Abstract
Multilocus sequence typing (MLST) is the gold standard genotyping technique for many microorganisms. This classification approach satisfies the requirements for a high-resolution, standardized, and archivable taxonomic system. Here, we describe the development of a novel MLST system to assist with the investigation of an unusual cluster of surgical site infections caused by Bipolaris spp. in postoperative cardiothoracic surgery (POCS) patients during January 2008 to December 2013 in the southeastern United States. We also used the same MLST system to perform a retrospective analysis on isolates from a 2012 Bipolaris endophthalmitis outbreak caused by a contaminated product. This MLST system showed high intraspecies discriminatory power for Bipolaris spicifera, B. hawaiiensis, and B. australiensis. Based on the relatedness of the isolates, the MLST data supported the hypothesis that infections in the POCS cluster were from different environmental sources while confirming that the endophthalmitis outbreak resulted from a point source, which was a contaminated medication.
INTRODUCTION
Dematiaceous fungi belonging to the genus Bipolaris are abundant in the environment. Many species in this genus are known to cause devastating disease in plants and staple crops around the world (1). In humans, they are common etiological agents of fungal sinusitis. Surgical site infections (SSIs) and deep tissue and invasive infections by members of this genus do occur, but they are extremely rare and typically associated with immunocompromised patients (2–7). The common disease-causing Bipolaris species in humans are Bipolaris spicifera, B. hawaiiensis, and, occasionally, B. australiensis (3). A recent proposal transfers these species to the genus Curvularia (1), but this new name has not yet been widely accepted.
In the clinical setting, Bipolaris infections are usually diagnosed using microscopy to distinguish the morphological characteristics of the fungus following culture. In recent years, the sequence comprising the internal transcribed spacer (ITS) regions and the 5.8S nuclear rRNA genes of the ribosomal cistron has been exploited for species assignment of dematiaceous fungi (8, 9). In most cases, ITS-based sequence identification is sufficient for determining species. However, ITS-based sequencing is limited by its dependence on the completeness and accuracy of available DNA sequence databases. A DNA typing system employing two or more loci, such as multilocus sequence typing (MLST), can be used when it is necessary to distinguish between isolates within a species (10, 11). MLST has been successfully implemented for species identification within a species complex as well as strain differentiation within a species for various fungal pathogens of humans such as Aspergillus fumigatus, Candida albicans, Candida glabrata, Candida tropicalis, Cryptococcus gattii, Cryptococcus neoformans, and Fusarium sp. (12–18). The recent investigations of infections caused by dematiaceous fungi (5, 19) have highlighted the importance of high-resolution, standardized, and archivable typing system(s) for this group of fungi.
In November 2013, the CDC was contacted by the Texas Department of State Health Services about a cluster of SSIs caused by Bipolaris spp. among patients who had recently undergone cardiothoracic surgery. Upon further investigation, 21 cases in postoperative cardiothoracic surgery (POCS) patients were identified from 10 different hospitals in three states (Texas, Arkansas, and Florida) during 2008-2013. Three compelling epidemiological features emerged during the investigation: (i) all patients had a recent history of cardiothoracic surgery and had SSIs caused by Bipolaris spp., (ii) we could not find cases among patients who had undergone other invasive surgery procedures, and (iii) the cases were clustered together temporally (more than half of the cases occurred in 2013) and geographically (cases were found in the southeastern United States). Exposure to a common contaminated product used during surgery was initially suspected, but common medical products or devices unique to the case patients were not identified in the epidemiologic investigation (A. Purfield, S. S. Vallabhaneni, K. Benedict, U. Luvsansharav, S. R. Lockhart, C. D. Pham, A. Laufer, N. Pascoe, G. Heseltine, W. Chung, E. Hall, K. B. Brust, C. F. Wheeler, S. Chideya, and B. J. Park, unpublished data). The investigation concluded that patients likely acquired the infections from separate environmental sources. In such cases, investigators often rely on laboratory-based techniques to understand the relatedness of patient isolates for validating the conclusions of the epidemiologic investigation. Although 10 patient isolates were available for analysis, a reliable method for strain typing did not exist at the time of the outbreak.
Here, we describe the development and implementation of a novel MLST system using six different DNA loci to differentiate B. spicifera, B. hawaiiensis, and B. australiensis isolates from case patients, controls, and the environment, collected during the investigation of the cluster of fungal SSIs among POCS patients as well as from a preexisting culture collection at the CDC, including Bipolaris isolates from a previous outbreak of endophthalmitis (5).
MATERIALS AND METHODS
Fungal collection, growth conditions, and DNA purification.
Sixty Bipolaris species isolates were included in this study (Table 1). Thirty-eight isolates were collected as part of the investigation into a cluster of fungal infections among POCS patients: 10 isolates obtained from surgical sites from POCS case patients, 15 isolates from respiratory tract or skin from patients who presented with Bipolaris infections that were not SSIs to hospitals where Bipolaris SSI cases were identified, and 12 isolates obtained through environmental sampling in hospitals with Bipolaris SSI cases. Twenty-two isolates used as unrelated controls were obtained from the CDC collection (n = 16) or the Centraalbureau voor Schimmelcultures (CBS) (http://www.cbs.knaw.nl/Collections/) collection (n = 6, including the type strain for each species). The CDC collection isolates included six B. hawaiiensis isolates which were collected as part of an investigation into a 2012 Bipolaris endophthalmitis outbreak linked to contaminated medication (11).
TABLE 1.
Isolate collection information and MLST-based allelic and sequence-type assignment
| Isolate collection information |
MLST locus allele no. |
MLST typing |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Name | Source | Year | BRN1 | EF1α | GPDH | RPB1 | RPB2 | SAL1 | ST | Species |
| B10583a | CSb patient | 2013 | 4 | 1 | 1 | 1 | 3 | 3 | 1 | B. spicifera |
| B10588a | Environment | 2013 | 2 | 1 | 4 | 3 | 4 | 5 | 2 | B. spicifera |
| B10582a | CS patient | 2013 | 3 | 1 | 3 | 3 | 8 | 2 | 3 | B. spicifera |
| CBS125738 | Sinus | Unknown | 1 | 1 | 8 | 4 | 2 | 3 | 4 | B. spicifera |
| B10680a | Sinus | 2014 | 3 | 1 | 5 | 2 | 4 | 5 | 5 | B. spicifera |
| B4388 | Sinus | 1987 | 3 | 1 | 7 | 4 | 4 | 1 | 6 | B. spicifera |
| B10575a | CS patient | 2013 | 3 | 1 | 4 | 3 | 6 | 5 | 7 | B. spicifera |
| B10586a | Environment | 2013 | 3 | 1 | 7 | 3 | 3 | 5 | 8 | B. spicifera |
| B10730a | Sinus | 2014 | 3 | 1 | 7 | 3 | 4 | 5 | 9 | B. spicifera |
| B6138 | Environment | 2001 | 3 | 1 | 5 | 3 | 4 | 5 | 10 | B. spicifera |
| B10681a | Sputum | 2014 | 3 | 1 | 5 | 2 | 7 | 5 | 11 | B. spicifera |
| B10578a | CS Patient | 2013 | 5 | 1 | 6 | 4 | 4 | 5 | 12 | B. spicifera |
| B10574a | CS patient | 2013 | 4 | 3 | 5 | 6 | 3 | 5 | 13 | B. spicifera |
| B10585a | Environment | 2013 | 3 | 1 | 6 | 3 | 8 | 5 | 14 | B. spicifera |
| B10581a | Environment | 2012 | 3 | 1 | 6 | 3 | 8 | 5 | 14 | B. spicifera |
| B10590a | Environment | 2013 | 3 | 1 | 4 | 6 | 3 | 7 | 15 | B. spicifera |
| B10788a | Nose | 2014 | 6 | 1 | 4 | 6 | 4 | 5 | 16 | B. spicifera |
| B10677a | Sinus | 2014 | 5 | 1 | 7 | 4 | 4 | 5 | 17 | B. spicifera |
| ATCC28335 | Alfalfa seed | 1973 | 7 | 3 | 3 | 6 | 1 | 5 | 18 | B. spicifera |
| CBS274.52c | Mandarin orange | Unknown | 7 | 3 | 3 | 6 | 3 | 5 | 19 | B. spicifera |
| B10584a | Environment | 2013 | 3 | 1 | 6 | 4 | 8 | 5 | 20 | B. spicifera |
| B10621 | Breast | 2013 | 3 | 1 | 4 | 3 | 9 | 5 | 21 | B. spicifera |
| B10791a | Sputum | 2014 | 6 | 1 | 4 | 3 | 4 | 7 | 22 | B. spicifera |
| B10794a | Sinus | 2014 | 5 | 1 | 7 | 4 | 3 | 6 | 23 | B. spicifera |
| B10580a | CS patient | 2012 | 3 | 1 | 4 | 4 | 9 | 5 | 24 | B. spicifera |
| B10589a | Environment | 2013 | 3 | 1 | 5 | 6 | 8 | 5 | 25 | B. spicifera |
| B10576a | CS patient | 2013 | 5 | 1 | 6 | 3 | 8 | 6 | 26 | B. spicifera |
| B10618a | Environment | 2013 | 6 | 1 | 6 | 3 | 8 | 5 | 27 | B. spicifera |
| B10619a | Environment | 2013 | 6 | 1 | 6 | 3 | 8 | 5 | 27 | B. spicifera |
| B10617a | CS patient | 2013 | 3 | 2 | 7 | 3 | 4 | 8 | 28 | B. spicifera |
| B10793a | Tracheal aspirate | 2014 | 3 | 1 | 4 | 3 | 8 | 8 | 29 | B. spicifera |
| B10579a | CS patient | 2011 | 5 | 1 | 6 | 2 | 8 | 8 | 30 | B. spicifera |
| B10682a | Sinus | 2013 | 8 | 1 | 5 | 5 | 5 | 5 | 31 | B. spicifera |
| B11060 | Calcium gluconate | 2013 | 3 | 1 | 8 | 5 | 8 | 5 | 32 | B. spicifera |
| B10594a | Sinus | 2013 | 6 | 1 | 4 | 4 | 8 | 7 | 33 | B. spicifera |
| B10587a | Environment | 2013 | 9 | 3 | 2 | 5 | 8 | 5 | 34 | B. spicifera |
| MSG06003 | Human, source unknown | 2014 | 6 | 1 | 6 | 5 | 8 | 5 | 35 | B. spicifera |
| B10592a | Environment | 2013 | 5 | 1 | 6 | 6 | 8 | 8 | 36 | B. spicifera |
| B3515 | Human, source unknown | 1981 | 7 | 1 | 8 | 5 | 9 | 5 | 37 | B. spicifera |
| B4432 | Corneal scraping | 1986 | 1 | 4 | 1 | 1 | 2 | 1 | 1 | B. hawaiiensis |
| B10790a | Bronchial wash | 2014 | 2 | 1 | 2 | 2 | 1 | 2 | 2 | B. hawaiiensis |
| B10620a | CS patient | 2013 | 4 | 2 | 3 | 5 | 3 | 3 | 3 | B. hawaiiensis |
| B10336 | Environment | 2013 | 3 | 3 | 3 | 6 | 4 | 4 | 4 | B. hawaiiensis |
| B10795a | Sinus | 2014 | 5 | 3 | 3 | 4 | 3 | 5 | 5 | B. hawaiiensis |
| B9526d | Endophthalmitis | 2012 | 3 | 3 | 3 | 6 | 4 | 9 | 6 | B. hawaiiensis |
| B9573d | Endophthalmitis | 2012 | 3 | 3 | 3 | 6 | 4 | 9 | 6 | B. hawaiiensis |
| B9574d | Endophthalmitis | 2012 | 3 | 3 | 3 | 6 | 4 | 9 | 6 | B. hawaiiensis |
| B9575d | Endophthalmitis | 2012 | 3 | 3 | 3 | 6 | 4 | 9 | 6 | B. hawaiiensis |
| B9525d | Endophthalmitis | 2012 | 3 | 3 | 3 | 6 | 4 | 9 | 6 | B. hawaiiensis |
| B9528# | Endophthalmitis | 2012 | 3 | 3 | 3 | 6 | 4 | 9 | 6 | B. hawaiiensis |
| B10789a | Bronchial wash | 2014 | 7 | 5 | 4 | 3 | 5 | 6 | 7 | B. hawaiiensis |
| B10679a | Sputum | 2013 | 6 | 5 | 4 | 3 | 5 | 7 | 8 | B. hawaiiensis |
| B10678a | Sinus | 2013 | 6 | 5 | 4 | 3 | 5 | 7 | 8 | B. hawaiiensis |
| CBS127091 | Unknown | Unknown | 6 | 5 | 4 | 3 | 5 | 8 | 9 | B. hawaiiensis |
| CBS173.57c | Asian rice | 1957 | 6 | 5 | 4 | 3 | 5 | 8 | 9 | B. hawaiiensis |
| CBS126975 | Unknown | Unknown | 1 | 2 | 1 | 1 | 1 | 3 | 1 | B. australiensis |
| B10792a | Leg abrasion | 2014 | 2 | 1 | 3 | 2 | 3 | 1 | 2 | B. australiensis |
| CBS172.57c | Asian rice | 1957 | 3 | 3 | 2 | 3 | 2 | 2 | 3 | B. australiensis |
| B4361 | Cat granuloma | 1986 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | B. ellisii |
| B10591a | Environment | 2013 | - | - | - | - | - | - | - | B. cynodontis |
Isolate from the POCS investigation.
CS, cardiothoracic surgery.
Isolate from the endophthalmitis.
Type strain from CBS.
Fungal isolates were propagated on Sabouraud dextrose agar. All isolates were maintained at 25°C. Total genomic DNA was purified using bead agitation and reagents in the DNeasy blood and tissue kit (Qiagen; Valencia, CA, USA) as previously described (20).
Molecular investigations of the ribosomal DNA and MLST loci.
PCR and DNA sequencing of both strands with BigDye Terminator (Life Technologies, Grand Island, NY, USA) were performed as described previously (21). The same primer sets were employed for both PCR and DNA sequencing at each locus. Locus-specific primers were designed as described previously (22) (Table 2). The sequences for melanin reductase (BRN1) (23), glyceraldehyde-3-phosphate dehydrogenase (GPDH), transcription elongation factor-1 alpha (EF1α), RNA polymerase II subunit 1-like (RPB1), RNA polymerase II subunit 1-like (RPB2), and scytalone dehydratase (SAL1) (24) were from the National Center for Biotechnology Information (Table 1). The β-tubulin primers were designed using the β-tubulin gene sequences from B. spicifera (n = 3) and Curvularia sp. (n = 1). PCR programs were as follows: 94°C for 5 min; 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and a final elongation step at 72°C for 2 min.
TABLE 2.
MLST loci information
| Locus | GenBank accession no.a | Primer typeb | Primer sequence (5′ to 3′) | No. of bp analyzed | Analyzed sequence fragment start and end points |
|---|---|---|---|---|---|
| βTUB | NA | F | AAATCGGTGCTGCTTTCTGGC | 287−296 | 5′-CCAT(T/C)TC(C/T)......TGGGCCAA-3′ |
| R | GTAGTGACCCTTGGCCCAGTT | ||||
| BRN1 | AB011638.1 | F | TATTGGAAAGGCTATGGCCA | 485 | 5′-TACGCCAA......AGAAG(G/A)TC-3′ |
| R | GGCAGACAGCGTGGTACAT | ||||
| EF1a | JN601010.1 | F | TCGGTGTCAAGCAGCTCAT | 569 | 5′-GAGGAGCG......CAGGTCGG-3′ |
| R | AATCTTCTCGAGGAGCTCGG | ||||
| GPDH | JN601036.1 | F | GTCTCGCATGCGTAGGTGT | 448–453 | 5′-TCCATTGA ......CGTCATGG-3′ |
| R | AGTGGTTGTGCAGGAGGC | ||||
| RPB1 | JQ965141.1 | F | TACAACCTGGCTACCCCG | 556 | 5′-TTCTCC(T/A)G......C(G/A)TT(C/T)TTC-3′ |
| R | CAAGAGCCTGCTTCTTCTTGTT | ||||
| RPB2 | JQ585695.1 | F | GTGGAAAACAACCAAGACTTCAA | 437 | 5′-CGGC(C/T)TGA......(A/G)GT(C/T)GTGC-3′ |
| R | ATATCACGAATCAAACTCATCTCGT | ||||
| SAL1 | AB587805.1 | F | CATGGCTAACTAGTTTGCAGATGT | 275 | 5′-G(G/T)CGGC(A/G/C)T......TCA(C/T)CA (G/A)C-3′ |
| R | CCGCGACTCATCCGTGTAT |
Accession number of gene sequence from which the primers were derived.
F, forward; R, reverse.
MLST marker stability and reproducibility were determined by performing sequential DNA analysis on two unrelated B. hawaiiensis isolates and one B. spicifera isolate. These isolates were subcultured every 2 weeks over a period of 6 weeks, followed by DNA extraction and sequencing of the MLST loci to monitor for genetic instability.
Molecular computational phylogenetic analysis.
Geneious (Biomatters Limited, San Francisco, CA, USA) was employed for visualizing and annotating DNA sequences and for phylogenetic analysis. The species assignment of fungal isolates was accomplished by using the ITS DNA sequence and the Basic Local Alignment Search Tool (BLAST) on the CBS fungal database website (http://www.cbs.knaw.nl/Collections/BioloMICSSequences.aspx?file=all). For phylogenetic analysis, single-locus sequences or concatenated multilocus sequences of all Bipolaris isolates were pool aligned using MUSCLE (or otherwise specified) prior to the construction of a phylogram. All phylograms were generated using the neighbor-joining model and rooted on Bipolaris ellisii. The genetic distance and reproducibility of the phylograms were calculated according to Tamura-Nei parameters and 1,000 bootstrap replicates, respectively. Simpson's index (SI) of diversity was calculated as previously described by Hunter and Gaston (25).
Database hosting.
The typing systems for B. spicifera, B. hawaiiensis, and B. australiensis with allele number and sequence type assignment are publicly available and hosted at the Fungal MLST Database site (http://mlst.mycologylab.org/DefaultInfo.aspx?Page=Home).
RESULTS
Morphological and ribosomal DNA sequence characteristics of mold isolates.
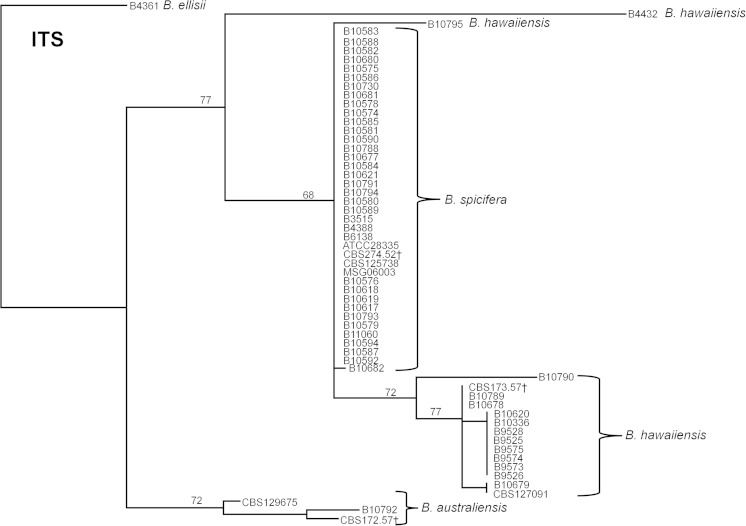
As part of the Bipolaris SSI cluster investigation, 38 Bipolaris isolates were received. By BLAST analysis of the ITS sequence, the Bipolaris isolates were further classified into B. spicifera (n = 30), B. hawaiiensis (n = 6), B. australiensis (n = 1), and B. cynodontis (n = 1). ITS sequences of the 22 isolates of Bipolaris used as unrelated controls for the development of the MLST system confirmed them as B. australiensis (n = 2), B. ellisii (n = 1), B. hawaiiensis (n = 10), and B. spicifera (n = 9). The B. spicifera isolates exhibited high ITS sequence identity; 38 of 39 shared identical ITS sequences (Fig. 1). The ITS sequences of B. australiensis and B. hawaiiensis, including their respective type strains, showed higher intraspecies diversity with Simpson's index (SI) values of 1 and 0.68, respectively.
FIG 1.
Dendrogram showing the discrimination of B. australiensis, B. hawaiiensis, and B. spicifera isolates inferred from ITS sequence data. The dendrogram was constructed with the neighbor-joining method using CLUSTALW alignment and rooted at B. ellisii. The genetic distance and topology reliability were calculated using the Tamura-Nei model and 1,000 bootstrap replicates, respectively. †Type strain from CBS.
Validation of genetic stability and amplification reproducibility for the genetic markers employed to differentiate Bipolaris species.
We investigated seven different loci (βTUB, BRN1, GPDH, EF1α, RPB1, RPB2, and SAL1) for the development of the MLST system. All primer sets generated robust bands for B. australiensis, B. ellisii, B. hawaiiensis, and B. spicifera. Not all of the loci were amplified for the B. cynodontis isolate, so it was dropped from the analysis. DNA sequences for all seven loci were stable in isolates that were subcultured three times over a period of 6 weeks. Analysis of the DNA sequences revealed a highly variable intron in the βTUB marker which is similar to introns observed in the βTUB gene of various other fungal species (26). The βTUB intron is approximately 46 to 55 bp long and contains 36 single nucleotide polymorphisms (SNPs). Upon further analysis of the βTUB locus, it was determined that the βTUB primers amplified two tubulin paralogs. As a result, βTUB was not included in the final MLST system.
Characteristics of the DNA markers used for differentiating B. australiensis, B. hawaiiensis, and B. spicifera.
Partial fragments of the BRN1, EF1α, GPDH, RPB1, RPB2, and SAL1 genes were combined to form the new MLST system (Table 3). No insertions or deletions were observed in any of the MLST loci for B. australiensis, B. hawaiiensis, and B. spicifera. The BRN1 and the GPDH loci both contained introns. The BRN1 locus contained an intron of 53 bp that contained 15 unique SNPs. The GPDH locus used had one full intron and part of another. The full intron was 62 bp in length and contained 29 unique SNPs. The partial intron (truncated by the end of the sequence used for analysis) was 15 bp and contained two SNPs. The predicted coding regions of BRN1 and GPDH loci also contained a high number of polymorphic sites. The majority of those sites were synonymous mutations; GPDH only had synonymous mutations while BRN1 also possessed five nonsynonymous mutations, an indication of possible selective pressure at this locus.
TABLE 3.
Allelic information at each MLST locusa
| Species | Allele no. | BRN1 | EF1α | GPDH | RPB1 | RPB2 | SAL1 | ITS |
|---|---|---|---|---|---|---|---|---|
| B. spicifera | ||||||||
| No. of isolates per allele | 1 | 1 | 34 | 1 | 1 | 1 | 1 | 38 |
| 2 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | |
| 3 | 18 | 4 | 3 | 15 | 6 | 1 | 0 | |
| 4 | 2 | 0 | 9 | 8 | 10 | 1 | 0 | |
| 5 | 6 | 0 | 6 | 5 | 1 | 26 | 0 | |
| 6 | 6 | 0 | 10 | 7 | 1 | 2 | 0 | |
| 7 | 3 | 0 | 6 | 0 | 1 | 3 | 0 | |
| 8 | 1 | 0 | 3 | 0 | 15 | 4 | 0 | |
| 9 | 1 | 0 | 0 | 0 | 3 | 0 | 0 | |
| SI | 0.75 | 0.23 | 0.84 | 0.77 | 0.77 | 0.55 | 0.05 | |
| B. hawaiiensis | ||||||||
| No. of isolates per allele | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| 3 | 7 | 8 | 9 | 5 | 2 | 1 | 1 | |
| 4 | 1 | 1 | 5 | 1 | 7 | 1 | 5 | |
| 5 | 1 | 5 | 0 | 1 | 5 | 1 | 8 | |
| 6 | 4 | 0 | 0 | 7 | 0 | 1 | 0 | |
| 7 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | |
| 8 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | |
| 9 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | |
| SI | 0.78 | 0.68 | 0.62 | 0.77 | 0.73 | 0.86 | 0.68 | |
| B. australiensis | ||||||||
| No. of isolates per allele | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| SI | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Total no. of isolates | 58 | 58 | 58 | 58 | 58 | 58 | 58 | |
| Composite SI | 0.87 | 0.79 | 0.90 | 0.88 | 0.87 | 0.79 | 0.55 |
Allele information will be stored at http://mlst.mycologylab.org/DefaultSlideUD.aspx?Page=Home.
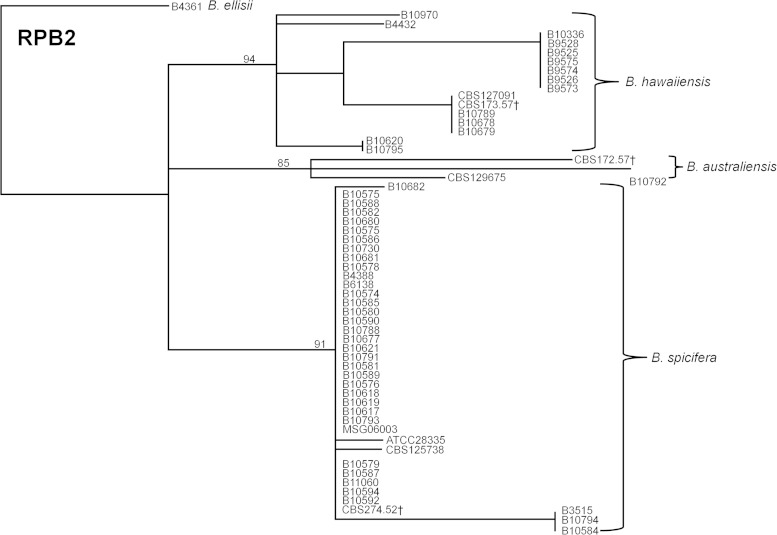
None of the three species (B. australiensis, B. spicifera, and B. hawaiiensis) shared an identical allele for any of the MLST markers. The ITS locus was able to differentiate both B. australiensis and B. spicifera, but suggested that B. hawaiiensis was polyphyletic (Fig. 1); two isolates, B4432 and B10795, did not cluster with the type isolate. Of the six markers used, RPB2 most reliably segregated all of the isolates to specific species clusters (Fig. 2).
FIG 2.
Dendrogram showing discrimination of B. australiensis, B. hawaiiensis, and B. spicifera isolates by the RPB2 sequence, which was constructed with the neighbor-joining method and rooted at B. ellisii. The genetic distance and topology reliability were calculated using the Tamura-Nei model and 1,000 bootstrap replicates, respectively. †Type strain from CBS.
Discriminatory power of the MLST system.
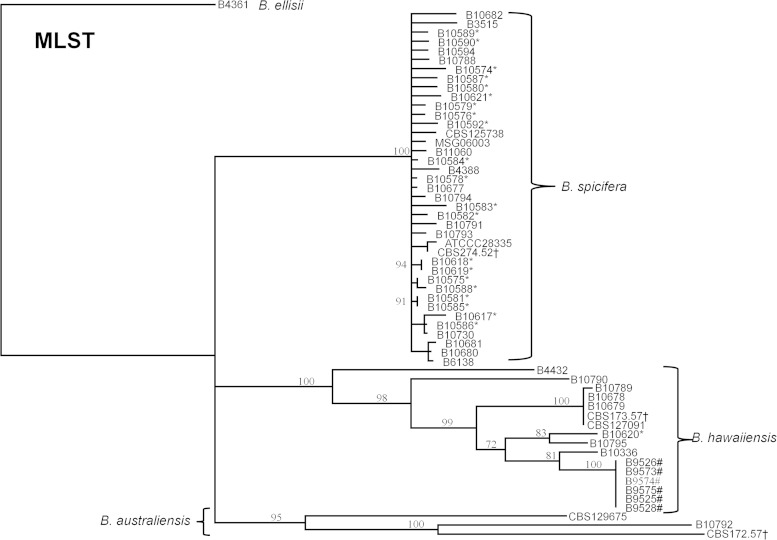
The MLST system displayed strong interspecies discriminatory power for the Bipolaris species investigated. It was able to segregate isolates of the same ITS-assigned species into distinct clades in concordance with the type strains, with bootstrapping values of >95% (Fig. 3). The MLST system also displayed good intraspecies differentiation (Table 1). For B. spicifera, GPDH displayed the highest discriminatory power with an SI of 0.84 while EF1α displayed the lowest discriminatory power with an SI of 0.23. Each MLST marker alone was able to divide the 39 B. spicifera isolates into multiple allele types, ranging between three and nine alleles per locus. For B. hawaiiensis, SAL1 displayed the highest discriminatory power with an SI of 0.86, while GPDH displayed the lowest discriminatory power with an SI of 0.62. There were too few isolates of B. australiensis to adequately calculate the discriminatory power.
FIG 3.
Dendrogram inferring the relatedness of 59 Bipolaris isolates using MLST. The dendrogram was constructed with the neighbor-joining method and rooted at B. ellisii. The genetic distance and topology reliability were calculated using the Tamura-Nei model and 1,000 bootstrap replicates, respectively. *Isolate from the POCS investigation; †type strain from CBS; #isolate from the endophthalmitis outbreak.
The MLST system divided the 39 B. spicifera isolates into 37 unique sequence types (STs). Isolates collected during the Bipolaris SSI investigation had unique STs except for two pairs of environmental isolates that belonged to the same ST; isolates B10618 and B10619 came from two different swabs of a medical instrument associated with a case-patient's care and shared an ST, and isolates B10581 and B10585 were cultured about 1 year apart from the environment of a hospital and shared an ST. The 16 B. hawaiiensis isolates were separated into 9 different STs. All isolates from a previous Bipolaris endophthalmitis outbreak (5) had identical STs, consistent with the epidemiological conclusion of a point source from a contaminated medication. The three B. australiensis isolates were divided into three unique STs.
DISCUSSION
In November 2013, the CDC was asked to assist in the investigation of a cluster of infections in POCS patients caused by the mold Bipolaris. One hypothesis was that this was a point source outbreak caused by a contaminated product. In addition to an epidemiologic investigation looking for common exposures, such a hypothesis can be investigated by DNA typing of the isolates and looking for genetic homology or heterogeneity. However, no typing system existed for Bipolaris at the time of the outbreak. We designed an MLST system using six distinct loci, all of which were informative. This new typing system was used to provide support against a point source in the POCS cluster and to corroborate the hypothesis of a contaminated product from an older outbreak.
This novel Bipolaris MLST system revealed a high degree of heterogeneity among the B. spicifera isolates. The 39 B. spicifera isolates separated into 37 unique STs. Two isolates collected a year apart at a hospital displayed identical MLST patterns. Together with the genetic stability experiments, this suggests that the MLST loci are stable. We also used this MLST system to analyze 16 B. hawaiiensis isolates which revealed 9 different STs. Although our isolate numbers were low, this MLST system also seemed to work for B. australiensis and B. ellisii; the MLST primers were able to amplify all six genes in both species. The typing system did not work for the species B. cynodontis.
Using the new MLST system, we were able to confirm that the B. spicifera isolates from the Bipolaris POCS investigation were highly heterogeneous with no identical patterns among isolates from the case patients. This strongly supported the epidemiological data suggesting that the source was likely not a single-source contaminated product but rather that the likely sources of the infections were environmental. In contrast, the B. hawaiiensis isolates implicated in the 2012 endophthalmitis outbreak were clonal to the extent that was shown with this typing system, which confirms the epidemiological data suggesting that the mode of transmission was likely due to contaminated triamcinolone steroid injected into the eyes of patients (5).
Although fungal outbreaks are relatively uncommon, there have been a number of high-profile fungal-related outbreaks in recent years (27). Fungal typing systems were not available for the majority of these clusters, and typing was performed in most cases after the epidemiologic investigation was completed. We show here that MLST high-resolution typing systems can be developed when only a minimum number of sequences are available for the species of interest. With the increasing number of fungal whole genomes becoming available for loci identification, fungal MLST systems can be developed quickly for immediate use in public health investigations. While whole genomes or a next-generation typing system like NGMLST (28) will likely be the typing tools of the future, the comparative speed and cost of MLST make it a welcome alternative for current fungal epidemiologic investigations.
ACKNOWLEDGMENTS
We thank Joyce Peterson, Carol Bolden, and Ngoc Le of the Fungal Reference Laboratory at the CDC, the staff at hospitals involved in the recent cluster and staff at Texas Department of State Health Services, and the Dallas, Tarrant, and Bell county health departments for their contributions to this investigation. We also thank Snigdha Vallabhaneni, Jordan Peart, Alex Kallen, Gary Heseltine, Wendy Chung, Karen Brust, Charlotte Wheeler, Emily Hall, Kaitlin Benedict, Ulzii Luvsansharav, Sekai Chideya, Alison Halpin, and Benjamin Park for their role in the outbreak and cluster investigation.
The findings and conclusions of this article are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
REFERENCES
- 1.Manamgoda DS, Rossman AY, Castlebury LA, Crous PW, Madrid H, Chukeatirote E, Hyde KD. 2014. The genus Bipolaris. Stud Mycol 79:221–288. doi: 10.1016/j.simyco.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandt ME, Warnock DW. 2003. Epidemiology, clinical manifestations, and therapy of infections caused by dematiaceous fungi. J Chemother 15(Suppl 2):S36–S47. [DOI] [PubMed] [Google Scholar]
- 3.da Cunha KC, Sutton DA, Fothergill AW, Cano J, Gene J, Madrid H, De Hoog S, Crous PW, Guarro J. 2012. Diversity of Bipolaris species in clinical samples in the United States and their antifungal susceptibility profiles. J Clin Microbiol 50:4061–4066. doi: 10.1128/JCM.01965-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yew SM, Chan CL, Lee KW, Na SL, Tan R, Hoh CC, Yee WY, Ngeow YF, Ng KP. 2014. A five-year survey of dematiaceous fungi in a tropical hospital reveals potential opportunistic species. PLoS One 9:e104352. doi: 10.1371/journal.pone.0104352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mikosz CA, Smith RM, Kim M, Tyson C, Lee EH, Adams E, Straif-Bourgeois S, Sowadsky R, Arroyo S, Grant-Greene Y, Duran J, Vasquez Y, Robinson BF, Harris JR, Lockhart SR, Torok TJ, Mascola L, Park BJ. 2014. Fungal endophthalmitis associated with compounded products. Emerg Infect Dis 20:248–256. doi: 10.3201/eid2002.131257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pauzner R, Goldschmied-Reouven A, Hay I, Vered Z, Ziskind Z, Hassin N, Farfel Z. 1997. Phaeohyphomycosis following cardiac surgery: case report and review of serious infection due to Bipolaris and Exserohilum species. Clin Infect Dis 25:921–923. doi: 10.1086/597638. [DOI] [PubMed] [Google Scholar]
- 7.Teran CG, Downes K, Medows M. 2014. Fatal Bipolaris spicifera infection in an immunosuppressed child. BMJ Case Rep 2014. doi: 10.1136/bcr-2013-009703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci U S A 109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Irinyi L, Serena C, Garcia-Hermoso D, Arabatzis M, Desnos-Ollivier M, Vu D, Cardinali G, Arthur I, Normand AC, Giraldo A, da Cunha KC, Sandoval-Denis M, Hendrickx M, Nishikaku AS, de Azevedo Melo AS, Merseguel KB, Khan A, Parente Rocha JA, Sampaio P, da Silva Briones MR, e Ferreira RC, de Medeiros Muniz M, Castanon-Olivares LR, Estrada-Barcenas D, Cassagne C, Mary C, Duan SY, Kong F, Sun AY, Zeng X, Zhao Z, Gantois N, Botterel F, Robbertse B, Schoch C, Gams W, Ellis D, Halliday C, Chen S, Sorrell TC, Piarroux R, Colombo AL, Pais C, de Hoog S, Zancope-Oliveira RM, Taylor ML, Toriello C, de Almeida Soares CM, et al. 2015. International Society of Human and Animal Mycology (ISHAM)-ITS reference DNA barcoding database-the quality controlled standard tool for routine identification of human and animal pathogenic fungi. Med Mycol 53:313–337. doi: 10.1093/mmy/myv008. [DOI] [PubMed] [Google Scholar]
- 10.Schubert K, Groenewald JZ, Braun U, Dijksterhuis J, Starink M, Hill CF, Zalar P, de Hoog GS, Crous PW. 2007. Biodiversity in the Cladosporium herbarum complex (Davidiellaceae, Capnodiales), with standardisation of methods for Cladosporium taxonomy and diagnostics. Stud Mycol 58:105–156. doi: 10.3114/sim.2007.58.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Donnell K, Sutton DA, Rinaldi MG, Sarver BA, Balajee SA, Schroers HJ, Summerbell RC, Robert VA, Crous PW, Zhang N, Aoki T, Jung K, Park J, Lee YH, Kang S, Park B, Geiser DM. 2010. Internet-accessible DNA sequence database for identifying fusaria from human and animal infections. J Clin Microbiol 48:3708–3718. doi: 10.1128/JCM.00989-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tavanti A, Gow NA, Senesi S, Maiden MC, Odds FC. 2003. Optimization and validation of multilocus sequence typing for Candida albicans. J Clin Microbiol 41:3765–3776. doi: 10.1128/JCM.41.8.3765-3776.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dodgson AR, Pujol C, Denning DW, Soll DR, Fox AJ. 2003. Multilocus sequence typing of Candida glabrata reveals geographically enriched clades. J Clin Microbiol 41:5709–5717. doi: 10.1128/JCM.41.12.5709-5717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tavanti A, Davidson AD, Johnson EM, Maiden MC, Shaw DJ, Gow NA, Odds FC. 2005. Multilocus sequence typing for differentiation of strains of Candida tropicalis. J Clin Microbiol 43:5593–5600. doi: 10.1128/JCM.43.11.5593-5600.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bain JM, Tavanti A, Davidson AD, Jacobsen MD, Shaw D, Gow NA, Odds FC. 2007. Multilocus sequence typing of the pathogenic fungus Aspergillus fumigatus. J Clin Microbiol 45:1469–1477. doi: 10.1128/JCM.00064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer W, Aanensen DM, Boekhout T, Cogliati M, Diaz MR, Esposto MC, Fisher M, Gilgado F, Hagen F, Kaocharoen S, Litvintseva AP, Mitchell TG, Simwami SP, Trilles L, Viviani MA, Kwon-Chung J. 2009. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med Mycol 47:561–570. doi: 10.1080/13693780902953886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Debourgogne A, Gueidan C, de Hoog S, Lozniewski A, Machouart M. 2012. Comparison of two DNA sequence-based typing schemes for the Fusarium solani species complex and proposal of a new consensus method. J Microbiol Methods 91:65–72. doi: 10.1016/j.mimet.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 18.O'Donnell K, Humber RA, Geiser DM, Kang S, Park B, Robert VA, Crous PW, Johnston PR, Aoki T, Rooney AP, Rehner SA. 2012. Phylogenetic diversity of insecticolous fusaria inferred from multilocus DNA sequence data and their molecular identification via FUSARIUM-ID and Fusarium MLST. Mycologia 104:427–445. doi: 10.3852/11-179. [DOI] [PubMed] [Google Scholar]
- 19.Smith RM, Schaefer MK, Kainer MA, Wise M, Finks J, Duwve J, Fontaine E, Chu A, Carothers B, Reilly A, Fiedler J, Wiese AD, Feaster C, Gibson L, Griese S, Purfield A, Cleveland AA, Benedict K, Harris JR, Brandt ME, Blau D, Jernigan J, Weber JT, Park BJ. 2013. Fungal infections associated with contaminated methylprednisolone injections. N Engl J Med 369:1598–1609. doi: 10.1056/NEJMoa1213978. [DOI] [PubMed] [Google Scholar]
- 20.Lockhart SR, Pham CD, Gade L, Iqbal N, Scheel CM, Cleveland AA, Whitney AM, Noble-Wang J, Chiller TM, Park BJ, Litvintseva AP, Brandt ME. 2013. Preliminary laboratory report of fungal infections associated with contaminated methylprednisolone injections. J Clin Microbiol 51:2654–2661. doi: 10.1128/JCM.01000-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gade L, Scheel CM, Pham CD, Lindsley MD, Iqbal N, Cleveland AA, Whitney AM, Lockhart SR, Brandt ME, Litvintseva AP. 2013. Detection of fungal DNA in human body fluids and tissues during a multistate outbreak of fungal meningitis and other infections. Eukaryot Cell 12:677–683. doi: 10.1128/EC.00046-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pham CD, Bolden CB, Kuykendall RJ, Lockhart SR. 2014. Development of a Luminex-based multiplex assay for detection of mutations conferring resistance to echinocandins in Candida glabrata. J Clin Microbiol 52:790–795. doi: 10.1128/JCM.03378-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimizu K, Tanaka C, Peng YL, Tsuda M. 1998. Phylogeny of Bipolaris inferred from nucleotide sequences of Brn1, a reductase gene involved in melanin biosynthesis. J Gen Appl Microbiol 44:251–258. doi: 10.2323/jgam.44.251. [DOI] [PubMed] [Google Scholar]
- 24.Saitoh Y, Izumitsu K, Morita A, Shimizu K, Tanaka C. 2012. Cloning of SalI, a scytalone dehydratase gene involved in melanin biosynthesis in Cochliobolus heterostrophus. Mycoscience 53:330–334. doi: 10.1007/S10267-011-0162-Z. [DOI] [Google Scholar]
- 25.Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol 26:2465–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Begerow D, John B, Oberwinkler F. 2004. Evolutionary relationships among beta-tubulin gene sequences of basidiomycetous fungi. Mycol Res 108:1257–1263. doi: 10.1017/S0953756204001066. [DOI] [PubMed] [Google Scholar]
- 27.Litvintseva AP, Brandt ME, Mody RK, Lockhart SR. 2015. Investigating fungal outbreaks in the 21st century. PLoS Pathog 11:e1004808. doi: 10.1371/journal.ppat.1004808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y, Frazzitta AE, Litvintseva AP, Fang C, Mitchell TG, Springer DJ, Ding Y, Yuan G, Perfect JR. 2015. Next generation multilocus sequence typing (NGMLST) and the analytical software program MLSTEZ enable efficient, cost-effective, high-throughput, multilocus sequencing typing. Fungal Genet Biol 75:64–71. doi: 10.1016/j.fgb.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]