Abstract
Purpose
Next-generation sequencing (NGS) based methods are being adopted broadly for genetic diagnostic testing, but the performance characteristics of these techniques have not been fully defined with regard to test accuracy and reproducibility.
Methods
We developed a targeted enrichment and NGS approach for genetic diagnostic testing of patients with inherited eye disorders, including inherited retinal degenerations, optic atrophy and glaucoma. In preparation for providing this Genetic Eye Disease (GEDi) test on a CLIA-certified basis, we performed experiments to measure the sensitivity, specificity, reproducibility as well as the clinical sensitivity of the test.
Results
The GEDi test is highly reproducible and accurate, with sensitivity and specificity for single nucleotide variant detection of 97.9% and 100%, respectively. The sensitivity for variant detection was notably better than the 88.3% achieved by whole exome sequencing (WES) using the same metrics, due to better coverage of targeted genes in the GEDi test compared to commercially available exome capture sets. Prospective testing of 192 patients with IRDs indicated that the clinical sensitivity of the GEDi test is high, with a diagnostic rate of 51%.
Conclusion
The data suggest that based on quantified performance metrics, selective targeted enrichment is preferable to WES for genetic diagnostic testing.
Keywords: Genetic diagnostic testing, next-generation sequencing, sensitivity, specificity, reproducibility
INTRODUCTION
Next-generation sequencing (NGS) based testing methods are increasingly being used for genetic diagnostic testing. This is especially true for genetically heterogeneous disorders, such as inherited retinal degenerations (IRDs), hearing loss, cardiomyopathies, mitochondrial disorders and cancer 1-4. There are multiple advantages to these approaches, including the ability to simultaneously sequence many genes and to quantify allele frequency 1,3,5,6. The Next-generation Sequencing: Standardization of Clinical Testing (Nex-StoCT) workgroup of the CDC and the American College of Medical Genetics and Genomics (ACMG) have issued guidelines for clinical laboratory standards for NGS-based testing methods, which include determination of test accuracy, analytical sensitivity and specificity, reproducibility and repeatability 7,8. Defining these test characteristics is important, and despite the increased use of NGS-based tests in Clinical Laboratory Improvement Amendments (CLIA) and/or College of American Pathologists (CAP) certified laboratories 3,6,9, the sensitivity, specificity and reproducibility of these techniques have been defined for only a small subset of tests 1,5,10.
Inherited eye disorders are important causes of vision loss. IRDs are among the most common causes of blindness in working age people 11, and glaucoma is a leading cause of irreversible blindness worldwide 12,13. Genetic diagnostic testing for these disorders is challenging due to their genetic heterogeneity. For example, mutations in over 200 genes can cause IRDs, and multiple genes are known to underlie inherited forms of glaucoma and optic atrophy (complete gene list available at: https://sph.uth.edu/retnet/) 14-16. It is increasingly desirable to obtain genetic diagnoses for patients with these disorders, as this information can influence patient care by both informing genetic risk assessment and identifying patients who would benefit from novel gene-based therapies 17-21.
Recently, several groups have reported the use of NGS techniques for genetic diagnostic testing of patients with IRDs 22-26. These reports demonstrate that NGS combined with targeted enrichment approaches is a superior method for genetic diagnostic testing for patients with IRDs that improved diagnostic rates and reduced cost compared to traditional sequencing methods. In the reports published to date, however, the accuracy, sensitivity and specificity, reproducibility and repeatability of the NGS approaches used have not been completely defined.
We developed a targeted enrichment and NGS approach for genetic diagnostic testing of patients with inherited eye disorders, including IRDs, optic atrophy and glaucoma. In preparation for providing this Genetic Eye Disease (GEDi) test on a CLIA-certified basis, we sought to determine the accuracy, sensitivity and specificity, reproducibility and repeatability and clinical sensitivity of this testing approach compared with whole exome sequencing (WES).
MATERIALS and METHODS
Patient Samples
The clinical study was approved by the institutional review boards of the University of Pennsylvania and the Massachusetts Eye and Ear Infirmary, and conformed to the tenets of the Declaration of Helsinki. Informed consent was obtained from all participants, who were recruited after having been identified to have a form of inherited retinal degeneration following clincial evaluation by EAP or ABF at Mass Eye and Ear or Children's Hospital Boston, respectively. Genomic DNA (gDNA) was extracted from patient blood using the PreAnalytiX (QIAGEN / BD Biosciences; Valencia, CA) PAXgene Blood DNA Kit (PAXgene Blood DNA Kit Handbook, 10/2009) or DNAzol (Life Technologies, Carlsbad, CA).
Targeted Enrichment
i. Targeted Enrichment Bait Library Design
The custom SureSelect targeted enrichment GEDi capture kit (Agilent Technologies, Inc., Santa Clara, CA) was designed to capture and enrich coding exons, 5'-/3'-UTRs, and select deep intronic regions known to harbor pathogenic mutations, associated with the 214 known IRD disease genes described in the Retinal Information Network database (RetNet; https://sph.uth.edu/Retnet/) up to April 2013, as well as 8 early-onset glaucoma and optic atrophy genes, using Agilent Technologies’ eArray web design tool (https://earray.chem.agilent.com/earray/). The GEDi capture kit also includes 24 candidate IRD disease genes, 9 age-related macular degeneration risk factor genes, and 1 non-syndromic hearing loss gene. Additional information regarding the parameters used for GEDi capture kit design is available in Supplemental Materials. The custom mitochondrial genome targeted enrichment baits we designed previously were also included as part of the GEDi capture kit 27. Complete lists of the GEDi targeted genes and intronic regions are shown in Tables S1a and S1b, respectively.
ii. Capture Library Sample Preparation
Illumina-compatible paired-end/multiplexable GEDi targeted enrichment capture libraries were generated as described, using the following parameters: a) no less than 1.5 mcg of sheared gDNA was used for pre-capture library generation; b) 5-cycles of pre-capture PCR were used for all samples; c) no less than 400 ng of pre-capture library was used during bait hybridizations; d) 14-cycles of post-capture PCR were used to generate all capture libraries; e) all samples were post-capture indexed; and f) sample multiplex ratios were determined based on post-capture indexed sample concentrations (Agilent methods, part no.:G7530-90000; Protocol v2.1, May 2011).
NGS Analysis
GEDi targeted enrichment sample sequencing was performed on an Illumina MiSeq NGS platform (Illumina, Inc., San Diego, CA). A 12X patient sample multiplex was clustered to an average cluster density of between 750 - 900 K clusters per mm2 and 121 × 6 × 121 bp indexed/paired-end analyzed using Illumina's 300 cycle MiSeq Reagent Kit V2.
Whole Exome Sequencing
Whole exome capture and sequencing were performed as described in Supplementary Methods.
Informatics Analyses
Analysis of the sequence data obtained was performed using a combination of publically available and custom software tools, as described 28. Briefly, BWA (version 0.6.2-r126) was used to align the sequence reads to the human reference genome used by the 1000 Genomes Project . SAMtools (version 0.1.18 or r982:295) was used to remove potential duplicates, and make initial SNP and indel calls, which were refined using a custom program 28. A coverage depth cutoff of 10X was applied. Resulting variant calls were annotated using our custom human bp codon resource 28. Custom scripts were also developed and used to identify candidate variants that fit different filtering criteria, such as genetic models. Variants that fit the appropriate inheritance patterns, and were rare based on data from the 1000 Genomes Project, the NHLBI Exome Sequencing Project Exome Variant Server, and our own internal controls were considered to be potentially pathogenic. See the Supplementary Methods section for addititional information.
RESULTS
GEDi Test Design
Probes were designed for 257 genes targeted by the GEDi selective capture system as well as the mitochondrial genome, since retinal degeneration and optic atrophy can accompany mitochondrial disease 27. Probes for previously identified deep intronic mutations in CEP290, OFD1, and USH2A have also been included in the GEDi probe set 29-31. The targeted regions constitute 1,210,190 bp in total (703,980 bp coding sequence), and are listed in Table S1.
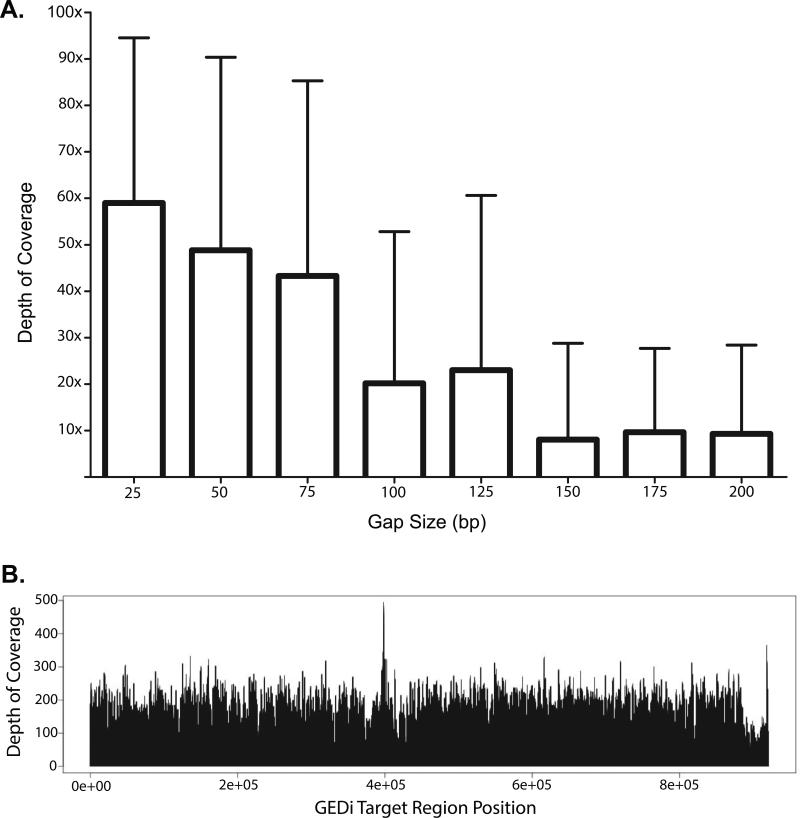
Probes for some of the targeted regions could not be designed due to the presence of repetitive or non-unique sequence elements. In total, there were 688 such design gaps ranging from 1-2,031 bp in length with an average length of 112 bp, accounting for a total of 76,980 bp (9,220 bp coding sequence). Analysis of empiric GEDi data shows that design gaps ≤ 75 bp (67% of gaps) were relatively well covered by “near-target” capture (Figure 1A).
Figure 1.
(A) Analysis of empiric GEDi data shows that design gaps ≤ 75 bp were relatively well covered by “near-target” capture. (B) Representative Depth-of-Coverage (DoC) plot for a 12x-multiplexed capture sample using the GEDi targeted enrichment kit and 2 × 121 bp paired-end sequenced using an Illumina MiSeq.
NGS Metrics
Figure 1B shows a representative Depth-of-Coverage (DoC) plot for a 12x-multiplexed sample captured using the GEDi targeted enrichment kit and sequenced using an Illumina MiSeq. The data shows relatively uniform coverage of the target regions. The average percentages of the target regions covered at 1X (99.8%), 10X (98.6%) and 20X (96.4%) DoC were also relatively constant for all of the sequencing analyses. The 1.4% of target regions which were not covered with ≥ 10X read depth included part or all of 14 exons, The overall average DoC for all samples analyzed was 98.8X ± 14.5X.
Test Performance Metrics
The Nex-StoCT and ACMG recommend that validation of an NGS-based diagnostic test include performance test characteristics for assay accuracy, analytical sensitivity and specificity, reproducibility and repeatability 7,8. To measure these parameters for the GEDi capture and sequencing test, 4 samples (three randomly selected patient samples and the NA12878 HapMap sample) were prepared and sequenced in triplicate on each of three separate days. We also performed WES and SNP array genotyping analyses of these 4 samples using Agilent V4+UTR whole exome enrichment kit and Illumina Omni 2.5 SNP arrays, respectively (see Supplemental Methods). The HapMap sample was included as an internal control for establishing QC metrics, and is included in all diagnostic runs to evaluate each diagnostic capture and sequencing run.
Sensitivity and Specificity
To assess the sensitivity and specificity of the GEDi test, we used the 2,443 SNPs located in GEDi genes that are represented on Omni 2.5 SNP array, using the Omni 2.5 data as the “gold standard.” For these analyses, sensitivity was calculated as the ability of the GEDi test to correctly identify a SNP when it was identified in the Omni 2.5 data. Similarly, the specificity was calculated as the ability of the GEDi test to correctly identify the lack of a variant at a given position when reference was detected by the Omni 2.5 array 5 (Table 1). For example, 495 ± 1 SNPs identified in the 9 GEDi replicates for the OGI-132-357 sample (range 492-497) were also identified in the Omni 2.5 data, and these were scored as true positives (Table 1). The GEDi test did not identify variants at 10 ± 1.4 positions where variants were identified in the Omni 2.5 data for OGI-132-357, and these were scored as false negatives, giving a sensitivity of 0.98 for variant detection. The GEDi test did not identify variants at any of the 1,919 SNPs with reference genotypes in the Omni 2.5 data, for a specificity of 1 (Table 1). The average sensitivity of the GEDi test, including data from the 9 replicates of all 4 samples, was 0.979 ± 0.007, and the specificity was 1 ± 0.
Table 1.
Sensitivity and specificity calculations for GEDi vs. Omni 2.5 SNP data.
| 2 × 2 Contingency Table | ||
|---|---|---|
| Omni + (SNP) | Omni - (REF) | |
| GEDi + (SNP) | True Positive | False Positive |
| GEDi - (REF) | False Negative | True Negative |
| Sensitivity = True Pos/(True Pos + False Neg) | Specificity = True Neg/(True Neg + False Pos) | |
| NA12878 | ||
|---|---|---|
| Omni + (SNP) | Omni - (REF) | |
| GEDi + (SNP) | 508 (503-510 SD:2) | 0 (0-0 SD:0) |
| GEDi - (REF) | 12 (8-20 SD:4) | 1933 (1919-1944 SD:13) |
| Sensitivity: 0.977 (0.967-0.981 SD:0.004) | Specificity: 1 (1-1 SD:0) | |
| OGI-281-608 | ||
|---|---|---|
| Omni + (SNP) | Omni - (REF) | |
| GEDi + (SNP) | 469 (462-472 SD:3) | 0 (0-0 SD:0) |
| GEDi - (REF) | 13 (10-20 SD:3) | 1944 (1944-1944 SD:0) |
| Sensitivity: 0.973 (0.959-0.979 SD:0.007) | Specificity: 1 (1-1 SD:0) | |
| OGI-132-357 | ||
|---|---|---|
| Omni + (SNP) | Omni - (REF) | |
| GEDi + (SNP) | 495 (492-497 SD:1) | 0 (0-0 SD:0) |
| GEDi - (REF) | 10 (8-13 SD:1.4) | 1919 (1919-1919 SD:0) |
| Sensitivity: 0.981 (0.974-0.984 SD:0.003) | Specificity: 1 (1-1 SD:0) | |
| OGI-307-717 | ||
|---|---|---|
| Omni + (SNP) | Omni - (REF) | |
| GEDi + (SNP) | 508 (506-511 SD:1.716) | 0 (0-0 SD:0) |
| GEDi - (REF) | 7 (4-9 SD:2) | 1917 (1917-1917 SD:0) |
| Sensitivity: 0.986 (0.983-0.992 SD:0.003) | Specificity: 1 (1-1 SD:0) | |
For each DNA sample, the number of positions at which variants (SNP) or reference (REF) were detected by the Omni 2.5 SNP arrays and GEDi test are indicated. For the GEDi data, the ranges derived from the 9 replicates for each sample tested are shown. The average sensitivity and specificity for each DNA sample are shown, with the ranges and standard deviation (SD) included in parentheses. The overall sensitivity and specificity reported in the text are the averages of these data for all 4 samples. A standard 2×2 contingency table with definitions of sensitivity and specificity is shown for reference.
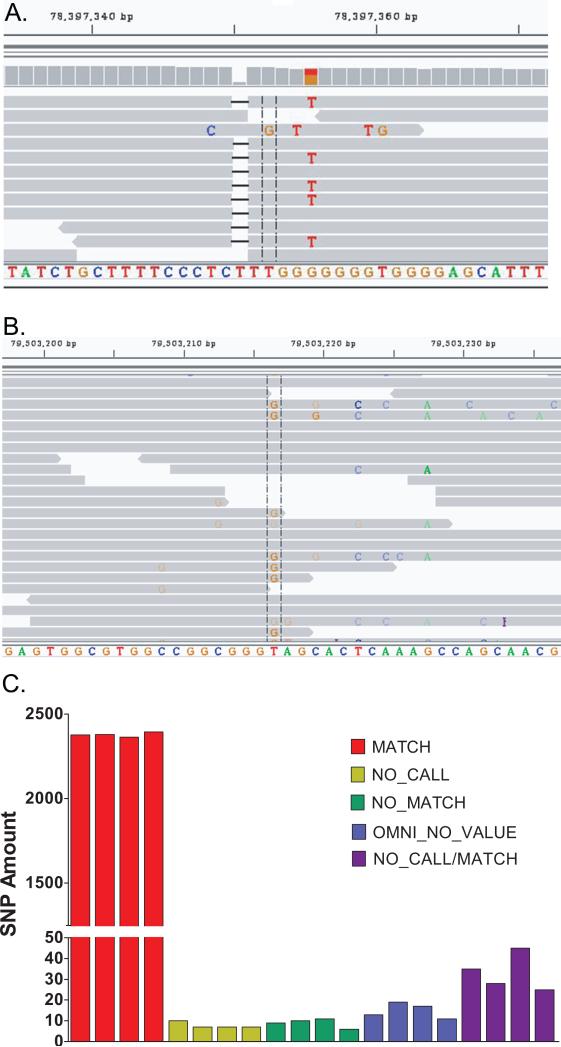
We investigated the false negative base calls in the GEDi data further and found that there were 7-11 discrepancies per sample identified between the GEDi and Omni 2.5 data (Table 2). In total, there were discrepancies detected at 23 positions that were predominantly related to the heterozygous vs. homozygous state of the same identified base, with a different base identified at only 1 position, chr15:78397352. The NGS data showed that at chr15:78397352 the Omni data were incorrect due to a single base deletion adjacent to the interrogated base, which shifted the base analyzed by the single-base extension method used in the Omni arrays (Figure 2A). Indels were associated with 4 additional GEDi vs. Omni discrepancies, and all but one of the remaining differences were due to low SNP quality scores in the SAMtools variant identification software (Table 2). At one position (chr4:6304087), the Omni data was incorrect (confirmed by Sanger sequencing), without any evident explanation. A small number (7-10) of bases were not called in the GEDi data across all replicates (No Call, Figure 2C), and Omni 2.5 SNP calls were not obtained for 11-19 positions (Omni No Value, Figure 2C). Bases were scored as “No Call/Match” if 1 or more replicates for each DNA sample had no call at that position, but all other replicates matched. There were 25-45 of these bases, with many of these (55/87 total = 63%) being due to no call in a single replicate (Table S2).
Table 2.
GEDi vs. Omni 2.5 Discrepancies Detected.
| NA12878 | OGI-132-357 | OGI-281-608 | OGI-307-717 | DISCREPANCY | |
|---|---|---|---|---|---|
| chr1:156146218 | 1/9 HOM/HET | 1 | |||
| chr1:213071341 | 1/9 HOM/HET | 1 | |||
| chr2:62052380 | 1/9 HOM/HET | 1 | |||
| chr2:166770120 | 3/8 HOM/HET | 1 | |||
| chr3:150645351 | 2/9 HOM/HET | 1 | |||
| chr3:193413502 | 1/5 HOM/HET | 1 | |||
| chr4:15982166 | 2/7 HOM/HET | 1 | |||
| chr5:178405941 | 2/9 HOM/HET | 1/9 HOM/HET | 1 | ||
| chr6:42932200 | 1/7 HOM/HET | 1/3 HOM/HET | 1/7 HOM/HET | 1 | |
| chr6:42932202 | 1/8 HOM/HET | 1/7 HOM/HET | 1 | ||
| chr9:102861613 | 1/9 HOM/HET | 1 | |||
| chr9:139327064 | 9/9 HOM/HET | 9/9 HOM/HET | 9/9 HOM/HET | 1 | |
| chr10:73461805 | 1/9 HOM/HET | 1 | |||
| chr10:85976966 | 9/9 HOM/HET | 9/9 HOM/HET | 1 | ||
| chr16:1265600 | 1/7 HOM/HET | 1 | |||
| chr16:1574863 | 1/9 HOM/HET | 1 | |||
| chr16:57937788 | 1/9 HOM/HET | 1 | |||
| chr17:11835331 | 1/9 HOM/HET | 1 | |||
| chr3:63986047 | 9/9 HOM/HET | 9/9 HOM/HET | 2 | ||
| chr4:6304087 | 9/9 HOM/HET | 5 | |||
| chr4:15981874 | 9/9 HOM/HET | 4 | |||
| chr4:15982166 | 9/9 HOM/HET | 9/9 HOM/HET | 4 | ||
| chr11:76895772 | 9/9 HOM/HET | 9/9 HOM/HET | 2 | ||
| chr15:78397352 | 9/9 DISCREP | 9/9 DISCREP | 9/9 DISCREP | 9/9 DISCREP | 3 |
Base positions of discrepancies detected in at least 1 GEDi sequence replicate and the Omni 2.5 SNP data for each sample analyzed are shown. The number of replicates with alternate results are indicated. Fewer than 9 replicates are indicated for some positions at which base calls were not made in some replicates. Discrepancies located in the top portion of the table were due to low SNP quality scores in SAMtools; the bottom portion of the table contains discrepancies specific to the GEDi vs. Omni data comparisons. The reasons identified for the discrepancies are indicated in the right column: 1 = low SNP quality score; 2 = heterozygous deletion of target base; 3 = homozygous deletion adjacent to target base; 4 = heterozygous insertion adjacent to target base; 5 = Omni incorrect, reason uncertain. Note that position chr4:15982166 is included in both halves of the table.
Figure 2.
(A) Integrative Genomics Viewer (IGV) screenshot of representative GEDi NGS validation data at chr15:78397352. The Omni 2.5 SNP data were determined to be incorrect in all samples due to a single base deletion adjacent to the interrogated base that shifted the analyzed base. (B) IGV screenshot of the putative c.1028T>G mutation of FSCN2 in OGI-267-573, clarifying the false positive variant call was due to mis-alignment of some NGS sequencing reads. (C) GEDi vs. Omni 2.5 concordance histogram plot corresponding to the 2,443 shared SNPs between the GEDi design and Omni 2.5 SNP for all 36 replicates of the 4 validation samples used in this study. KEY: MATCH – All GEDi NGS replicates matched Omni 2.5 SNP data; NO CALL – no NGS result; NO MATCH – ≥ 1 NGS replicate did not match Omni 2.5 SNP data; OMNI NO VALUE – no Omni 2.5 SNP result; NO CALL/MATCH – ≥ 1 NGS replicate had no result; all other NGS replicates matched Omni 2.5 SNP data.
The accuracy of the GEDi test was also supported by comparison of the GEDi sequence data for the HapMap sample NA12878 with publically available Platinum 200X average depth WGS data for NA12878 from Illumina (http://www.illumina.com/platinumgenomes). Within the 1,197,667 bp in the GEDi capture regions, excluding the mitochondrial chromosome, there were 962 SNPs and 89 indels identified in the NA12878 WGS data by Illumina. The accuracy of the GEDi test to identify both the SNPs and indels was 99.9%. The sensitivity and specificity for SNP detection were 96.4% and 99.9%, and for indel detection were 91.6% and 99.9%, respectively. It is likely that the sensitivity of the GEDi test for SNP detection is even higher, as we identified 47 SNPs called in the Illumina Platinum data that are located in a highly repetitive 11 kb chr17 region (chr17:21311917-21323163) in the gene KCNJ12 that are likely to be incorrect due to poor read alignment (Figure S1).
GEDi vs. WES
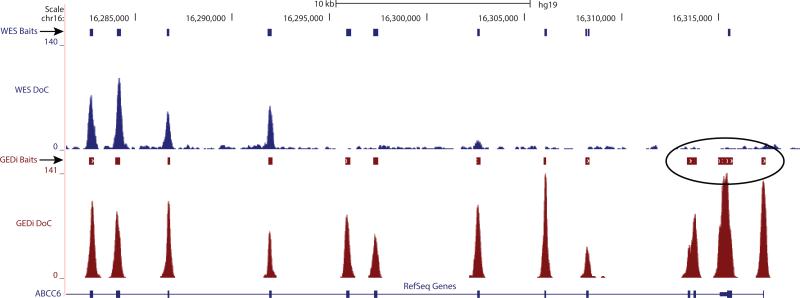
The GEDi test performance was compared to WES by analyzing the WES data of the same 4 validation samples for the 2,443 Omni 2.5 SNPs in the GEDi gene set. The average depth of coverage achieved by WES in these experiments was 100X, with 98% of the targeted regions covered at 10X sequence depth. Using the Omni 2.5 data as the “gold standard,” the sensitivity of WES was 0.883 ± 0.004, and the specificity of WES was 0.9998 ± 0.0003. While both GEDi and WES have excellent specificity, this comparison shows that WES is approximately 10% less sensitive than the GEDi test. Analysis shows that this is due to lack of sequence coverage in the WES data, with approximately 10% of the 2,443 positions interrogated in these analyses having insufficient coverage (≥10X) to make an accurate base call (Table S3). The majority of these positions (76%) were common in all 4 samples, suggesting that these positions were covered less efficiently in the WES capture design. Comparison of the WES and GEDi capture baits at these positions confirmed this hypothesis, and showed that 88% of the positions without coverage in the WES data had no baits in the V4+UTR capture set, whereas the GEDi capture set had at least 1 bait at these positions (Table S3). An example of one of these regions in shown in Figure 3. Further, there are 947 mutations in IRD disease genes reported in HGMD, ClinVar and Ensemble that would be detected by GEDi sequencing which fall in regions that are not covered in the Agilent V4+UTR WES capture set (Table S4). Conversely, WES did detect bases at an average of 5.5 positions out of 2,443 (0.22%) for which GEDi sequencing provided no call.
Figure 3.
Comparison of V4+UTR WES and GEDi capture baits at the 5′-end of ABCC6. The ABCC6 reference used is a “collapsed” reference that accounts for all known gene isoforms.
Reproducibility and Repeatability
The reproducibility of variant detection by GEDi was assessed by comparing the detection rates for the 2,443 common SNPs in all 9 GEDi datasets from each of the 4 samples. Bases that were discrepant in 1 or more of the 9 datasets for each sample in this “GEDi vs. GEDi” comparison were identified. GEDi capture followed by Illumina sequencing is highly reproducible, with only 4-6 discrepancies detected between the replicates for each DNA sample. In each case, the discrepancies related to the heterozygous vs. homozygous state of the same identified base. In the majority of cases, one out of the 9 sequence runs performed for each DNA sample contained the discrepancy (Table S5). Further analysis of the data for each of the 17 total discrepancies showed that they were due to low SNP quality score in the SAMtools variant identification software. Sixteen of these are the same as those detected in the GEDi vs. Omni 2.5 comparisons described above (Table S5).
The repeatability and reproducibility of the GEDi test was also evaluated using the kappa statistic, or kappa coefficient of agreement 32. For GEDi replicates performed on the same day the kappa statistic was 0.83088, indicating almost perfect agreement between the data obtained in the three replicates for each DNA sample analyzed 32. For GEDi tests performed for the 4 individual samples on each of the three separate days the kappa statistic was 0.76366, indicating excellent reproducibility 32.
Mutation Detection
The GEDi test correctly identified mutations in 17/18 patient samples with known IRDs, glaucoma and optic atrophy variants, including 10 indels (Table S6). GEDi testing did not correctly identify the pathogenic FOXC1 indel mutation in a patient with glaucoma; however, analysis showed a design gap in FOXC1 where this pathogenic mutation is located.
Clinical Sensitivity
GEDi clinical sensitivity was analyzed using samples from 192 probands with diagnoses of isolated or syndromic IRD, albinism or microphthalmos (Table S7). Analyses of the sequence data identified genetic diagnoses for 98 of the probands, representing a clinical sensitivity of 51%, consistent with findings from other studies (Table S7) 22-26. The majority of these diagnoses were consistent with the subject's clinical presentation and family history. Two subjects without a family history of disease were found to have mutations in the known dominant IRD disease genes PRPH2 and IMPDH1, consistent with identification of de novo mutations in the affected individuals; segregation analyses confirmed de novo mutations in these two subjects (OGI-301-703 and OGI-274-582, Table S7). While de novo mutations have been reported as the cause of dominant RP, de novo mutations in the PRPH2 and IMPDH1 genes have not been previously reported. The majority of subjects that were not diagnosed genetically by GEDi testing had non-syndromic RP (52/89 = 58%; Table S8).
Mutation Validation
In total, we identified 147 likely pathogenic mutations by GEDi capture and NGS sequencing, and all but four of these were validated by PCR and Sanger sequencing (Supplemental methods and Table S9). Review of the NGS data for the 4 putative mutations that were not validated by Sanger sequencing showed that 3 of the 4 mutations, corresponding to 2 probands (OGI-040-100 and OGI-271-579) were detected by less than 10 reads (Table S9). The 4th putative mutation not detected by Sanger sequencing, had excellent DoC (Table S9); however, the heterozygous G base call was due to mis-alignment of some of the NGS sequence reads, resulting in a false positive variant call (Figure 2B).
Missed Diagnoses
GEDi capture and sequencing did not initially identify a genetic cause of disease in 5 patients for whom genetic diagnoses were ultimately obtained (Table S10). These cases are instructive, and information from them has been used to iteratively improve the GEDi test. For example, in two cases, OGI-147-394 and OGI-387-839, GEDi sequencing identified a single potentially pathogenic variant in ABCA4 and USH2A, respectively, but the second mutant allele was not initially detected (Table S10). The second alleles were subsequently identified by Sanger sequencing, both being deep intronic mutations known to alter splicing 31,33. Probes for the relevant intronic region for USH2A have been added to subsequent versions of the GEDi capture set (Table S1b), and those corresponding to deep intronic ABCA4 mutations will be added to the next version of GEDi 33. Information regarding the remaining 3 cases listed in Table S10 is included in Supplementary Information.
Improved Diagnoses
Of note, seven of the subjects studied were found to have mutations in genes that are not primarily associated with their phenotypes (denoted with * in Table S7). Specific examples include cone dystrophy due to mutations in the ORF15 region of RPGR, NRL mutations in Chorio-retinal atrophy, and TMEM67 mutations in Senior-Loken syndrome (additional details in Supplemental Information).
DISCUSSION
Our results suggest that selective targeted enrichment and NGS is the preferred method for diagnostic testing, especially for genetically heterogeneous disorders such as IRDs. The GEDi test has improved sensitivity when compared to WES while maintaining nearly perfect specificity. Our results show that the higher sensitivity of the GEDi test is due to improved probe design compared to commercially available exome capture sets where probes were missing for approximately 10% of the regions targeted by the GEDi test. While the concept that targeted sequencing can out-perform standard exome sequencing based on better coverage has been discussed in reviews and commentaries regarding genetic diagnostic testing, only limited empiric comparisons of these two approaches to genetic diagnostic testing have been reported previously 3,10,34. Thus, while WES is now available as a clinical diagnostic test at some centers, and reports of using WES for diagnostic testing have been published, quantitation of the performance characteristics of the GEDi test makes it possible to identify and quantify the advantages of selective targeted enrichment over WES 6,35,36.
There are additional advantages of selective enrichment or panel tests over WES for diagnostic testing. The turn-around-time for the GEDi “panel” test run on a MiSeq NGS platform is approximately 1 day, which is considerably less than WES samples run on a HiSeq 2000 instrument (~12 days). The current costs of selective exon capture tests are also lower than WES, although it is likely that this difference will continue to diminish over time. At present (8/2014), the cost of the materials needed for GEDi testing per patient is approximately $430, compared to $1,325 per patient for WES using the sequence depth described. In addition, panel testing has a higher pre-test probability of finding a meaningful result, and reduces the potential for making incidental sequence findings, which can be challenging for both health care providers and patients 37,38.
While multiple characteristics make selective targeted enrichment a preferable method for genetic diagnostic testing, there are some drawbacks to this approach. First, hybridization-based capture approaches are limited by “design gaps”, regions where it is not possible to design targeted enrichment probes. Specifically, genome regions with high GC content and/or repetitive elements can be resistant to accurate capture probe design 3. Fortunately, based on the data obtained for the GEDi test, near-target sequence coverage limits this problem to gaps larger than 75 bp, which reduces, although does not, eliminate this problem. It is also possible to use alternative approaches to capture regions in hybridization design gaps, including amplification-based strategies such as Agilent Technologies’ HaloPlex technique 39.
Sufficient sequence depth is also needed to make accurate base calls from the NGS data. For GEDi we showed that a minimum depth of coverage of 10X gave a specificity of 100%. Even with sufficient depth of coverage, mis-alignment of short NGS reads can lead to incorrect base identification, especially for repetitive regions or genes with paralogous copies elsewhere in the genome, which we observed in both the GEDi and WES data. Until longer sequencing reads become routinely available, this is likely to remain a problem; however, familial segregation studies and Sanger validation of pathogenic alleles can be useful to resolve these discrepancies.
We carefully evaluated the overall performance characteristics of the GEDi test and showed that the test is both sensitive and specific and is highly reproducible and accurate. Thorough analyses of these test characteristics have been reported for one other NGS-based diagnostic test, a targeted enrichment and NGS-based test for 25 genes associated with cancer called the WUCaMP assay 5. The sensitivity and specificity of the WUCaMP assay were determined by comparing test data with WGS data from Complete Genomics for HapMap sample NA19240. For these studies, the test samples were sequenced to a high depth of coverage, with 96.9% of the targeted regions covered at ≥ 50X depth. The reported sensitivity and specificity for detecting SNVs were 98.3% and 100%, respectively 5. The sensitivity and specificity of the GEDi test for SNV detection are comparable, at 96.4% to 97.9% and 99.9% to 100%, respectively, with an overall accuracy for both the SNPs and indels of 99.9%. We also showed that the GEDi test is highly repeatable and reproducible, with kappa statistics of 0.83088 and 0.76366, respectively, indicating excellent agreement between the data obtained in the replicate testing of the 4 individual DNA samples 32.
The clinical sensitivity of the GEDi test was 51% in patients with IRDs, a rate that is consistent with prior reports 22-26. It is hypothesized that subjects without mutations in GEDi target genes must have mutations in novel disease genes, or in non-coding portions of the currently identified IRD disease genes. Exome and genome sequencing will be required for identifications of these mutations. It is also possible that some subjects have mutations that cannot be readily detected by sequencing-based approaches, such as CNVs 25,40.
Comprehensive genetic diagnostic testing for genetically and phenotypically heterogeneous disorders such as IRDs can also lead to diagnoses outside of the reported genotype-phenotype relationships. Seven of the patients with genetic diagnoses had atypical phenotypic features confirming that it can be difficult to predict the genetic cause of disease based on clinical findings alone 26.
In summary, the GEDi test offers a number of advantages as a clinical diagnostic test for patients with inherited eye disorders. Given the potential for gene-based therapies for inherited disorders in general, and inherited eye disorders specifically, genetic diagnostic testing will increasingly be necessary for optimal care of patients with genetic diseases. Further, the GEDi test statistics make a strong case for the use of targeted tests in the clinical setting, as they are highly accurate, reproducible and have better overall performance than more general tests such as conventional WES analyses.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful for the technical contribution of Aliete E. Langsdorf, and to Dr. Kendrick Goss for assistance with performing the PPT1 assays. This work was supported by grants from the National Institutes of Health [EY012910 (E.A.P.), and P30EY014104 (MEEI core support)], the March of Dimes (J.L.W.) and the Foundation Fighting Blindness USA (E.A.P., Q.L.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding organizations or the National Institutes of Health.
Footnotes
Contributor statement
Experiments were designed by M.B.C., X.G., J.L.W. and E.A.P. Subject, family and control samples were provided by E.M.P., A.B.F., J.L.W. and E.A.P. GEDi sequencing, WES and Omni 2.5 array analyses were performed by M.B.C., D.G.T., E.D.A., M.J. and D.Y.W. Bioinformatic pipeline development and data analyses were performed by X.G., D.N.G., M.B.C., E.M.P. and Q.L. Validation sequencing and segregation analyses were performed by M.B.C., D.G.T., M.J., D.Y.W.,Z.D. F-K., E.D.A., M.E.S and K.M.B. Assays of PPT1 activity were performed and analyzed by K.B.S. and D.A.S. The manuscript was written by M.B.C., E.M.P., K.M.B, X.G., J.L.W. and E.A.P.
Competing Interests
The authors have no conflicts of interest to disclose.
Data Access
GEDi sequence data, WES sequence data and Omni 2.5 SNP data for the 4 samples used to quantify the performance of the GEDi test have been submitted to dbGAP; study accession phs000798.v1.p1. Similarly, The variants listed in Table S7 have been submitted to the ClinVar database.
Supplementary Information is available at the Genetics in Medicine website.
REFERENCES
- 1.Calvo SE, Compton AG, Hershman SG, et al. Molecular diagnosis of infantile mitochondrial disease with targeted next-generation sequencing. Sci Transl Med. 2012 Jan 25;4(118):118ra110. doi: 10.1126/scitranslmed.3003310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sommen M, Van Camp G. Genetic diagnostics of early childhood hearing loss: better testing with next-generation DNA sequencing. B-Ent. 2013;(Suppl 21):51–56. [PubMed] [Google Scholar]
- 3.Rehm HL. Disease-targeted sequencing: a cornerstone in the clinic. Nat Rev Genet. 2013 Apr;14(4):295–300. doi: 10.1038/nrg3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pugh TJ, Kelly MA, Gowrisankar S, et al. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genetics In Medicine. 2014 Feb 6; doi: 10.1038/gim.2013.204. [DOI] [PubMed] [Google Scholar]
- 5.Cottrell CE, Al-Kateb H, Bredemeyer AJ, et al. Validation of a next-generation sequencing assay for clinical molecular oncology. Journal Molecular Diagnostics. 2014 Jan;16(1):89–105. doi: 10.1016/j.jmoldx.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levenson D. Whole-exome sequencing emerges as clinical diagnostic tool: Testing method proves useful for diagnosing wide range of genetic disorders. American Journal Medical Genetics. 2014 Jan;164(1):ix–x. doi: 10.1002/ajmg.a.36385. [DOI] [PubMed] [Google Scholar]
- 7.Gargis AS, Kalman L, Berry MW, et al. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nature Biotechnology. 2012 Nov;30(11):1033–1036. doi: 10.1038/nbt.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rehm HL, Bale SJ, Bayrak-Toydemir P, et al. ACMG clinical laboratory standards for next-generation sequencing. Genetics In Medicine. 2013 Sep;15(9):733–747. doi: 10.1038/gim.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. New England Journal Medicine. 2013 Oct 17;369(16):1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shearer AE, Black-Ziegelbein EA, Hildebrand MS, et al. Advancing genetic testing for deafness with genomic technology. Journal Medical Genetics. 2013 Sep;50(9):627–634. doi: 10.1136/jmedgenet-2013-101749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buch H, Vinding T, La CM, Appleyard M, Jensen GB, Nielsen NV. Prevalence and causes of visual impairment and blindness among 9980 Scandinavian adults: the Copenhagen City Eye Study. Ophthalmology. 2004;111(1):53–61. doi: 10.1016/j.ophtha.2003.05.010. [DOI] [PubMed] [Google Scholar]
- 12.Congdon N, O'Colmain B, Klaver CC, et al. Causes and prevalence of visual impairment among adults in the United States. Archives Ophthalmology. 2004 Apr;122(4):477–485. doi: 10.1001/archopht.122.4.477. [DOI] [PubMed] [Google Scholar]
- 13.Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. British Journal Ophthalmology. 2006 Mar;90(3):262–267. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan BJ, Wiggs JL. Glaucoma: genes, phenotypes, and new directions for therapy. Journal Clinical Investigation. 2010 Sep;120(9):3064–3072. doi: 10.1172/JCI43085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gemenetzi M, Yang Y, Lotery AJ. Current concepts on primary open-angle glaucoma genetics: a contribution to disease pathophysiology and future treatment. Eye. 2012 Mar;26(3):355–369. doi: 10.1038/eye.2011.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neuhann T, Rautenstrauss B. Genetic and phenotypic variability of optic neuropathies. Expert Review Neurotherapeutics. 2013 Apr;13(4):357–367. doi: 10.1586/ern.13.19. [DOI] [PubMed] [Google Scholar]
- 17.Maguire AM, Simonelli F, Pierce EA, et al. Safety and efficacy of gene transfer for Leber's congenital amaurosis. New England Journal of Medicine. 2008 May 22;358(21):2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bainbridge JW, Smith AJ, Barker SS, et al. Effect of gene therapy on visual function in Leber's congenital amaurosis. New England Journal of Medicine. 2008;358(21):2231–2239. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- 19.Cideciyan AV, Aleman TS, Boye SL, et al. Human gene therapy for RPE65 isomerase deficiency activates the retinoid cycle of vision but with slow rod kinetics. Proceedings National Academy Sciences USA. 2008 Sep 30;105(39):15112–15117. doi: 10.1073/pnas.0807027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacLaren RE, Groppe M, Barnard AR, et al. Retinal gene therapy in patients with choroideremia: initial findings from a phase 1/2 clinical trial. Lancet. 2014 Mar 29;383(9923):1129–1137. doi: 10.1016/S0140-6736(13)62117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiggs JL, Pierce EA. Genetic testing for inherited eye disease: who benefits? JAMA Ophthalmol. 2013 Oct;131(10):1265–1266. doi: 10.1001/jamaophthalmol.2013.4509. [DOI] [PubMed] [Google Scholar]
- 22.Song J, Smaoui N, Ayyagari R, et al. High-throughput retina-array for screening 93 genes involved in inherited retinal dystrophy. Investigative Ophthalmology & Visual Science. 2011;52(12):9053–9060. doi: 10.1167/iovs.11-7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Audo I, Bujakowska KM, Leveillard T, et al. Development and application of a next-generation-sequencing (NGS) approach to detect known and novel gene defects underlying retinal diseases. Orphanet Journal Rare Diseases. 2012;7:8. doi: 10.1186/1750-1172-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neveling K, Collin RW, Gilissen C, et al. Next-generation genetic testing for retinitis pigmentosa. Human Mutation. 2012 Jun;33(6):963–972. doi: 10.1002/humu.22045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eisenberger T, Neuhaus C, Khan AO, et al. Increasing the Yield in Targeted Next-Generation Sequencing by Implicating CNV Analysis, Non-Coding Exons and the Overall Variant Load: The Example of Retinal Dystrophies. PLoS One. 2013;8(11):e78496. doi: 10.1371/journal.pone.0078496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang F, Wang H, Tuan HF, et al. Next generation sequencing-based molecular diagnosis of retinitis pigmentosa: identification of a novel genotype-phenotype correlation and clinical refinements. Human Genetics. 2014 Mar;133(3):331–345. doi: 10.1007/s00439-013-1381-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falk MJ, Pierce EA, Consugar M, et al. Mitochondrial disease genetic diagnostics: optimized whole-exome analysis for all MitoCarta nuclear genes and the mitochondrial genome. Discov Med. 2012 Dec;14(79):389–399. [PMC free article] [PubMed] [Google Scholar]
- 28.Falk MJ, Zhang Q, Nakamaru-Ogiso E, et al. NMNAT1 mutations cause Leber congenital amaurosis. Nature Genetics. 2012 Jul 29;44(9):1040–1045. doi: 10.1038/ng.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.den Hollander AI, Koenekoop RK, Yzer S, et al. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. American Journal of Human Genetics. 2006 Sep;79(3):556–561. doi: 10.1086/507318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Webb TR, Parfitt DA, Gardner JC, et al. Deep intronic mutation in OFD1, identified by targeted genomic next-generation sequencing, causes a severe form of X-linked retinitis pigmentosa (RP23). Human Molecular Genetics. 2012 Aug 15;21(16):3647–3654. doi: 10.1093/hmg/dds194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vache C, Besnard T, le Berre P, et al. Usher syndrome type 2 caused by activation of an USH2A pseudoexon: implications for diagnosis and therapy. Human Mutation. 2012 Jan;33(1):104–108. doi: 10.1002/humu.21634. [DOI] [PubMed] [Google Scholar]
- 32.Viera AJ, Garrett JM. Understanding interobserver agreement: the kappa statistic. Family Medicine. 2005 May;37(5):360–363. [PubMed] [Google Scholar]
- 33.Braun TA, Mullins RF, Wagner AH, et al. Non-exomic and synonymous variants in ABCA4 are an important cause of Stargardt disease. Human Molecular Genetics. 2013 Dec 20;22(25):5136–5145. doi: 10.1093/hmg/ddt367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Redin C, Le Gras S, Mhamdi O, et al. Targeted high-throughput sequencing for diagnosis of genetically heterogeneous diseases: efficient mutation detection in Bardet-Biedl and Alstrom syndromes. Journal Medical Genetics. 2012 Aug;49(8):502–512. doi: 10.1136/jmedgenet-2012-100875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flintoft L. Clinical genetics: exomes in the clinic. Nat Rev Genet. 2013 Dec;14(12):824. doi: 10.1038/nrg3620. [DOI] [PubMed] [Google Scholar]
- 36.Delanty N, Goldstein DB. Diagnostic exome sequencing: a new paradigm in neurology. Neuron. 2013 Nov 20;80(4):841–843. doi: 10.1016/j.neuron.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 37.Dorschner MO, Amendola LM, Turner EH, et al. Actionable, pathogenic incidental findings in 1,000 participants' exomes. American Journal Human Genetics. 2013 Oct 3;93(4):631–640. doi: 10.1016/j.ajhg.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Platt J, Cox R, Enns GM. Points to Consider in the Clinical Use of NGS Panels for Mitochondrial Disease: An Analysis of Gene Inclusion and Consent Forms. Journal Genetic Counseling. 2014 Jan 8; doi: 10.1007/s10897-013-9683-2. [DOI] [PubMed] [Google Scholar]
- 39.Berglund EC, Lindqvist CM, Hayat S, et al. Accurate detection of subclonal single nucleotide variants in whole genome amplified and pooled cancer samples using HaloPlex target enrichment. BMC Genomics. 2013;14:856. doi: 10.1186/1471-2164-14-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fromer M, Moran JL, Chambert K, et al. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. American Journal Human Genetics. 2012 Oct 5;91(4):597–607. doi: 10.1016/j.ajhg.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.