Abstract
G protein-coupled receptors are vital membrane proteins that allosterically transduce biomolecular signals across the cell membrane. However, the process by which ligand binding induces protein conformation changes is not well understood biophysically. Rhodopsin, the mammalian dim-light receptor, is a unique test case for understanding these processes because of its switch-like activity; the ligand, retinal, is bound throughout the activation cycle, switching from inverse agonist to agonist after absorbing a photon. By contrast, the ligand-free opsin is outside the activation cycle and may behave differently. We find that retinal influences rhodopsin dynamics using an ensemble of all-atom molecular dynamics simulations that in aggregate contain 100 μs of sampling. Active retinal destabilizes the inactive state of the receptor, whereas the active ensemble was more structurally homogenous. By contrast, simulations of an active-like receptor without retinal present were much more heterogeneous than those containing retinal. These results suggest allosteric processes are more complicated than a ligand inducing protein conformational changes or simply capturing a shifted ensemble as outlined in classic models of allostery.
Introduction
G protein-coupled receptors (GPCRs) are integral membrane proteins that serve wide-ranging functions from olfaction (1,2) to modulating cardiac response (3). There are more than 800 GPCRs in humans (4), and they are targeted by a remarkable number of drugs (5–7). These proteins transmit signals across the cell membrane using an allosteric mechanism; ligands binding in the canonical orthosteric site modulate receptor conformations such that G proteins inside the cell can bind the cytoplasmic face of the GPCR. Rhodopsin, the mammalian dim-light receptor, was the first GPCR to be reported at high resolution (8) and has served as a useful model for understanding the dynamics and function of other proteins. However, the process by which ligand conformational change is communicated to the protein’s active site remains poorly understood despite vast biochemical and biophysical efforts (9,10). Some hypotheses suggest that highly conserved motifs in the protein may serve as functional microswitches, making concerted transitions upon ligand binding (10–17), but the details of any such process remain elusive (15,18,19).
Early discussions of GPCR dynamics were generally framed in terms of two states (active and inactive) connected by a single path. Although this view has yielded to a more complex understanding of GPCR dynamics, a simplified single pathway description is still prevalent, despite mounting evidence that suggests this idea is flawed (20–22). The complexity of GPCR activation is only beginning to be understood (20–24). Experimental evidence suggests that multiple states and multiple activation pathways may exist (10,20,21,25,26). Additionally, GPCR activity may be modulated by external factors, such as dimerization (27), membrane environment (28–31), and pH (16,32–34). Even ligands that evoke a functionally similar response may have a varied impact on specific residues (21). For example, the adrenergic receptor () can sample a diverse range of conformational states with variable populations and lifetime shifts that depend on the environment and presence of ligands (25).
The subtlety of GPCRs’ functional dynamics presents a major opportunity for contribution from computational methods. Although experimental techniques can probe a small subset of interesting degrees of freedom, molecular dynamics (MD) simulations capture atomistic-resolution details of the system. As a result, MD simulations can unambiguously characterize the structural ensemble accessible to GPCRs under different conditions, without the perturbations of adding probes such as fluorescent dyes.
However, all-atom simulations are so computationally costly that supercomputers or specialized hardware are often needed, and even then acquiring sufficient statistical sampling is a serious problem. This problem is often underestimated, because even degrees of freedom that interconvert rapidly can be linked to the much slower dynamics of the entire protein (35–37). Enhanced sampling methods can improve the situation, but most advanced methods require the choice of an informative reaction coordinate, which can be very difficult to obtain. On the other hand simplified models can be quite useful (19), but often lack the resolution to answer very specific scientific questions. Because of the large datasets and their biomedical relevance, understanding allostery in the activation of GPCRs is a topic of intense interest.
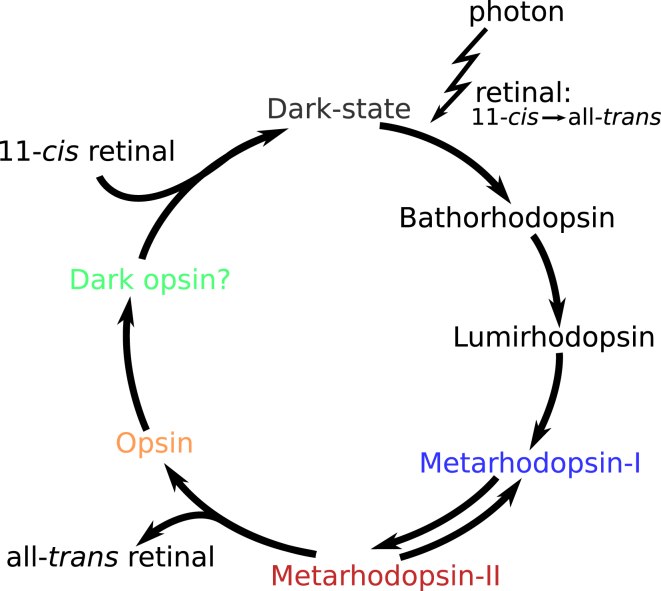
In this study, we use rhodopsin as a platform to better understand allostery, exploring the effects of the presence or absence of retinal on its conformational ensemble. Rhodopsin is unique among GPCRs because of its switch-like behavior; retinal undergoes an 11-cis to all-trans bond isomerization upon absorption of a photon, making an in situ switch from inverse agonist to agonist and allowing protein activation to proceed. Rhodopsin, together with its apo-form, opsin, makes an effective case for ligand effects on protein dynamics because, unlike most ligand-binding proteins, the apo form lies outside of this functional cycle (Fig. 1).
Figure 1.
Rhodopsin activation cycle. 11-cis retinal is prebound in the inactive dark-state. Absorption of a photon causes 11-cis to all-trans photoisomerization, and changes retinal from an inverse agonist to agonist. Activation then proceeds through several trappable nonequilibrium intermediates (not all shown) before reaching Metarhodopsin-I (Meta-I). Meta-I exists in equilibrium with the fully active Metarhodopsin-II (Meta-II). The retinal-rhodopsin bond is cleaved and retinal is able to leave, resulting in the apo-protein opsin. Upon rebinding of an 11-cis retinal this cycle is then ready to be repeated. To see this figure in color, go online.
Crystal structures of opsin show a very active-like conformation (38,39), in distinct contrast to functional experiments showing that opsin does not efficiently induce downstream G protein signaling (40). To rationalize this apparent contradiction, we used MD simulations to produce ensembles of conformations for both opsin and rhodopsin using active-like and inactive-like starting structures. Specifically, we simulated the Metarhodopsin-I (Meta-I) and Metarhodopsin-II (Meta-II) states of rhodopsin, and opsin in both its active-like crystal structure and a hypothesized inactive-like dark opsin (see Fig. 1 and Table 1). Our results suggest that opsin is able to sample a superset of those conformations accessible to Meta-II rhodopsin. Remarkably, these simulations capture ligand-dependent changes to the structural ensemble. In Meta-I simulations, both the ligand and the protein are more dynamic.
Table 1.
Simulation details
| System | PDB ID | Notes | Simulation Time (ns) |
|---|---|---|---|
| Dark opsin | 1U19 | retinal removed | 1 3,000, 5 4,000 |
| Opsin | 3CAP | 3 4,000, 3 4,200 | |
| Meta-I | Meta-I | (from 45) | 6 4,800 |
| Meta-II | 3PXO | 3 4,000, 3 4,200 | |
| Total | 101,000 ns |
An exhaustive list is available in Table S1.
In the context of allostery, the results are quite nuanced; whereas an active-like apo-protein is more conformationally heterogeneous, an inactive protein bound to activating ligand is destabilized. In rhodopsin, it appears that the ligand has a role beyond that outlined by the conformational equilibrium model of allostery: it can stabilize one ensemble of conformations and destabilize another, as suggested by more recent allosteric hypotheses (41–44).
Materials and Methods
Simulation details
Four different protein starting structures were used in the current study. Three were obtained from x-ray crystal structure data; opsin and Meta-II were used as is, whereas dark opsin was built by removing the retinal from a crystal structure of dark-state rhodopsin. The fourth (Meta-I) was taken from the final coordinates of the complex-counterion simulation conducted in a previous study (45–47); we assert that this structure resembles the Meta-I state of the protein based on the behavior of the Schiff base, as well as the match between the theoretical NMR spectra calculated from this trajectory and experiment (46). Six independent systems were then built for each structure, for a total of 24 trajectories (see Table 1). Each structure was embedded in a randomized lipid bilayer containing 123 SDPE (1-stearoyl-2-docosahexaenoyl-phosphatidylethanolamine) molecules, 8200 waters, and 100 mM NaCl (beyond that needed to neutralize the system). SDPE lipids were chosen because polyunsaturated fatty acids and phosphatidylethanolamine headgroups were experimentally shown to favor the active Meta-II conformation (28–31,48,49). We also simulated low-pH conditions, protonating Glu1343.49 and Glu181EL2. This decision was based on the work of multiple investigators (16,32–34,50,51), which suggested such conditions favor activation. Six replicates were run for each protein starting structure, ranging from 3 to 4 μs each. Each replicate for a given starting state was assembled using the same protein structure, but the lipid and water coordinates were generated independently using a previously published protocol (52).
The system was simulated in the NPγT ensemble using the CHARMM36 lipid parameters (53) and CHARMM22 protein parameters (54) with CMAP corrections (55,56). These simulations were begun before SDPE was specifically parametrized (see 57), and we found that a lateral tension of 30 dyn/cm was needed to obtain the correct area per lipid (data not shown). Retinal parameters were obtained from Scott Feller (see 58).
This system was then subjected to MD using NAMD 2.8 (59), with the Velocity Verlet integrator and a 2 fs timestep (60). Bonds to hydrogen were constrained using the RATTLE algorithm (61). The Langevin piston barostat was combined with Langevin dynamics to produce the correct isothermal-isobaric ensemble at 310 K (62,63). The nonbonded cutoff was set to 10 Å, whereas long-range electrostatics were calculated using smooth particle-mesh Ewald summation (64) with a 75 75 96 cubic grid ( 1 Å/grid point). These simulations were run on the University of Rochester’s BlueGene/P and BlueGene/Q computers. The trajectory details can be found in Table 1.
Simulation analysis
All simulation analyses were implemented in LOOS (version 2.0.5, loos.sourceforge.net), an open-source simulation development library maintained by the Grossfield lab (65). Images were rendered using PyMol (version 1.6, www.pymol.org), and data was plotted with gnuplot (version 4.6, www.gnuplot.info).
Results
We computed an ensemble of conformations that represent different functional states of rhodopsin, finding that retinal plays an important role in modulating the protein’s behavior. Somewhat surprisingly, the ligand’s interaction with the protein was quite promiscuous. These results provide evidence that the rhodopsin system behaves in a manner that cannot be explained by the classic allosteric model of conformational selection (66).
Dynamics of experimentally interesting degrees of freedom
Retinal
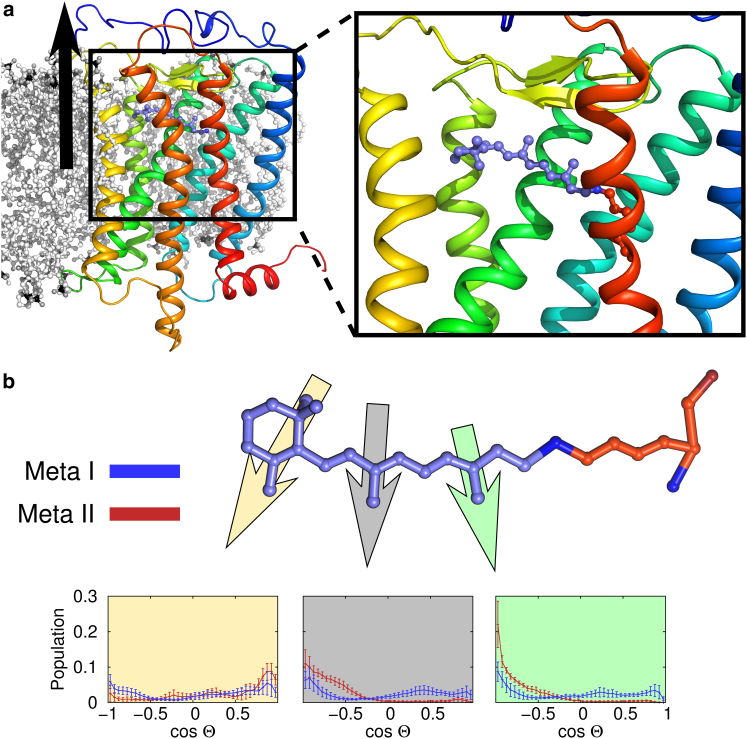
To understand how retinal impacts rhodopsin’s conformational ensemble, we first investigated how the protein’s starting state correlated with ligand dynamics. Our previous work showed that after the 11-cis to all-trans isomerization retinal became more flexible (46,47,67). Here we extend that view to understand how all-trans retinal behaves in the active Meta-II state versus the preactive Meta-I state (Fig. 2) using the new ensemble of simulations outlined in Table 1.
Figure 2.
Retinal dynamics distinguish the Meta-I and Meta-II ensembles. (a) Illustration showing rhodopsin in a lipid bilayer. The protein is shown as a rainbow colored cartoon with the N-terminus colored blue. Lipids are shown in a black/white ball-and-stick representation (note several lipids are removed for clarity). The black arrow denotes the membrane normal. The inset shows a close-up view of retinal (purple ball-and-sticks) in the binding pocket. It is covalently attached to Lys2967.43 (orange) by a Schiff-base linked nitrogen. (b) Retinal orientations. A morphed retinal is shown colored as in (a). Its orientation was monitored using the three vectors illustrated by yellow, gray, and green arrows. Specifically, we computed the dot-product between a vector drawn from either: the C1–C5 atoms (yellow), the C9–C19 atoms (gray), or the C13–C20 atoms (green) and the membrane normal (black arrow in a). This angle was histogrammed over the entire Meta-I (blue data) or Meta-II (red data) ensemble of trajectories. The average histograms are shown below, with error bars representing the standard error. To see this figure in color, go online.
Previous efforts by several labs have shown that retinal makes state-specific protein-ligand interactions (68–72). We monitored retinal using three regions of the ligand equivalent to those identified by Brown and coworkers, the orientations of the C5-, C9-, and C13-methyl groups (46,47,73–76). In this analysis, the vectors denoted by the three arrows in Fig. 2 b (see caption for details) were compared with the membrane normal (black vector in Fig. 2 a) using a simple dot product. (Retinal orientations for individual trajectories are available in Figs. S1–S6 in the Supporting Material.)
The ligand-binding pocket
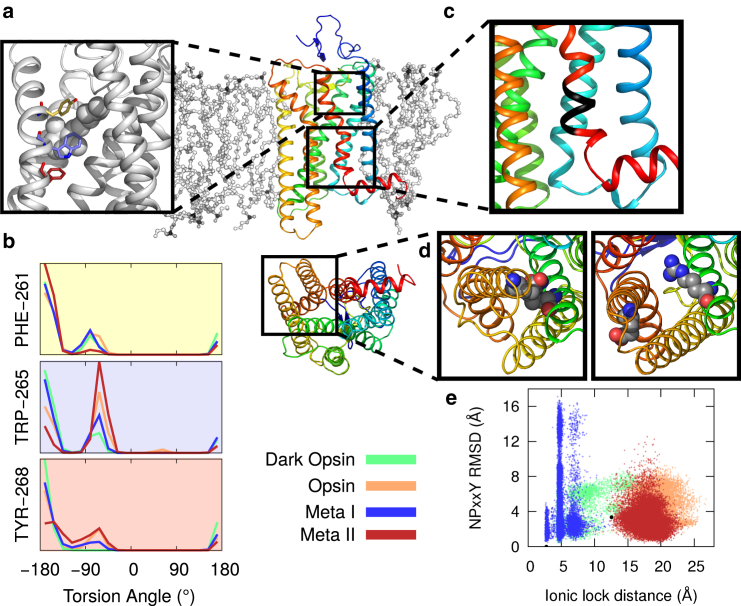
Because the orientation of retinal’s β-ionone ring was quite varied whereas the dynamics closer to the covalently bound Lys2967.43 depended on the protein’s state, it is logical to examine the behavior of nearby aromatic side-chains that interact with retinal, coupling it to the rest of the protein. These particularly include Phe2616.44, Trp2656.48, and Tyr2686.51, the rotamer toggle switch residues. Phe2616.44 shows no discernible trend, but in the Meta-II ensemble the gauche- rotamer had a much smaller population than in the other simulations (Fig. 3 b), which may be an impact of the stable ligand conformation (see the previous section). In Trp2656.48 and Tyr2686.51 the active-like ensembles (opsin and Meta-II) showed a different preferred orientation to the inactive-like ensembles (dark opsin and Meta-I). Trp2656.48 (Fig. 3 b, middle) showed a stronger preference for the gauche- rotamer in opsin and Meta-II, whereas dark opsin and Meta-I occupied a primarily trans rotamer. As we will show below (see also Fig. 4) this same population ordering—dark opsin, Meta-I, opsin, Meta-II—was seen in measures of whole-protein activity. Tyr2686.51 (Fig. 3 b, bottom) also showed a distinction between active-like and inactive-like ensembles.
Figure 3.
Dynamics of biologically distinguished structural motifs. (Center) Rhodopsin is shown as a rainbow cartoon embedded in a lipid bilayer. The bottom view shows the protein’s cytoplasmic face. (a) Illustration showing retinal (white spheres) in its binding pocket and rotamer toggle residues: Phe2616.44 (pale yellow), Trp2656.48 (pale violet), and Tyr2686.51 (dark red). (b) Histograms of the population of torsion angles for each of the three toggle switch residues. Data is colored by the simulation ensemble. (Note: a version of this figure with error bars is depicted in Fig. S8.) (c) Close-up view of the NPxxY motif. (d) The ionic lock. Arg1353.50 and Glu2476.30 are shown forming a salt bridge (left) as in the inactive crystal structure (PDB ID: 1U19) or in an open conformation (right) as in the active crystal structure (PDB ID: 3PXO). (e) NPxxY motif versus ionic lock dynamics. The NPxxY RMSD from the inactive crystal structure (y axis) is shown versus the ionic lock distance (x axis). Every frame (sampled at a rate of 1 ns) is shown colored according to ensemble. To see this figure in color, go online.
Figure 4.
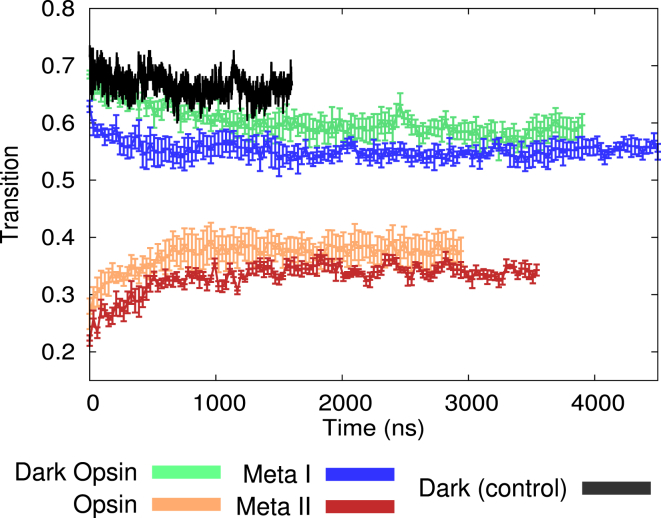
Contact-based reaction coordinate shows opsin and Meta-I ensembles are more heterogeneous. The average transition is shown for all 24 trajectories as a function of simulation time. This value is calculated using Eq. 1, where contacts are computed on a per-residue basis using side-chain centers of mass. Data is averaged over the six independent simulations in each of the four ensembles with error bars representing the standard deviation. The single dark-state simulation from Grossfield et al. (45) is shown for reference. To see this figure in color, go online.
Cytoplasmic regions
We also monitored the dynamics of two conserved cytoplasmic motifs: the ionic lock (a salt bridge between Glu2476.30 and Arg1353.50) and the NPxxY sequence in TM7 (Fig. 3, c and d). Interresidue distances for the ionic lock are plotted as a function of simulation time in Fig. S7. There was only a single transition in this coordinate, in one of the opsin simulations, although there was one short-lived opening event in one of the dark opsin runs. If we monitor the ionic lock distance versus the NPxxY motif root mean square deviation (RMSD) from the inactive structure, as in previous work (18,77), the picture is very different. We found that the NPxxY region is quite flexible, especially in the Meta-I ensemble, even though there are no transitions in the ionic lock (Fig. 3 e). By contrast, the lock flickers in dark opsin simulations, but the NPxxY region is not as flexible. In opsin, there was a single lock closing event, which did coincide with a decrease in NPxxY RMSD, but the Meta-II ensemble produced NPxxY conformations just as close to the inactive structure even though the lock never transitioned.
Whole protein dynamics reveal heterogeneity in opsin and Meta-I
We found that probing only experimentally motivated degrees of freedom gave an incomplete picture of rhodopsin’s dynamics and thus also explored protein dynamics using data-derived methods. First, we introduced a reaction coordinate based on the interresidue contacts formed and broken in our simulations. Instead of analyzing all contacts, we limit this analysis only to those contacts present in either the inactive and active crystal structures, but not both. A smooth tanh function was used to define connectivity between two residues, i and j, as in the following:
| (1) |
where is a switching distance of 8 Å and is the distance between the two side chains. The contact values were then summed and normalized such that a value of 1 is obtained when the contacts are exactly those found in the inactive crystal, and 0 when they are those found in the active structure. This quantity, the transition value, was monitored as a function of simulation time, t, as depicted in Fig. 4. Because of thermal fluctuations, values near 1 or 0 are rarely sampled.
Fig. 4 contains data from a previously published (45) control simulation of inactive rhodopsin (black curve); because 11-cis retinal totally deactivates rhodopsin, this serves as a base level of conformational heterogeneity. By the time this control system was equilibrated it had already made an 0.3 transition along the reaction coordinate (from 1.0 to 0.7) simply because of thermal fluctuations.
Considering the two active-like starting structures—opsin and Meta-II—opsin appears to progress to a slightly more inactive-like state than Meta-II, only in part because of the single trajectory that underwent a deactivation event. This makes sense given that a range of experimental evidence suggests that opsin shows much lower activity than Meta-II rhodopsin.
Opsin samples a superset of Meta-II conformations
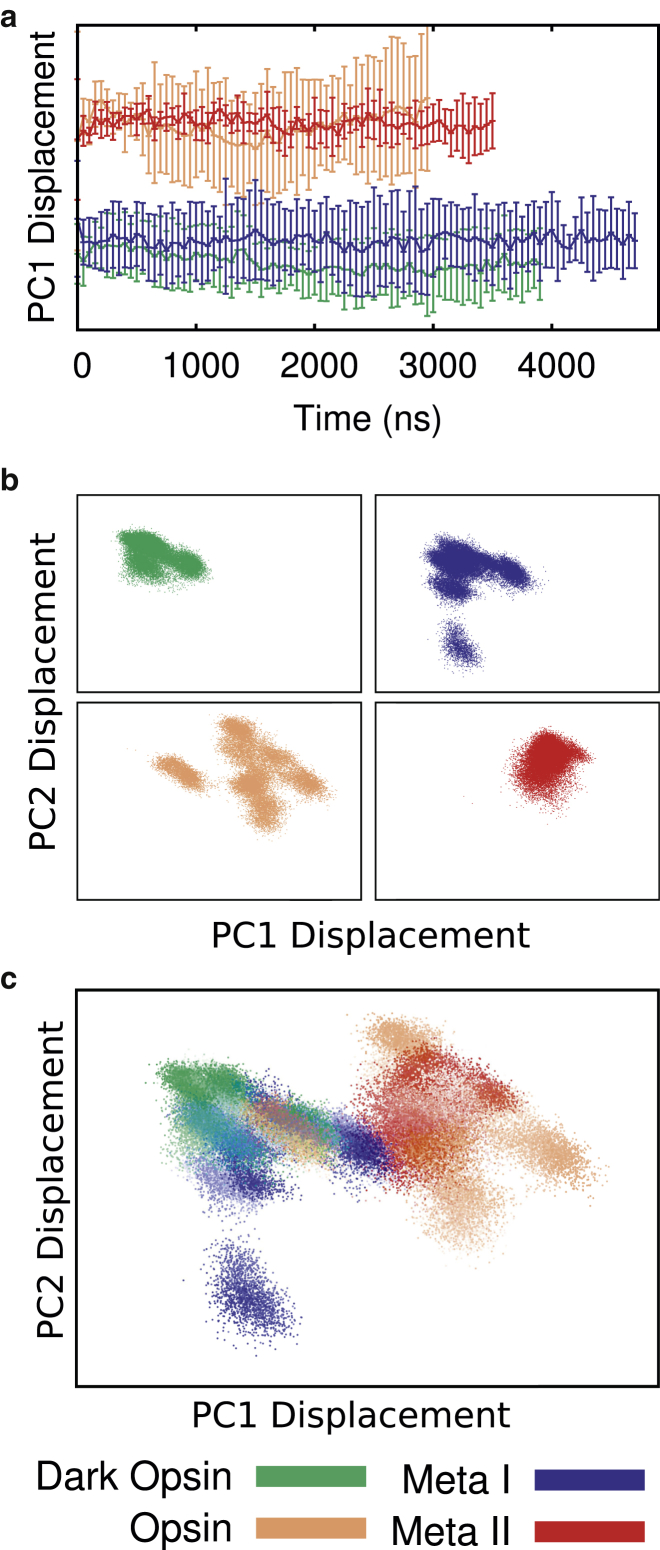
To better understand the conformational heterogeneity in our simulations, we next calculated the principal components of motion using only the transmembrane α-carbons. (Fig. 5). For this calculation we grouped all 24 trajectories together, so that the resulting eigenvectors form a common basis set describing the fluctuations across all four ensembles. Fig. 5 a shows the average projection of each simulation onto the first principal component. Each curve depicts the average of all six trajectories from the four different starting states.
Figure 5.
Ensemble-based principal component analysis was conducted on the transmembrane α-carbons of all 24 trajectories in aggregate, so the results depict how each simulation is projected onto the same basis set. Each of the trajectories used in this analysis is labeled according to its ensemble as shown in the key at the bottom of the figure. (a) Average projection of trajectory time course onto principal component one. Simulations are grouped by the four starting structures outlined in Table 1. Curves show the average across all six trajectories for each ensemble, with error bars showing the average of the standard deviations in each dataset, to emphasize which trajectories are more heterogeneous. (b and c) Projection onto the first two principal components. Each data point represents a structure from the ensemble of trajectories (sampled at 1 ns a piece) and colored according to ensemble. The x axis represents displacement along PC1, the y axis displacement along PC2. (b) Each ensemble is shown separately on the same scale. (c) Merged view of the result from (b). Opacity of the points shows the time evolution of each independent trajectory. To see this figure in color, go online.
Principal component 1 (Fig. 5 a) distinguished the active and inactive structures: the active-like opsin and Meta-II ensembles have similar average displacements, as do the inactive-like dark opsin and Meta-I. However, the opsin ensemble had a much larger standard deviation than Meta-II, indicating more disparity of structures in that dataset, driven at least in part by the simulation where the ionic lock closed. Along the same lines, dark opsin shows smaller fluctuations in this dimension than Meta-I, again consistent with Fig. 4. Meta-II trajectories also sampled a narrower range of conformations. In Fig. 5, b and c we expanded this analysis to the first two principal components. Every frame (sample rate = 1 ns) from the 24 simulations was plotted. Opsin (Fig. 5 b, lower left) sampled a superset of those conformations accessible to the fully active Meta-II (Fig. 5 b, lower right). Moreover, opsin had more dynamic range in PC1 and PC2 and some structures produced by opsin simulations overlapped with the inactive ensembles (Meta-I and dark opsin). Meta-I was also very dynamic—it accounted for the largest displacement in PC2, and produced conformations with displacements that adjoin the active-like ensembles. These results are also corroborated by projections onto the first three principal components (see Movie S1).
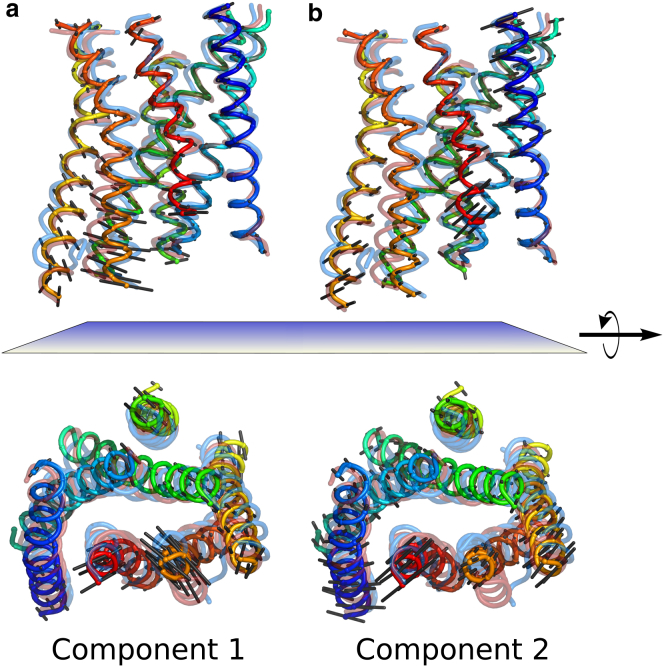
Although this analysis provided an intuitive way to understand the structural variation in our dataset, the biological meaning of these reduced coordinates was less clear. Therefore, we mapped the first two principal component eigenvectors back onto the structures themselves (Fig. 6). The dark-state (marine) and Meta-II (dark red) crystal structures are also shown for reference. The largest contribution to PC1 (Fig. 6 a) comes from the cytoplasmic portion of TM5 and TM6. The vectors around TM6 in particular appear to connect the inactive and active structures. By contrast, PC2 (Fig. 6 b) captures motion reminiscent of TM5 unwinding and the large variation in the NPxxY region. In contrast to AR, the motions of TM6 and the NPxxY region may be decoupled in rhodopsin activation.
Figure 6.
Mapping principal components back onto structures. (a) Illustration showing PC1. The first eigenvector is projected back onto a cartoon representation of the average structure from all 24 simulations. Rhodopsin is shown as a rainbow, with blue indicating the N-terminus. Black sticks project from the cα position in the direction of the first principal component, with a length proportional to their contribution to the motion. Transparent overlays show the Meta-II (78) (dark red) and dark-state (72) (marine) crystal structures. The top view shows rhodopsin in the plane of the membrane, the bottom view is normal to the membrane from the cytoplasmic side. (b) As in (a), except with sticks representing the second principal component of motion. To see this figure in color, go online.
Opsin and Meta-I explore conformations more rapidly
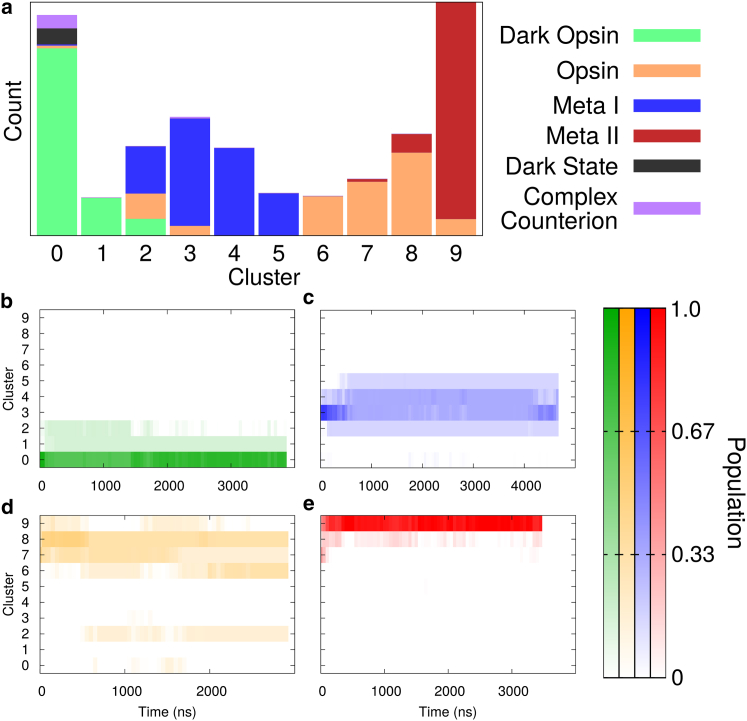
Visual inspection of Movie S1 shows that opsin and Meta-I were not only filling larger volumes in conformational space, but at the end of the simulations they were still spreading out, whereas dark opsin and Meta-II were more contained. This would suggest that our opsin and Meta-I simulations were not only more heterogeneous, but were also expanding more rapidly. To better understand this subtle difference, we used a structural clustering algorithm (Fig. 7), where the simulations were separated into k clusters based on RMSD of the transmembrane α-carbons. Results are shown using k = 10 clusters, but were robust when varying k from 5 to 50. We included our whole dataset of 24 trajectories, with two additional previously published simulations: the dark-state simulation and a transition simulation covering dark-state to Meta-I (referred to as the complex counterion trajectory in Grossfield et al. (45)).
Figure 7.
Structure-based clustering reveals a heterogeneity of substates in opsin and Meta-I. A k-means clustering algorithm based on RMSD was used on all 24 simulations in aggregate plus the dark-state and complex counterion simulations from Grossfield et al. (45). (a) Histogram showing the raw counts for each cluster. Data is colored according to starting structure. Cluster numbering is arbitrary. (b–e) Time course of clustering data for all six simulations with the same starting structure. The color intensity represents population, which is plotted for each cluster (y axis) as a function of simulation time (x axis). The data is averaged in 50 ns windows across all six trajectories for (b) dark opsin, (c) Meta-I, (d) opsin, and (e) Meta-II. To see this figure in color, go online.
Fig. 7 a shows a histogram of the population of each cluster, color-coded by the starting protein structure. Once again, this analysis identifies opsin and Meta-I as very heterogeneous. Structures produced by the opsin simulations (orange) occupy seven different bins, and those produced in the Meta-I simulations (blue) occupy five bins. By contrast, Meta-II and dark opsin occupy four and two clusters, respectively; they produced a more homogeneous ensemble of structures.
The time course of these conformational changes was even more revealing (Fig. 7, b–e). Here cluster population is shown as a function of simulation time, with each of the four ensembles depicted separately, but summed over the six replicas of each system. Because all simulations within an ensemble had the same starting structure, we expect them to occupy the same initial cluster and diverge from there, as can be seen in all four panels. Dark opsin (Fig. 7 b) occupied three bins, (nominally clusters 0, 1, and 2). The simulations began producing structures that fell into cluster 0, but very quickly there was some divergence into clusters 1 and 2. This population distribution then persisted for the first 1500 ns, until cluster 2 became unoccupied. However, instead of diverging further, the population of cluster 0 increased during the second half of the simulation time. In other words, the dark opsin ensemble was sampling structures more similar to its starting conformation after 1500 ns of simulation. We also saw that Meta-I and opsin were not only occupying more clusters, but their original clusters (numbers 3 and 8, respectively) were diminished at the end of the simulations (Fig. 7, c and d) and these trajectories continued to diverge. Meta-II (Fig. 7 e) simulations occupied three clusters, but the vast majority of this data fell into a single cluster, whereas the other two were only occupied transiently. This result suggests the more heterogeneous ensembles are continuing to diverge more rapidly, even after 3 to 4 μs of sampling.
Discussion
The results presented in this study indicate that retinal has a dramatic and complex influence on the conformational ensemble available to rhodopsin. All-trans retinal serves both to stabilize the active conformational ensemble and destabilize the inactive ensemble. Alternatively, without retinal opsin is more structurally diverse, even spontaneously sampling inactive-like structures (Figs. 3 e, 5 c, and 7 a).
In both retinal-containing ensembles, the β-ionone ring was quite dynamic (Fig. 2 b, left); this flexibility can also be seen in the individual trajectories (Figs. S1–S4). However, there are differences between Meta-I and Meta-II closer to the Schiff-base linkage. The C9-methyl (Fig. 2 b, middle) and C13-methyl (Fig. 2 b, right) adopted multiple different configurations in Meta-I simulations (see Fig. S5). An analogous result was observed by Lee and Lyman in the A2A adenosine receptor, suggesting that GPCR ligands are quite promiscuous within the binding pocket (79). We also observed heterogeneity closer to Lys2967.43 in previous simulations of Meta-I formation (47). However, in the Meta-II ensemble, the C13-methyl tended to point toward the cytoplasm () as seen in Figs. 2 b and S6. The portion of retinal linked to Lys2967.43 was less flexible in the Meta-II ensemble of simulations compared with Meta-I. This ligand stability correlated with the overall protein dynamics in the Meta-II ensemble, whereas Meta-I sampled a more diverse set of structures.
Interestingly, the residues comprising the rotamer toggle switch did not show a preference that segregates active-like and inactive-like ensembles; rather, multiple rotamer angles were sampled without leading to large-scale conformation changes in TM6. Although these highly conserved residues are implicated in GPCR activation, our simulations are consistent with recent work that casts doubt on any simplistic mechanism describing their action (10–18,71,77,80). We speculate that these residues may act through clutches as in a car’s transmission: when engaged, the clutches link these residues to a larger network that facilitates conformational change. When not engaged—the far more probable scenario from our limited data—rotameric transitions are not coupled to large-scale motions. Our present dataset is not sufficient to support this idea beyond speculation. Future work will be required to substantiate this claim and determine what precisely these clutches are, and how they are engaged.
The simulations presented here represents one of the larger collections of all-atom GPCR trajectories published, but it is clear these simulations have not converged to the full ensemble of conformations accessible to rhodopsin; the two opsin setups are chemically identical, as are the Meta-II and Meta-I simulations, and yet the different starting protein structures are enough to yield dramatically different ensembles. In essence, the simulations are long enough to sample well within a given state, but not long enough to explore transitions between states in a statistically significant way. This means that our efforts are more productively focused on understanding the dynamics of each state, so that we can connect the differences between states to protein function. The results suggest that the presence of retinal stabilizes the protein in a more active-like ensemble of conformations, which may be because of the specific retinal orientation discussed earlier (Figs. 2 b and S6, b and c). In effect, all-trans retinal may discourage rhodopsin from taking inactive structures—selecting for these active conformations. By contrast, Meta-I is consistently more active-like than dark opsin. Because all-trans retinal appears in situ within the binding pocket, it may serve a dual role, destabilizing the Meta-I ensemble as well as stabilizing Meta-II, playing a role similar to that suggested previously (41,42).
This is evident in the heterogeneity of Meta-I and the relative homogeneity of Meta-II (Figs. 4, 5, and 7). The opposite trend was seen when retinal was absent: inactive-like conformations were preferred. Interestingly, opsin was able to transition from an active-like conformation to an inactive one. Rather than thinking in terms of retinal inducing this conformational change or selecting/capturing a preferred conformation, we suggest thinking in terms of the underlying energy landscape. The presence (and state) of retinal is simply shifting the free energy minima. After all, we know that all conformations are possible—some just have vanishingly small probabilities.
From this data, it appears the ligand is not only changing the conformational ensemble accessible to the protein, but also how rapidly the protein explores the free energy landscape. It may be that ligand presence is altering the landscape, in this case effectively smoothing barriers (or increasing the attempt rate) to activation. This result argues in favor of the more recent views of allostery (e.g., 41–44).
Conclusions
Our results suggest that retinal has a substantial impact on rhodopsin dynamics—even this functionally switch-like protein is quite plastic at the molecular level. This is consistent with single-molecule experiments on , suggesting that there are diverse states sampled along the activation-deactivation continuum (21,25).
Rather than thinking in terms of a discrete set of substates, the emerging view is that proteins are more dynamic and sample a diverse ensemble of configurations. Although some of the biologically important motifs studied coincided with overall protein dynamics, others did not appear to correlate with any functionally related motion. One analogy to describe how allosteric information might be communicated is to think of these regions as a series of gears that are connected by (dis)engageable clutches. In this view, a motif can move freely—sampling many conformations without affecting the functional state—so long as the clutch connecting it to the protein at large is disengaged. Once engaged, that same motif has a vital impact on the overall structure of the protein. In this view, the purpose of the ligand would be to influence the position of these clutches. Understanding how these gears interconnect in the context of the protein’s free energy landscape will be a topic for much future work. This will need to be understood in the realm of statistical mechanics, going beyond the early phenomenological models of allostery. However, the picture of allostery is more complicated than solely selecting one conformation or directly inducing a conformation switch; the ligand can serve both functions.
Author Contributions
N.L. and A.G. designed the research, N.L., T.D.R., and S.A.D. performed the research, N.L. and A.G. wrote the article.
Acknowledgments
The authors thank Dejun Lin, Mark Dumont, and Jessi Leioatts for their critical review and suggestions to the manuscript. We also thank the University of Rochester Center for Integrated Research Computing and Health Sciences Center for Computational Innovation for the computational resources to run these simulations. This work was supported by NIH project number R01GM095496 (A.G.) and T32GM068411 (N.L.).
Editor: Nathan Baker
Footnotes
Nicholas Leioatts’s present address is Department of Theoretical and Computational Biophysics, Max Planck Institute for Biophysical Chemistry, Am Fassberg 11, 37077 Göttingen, Germany.
Eight figures, one table, and one movie are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00655-4.
Supporting Material
This movie shows the average displacement of simulations along the first three principal components (also called right singular vectors). All 24 simulations are shown colored according to the starting structure. The movie shows the time progression of each simulation. Dark opsin simulations, green; opsin simulations, orange; Meta-I simulations, blue; and Meta-II simulations, red.
References
- 1.Mombaerts P. Seven-transmembrane proteins as odorant and chemosensory receptors. Science. 1999;286:707–711. doi: 10.1126/science.286.5440.707. [DOI] [PubMed] [Google Scholar]
- 2.Firestein S. The good taste of genomics. Nature. 2000;404:552–553. doi: 10.1038/35007167. [DOI] [PubMed] [Google Scholar]
- 3.Granier S., Kobilka B. A new era of GPCR structural and chemical biology. Nat. Chem. Biol. 2012;8:670–673. doi: 10.1038/nchembio.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fredriksson R., Lagerström M.C., Schiöth H.B. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 5.Bahar I., Lezon T.R., Shrivastava I.H. Normal mode analysis of biomolecular structures: functional mechanisms of membrane proteins. Chem. Rev. 2010;110:1463–1497. doi: 10.1021/cr900095e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Overington J.P., Al-Lazikani B., Hopkins A.L. How many drug targets are there? Nat. Rev. Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 7.Lagerström M.C., Schiöth H.B. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 8.Palczewski K., Kumasaka T., Miyano M. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 9.Lefkowitz R.J. Historical review: a brief history and personal retrospective of seven-transmembrane receptors. Trends Pharmacol. Sci. 2004;25:413–422. doi: 10.1016/j.tips.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 10.Nygaard R., Frimurer T.M., Schwartz T.W. Ligand binding and micro-switches in 7TM receptor structures. Trends Pharmacol. Sci. 2009;30:249–259. doi: 10.1016/j.tips.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Crocker E., Eilers M., Smith S.O. Location of Trp265 in metarhodopsin II: implications for the activation mechanism of the visual receptor rhodopsin. J. Mol. Biol. 2006;357:163–172. doi: 10.1016/j.jmb.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 12.Elling C.E., Frimurer T.M., Schwartz T.W. Metal ion site engineering indicates a global toggle switch model for seven-transmembrane receptor activation. J. Biol. Chem. 2006;281:17337–17346. doi: 10.1074/jbc.M512510200. [DOI] [PubMed] [Google Scholar]
- 13.Schwartz T.W., Frimurer T.M., Elling C.E. Molecular mechanism of 7TM receptor activation—a global toggle switch model. Annu. Rev. Pharmacol. Toxicol. 2006;46:481–519. doi: 10.1146/annurev.pharmtox.46.120604.141218. [DOI] [PubMed] [Google Scholar]
- 14.Valentin-Hansen L., Groenen M., Schwartz T.W. The arginine of the DRY motif in transmembrane segment III functions as a balancing micro-switch in the activation of the β2-adrenergic receptor. J. Biol. Chem. 2012;287:31973–31982. doi: 10.1074/jbc.M112.348565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trzaskowski B., Latek D., Filipek S. Action of molecular switches in GPCRs—theoretical and experimental studies. Curr. Med. Chem. 2012;19:1090–1109. doi: 10.2174/092986712799320556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahalingam M., Martínez-Mayorga K., Vogel R. Two protonation switches control rhodopsin activation in membranes. Proc. Natl. Acad. Sci. USA. 2008;105:17795–17800. doi: 10.1073/pnas.0804541105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi L., Liapakis G., Javitch J.A. Beta2 adrenergic receptor activation. Modulation of the proline kink in transmembrane 6 by a rotamer toggle switch. J. Biol. Chem. 2002;277:40989–40996. doi: 10.1074/jbc.M206801200. [DOI] [PubMed] [Google Scholar]
- 18.Dror R.O., Arlow D.H., Shaw D.E. Activation mechanism of the β2-adrenergic receptor. Proc. Natl. Acad. Sci. USA. 2011;108:18684–18689. doi: 10.1073/pnas.1110499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leioatts N., Suresh P., Grossfield A. Structure-based simulations reveal concerted dynamics of GPCR activation. Proteins. 2014;82:2538–2551. doi: 10.1002/prot.24617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park P.S.-H., Lodowski D.T., Palczewski K. Activation of G protein-coupled receptors: beyond two-state models and tertiary conformational changes. Annu. Rev. Pharmacol. Toxicol. 2008;48:107–141. doi: 10.1146/annurev.pharmtox.48.113006.094630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kahsai A.W., Xiao K., Lefkowitz R.J. Multiple ligand-specific conformations of the β2-adrenergic receptor. Nat. Chem. Biol. 2011;7:692–700. doi: 10.1038/nchembio.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nygaard R., Zou Y., Kobilka B.K. The dynamic process of β(2)-adrenergic receptor activation. Cell. 2013;152:532–542. doi: 10.1016/j.cell.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Onaran H.O., Costa T. Where have all the active receptor states gone? Nat. Chem. Biol. 2012;8:674–677. doi: 10.1038/nchembio.1024. [DOI] [PubMed] [Google Scholar]
- 24.Perez D.M., Karnik S.S. Multiple signaling states of G-protein-coupled receptors. Pharmacol. Rev. 2005;57:147–161. doi: 10.1124/pr.57.2.2. [DOI] [PubMed] [Google Scholar]
- 25.Bockenhauer S., Fürstenberg A., Moerner W.E. Conformational dynamics of single G protein-coupled receptors in solution. J. Phys. Chem. B. 2011;115:13328–13338. doi: 10.1021/jp204843r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zocher M., Fung J.J., Müller D.J. Ligand-specific interactions modulate kinetic, energetic, and mechanical properties of the human β2 adrenergic receptor. Structure. 2012;20:1391–1402. doi: 10.1016/j.str.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bouvier M. Oligomerization of G-protein-coupled transmitter receptors. Nat. Rev. Neurosci. 2001;2:274–286. doi: 10.1038/35067575. [DOI] [PubMed] [Google Scholar]
- 28.Soubias O., Gawrisch K. The role of the lipid matrix for structure and function of the GPCR rhodopsin. Biochim. Biophys. Acta. 2011;1818:234–240. doi: 10.1016/j.bbamem.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soubias O., Teague W.E., Jr., Gawrisch K. Contribution of membrane elastic energy to rhodopsin function. Biophys. J. 2010;99:817–824. doi: 10.1016/j.bpj.2010.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feller S.E., Gawrisch K., Woolf T.B. Rhodopsin exhibits a preference for solvation by polyunsaturated docosohexaenoic acid. J. Am. Chem. Soc. 2003;125:4434–4435. doi: 10.1021/ja0345874. [DOI] [PubMed] [Google Scholar]
- 31.Brown M.F. Modulation of rhodopsin function by properties of the membrane bilayer. Chem. Phys. Lipids. 1994;73:159–180. doi: 10.1016/0009-3084(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 32.Lüdeke S., Beck M., Vogel R. The role of Glu181 in the photoactivation of rhodopsin. J. Mol. Biol. 2005;353:345–356. doi: 10.1016/j.jmb.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 33.Vogel R., Siebert F., Sheves M. Modulating rhodopsin receptor activation by altering the pKa of the retinal Schiff base. J. Am. Chem. Soc. 2006;128:10503–10512. doi: 10.1021/ja0627848. [DOI] [PubMed] [Google Scholar]
- 34.Sato K., Morizumi T., Shichida Y. Direct observation of the pH-dependent equilibrium between metarhodopsins I and II and the pH-independent interaction of metarhodopsin II with transducin C-terminal peptide. Biochemistry. 2010;49:736–741. doi: 10.1021/bi9018412. [DOI] [PubMed] [Google Scholar]
- 35.Lyman E., Zuckerman D.M. Ensemble-based convergence analysis of biomolecular trajectories. Biophys. J. 2006;91:164–172. doi: 10.1529/biophysj.106.082941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grossfield A., Zuckerman D.M. Quantifying uncertainty and sampling quality in biomolecular simulations. Annu. Rep. Comput. Chem. 2009;5:23–48. doi: 10.1016/S1574-1400(09)00502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neale C., Hsu J.C.Y., Pomès R. Indolicidin binding induces thinning of a lipid bilayer. Biophys. J. 2014;106:L29–L31. doi: 10.1016/j.bpj.2014.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park J.H., Scheerer P., Ernst O.P. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 39.Scheerer P., Park J.H., Ernst O.P. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 40.Kono M., Crouch R.K. In vitro assays of rod and cone opsin activity: retinoid analogs as agonists and inverse agonists. Methods Mol. Biol. 2010;652:85–94. doi: 10.1007/978-1-60327-325-1_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nussinov R., Tsai C.-J. Allostery in disease and in drug discovery. Cell. 2013;153:293–305. doi: 10.1016/j.cell.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 42.Tsai C.-J., Nussinov R. A unified view of “how allostery works.”. PLOS Comput. Biol. 2014;10:e1003394. doi: 10.1371/journal.pcbi.1003394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Motlagh H.N., Wrabl J.O., Hilser V.J. The ensemble nature of allostery. Nature. 2014;508:331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Motlagh H.N., Hilser V.J. Agonism/antagonism switching in allosteric ensembles. Proc. Natl. Acad. Sci. USA. 2012;109:4134–4139. doi: 10.1073/pnas.1120519109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grossfield A., Pitman M.C., Gawrisch K. Internal hydration increases during activation of the G-protein-coupled receptor rhodopsin. J. Mol. Biol. 2008;381:478–486. doi: 10.1016/j.jmb.2008.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martínez-Mayorga K., Pitman M.C., Brown M.F. Retinal counterion switch mechanism in vision evaluated by molecular simulations. J. Am. Chem. Soc. 2006;128:16502–16503. doi: 10.1021/ja0671971. [DOI] [PubMed] [Google Scholar]
- 47.Leioatts N., Mertz B., Brown M.F. Retinal ligand mobility explains internal hydration and reconciles active rhodopsin structures. Biochemistry. 2014;53:376–385. doi: 10.1021/bi4013947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baldwin P.A., Hubbell W.L. Effects of lipid environment on the light-induced conformational changes of rhodopsin. 2. Roles of lipid chain length, unsaturation, and phase state. Biochemistry. 1985;24:2633–2639. doi: 10.1021/bi00332a007. [DOI] [PubMed] [Google Scholar]
- 49.Feller S.E., Gawrisch K. Properties of docosahexaenoic-acid-containing lipids and their influence on the function of rhodopsin. Curr. Opin. Struct. Biol. 2005;15:416–422. doi: 10.1016/j.sbi.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 50.Yan E.C.Y., Kazmi M.A., Mathies R.A. Retinal counterion switch in the photoactivation of the G protein-coupled receptor rhodopsin. Proc. Natl. Acad. Sci. USA. 2003;100:9262–9267. doi: 10.1073/pnas.1531970100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim J.-M., Altenbach C., Khorana H.G. Structural origins of constitutive activation in rhodopsin: role of the K296/E113 salt bridge. Proc. Natl. Acad. Sci. USA. 2004;101:12508–12513. doi: 10.1073/pnas.0404519101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grossfield A., Feller S.E., Pitman M.C. A role for direct interactions in the modulation of rhodopsin by omega-3 polyunsaturated lipids. Proc. Natl. Acad. Sci. USA. 2006;103:4888–4893. doi: 10.1073/pnas.0508352103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.MacKerell A.D., Jr., Bashford D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 55.Mackerell A.D., Jr., Feig M., Brooks C.L., 3rd Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 56.MacKerell A.D., Jr., Feig M., Brooks C.L., 3rd Improved treatment of the protein backbone in empirical force fields. J. Am. Chem. Soc. 2004;126:698–699. doi: 10.1021/ja036959e. [DOI] [PubMed] [Google Scholar]
- 57.Huang J., MacKerell A.D., Jr. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu S., Brown M.F., Feller S.E. Retinal conformation governs pKa of protonated Schiff base in rhodopsin activation. J. Am. Chem. Soc. 2013;135:9391–9398. doi: 10.1021/ja4002986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Voet D., Voet J. 3rd ed. Wiley; Hoboken, NJ: 2004. Biochemistry. [Google Scholar]
- 61.Ryckaert J.-P., Ciccotti G., Berendsen H.J. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 62.Feller S.E., Zhang Y., Brooks B.R. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 63.Schneider T., Stoll E. Molecular-dynamics study of a three-dimensional one-component model for distortive phase transitions. Phys. Rev. B. 1978;17:1302–1322. [Google Scholar]
- 64.Essmann U., Perera L., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 65.Romo T.D., Leioatts N., Grossfield A. Lightweight object oriented structure analysis: tools for building tools to analyze molecular dynamics simulations. J. Comput. Chem. 2014;35:2305–2318. doi: 10.1002/jcc.23753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Changeux J.-P. The feedback control mechanisms of biosynthetic L-threonine deaminase by L-isoleucine. Cold Spring Harb. Symp. Quant. Biol. 1961;26:313–318. doi: 10.1101/sqb.1961.026.01.037. [DOI] [PubMed] [Google Scholar]
- 67.Lau P.-W., Grossfield A., Brown M.F. Dynamic structure of retinylidene ligand of rhodopsin probed by molecular simulations. J. Mol. Biol. 2007;372:906–917. doi: 10.1016/j.jmb.2007.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fukada Y., Okano T., Liu R.S. Comparative study on the chromophore binding sites of rod and red-sensitive cone visual pigments by use of synthetic retinal isomers and analogues. Biochemistry. 1990;29:3133–3140. doi: 10.1021/bi00464a033. [DOI] [PubMed] [Google Scholar]
- 69.Srinivasan S., Ramon E., Garriga P. Binding specificity of retinal analogs to photoactivated visual pigments suggest mechanism for fine-tuning GPCR-ligand interactions. Chem. Biol. 2014;21:369–378. doi: 10.1016/j.chembiol.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 70.Ahuja S., Crocker E., Smith S.O. Location of the retinal chromophore in the activated state of rhodopsin∗. J. Biol. Chem. 2009;284:10190–10201. doi: 10.1074/jbc.M805725200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hornak V., Ahuja S., Smith S.O. Light activation of rhodopsin: insights from molecular dynamics simulations guided by solid-state NMR distance restraints. J. Mol. Biol. 2010;396:510–527. doi: 10.1016/j.jmb.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Okada T., Sugihara M., Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. J. Mol. Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 73.Struts A.V., Salgado G.F.J., Brown M.F. Structural analysis and dynamics of retinal chromophore in dark and meta I states of rhodopsin from 2H NMR of aligned membranes. J. Mol. Biol. 2007;372:50–66. doi: 10.1016/j.jmb.2007.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Salgado G.F.J., Struts A.V., Brown M.F. Deuterium NMR structure of retinal in the ground state of rhodopsin. Biochemistry. 2004;43:12819–12828. doi: 10.1021/bi0491191. [DOI] [PubMed] [Google Scholar]
- 75.Struts A.V., Salgado G.F.J., Brown M.F. Solid-state 2H NMR relaxation illuminates functional dynamics of retinal cofactor in membrane activation of rhodopsin. Proc. Natl. Acad. Sci. USA. 2011;108:8263–8268. doi: 10.1073/pnas.1014692108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salgado G.F.J., Struts A.V., Brown M.F. Solid-state 2H NMR structure of retinal in metarhodopsin I. J. Am. Chem. Soc. 2006;128:11067–11071. doi: 10.1021/ja058738+. [DOI] [PubMed] [Google Scholar]
- 77.Miao Y., Nichols S.E., McCammon J.A. Activation and dynamic network of the M2 muscarinic receptor. Proc. Natl. Acad. Sci. USA. 2013;110:10982–10987. doi: 10.1073/pnas.1309755110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Choe H.-W., Kim Y.J., Ernst O.P. Crystal structure of metarhodopsin II. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 79.Lee J.Y., Lyman E. Agonist dynamics and conformational selection during microsecond simulations of the A(2A) adenosine receptor. Biophys. J. 2012;102:2114–2120. doi: 10.1016/j.bpj.2012.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Holst B., Nygaard R., Schwartz T.W. A conserved aromatic lock for the tryptophan rotameric switch in TM-VI of seven-transmembrane receptors. J. Biol. Chem. 2010;285:3973–3985. doi: 10.1074/jbc.M109.064725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This movie shows the average displacement of simulations along the first three principal components (also called right singular vectors). All 24 simulations are shown colored according to the starting structure. The movie shows the time progression of each simulation. Dark opsin simulations, green; opsin simulations, orange; Meta-I simulations, blue; and Meta-II simulations, red.