Abstract
Chronic progressive external ophthalmoplegia (CPEO) is common in mitochondrial disorders and is frequently associated with multiple mtDNA deletions. The onset is typically in adulthood, and affected subjects can also present with general muscle weakness. The underlying genetic defects comprise autosomal-dominant or recessive mutations in several nuclear genes, most of which play a role in mtDNA replication. Next-generation sequencing led to the identification of compound-heterozygous RNASEH1 mutations in two singleton subjects and a homozygous mutation in four siblings. RNASEH1, encoding ribonuclease H1 (RNase H1), is an endonuclease that is present in both the nucleus and mitochondria and digests the RNA component of RNA-DNA hybrids. Unlike mitochondria, the nucleus harbors a second ribonuclease (RNase H2). All affected individuals first presented with CPEO and exercise intolerance in their twenties, and these were followed by muscle weakness, dysphagia, and spino-cerebellar signs with impaired gait coordination, dysmetria, and dysarthria. Ragged-red and cytochrome c oxidase (COX)-negative fibers, together with impaired activity of various mitochondrial respiratory chain complexes, were observed in muscle biopsies of affected subjects. Western blot analysis showed the virtual absence of RNase H1 in total lysate from mutant fibroblasts. By an in vitro assay, we demonstrated that altered RNase H1 has a reduced capability to remove the RNA from RNA-DNA hybrids, confirming their pathogenic role. Given that an increasing amount of evidence indicates the presence of RNA primers during mtDNA replication, this result might also explain the accumulation of mtDNA deletions and underscores the importance of RNase H1 for mtDNA maintenance.
Main Text
Accumulation of multiple mtDNA deletions (ΔmtDNAs) in skeletal muscle and the brain is the molecular hallmark of a group of autosomal-recessive or -dominant mitochondrial encephalomyopathies, characterized by chronic progressive external ophthalmoplegia (CPEO), caused by nuclear genetic mutations affecting the integrity of the mitochondrial genome. This condition is part of a spectrum of clinically and genetically heterogeneous disorders of intergenomic communication; this spectrum also includes mtDNA depletion syndromes. An increasing number of genes, encoding components of the mtDNA replisome, mitochondrial deoxynucleotide supply, mitochondrial dynamics and quality control, and also proteins of still-unknown function,1,2 have been associated with this category of mitochondrial disorders. Recently, mutations in DNA2 (MIM: 601810)3 and MGME1 (MIM: 615076),4 encoding exonucleases involved in mtDNA repair or maturation, have been described in subjects presenting with CPEO associated with ΔmtDNA (CPEO/ΔmtDNA). We show here that mutations in RNASEH1 (MIM: 604123), encoding an endonuclease specific to RNA-DNA double-stranded hybrids,5 cause the same syndrome.
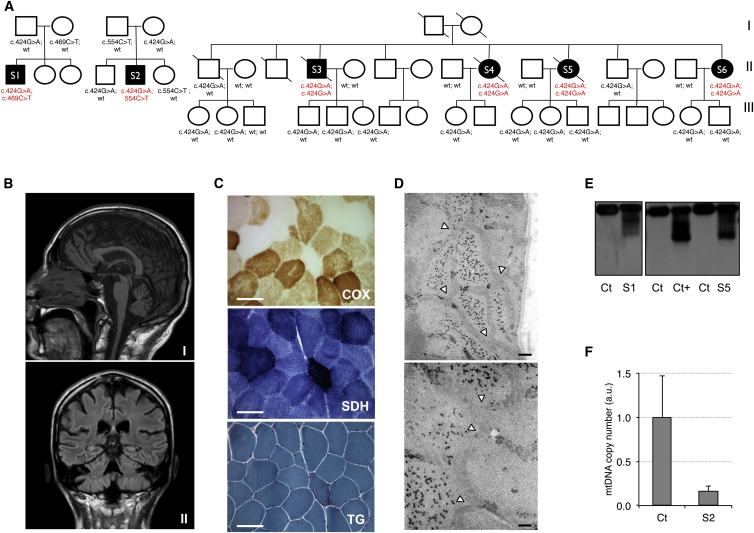
We studied two singleton subjects (S1 and S2) and four affected siblings (S3–S6) (Figure 1A). S1 and S2 developed juvenile-onset CPEO, muscle weakness and wasting, peripheral sensory-motor neuropathy, progressive spino-cerebellar ataxia, and pyramidal signs. Involvement of respiratory muscles explains the severe nocturnal dyspnea and orthopnoea in S1. CPEO occurred later in siblings S3–S6, but their symptoms and progression were similar and included CPEO, dysphagia, and respiratory impairment. S3, S4, and S5 died at 60, 70, and 63 years of age, respectively; S6 is now 65 years old, wheelchair bound, fed by a percutaneous-endoscopic-gastrostomy tube, and under non-invasive ventilation. All affected individuals showed cerebellar involvement, secondary to MRI-proven cerebellar atrophy (Figure 1B). In addition to having CPEO, affected subjects displayed a number of peculiar features, e.g., exquisite involvement of respiratory muscles and progressive ataxia due to severe involvement of the spino-cerebellar pathways and cerebellar atrophy, which could be useful clues for diagnosis. Muscle biopsies of all six subjects showed numerous ragged-red, intensely succinate dehydrogenase (SDH)-positive, COX-negative fibers (Figure 1C). Electron microscopy on muscle from S2 showed extremely elongated, hyperfused mitochondria (Figure 1D). Spectrophotometric analysis of muscle homogenates from S1 and S2 and myoblasts from S2 showed partially reduced complex I and IV activities, whereas values were normal in fibroblasts from S1. Detailed clinical and biochemical descriptions of the affected individuals are reported as supplemental case reports (see Supplemental Data). Informed consent for participation in this study was obtained from all investigated subjects in agreement with the Declaration of Helsinki, and the study was approved by the ethical committees of the centers where biological samples were obtained. Southern blot analysis showed multiple ΔmtDNAs in muscle from S1–S4 and S6 (no muscle DNA was available for S5) (Figure 1E). The mtDNA copy number was unchanged (105% in S2) or increased (300% in S1) in skeletal muscle, whereas significant mtDNA depletion (16%) was detected by qPCR in myoblasts from S2 (Figure 1F).
Figure 1.
Clinical, Morphological, and Genetic Features of Individuals with Mutations in RNASEH1
(A) Pedigrees of individuals with mutations in RNASEH1. Black symbols designate affected subjects, who are numbered according to the main text. The mutation status of each analyzed family member is indicated and is based on the NCBI RefSeq RNASEH1 transcript.
(B) Brain MRI in S1. T1 sagittal (I) and cross (II) sections show cerebellar atrophy and mild bulbar atrophy in S1.
(C) Histological findings in muscle biopsy from S2. Abbreviations are as follows: COX, cytochrome c oxidase; SDH, succinate dehydrogenase; Gomori Trichrome, GT. GT staining shows one ragged-red COX-negative and SDH-positive fiber and several COX-negative fibers. Scale bars represent 25μm.
(D) Electron microscopy with a seamless mitochondrial network intermingled among myofibrils in a muscle biopsy from S2. Arrowheads indicate ramification of the mitochondrial syncytium. Scale bars represent 0.25 μm in the upper panel and 0.10 μm in the lower panel.
(E) Southern blot analysis of mtDNA obtained from muscle biopsies of affected individuals S1 and S5, of control subjects (Ct), and of an individual with POLG1 mutations (Ct+).
(F) mtDNA levels assessed by qPCR in myoblasts from S2 and three control subjects (Ct). The mean value of the mtDNA/nDNA ratio obtained in control subjects was set as 1. Error bars represent SDs.
In order to identify the underlying genetic cause in our probands, we first ruled out several genes known to be associated with CPEO/ΔmtDNA syndromes by Sanger sequencing. We then performed whole-exome sequencing on DNA from S1; after filtering (Tables S1 and S2), we searched for homozygous or compound-heterozygous variants that followed a recessive inheritance pattern according to the pedigree of S1’s family. From selected previously unreported or rare variants in genes encoding known or predicted mitochondrial proteins, we zeroed in on two changes in RNASEH1 (GenBank: NM_002936.4), c.424G>A (p.Val142Ile) on the paternal allele and a nonsense mutation, c.469C>T (p.Arg157∗), on the maternal allele. Next, we screened RNASEH1 in a cohort of 40 genetically undefined CPEO/ΔmtDNA-affected individuals and identified compound-heterozygous mutations c.424G>A (p.Val142Ile) and c.554C>T (p.Ala185Val) in S2. S3 was homozygous for the same variant, c.424G>A, found in S1 and S2; this mutation segregated with the disease, given that it was homozygous in affected family members (Figure 1A). All the identified RNASEH1 variants were absent or extremely rare (<0.01%) in the Exome Aggregation Consortium (ExAC) Browser (Table S3).
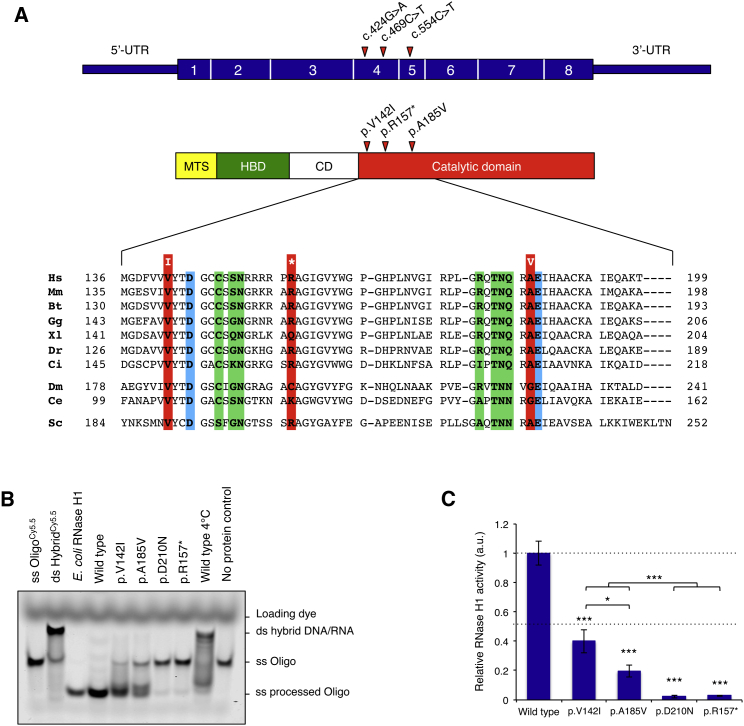
All amino acid changes map to the first third of the RNase H1 catalytic domain (Figure 2A). p.Arg157∗ affects the 22nd residue of the catalytic domain, leading to the synthesis of a truncated protein lacking catalytic activity, as proven by in vitro assay (see below). Substitutions p.Val142Ile and p.Ala185Val affect amino acids that are close to conserved residues, are part of the active site, or interact with RNA or DNA strands (Figure 2A).
Figure 2.
Characterization of the Structure and Enzymatic Activity of RNaseH1 Variants
(A) Human RNaseH1 (top) consists of four domains: a mitochondrial targeting sequence (MTS) that directs the protein to mitochondria and is cleaved after import, a hybrid binding domain (HBD) involved in recognition of DNA-RNA heteroduplexes, a catalytic domain responsible for the cleavage of the RNA component in the heteroduplex, and a flexible connection domain (CD) that links the last two domains. Phylogenetic alignment of the human protein region containing the substitutions found in affected individuals is shown below. Conserved residues that are altered in affected subjects are boxed in red and are found near conserved residues in the active site or interacting with DNA or RNA in the heteroduplex; these latter residues are boxed in blue or green, respectively. Abbreviations are as follows: Hs, H. sapiens; Mm, M. musculus; Bt, B. taurus; Gg, G. gallus; Xl, X. laevis; Dr, D. rerio; Ci, C. intestinalis; Dm, D. melanogaster; Ce, C. elegans; Sc, S. cerevisiae.
(B) RNase H1 activity of recombinant proteins. The portion corresponding to the mature protein, lacking the mitochondrial targeting signal (MTS), was cloned into a pET-28a bacterial expression vector (Novagen) containing an N-terminal His6 tag. Protein production was carried out in BL21(DE3)pLysS E. coli cells at 37°C in lysogeny broth medium. Proteins were loaded onto a Ni-NTA agarose column and eluted with 400 mM imidazole. 80 nmol of DNA-RNA heteroduplex oligonucleodies were incubated with E. coli RNase H1 or either wild-type or altered recombinant proteins for 1 hr.6 Reaction products were loaded onto 15% polyacrylamide gels, and gel imaging and quantification followed.
(C) Relative RNase H1 activity was obtained by quantification of the amount of processed oligonucleotide normalized by protein loading (based on western blotting; Figure S1C) and relative to wild-type protein. ∗p < 0.05, ∗∗∗p < 0.001 (two-tailed unpaired Student’s t test). n = 3 experiments. Error bars represent 1 SD.
RNASEH1 encodes mitochondrial and nuclear protein variants, depending on which initiation AUG codons, encoding Met1 or Met27, are adopted. An upstream AUG, encoding Met0, competes with initiation at Met1 (which begins the mitochondrial variant), so that in physiological conditions, the nuclear form (which starts with Met27) is predominant.7 An N-terminal MTS targets this protein to mitochondria and is eventually cleaved off by the mitochondrial matrix peptidase after translocation, resulting in a protein almost identical to the nuclear form. A second, exclusively nuclear ribonucleotidase affecting RNA-DNA hybrids is RNase H2, alterations in which are responsible for Aicardi-Goutieres syndrome (MIM: 610333, 610181). To circumvent potential cross-contamination and demonstrate the effect of the RNASEH1 mutations on protein activity, we affinity purified wild-type (WT) and altered forms of mature human RNase H1 from overexpressing bacterial strains (Figures 1A and 1B). The in vitro activity of the recombinant proteins (Figure 2B) was assessed on fluorescent-labeled oligonucleotides forming a 6-bp-long DNA-RNA heteroduplex (Figure S1B). Whereas the WT recombinant protein and E. coli RNase H1 were both able to fully cleave the RNA from the heteroduplex, the recombinant p.Val142Ile and p.Ala185Val proteins displayed partial activity and left a significant amount of intact single-stranded and incompletely cleaved oligonucleotides, similar to those obtained by incubating the WT protein with the heteroduplex on ice. Similar to the negative “no-protein” control, recombinant truncated p.Arg157∗ protein showed negligible activity, as did the p.Asp210Asn protein, previously reported as catalytically inactive8 (Figure 2C).
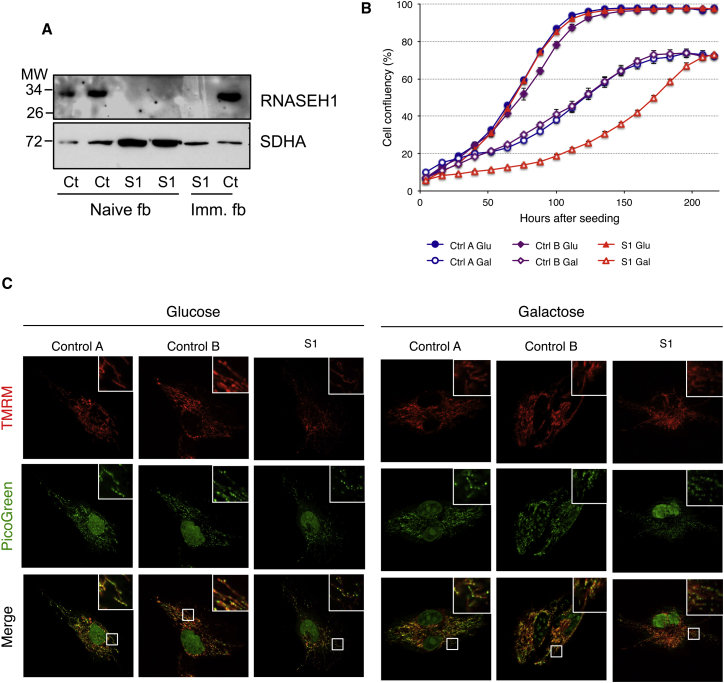
To further investigate the molecular consequences of RNASEH1 mutations, we characterized skin fibroblasts from S1 and two healthy control individuals in permissive glycolytic-prone medium (containing glucose as the sole carbon source) or oxidative phosphorylation (OXPHOS)-obligatory medium (containing galactose instead of glucose). RNASEH1 transcript levels in cells from S1 were decreased to 47% or 36% when grown in glucose or galactose, respectively (Figure S2), most likely as a result of nonsense-mediated mRNA decay of the transcript harboring the premature stop codon. Western blot analysis showed the virtual absence of RNase H1 in total cell lysates from S1 (Figure 3A), suggesting instability of both p.Arg157∗ and p.Val142Ile RNase H1 variants. Whereas glucose-grown cells showed no significant differences, cells from S1 grew more slowly in galactose than control cells did (Figure 3B), suggesting mitochondrial dysfunction. Accordingly, cell-cycle analysis revealed a significant increase in G1 and G0 phases and a decrease in S phases in galactose-grown cells from S1 (Figure S3). To further assess mitochondrial dysfunction, we analyzed mitochondrial morphology and membrane potential (ΔP) on live cells by using tetramethylrhodamine methyl ester (TMRM) and JC-1 immunofluorescence (Figure 3C). Glucose-grown cells from S1 displayed a mitochondrial network similar to that of control cells, albeit the intensity of the staining was significantly lower and thus indicated reduced ΔP (Figures 3C and S4A). Flow-cytometry analysis using JC-1 staining showed a 50% ΔP decrease in cells from S1 (Figure S4B). Whereas galactose-grown control cells showed neither alteration in mitochondrial morphology nor ΔP changes, cells from S1 displayed perinuclear aggregation of fragmented mitochondria (Figure 3C) and higher ΔP (Figure S4B).
Figure 3.
Growth and Mitochondrial Alterations in RNASEH1 Mutant Fibroblasts
(A) Western blot analysis, using antibodies against RNASEH1 (Abcam) and SDHA (Mitosciences-Invitrogen), of fibroblasts (fb) from S1 and control subjects (Ct); SDHA was taken as a loading control. Both naive and immortalized (Imm.) fibroblasts were used.
(B) Growth curves of control cells and RNASEH1 cells from S1 in glucose (high-glucose medium) and galactose (glucose-free medium supplemented with 50 mM galactose). Cell growth was monitored continuously by the IncuCyte live cell imager (Essen Bioscience). Data represent the average ± SD of three independent experiments.
(C) Mitochondria (red) were stained for 20 min with ΔP-dependent dye (20 nM TMRM), and DNA (green) was stained with PicoGreen (3 μl/ml) in live cells grown in glucose or galactose for 5 days. A digitally enhanced TMRM image from RNASEH1 mutant fibroblasts from S1 is shown in Figure S4A.
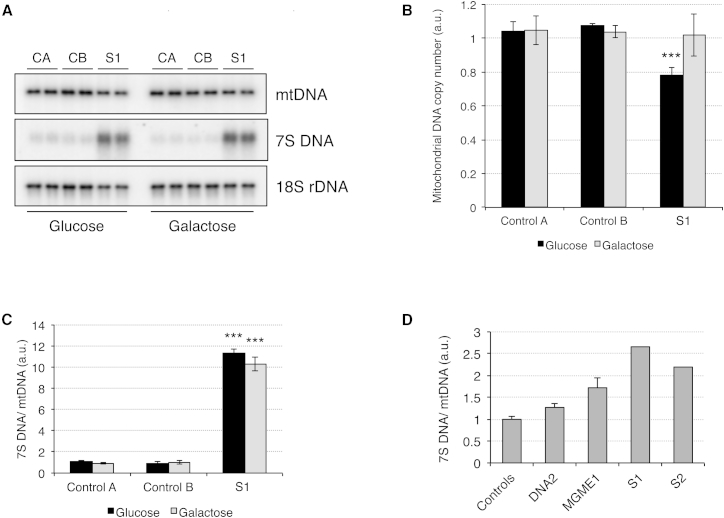
To investigate the effect of RNASEH1 mutations on mtDNA, we first analyzed nucleoid structure in live cells from control individuals and S1 by PicoGreen staining. Glucose-grown fibroblasts from S1 displayed fewer, smaller, and sharper nucleoids (Figures 3C and S5), whereas in galactose-grown cells, these differences were less prominent (Figure 3C), suggesting more active mtDNA metabolism in OXPHOS-obligatory conditions. Because PicoGreen staining is dependent on both the amount and topology of mtDNA,9 and we had observed mtDNA depletion in S2 myoblasts, we further investigated mtDNA copy number by Southern blot analysis. Fibroblasts from S1 showed mild (80% of control mean value) mtDNA depletion only in glucose, and no ΔmtDNA was detected (Figures 4A and 4B). These results were confirmed by qPCR (Figure S6). However, we observed a >10-fold increase in 7S DNA (Figure 4C), the third strand of the mtDNA displacement loop (D-loop); mutant muscle also displayed higher levels of 7S DNA (Figure 4D), suggesting aborted replication.
Figure 4.
Mitochondrial DNA Copy Number and 7S DNA Mutations in RNASEH1 Mutant Fibroblasts from S1and Muscle from S1 and S2
(A) Total DNA from control and RNASEH1 mutant fibroblasts was digested with PvuII, and mtDNA was analyzed by one-dimensional Southern blot. Radioactive probes (able to detect both linearized mtDNA and 7S DNA) specific to the human mtDNA D-loop region (nucleotide positions 16.341–16.151) and 18S ribosomal DNA were applied.
(B) The relative mtDNA copy number (mtDNA/18S rDNA ratio) was obtained after quantification of the Southern blot signal with PhosphorImager screens and normalization to control fibroblasts. ∗∗∗p < 0.001 (two-tailed Student’s t test). n = 2 experiments. Error bars represent 1 SD.
(C) 7S DNA levels (7S DNA/mtDNA ratio) were obtained after quantification of the Southern blot signal with PhosphorImager screens and normalization to control fibroblasts. ∗∗∗p < 0.001 (two-tailed Student’s t test). n = 2 experiments. Error bars represent 1 SD.
(D) 7S DNA levels (7S DNA/mtDNA ratio) in muscle from S1, S2, and subjects with mutations in MGME1 and DNA2 were assessed by qPCR. Error bars represent 1 SD.
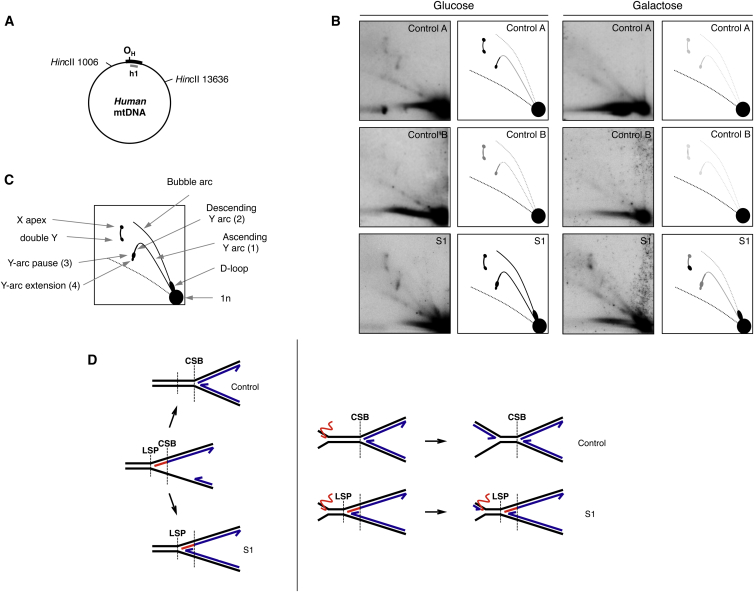
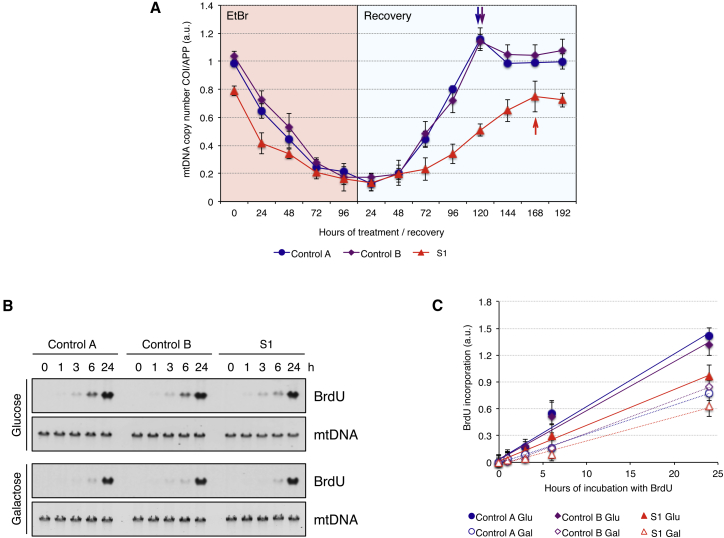
Because these results indicated that RNASEH1 mutations affected mtDNA replication, we studied replication intermediates by two-dimensional gel electrophoresis.10–12 Compared to control cells, glucose-grown fibroblasts from S1 displayed a 2- to 4-fold increased signal (Figures 5A–5C and S7A). New replication intermediates, Y-arc extensions, located at the end of the Y-arc pause (Figures 5B and 5C), were detected in fibroblasts from S1. This extension of the Y-arc might originate as a result of the presence of persisting RNA between the light-strand promoter and the conserved sequence block 1 or 2, which would be removed in control fibroblasts by proficient RNase H1 (Figure 5D).12–14 Fibroblasts from S1 also showed an increase in double-Y intermediates, indicating that replication paused at ribosomal genes, most likely because altered RNase H1 failed to efficiently remove rRNA transcripts (Figures 5B and 5D). A stronger signal in replication intermediates is consistent with replication slowdown, as also reported in MGME1 mutations,4 and HEK cells overexpressing catalytic alterations of the mtDNA helicase or polymerase.15 Galactose-grown cells showed decreased signals of almost all replication intermediates (Figure S7B). Finally, we performed mtDNA depletion and repopulation experiments. Glucose-grown cells exposed to the DNA intercalator EtBr for 96 hr showed a decrease of mtDNA amount to <20% in all cell lines (Figure 6A). Whereas control cells recovered to initial mtDNA levels in 120 hr, cells from S1 required 168 hr, indicating a slower replication rate. Mutations in other genes involved in mtDNA replication such as POLG1 (MIM: 174763), RRM2B (MIM: 604712), and MGME1 have also been associated with slow or virtually no recovery in human fibroblasts.4,16 Active replication was measured as the rate of BrdU incorporation in fibroblasts where nuclear replication was inhibited with aphidicolin (Figure 6B). Fibroblasts from S1 showed 30% less BrdU incorporation than did control cells in both glucose and galactose media, indicating slower mtDNA replication in fibroblasts from S1, most likely due to stalling caused by RNA tracts not being efficiently removed by altered RNase H1.14,17
Figure 5.
Effect of Mutant RNASEH1 Fibroblasts from S1 on mtDNA Replication
(A) Restriction enzyme sites and position of the probe are labeled. The origin of the H-strand replication (OH) is marked within the non-coding region (black bar).
(B) mtDNA replication intermediates analyzed by two-dimensional agarose gel electrophoresis and Southern blot. Explanatory cartoons are provided at the right of each image; shades of gray reflect the different intensities of the signal.
(C) A cartoon based on this and previous work10–14 shows different replication intermediates associated with this fragment and the non-replicating fragment, 1n.
(D) Proposed models that could explain the presence of a Y-arc extension (left) and increased double-Y (right) mtDNA replication intermediates from RNASEH1 mutant fibroblasts from S1. Parental DNA strands are in black, nascent DNA strands are in blue, and RNA is in red. The L-strand promoter (LSP) and conserved sequence block region (CSB) are marked as reference points.
Figure 6.
mtDNA Replication Is Slowed Down in Mutant RNASEH1 Fibroblasts from S1
(A) mtDNA copy number during depletion and recovery in control and RNASEH1 mutant fibroblasts. Depletion was achieved by the addition of 100 ng/ml EtBr to the culture medium for 96 hr, and recovery was followed up for 192 hr after the removal of the drug. mtDNA copy number was determined by qPCR, and each data point represents the mean values from two independent determinations ± SD. Arrowheads indicate the time point at which mtDNA was recovered at or close to initial values for each cell line.
(B) Detection of newly synthesized mtDNA. Cells were treated with 20 μM Aphidicolin (Sigma) for 6 hr to block nuclear DNA replication. Total DNA from control and RNASEH1 mutant fibroblasts subjected to pulse labeling of mtDNA with BrdU (100 μM) for 0, 1, 3, 6, and 24 hr was digested with NaeI and analyzed by one-dimensional southwestern blot. The BrdU signal was immonodetected with anti-BrdU antibody (Becton Dickinson) and was followed by mtDNA detection using a radioactive probe specific to the human mtDNA D-loop region.
(C) Plot of BrdU incorporation (normalized to mtDNA loading) over time; the linear regression for each cell line is shown. Each data point represents the mean of two replicates ± SD.
Our findings show that RNase H1 is required for effective mtDNA replication in humans;18 this has also been suggested by the embryolethality of an Rnaseh1−/− mouse model, which becomes virtually ρ0 by p.c. 8.5.19 Deleterious RNASEH1 mutations slow down and stall mtDNA replication, causing both mtDNA depletion and deletions and ultimately leading to clinically overt mitochondrial disease. Unlike MGME1, which is exclusively mitochondrial, and similar to DNA2, RNase H1 is localized in both the nucleus and mitochondria. Nevertheless, its impairment determines a virtually exclusive mtDNA damage, most likely because of the compensatory role of nuclear RNase H2. All three enzymes are involved in maturation of mtDNA and possibly in its repair, although RNase H1 seems to play a direct important role in mtDNA replication by processing the RNA primers of nascent strands. This could also explain the accumulation of 7S DNA species, given that defective RNase H1 increases the proportion of intermediates with the 7S RNA still attached to the nascent H strand. This kind of molecule is poorly processed by MGME1,4 and that could result in aborted replication. FENMIT is an exclusively mitochondrial isoform of FEN1 and also interacts with RNA-DNA hybrids.20 No mitochondrial diseases have been identified for FENMIT to date, and the degree of overlap with RNase H1 is not known. However, we could speculate that FENMIT could replace RNase H1 in some instances and therefore contribute to the mild phenotype in subjects with RNase H1 alterations. According to the analysis of the replication intermediates by two-dimensional gel electrophoresis, altered RNase H1 is associated with increased pausing of the replication fork. Excessive pausing has been proposed as a mechanism leading to the accumulation of multiple deleted species and impairment of the replisome machinery either through breakage or recombination,21,22 which could also explain partial mtDNA depletion and slower recovery of mtDNA copy number associated with RNASEH1 mutations.
Acknowledgments
We acknowledge the “Cell Lines and DNA Bank of Paediatric Movement Disorders and Neurodegenerative Diseases” of the Telethon Network of Genetic Biobanks (grant GTB12001J) and the EurobiobanK Network. This work was supported by the Medical Research Council, the Pierfranco and Luisa Mariani Foundation, Telethon grant GGP11011, the Italian Ministry of Health (grant GR2010-2316392), and European Research Council advanced grant FP7-322424.
Published: June 18, 2015
Footnotes
Supplemental Data include supplemental case reports, seven figures, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.05.013.
Contributor Information
Daniele Ghezzi, Email: daniele.ghezzi@istituto-besta.it.
Massimo Zeviani, Email: mdz21@mrc-mbu.cam.ac.uk.
Web Resources
The URLs for data presented herein are as follows:
Exome Aggregation Consortium (ExAC) Browser, http://exac.broadinstitute.org/
OMIM, http://www.omim.org/
Supplemental Data
References
- 1.Lamperti C., Zeviani M. Encephalomyopathies caused by abnormal nuclear-mitochondrial intergenomic cross-talk. Acta Myol. 2009;28:2–11. [PMC free article] [PubMed] [Google Scholar]
- 2.Copeland W.C. Defects in mitochondrial DNA replication and human disease. Crit. Rev. Biochem. Mol. Biol. 2012;47:64–74. doi: 10.3109/10409238.2011.632763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ronchi D., Di Fonzo A., Lin W., Bordoni A., Liu C., Fassone E., Pagliarani S., Rizzuti M., Zheng L., Filosto M. Mutations in DNA2 link progressive myopathy to mitochondrial DNA instability. Am. J. Hum. Genet. 2013;92:293–300. doi: 10.1016/j.ajhg.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kornblum C., Nicholls T.J., Haack T.B., Schöler S., Peeva V., Danhauser K., Hallmann K., Zsurka G., Rorbach J., Iuso A. Loss-of-function mutations in MGME1 impair mtDNA replication and cause multisystemic mitochondrial disease. Nat. Genet. 2013;45:214–219. doi: 10.1038/ng.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerritelli S.M., Crouch R.J. Cloning, expression, and mapping of ribonucleases H of human and mouse related to bacterial RNase HI. Genomics. 1998;53:300–307. doi: 10.1006/geno.1998.5497. [DOI] [PubMed] [Google Scholar]
- 6.Alla N.R., Nicholson A.W. Evidence for a dual functional role of a conserved histidine in RNA·DNA heteroduplex cleavage by human RNase H1. FEBS J. 2012;279:4492–4500. doi: 10.1111/febs.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki Y., Holmes J.B., Cerritelli S.M., Sakhuja K., Minczuk M., Holt I.J., Crouch R.J. An upstream open reading frame and the context of the two AUG codons affect the abundance of mitochondrial and nuclear RNase H1. Mol. Cell. Biol. 2010;30:5123–5134. doi: 10.1128/MCB.00619-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu H., Lima W.F., Crooke S.T. Investigating the structure of human RNase H1 by site-directed mutagenesis. J. Biol. Chem. 2001;276:23547–23553. doi: 10.1074/jbc.M009676200. [DOI] [PubMed] [Google Scholar]
- 9.He J., Cooper H.M., Reyes A., Di Re M., Kazak L., Wood S.R., Mao C.C., Fearnley I.M., Walker J.E., Holt I.J. Human C4orf14 interacts with the mitochondrial nucleoid and is involved in the biogenesis of the small mitochondrial ribosomal subunit. Nucleic Acids Res. 2012;40:6097–6108. doi: 10.1093/nar/gks257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reyes A., Yasukawa T., Cluett T.J., Holt I.J. Analysis of mitochondrial DNA by two-dimensional agarose gel electrophoresis. Methods Mol. Biol. 2009;554:15–35. doi: 10.1007/978-1-59745-521-3_2. [DOI] [PubMed] [Google Scholar]
- 11.Di Re M., Sembongi H., He J., Reyes A., Yasukawa T., Martinsson P., Bailey L.J., Goffart S., Boyd-Kirkup J.D., Wong T.S. The accessory subunit of mitochondrial DNA polymerase gamma determines the DNA content of mitochondrial nucleoids in human cultured cells. Nucleic Acids Res. 2009;37:5701–5713. doi: 10.1093/nar/gkp614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holt I.J., Reyes A. Human mitochondrial DNA replication. Cold Spring Harb. Perspect. Biol. 2012;4:a012971. doi: 10.1101/cshperspect.a012971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasukawa T., Reyes A., Cluett T.J., Yang M.Y., Bowmaker M., Jacobs H.T., Holt I.J. Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J. 2006;25:5358–5371. doi: 10.1038/sj.emboj.7601392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reyes A., Kazak L., Wood S.R., Yasukawa T., Jacobs H.T., Holt I.J. Mitochondrial DNA replication proceeds via a ‘bootlace’ mechanism involving the incorporation of processed transcripts. Nucleic Acids Res. 2013;41:5837–5850. doi: 10.1093/nar/gkt196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wanrooij S., Goffart S., Pohjoismäki J.L., Yasukawa T., Spelbrink J.N. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007;35:3238–3251. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stewart J.D., Schoeler S., Sitarz K.S., Horvath R., Hallmann K., Pyle A., Yu-Wai-Man P., Taylor R.W., Samuels D.C., Kunz W.S., Chinnery P.F. POLG mutations cause decreased mitochondrial DNA repopulation rates following induced depletion in human fibroblasts. Biochim. Biophys. Acta. 2011;1812:321–325. doi: 10.1016/j.bbadis.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 17.Pohjoismäki J.L., Holmes J.B., Wood S.R., Yang M.Y., Yasukawa T., Reyes A., Bailey L.J., Cluett T.J., Goffart S., Willcox S. Mammalian mitochondrial DNA replication intermediates are essentially duplex but contain extensive tracts of RNA/DNA hybrid. J. Mol. Biol. 2010;397:1144–1155. doi: 10.1016/j.jmb.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruhanen H., Ushakov K., Yasukawa T. Involvement of DNA ligase III and ribonuclease H1 in mitochondrial DNA replication in cultured human cells. Biochim. Biophys. Acta. 2011;1813:2000–2007. doi: 10.1016/j.bbamcr.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerritelli S.M., Frolova E.G., Feng C., Grinberg A., Love P.E., Crouch R.J. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol. Cell. 2003;11:807–815. doi: 10.1016/s1097-2765(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 20.Kazak L., Reyes A., He J., Wood S.R., Brea-Calvo G., Holen T.T., Holt I.J. A cryptic targeting signal creates a mitochondrial FEN1 isoform with tailed R-Loop binding properties. PLoS ONE. 2013;8:e62340. doi: 10.1371/journal.pone.0062340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bailey L.J., Cluett T.J., Reyes A., Prolla T.A., Poulton J., Leeuwenburgh C., Holt I.J. Mice expressing an error-prone DNA polymerase in mitochondria display elevated replication pausing and chromosomal breakage at fragile sites of mitochondrial DNA. Nucleic Acids Res. 2009;37:2327–2335. doi: 10.1093/nar/gkp091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicholls T.J., Zsurka G., Peeva V., Schöler S., Szczesny R.J., Cysewski D., Reyes A., Kornblum C., Sciacco M., Moggio M. Linear mtDNA fragments and unusual mtDNA rearrangements associated with pathological deficiency of MGME1 exonuclease. Hum. Mol. Genet. 2014;23:6147–6162. doi: 10.1093/hmg/ddu336. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.