Abstract
Bacteremia due to Cloacibacillus species is poorly described. We present three cases involving either Cloacibacillus evryensis or Cloacibacillus porcorum. The isolates were identified by 16S rRNA gene sequencing and were susceptible to antibiotics commonly used for anaerobic infections. The clinical significance of these organisms as potential emerging pathogens is discussed.
CASE REPORT
Case 1 was that of a 90-year-old man who was admitted to the Centre Hospitalier Universitaire Dr. Georges-L.-Dumont (Moncton, NB, Canada) for severe proctitis following rectal radiotherapy. The patient was receiving oral palliative chemotherapy with capecitabine for a rectal adenocarcinoma with lung metastasis. The patient had developed rectal bleeding and pain associated with fecal incontinence 3 days before his admission. He had a pacemaker and a medical history of hemorrhoidectomy.
On admission, the patient was afebrile with rectal pain and redness. There were no signs or symptoms of infection. The patient was managed with analgesia and local topical corticotherapy. The complete blood count on admission was within the normal range, except for a hemoglobin level of 107 (normal, 132 to 170) g/liter. On day 16 of hospitalization, he developed a fever of 39.8°C with no other symptoms except for rectal pain. There was redness of the rectal area and pain upon rectal examination. Hematological analysis revealed leukocytosis with a white cell count of 23.9 × 109 (normal, 4.0 × 109 to 11.0 × 109)/liter, a hemoglobin level of 101 g/liter, and a platelet count of 185 × 109 (normal, 130 × 109 to 400 × 109)/liter. A computerized tomography (CT) scan of the chest, abdomen, and pelvis showed a small amount of right-side pleural effusion, two pulmonary nodules, and bladder and rectal wall thickening with prostate enlargement. Two sets of aerobic and anaerobic blood cultures were collected. Empirical antimicrobial therapy consisted of 3.375 g of intravenous piperacillin-tazobactam administered every 6 h. The patient defervesced within 24 h of the introduction of the intravenous antibiotic. On day 18 of hospitalization, the intravenous antibiotic was discontinued. Oral ciprofloxacin and metronidazole were prescribed for 10 days. After 141 h of incubation at 37°C in aerobic and anaerobic blood culture systems (BD Bactec; Becton, Dickinson, Sparks, MD, USA), one anaerobic blood culture bottle was positive for rare Gram-negative bacilli. Strictly anaerobic Gram-negative bacilli were later isolated on BD brucella blood agar (Becton, Dickinson) supplemented with hemin (5 μg/ml) and vitamin K (1 μg/ml) after 6 days of incubation at 37°C. Preliminary identification tests for anaerobes with special-potency disks showed an inhibition zone diameter of >10 mm for kanamycin (1,000 μg), whereas there was no inhibition zone for colistin (10 μg) or vancomycin (5 μg). The isolate could not be identified by the RapID ANA II system (Remel, Lenexa, KS, USA). The strictly anaerobic, slowly growing, Gram-negative bacillus was identified as Cloacibacillus evryensis on the basis of 16S rRNA gene nucleotide sequence identity with the species type strain (158T) and designated strain LSPQ-04215 (Laboratoire de Santé Publique du Québec strain 04215). No other organism was isolated from blood cultures. The patient was discharged on day 22 of hospitalization. One year later, he was living at home under oral palliative chemotherapy. He had no relapse of his bacteremia.
Case 2 was that of a 94-year-old community-dwelling woman who was admitted to the McGill University Health Centre (Montreal, QC, Canada) for presumed urosepsis complicated by acute delirium and hypovolemic hyponatremia. Her medical history included atrial fibrillation requiring anticoagulation, coronary artery disease, hypertension, dyslipidemia, osteoporosis, and penicillin allergy. On arrival at the hospital, the patient was delirious with a temperature of 36.8°C. Other vital signs were within normal parameters. Urinalysis revealed pyuria and the presence of urinary nitrates. A complete blood count revealed leukocytosis (white blood cell count, 17.8 × 109/liter), a hemoglobin concentration of 133 g/liter, and a platelet count of 313 × 109/liter. Chest radiography was unremarkable. A urine culture and one set each of aerobic and anaerobic blood cultures were performed (BD Bactec). Empirical intravenous antimicrobial therapy with 1 g of ceftriaxone every 12 h was initiated for presumed community-acquired urosepsis. The patient's condition improved rapidly, and on day 3 after admission, her antibiotherapy was changed to oral trimethoprim-sulfamethoxazole (TMP-SMX). The urine culture yielded Escherichia coli at >108 CFU/ml, whereas growth was detected in the anaerobic blood culture bottle at 91 h of incubation and yielded small Gram-negative bacilli. Slow growth of a strictly anaerobic Gram-negative bacillus was observed on 5% sheep blood agar. After 5 days of incubation, preliminary identification tests for anaerobes using special-potency disks showed an inhibition zone diameter of 14 mm for kanamycin (1,000 μg) and no inhibition zone for colistin (10 μg) or vancomycin (5 μg). Bile medium did not inhibit the growth of the organism. The isolate could not be identified by the RapID ANA II system. The strictly anaerobic, slowly growing, Gram-negative bacillus was identified as C. evryensis on the basis of 16S rRNA gene nucleotide sequence identity with the species type strain (158T) and was designated strain LSPQ-04216. No other organism was isolated from blood cultures. The patient was discharged on day 4 following admission with a prescription for TMP-SMX to complete 14 days of antibiotherapy.
Case 3.
A 54-year-old woman was admitted to the Centre Hospitalier de l'Université de Montréal-Hôpital Saint-Luc (Montreal, QC, Canada) for vomiting, right iliac fossa pain and tenderness, and a temperature of 39.5°C for the last 24 h. The patient was known for obesity (118 kg) and a medical history of cholecystectomy. The medical exam revealed a positive McBurney point. Hematological investigations revealed leukocytosis with a white cell count of 17.5 × 109/liter with 90% neutrophils, a hemoglobin level of 155 g/liter, a platelet count of 230 × 109/liter, a creatinine level of 62 μmol/liter, and a total bilirubin level of 29.6 (normal, 7 to 23) μmol/liter. The aspartate aminotransferase, alanine transaminase, and alkaline phosphatase levels were normal. The urine biochemical analysis and bacterial culture were normal and negative, respectively. Two sets of blood cultures were done (BD Bactec). An abdomino-pelvic CT scan showed a 9-cm uterine fibroma, hepatic steatosis, and inflamed fat with gas bubbles around the appendix, compatible with acute appendicitis. Empirical antimicrobial therapy consisted of 3.375 g of intravenous piperacillin-tazobactam every 6 h. The patient improved progressively and became afebrile with a decrease in abdominal pain. One anaerobic bottle out of the two blood cultures was positive for a slowly growing, anaerobic, Gram-negative rod that was later isolated on fastidious anaerobe agar (Oxoid, Basingstoke, United Kingdom). No other organism was isolated. The strictly anaerobic, slowly growing, Gram-negative bacillus was identified as Cloacibacillus porcorum on the basis of 16S rRNA gene nucleotide sequence identity with the species type strain (CL-84T) and was designated strain LSPQ-04226. On day 4, the white cell count was 8.6 × 109/liter with 73% neutrophils, the hemoglobin level at 115 g/liter, the platelet count was 269 × 109/liter, and the creatinine level was 62 μmol/liter. The intravenous antimicrobial agent was changed to oral amoxicillin-clavulanic acid at 875 mg every 12 h for 10 days. The patient continued to improve and was followed up at the outpatient clinic by the digestive surgeon. Three months later, a complete colonoscopy showed only slight sigmoid diverticular disease.
The genus Cloacibacillus was first described by Ganesan et al. (1) and comprises two validly described species, C. evryensis and C. porcorum (2). Cells of the genus Cloacibacillus are strictly anaerobic, Gram-negative bacilli that are nonmotile and were originally isolated from environmental sources. C. evryensis was isolated from an anaerobic digester of a wastewater treatment plant (1); C. porcorum was isolated from the mucosal lining of a pig cecum (2). Members of this genus can be identified by 16S rRNA gene sequencing, cellular fatty acid profiles, DNA G+C content, and metabolic end product analysis (1). The genus Cloacibacillus is a member of the family Synergistaceae in the phylum Synergistetes, which was described in 2009 (3). The phylum contains 12 genera, Aminiphilus, Aminobacterium, Aminomonas, Anaerobaculum, Cloacibacillus, Dethiosulfovibrio, Fretibacterium, Jonquetella, Pyramidobacter, Synergistes, Thermanaerovibrio, and Thermovirga (4). Previously, most of the 16S rRNA gene sequences available from this phylum were obtained from culture-independent studies (3, 5–11). A small number of isolates were shown to be present in human infections, including soft tissue infections, abscesses, blood, peritoneal fluid, and dental infections (5–11). However, the lack of identification of Synergistetes isolates at the genus level led to the generic grouping under the Synergistes group of organisms (SGO) (3, 7, 11–13). With the initial description of the genus Cloacibacillus in 2008, 16S rRNA gene sequencing and phylogenetic analysis are now available for the proper identification of Cloacibacillus species (1, 2). We report here the first clinical description of Cloacibacillus bacteremia in three patients from whom either C. evryensis or C. porcorum was isolated. The three isolates, LSPQ-04215, LSPQ-04216, and LSPQ-04226, were identified by 16S rRNA gene sequencing with the BigDye Terminator v3.1 Cycle Sequencing kit on an ABI3130 XL genetic analyzer (Applied Biosystems, Foster City, CA, USA) (14). To identify the taxonomic neighbors of the three clinical isolates, the 16S rRNA gene sequences were used for an initial BLAST search (http://www.ncbi.nlm.nih.gov/BLAST) against GenBank. Phylogenetic and molecular evolutionary analyses with genera of the phylum Synergistetes were performed on 1,136 nucleotides with MEGA version 5 (15). Isolates LSPQ-04215 and LSPQ-04216 showed 100% nucleotide sequence identity with C. evryensis strain 158T (GenBank accession no. CU463952). Likewise, LSPQ-04226 showed 100% nucleotide sequence identity with C. porcorum strain CL-84T (GenBank accession no. JQ809697). The two C. evryensis isolates, LSPQ-04215 and LSPQ-04216, showed 97% nucleotide sequence identity with the 16S rRNA gene sequences of both C. porcorum CL-84T and LSPQ-04226. A neighbor-joining phylogenetic tree showed that isolates LSPQ-04215 and LSPQ-04216 clustered with C. evryensis 158T while LSPQ-04226 clustered with C. porcorum CL-84T (Fig. 1). Interestingly, we found that several isolates previously named Synergistes sp., Synergistes bacterium, or simply “bacterium” in the features contained in their annotation sequences submitted to GenBank clearly belonged to the species C. evryensis or C. porcorum (Fig. 1). Several Cloacibacillus-like strains were isolated from human infections, but the use of older taxonomic classification positioned them on various branches of the SGO phylogeny (Fig. 1). In the three university hospitals where isolates LSPQ-04215, LSPQ-04216, and LSPQ-04226 originated, conventional anaerobic characterization was performed according to reference 16 and was based on growth under anaerobic conditions; Gram staining; commercially available anaerobe identification substrates; testing for susceptibility to kanamycin, colistin, and vancomycin; and growth on bile medium. Cloacibacillus species are obligate anaerobes that grow slowly (1, 2). The calculated doubling time previously reported varies from 8 h for C. porcorum to 15 h for C. evryensis (1, 2), which could explain the rare Gram-negative bacilli observed in blood cultures after more than 3 days of incubation in the cases reported here. All three isolates, LSPQ-04215, LSPQ-04216, and LSPQ-04226, are susceptible to kanamycin and resistant to colistin and vancomycin, like C. evryensis 158T and C. porcorum CL-84T (1, 2). The conventional phenotype-based methods for anaerobes failed to identify Cloacibacillus species. The three isolates could not be identified by Vitek MS matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (bioMérieux Canada, Saint-Laurent, QC, Canada). A single isolate, C. evryensis LSP-04215, was tested on a Bruker MALDI Biotyper (Bruker, Madison, WI, USA). It could not be identified. The genus Cloacibacillus was not included in Vitek MS IVD version 2.0 (bioMérieux Canada), a clinically relevant species database; SARAMIS (spectral archive and microbial identification system) version 4.1 (bioMérieux Canada); or in the Bruker MALDI Biotyper databases. New spectrum entries were added to the SARAMIS database by using cultures from three different times of incubation (3, 7, and 10 days) for each of the three isolates. On the basis of this newly updated library, the SARAMIS database was capable of identifying the three isolates and assigned them to either C. evryensis or C. porcorum, respectively. We are planning to add both type strains (C. evryensis 158T and C. porcorum CL-84T) to the SARAMIS database in the near future. The antimicrobial susceptibilities of the three isolates were determined by the agar dilution method after 48 h of anaerobic incubation on prereduced laked sheep blood agar supplemented with hemin (5 μg/ml) and vitamin K (1 μg/ml), as recommended by the Clinical and Laboratory Standards Institute (17). The MICs for each isolate are reported in Table 1. As human infections with both Cloacibacillus species have remained largely uncharacterized, we reviewed the patients' charts for symptoms of infection, inflammation parameters such as fever, leukocyte counts, monomicrobial bacteremia, and clinical diagnosis. We also compared the isolation sites and clinical diagnoses of our three cases to the previous cases of Cloacibacillus-like strains by using the features contained in the annotation of the sequence data submitted to GenBank (Table 2). The latter were isolated from the environment and from human blood, peritoneal fluid, and the digestive tract (Table 2). The patients in cases 1 and 3 had proctitis and appendicitis, respectively, indicating a probable intestinal origin of their bacteremia. The probable intestinal origin of the Cloacibacillus bacteremia in our three case reports is in accordance with the isolation sites of several Cloacibacillus-like strains reviewed in Table 2. The antimicrobial susceptibility testing of the three Cloacibacillus isolates showed susceptibility to antibiotics commonly used for anaerobic infections. The patients all improved clinically after initiation of antibiotherapy. The three cases presented suggest that Cloacibacillus species are low-virulence, opportunistic pathogens associated with elderly or otherwise debilitated hosts. It is noteworthy that the patient described in case 2 made a full clinical recovery despite receiving only ceftriaxone and TMP-SMX, two antibiotics with limited uses against anaerobes. None of the patients described relapsed with Cloacibacillus bacteremia, and none of the patients died within 30 days.
FIG 1.
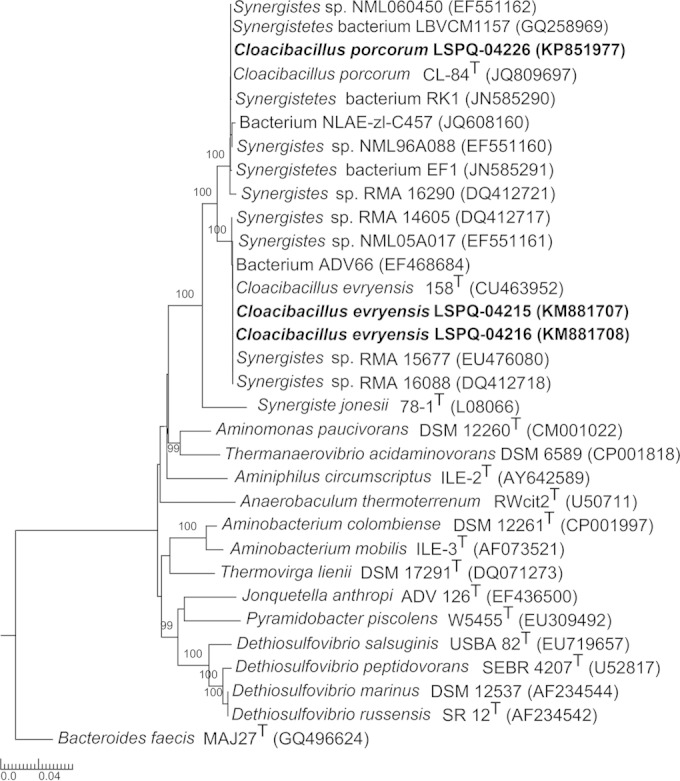
Neighbor-joining phylogenetic tree based on a comparison of partial 16S rRNA gene sequences (1,136 nucleotides) showing the relationships among strains LSPQ-04215, LSPQ-04216, and LSPQ-04226 and related taxa within the Cloacibacillus genus and other members of the Synergistetes phylum. Bacteroides faecis MAJ27T was used as an outgroup. Bootstrap percentages (based on 1,000 replications) higher than 90% are shown at branching points. Names of strains and accession numbers of 16S rRNA genes are as cited in GenBank. GenBank accession numbers for 16S rRNA gene sequences are shown in parentheses. The scale bar indicates the estimated number of substitutions per site. A superscript T indicates a type strain.
TABLE 1.
MICs of selected antibiotics for C. evryensis LSPQ-04215 and LSPQ-04216 and C. porcorum LSPQ-04226
| Antibiotic | MIC (μg/ml) |
||
|---|---|---|---|
| C. evryensis LSPQ-04215 | C. evryensis LSPQ-04216 | C. porcorum LSPQ-04226 | |
| Clindamycin | 0.25 | 0.5 | 0.5 |
| Penicillin | 0.25 | ≤0.125 | ≤0.125 |
| Cefoxitin | ≤2 | ≤2 | ≤2 |
| Meropenem | 0.125 | 0.03 | 0.03 |
| Metronidazole | 0.5 | 0.06 | 0.5 |
| Piperacillin-tazobactam | 2/4 | 1/4 | 2/4 |
TABLE 2.
Identification of Cloacibacillus and Cloacibacillus-like 16S rRNA sequences deposited in GenBank to the species level, their isolation sites, and relevant clinical data
| 16S rRNA gene sequence identification in GenBanka | Strain name (GenBank accession no.) | Identification based on ≥99% homology with type strain | Isolation site | Clinical diagnosis | Human clinical relevance |
|---|---|---|---|---|---|
| C. evryensis | 158T (CU463952) | C. evryensis | Environment | NAb | No |
| C. evryensis | LSPQ-04215 (KM881707) | C. evryensis | Blood | Bacteremia | Yes |
| C. evryensis | LSPQ-04216 (KM881708) | C. evryensis | Blood | Bacteremia | Yes |
| Synergistes sp. | RMA16088 (DQ412718) | C. evryensis | Peritoneal fluid | NA | Yes |
| Synergistes sp. | NML05A017 (EF551161) | C. evryensis | Blood | NA | Yes |
| Synergistes sp. | RMA 14605 (DQ412717) | C. evryensis | Peritoneal fluid | NA | Yes |
| Synergistes sp. | ADV66 (EF468684) | C. evryensis | NA | NA | NA |
| Synergistes sp. | RMA 15677 (EU476080) | C. evryensis | Peritoneal fluid | NA | Yes |
| C. porcorum | CL-84T (JQ809697) | C. porcorum | Environnment | NA | No |
| C. porcorum | LSPQ-04226 (KP851977) | C. porcorum | Blood | Bacteremia | Yes |
| Synergistes sp. | RMA 16290(DQ412721) | C. porcorum | Peritoneal fluid | NA | Yes |
| Synergistes sp. | NML96A088 (EF551160) | C. porcorum | Blood | NA | Yes |
| Synergistes sp. | NML060450 (EF551162) | C. porcorum | Blood | NA | Yes |
| Synergistetes bacterium | RK1 (JN585290) | C. porcorum | Digestive tract of red Kangaroo (Macropus rufus) | NA | No |
| Synergistetes bacterium | EF1 (JN585291) | C. porcorum | Emu (Dromaius novaehollandiae) feces | NA | No |
| Bacterium | NLAE-zl-C457 (JQ608160) | C. porcorum | Cow feces | NA | No |
| Synergistetes sp. | LBVCM1157 (GQ258969) | C. porcorum | Blood | NA | Yes |
| Synergistes jonesii | 78-1T (L08066) | C. porcorum | Rumen tract | NA | No |
The two C. evryensis strains and the C. porcorum strain isolated in this study are underlined.
NA, not available.
Physicians should be aware that Cloacibacillus species can be opportunistic human pathogens associated with intestinal infections.
Nucleotide sequence accession numbers.
The GenBank accession numbers for isolates LSPQ-04215, LSPQ-04216, and LSPQ-04226 are KM881707, KM881708, and KP851977, respectively.
ACKNOWLEDGMENTS
S.L. has a research agreement with bioMérieux Canada. All other authors declare that they have no conflicts of interest.
We thank Maureen Hastie and Jean-Charles Côté for critical reading of the manuscript. We also thank bioMérieux Canada for access to the Vitek MS MALDI-TOF system.
REFERENCES
- 1.Ganesan A, Chaussonnerie S, Tarrade A, Dauga C, Bouchez T, Pelletier E, Le Paslier D, Sghir A. 2008. Cloacibacillus evryensis gen. nov., sp. nov., a novel asaccharolytic, mesophilic, amino-acid-degrading bacterium within the phylum ‘Synergistetes’, isolated from an anaerobic sludge digester. Int J Syst Evol Microbiol 58(Pt 9):2003–2012. doi: 10.1099/ijs.0.65645-0. [DOI] [PubMed] [Google Scholar]
- 2.Looft T, Levine UY, Stanton TB. 2013. Cloacibacillus porcorum sp. nov., a mucin-degrading bacterium from the swine intestinal tract and emended description of the genus Cloacibacillus. Int J Syst Evol Microbiol 63:1960–1966. doi: 10.1099/ijs.0.044719-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jumas-Bilak E, Roudière L, Marchandin H. 2009. Description of ‘Synergistetes' phyl. nov. and emended description of the phylum ‘Deferribacteres’ and of the family Syntrophomonadaceae, phylum ‘Firmicutes’. Int J Syst Evol Microbiol 59:1028–1035. doi: 10.1099/ijs.0.006718-0. [DOI] [PubMed] [Google Scholar]
- 4.Euzéby JP. 2015. List of prokaryotic name with standing in nomenclature. http://www.bacterio.net/. Accessed 30 March 2015.
- 5.Hugenholtz P, Hooper SD, Kyrpides NC. 2009. Focus: Synergistetes. Environ Microbiol 11:1327–1329. doi: 10.1111/j.1462-2920.2009.01949.x. [DOI] [PubMed] [Google Scholar]
- 6.Munson MA, Pitt-Ford T, Chong B, Weightman A, Wade WG. 2002. Molecular and cultural analysis of the microflora associated with endodontic infections. J Dent Res 81:761–766. doi: 10.1177/154405910208101108. [DOI] [PubMed] [Google Scholar]
- 7.Horz HP, Citron DM, Warren YA, Goldstein EJ, Conrad G. 2006. Synergistes group organisms of human origin. J Clin Microbiol 44:2914–2920. doi: 10.1128/JCM.00568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Lillo A, Ashley FP, Palmer RM, Munson MA, Kyriacou L, Weightman AJ, Wade WG. 2006. Novel subgingival bacterial phylotypes detected using multiple universal polymerase chain reaction primer sets. Oral Microbiol Immunol 21:61–68. doi: 10.1111/j.1399-302X.2005.00255.x. [DOI] [PubMed] [Google Scholar]
- 9.Jumas-Bilak E, Carlier JP, Jean-Pierre H, Citron D, Bernard K, Damay A, Gay B, Teyssier C, Campos J, Marchandin H. 2007. Jonquetella anthropi gen. nov., sp. nov., the first member of the candidate phylum ‘Synergistetes’ isolated from man. Int J Syst Evol Microbiol 57:2743–2748. doi: 10.1099/ijs.0.65213-0. [DOI] [PubMed] [Google Scholar]
- 10.Downes J, Vartoukian SR, Dewhirst FE, Izard J, Chen T, Yu WH, Sutcliffe IC, Wade WG. 2009. Pyramidobacter piscolens gen. nov., sp. nov., a member of the phylum ‘Synergistetes’ isolated from the human oral cavity. Int J Syst Evol Microbiol 59:972–980. doi: 10.1099/ijs.0.000364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vartoukian SR, Palmer RM, Wade WG. 2010. Cultivation of a Synergistetes strain representing a previously uncultivated lineage. Environ Microbiol 12:916–928. doi: 10.1111/j.1462-2920.2009.02135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vartoukian SR, Palmer RM, Wade WG. 2007. The division Synergistes. Anaerobe 13:99–106. doi: 10.1016/j.anaerobe.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 13.Godon JJ, Morinière J, Moletta M, Gaillac M, Bru V, Delgènes JP. 2005. Rarity associated with specific ecological niches in the bacterial world: the Synergistes example. Environ Microbiol 7:213–224. doi: 10.1111/j.1462-2920.2004.00693.x. [DOI] [PubMed] [Google Scholar]
- 14.Bekal S, Gaudreau C, Laurence RA, Simoneau E, Raynal L. 2006. Streptococcus pseudoporcinus sp. nov., a novel species isolated from the genitourinary tract of women. J Clin Microbiol 44:2584–2586. doi: 10.1128/JCM.02707-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jousimies-Somer HR, Summanen P, Citron DM, Baron EJ, Wexler HM, Finegold SM. 2002. Wadsworth-KTL anaerobic bacteriology manual, 6th ed Star Publishing Co., Belmont, CA. [Google Scholar]
- 17.Clinical and Laboratory Standards Institute. 2012. Methods for antimicrobial susceptibility testing of anaerobic bacteria, 8th ed Approved standard M11-A8. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]


