Abstract
In gene expression studies, it is critical to normalize data using a stably expressed endogenous control gene in order to obtain accurate and reliable results. However, we currently do not have a universally applied endogenous control gene for normalization of data for gene expression studies, particularly those involving 60Co γ-ray-exposed human blood samples. In this study, a comparative assessment of the gene expression of six widely used housekeeping endogenous control genes, namely 18S, ACTB, B2M, GAPDH, MT-ATP6 and CDKN1A, was undertaken for a range of 60Co γ-ray doses (0.5, 1.0, 2.0 and 4.0 Gy) at 8.4 Gy min−1 at 0 and 24 h post-irradiation time intervals. Using the NormFinder algorithm, real-time PCR data obtained from six individuals (three males and three females) were analyzed with respect to the threshold cycle (Ct) value and abundance, ΔCt pair-wise comparison, intra- and inter-group variability assessments, etc. GAPDH, either alone or in combination with 18S, was found to be the most suitable endogenous control gene and should be used in gene expression studies, especially those involving qPCR of γ-ray-exposed human blood samples.
Keywords: human peripheral blood lymphocyte (HPBL), gene expression, qPCR, endogenous control gene, 60Co γ-rays
INTRODUCTION
In the post-genomic era and the shifting paradigms of radiation biology, studies in the domain of molecular radiobiology involve assessment of gene expression following irradiation by techniques such as Quantitative Real-Time PCR (qPCR), DNA Microarray, etc. Such assessments require a complex mathematical algorithm involving an endogenous control gene [1–11]. An ideal endogenous control gene ought to be constitutively expressed and invariant for a range of experimental conditions and interventions, subjects, tissues, etc. A housekeeping gene meets these criteria and, hence, is normally used as the endogenous control gene to normalize background gene expression levels. A glance through the published literature shows that a range of endogenous control, reference or normalizer genes have been used in various studies. It is obvious that the different reference genes would vary in their native and induced expressions in response to treatments or experimental conditions, as well as between subjects, tissues, etc. [3, 9, 11–20]. For these reasons, combining the results of the various studies (and interlab comparison of results) is difficult. An inappropriate reference or control gene may also lead to misinterpretation of the gene expression data. However, to date there is no consensus on a universal endogenous control gene. Thus, there is an urgent need to standardize the procedure by finding one or two of the most suitable endogenous control genes (by consensus) that exhibit minimal variation in gene expression results and permit comparison of the findings in the various studies and laboratories.
Some attempts have been made in the past to identify stable and convenient endogenous control genes in human studies [10, 16, 21]. The 18S and β-actin genes have been used in irradiated human blood as reference genes for normalization [21–24]. In some studies, PPIB [16] and a combination of the TRAP1, FPGS, DECR1 and PPIB [10] genes have been used as reference genes in studies involving human peripheral blood. In other studies, while the GAPDH, B2M and ACTB genes were shown to be reliable reference genes in peripheral blood mononuclear cells in post-traumatic stress disorder patients [25], β-actin and TUBB1 were used as the reference genes in human skin fibroblasts after UVB irradiation [26]. Similarly, the 18S gene alone was used as a normalizer gene in irradiated human fibroblasts [27]. On the other hand, many reports show that the reference genes used were not stable. For example, low-dose X-ray irradiation was reported to downregulate β-actin up to 17 h post-radiation in human peripheral blood lymphocytes in vitro [28]. The expression of the CDKN1A gene was also demonstrated to be upregulated in the blood of patients undergoing total body irradiation [22]. While the 18S and B2M genes were reported to be unstable under different radiation qualities in two human cell lines, the GAPDH and ATP6 genes were reportedly stable and, hence, were used as the reference genes [29]. For obvious reasons, unstable genes do not make good normalizer genes. As a consequence of this, gene expression analysis using qPCR also utilized normalization to intergenic and intragenic regions of candidate radiation-responsive genes for dose prediction as well as reduced interindividual variations in the absence of untreated basal gene expression [30]. Hence, it is apparent that there is currently no universal reference gene that is stably and abundantly expressed under various experimental conditions and able to serve as an ideal and common endogenous control gene [2, 20, 31].
To the best of our knowledge and belief, so far no comparative assessment has been made between the commonly used endogenous control genes in human blood exposed to 60Co γ-rays in order to find the most suitable normalizer gene for gene expression studies. Therefore, the primary goal of this study was to make a comparative analysis of the commonly used endogenous control genes for their suitability to use in gene expression studies. To achieve this goal, we have examined six housekeeping genes, namely 18S (ribosomal protein), ACTB (β-actin), B2M (β-2-microglobulin), GAPDH (glyceraldehyde-3-phosphate dehydrogenase), MT-ATP6 (mitochondrially encoded ATP synthase 6) and CDKN1A (cyclin-dependent kinase inhibitor 1A), in human whole blood by qPCR, either immediately (0 h group) or at 24 h post-irradiation period following exposure to a range of doses of 60Co γ-rays.
MATERIALS AND METHODS
Sample preparation
Approximately 5 ml of blood was collected from each of six consenting volunteers (three males and three females) in the age range of 25–30 years. Each sample was distributed into five equal parts in 1.5 ml Eppendorf tubes. Tube 1 served as the sham-exposed control, while tubes 2 to 5 were exposed ex vivo to 60Co γ-rays at 0.5 Gy, 1.0 Gy, 2.0 Gy and 4.0 Gy, respectively (LDBI 2000 purchased from BRIT, BARC, Mumbai; dose rate of ∼8.39 Gy min−1). Equal volumes (0.25 ml each) of sham-exposed and irradiated samples were aliquoted into two new tubes. To each of these tubes, 0.25 ml RPMI-1640 (Sigma–Aldrich) medium supplemented with 10% heat-inactivated fetal bovine serum (Hi-Media) was added [23]. Since the post-irradiation incubation periods of 0 (immediately after irradiation) and 24 h were chosen for the two sets of samples, 0.75 ml of TRI Reagent BD (Sigma–Aldrich) and 25 μl of 5N acetic acid were added immediately in one set (0 h post-irradiation group), then mixed and stored at −50°C. The second set (24 h post-irradiation group) was transferred to a CO2 incubator (Thermo Fisher). After 24 h in the CO2 incubator (15% CO2; ∼100% humidity; 37°C), equal volume of TRI Reagent BD and 5N acetic acid were added and mixed as before. The samples were stored at −50°C until further use.
RNA isolation and cDNA synthesis
Total RNA was isolated directly from the frozen whole blood–TRI Reagent BD mixture according to the manufacturer's (Sigma–Aldrich's) instructions. RNA concentration and purity were estimated using a NanoDrop 2000c (Thermo Fischer), and A260/A280 values of >1.8 were considered to be satisfactory. For cDNA synthesis, a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) was employed with 1.0 µg of the RNA template and random hexamer primers, according to the manufacturer's instructions. The conditions of reactions in the thermo cycler were 25°C for 10 min, 37°C for 120 min and 85°C for 5 min. In order to check for genomic DNA contamination, reactions without Reverse Transcriptase (RT) were also run to serve as ‘−RT’ controls. Primers containing two exon boundaries were also employed to avoid genomic DNA contamination. The cDNA samples were stored at −50°C until further use.
qPCR analysis
For gene expression analysis of all six housekeeping genes (18S, ACTB, B2M, GAPDH, MT-ATP6 and CDKN1A), TaqMan Gene Expression Assays (Applied Biosystems) were employed (Table 1). The qPCR was carried out with an Applied Biosystems 7500 Fast Real-Time PCR system with 5 µl TaqMan Fast Universal PCR Master Mix in a 10-µl reaction volume. The optimized thermal cycling conditions in Fast Mode were 95°C for 2 min, 40 cycles at 95°C for 3 s each and 60°C for 30 s.
Table 1.
List of endogenous genes selected for this study
| Gene symbol | Gene name | Function | Assay ID |
|---|---|---|---|
| 18S | Eukaryotic 18S ribosomal RNA | Component of ribosomal subunit (40S) | Hs99999901_s1 |
| ACTB | Beta-actin | Cell motility, structure and integrity | Hs99999903_m1 |
| B2M | Beta 2-microglobulin | Component of MHC I on all nucleated cells, protein binding, antigen presentation | Hs99999907_m1 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | Glycolytic enzyme involved in the breakdown of glucose | Hs03929097_g1 |
| MT-ATP6 | Mitochondrially encoded ATP synthase 6 | Component of ATP synthase complex V, ATP production via oxidative phosphorylation | Hs02596862_g1 |
| CDKN1A | cyclin-dependent kinase inhibitor 1A | Regulatory enzyme in cell cycle progression | Hs00355782_m1 |
Data analysis and statistics
The threshold cycle (Ct) value, which is inversely proportional to the target mRNA abundance, was used to estimate the level of gene expression. The inverse of the Ct value (that is, 1/Ct), therefore, gives the abundance value of the mRNA. Relative stability was determined by the ΔCt method [18], comparing all possible gene combinations. The level of variability was indicated by the range of the standard deviation of the Ct values (StdDev) across samples. In this method, comparison of the ΔCt values of the different genes provides information on which pairs show less variability and hence which genes are stably expressed among the samples tested. A relatively large panel of genes can be compared against one another and either chosen or discarded on the basis of ΔCt. The average ΔCt is derived by dividing the ΔCt of one gene with that of another, and the average standard deviation is a measure of the gene expression variability. Further data analysis was carried out using NormFinder software [32]. NormFinder provides intra- and inter-variability, the best endogenous control, and also the best combination of two endogenous controls. The NormFinder applies a mathematical model to separate the analysis of the sample subgroups, estimates both the intra- and the intergroup expression variations, and calculates the stability value of a candidate gene. It works on a Microsoft Excel platform that automatically calculates the stability value for all candidate normalization genes containing any number of samples arranged in any number of groups. This approach ranks the best candidate gene with the minimal estimated intra- and intergroup variation, whereas the pair-wise comparison approach tends to select those genes with the highest degree of similarity across the sample sets. In the pair-wise comparison approach, the gene with the minimum expression variation does not necessarily get chosen as the best candidate gene. The most stable gene expression is indicated by the lowest average expression stability value. All data are shown as mean ± SD. One-way ANOVA was employed to determine the statistical significance of Ct values. Differences of P < 0.05 were considered statistically significant.
RESULTS AND DISCUSSION
Genome-wide studies have provided an insight into possible perturbations of biological functions in human peripheral blood lymphocytes (HPBLs) following γ-irradiation [33–35]. Exposure of HPBLs to environmental stresses, including ionizing radiation, is known to activate multiple signal transduction pathways, and rapidly results in complex patterns of gene expression change. As a biological material, human whole blood offers a great advantage, since circulating lymphocytes are both sensitive to early radiation injury and also highly responsive in terms of induced gene expression changes. As they are also relatively easily biopsied, non-stimulated HPBLs provide an ideal model for development of a gene expression biodosimeter for radiation exposure. qPCR is one of the most sensitive and reproducible relative quantification methods for gene expression analysis and provides simultaneous measurement of gene expression in many different samples. In qPCR, selection of an ideal housekeeping gene is an important criterion for a reliable and accurate interpretation of results. Therefore, any candidate housekeeping gene for the purpose of differential gene expression analysis should remain stably expressed between samples taken from different timepoints and under different experimental conditions [18]. The most commonly used housekeeping genes, such as 18S, GAPDH and ACTB are known to vary considerably in their transcriptional levels between different individuals, different cell types, different developmental stages and under different experimental conditions [19, 20, 36]. Even though the level of ribosomal 18S is not a direct indicator of mRNA level or gene expression, it is also used widely in gene expression analysis.
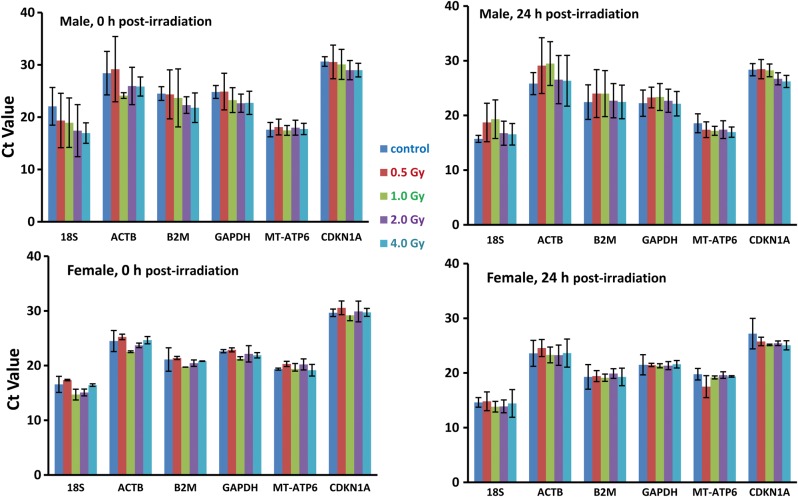
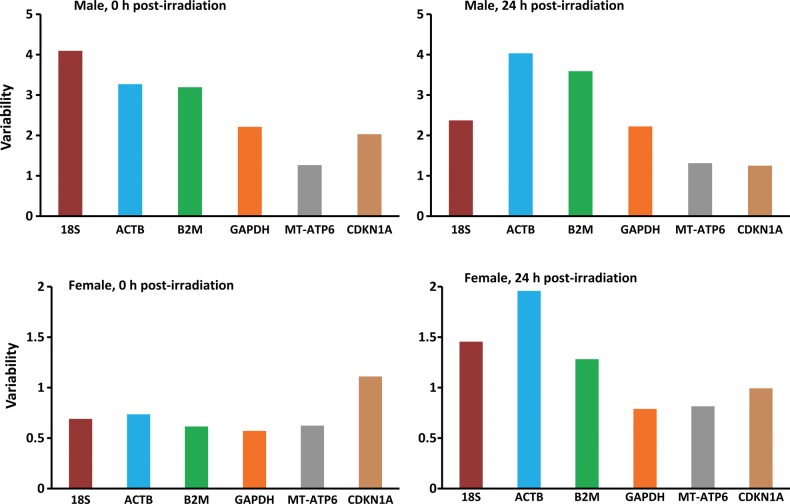
In this study, we first compared the gene expression levels of the six chosen genes by a direct Ct method, which gave some indication of the overall expression variations [25]. To analyze whether or not the gene expression was affected by γ-irradiation, the average Ct values for each group were compared (Fig. 1). The mean Ct value ranged from 15.71 to 30.65 in males (Fig. 1, top panels) and 13.83 to 30.5 in females (Fig. 1, bottom panels). It ranged between 14.7 and 30.5 at 0 h post-irradiation incubation (Fig. 1, left panels), whereas it ranged between 13.8 and 28.4 at 24 h post-irradiation incubation (Fig. 1, right panels). All six housekeeping genes exhibited essentially similar trends in both genders and post-irradiation incubation groups. We further analyzed the data to look for statistically significant difference between the two genders by one-way ANOVA between pairs of gender groups (Male Control vs Female Control; Male 0.5 Gy vs Female 0.5 Gy, etc.) The results suggest that there is no significant difference between the two (Supplementary Table 1). With the exception of MT-ATP6 for the 1.0, 2.0 and 4.0 Gy groups, the P values are greater than 0.05 and hence fail to reject the null hypothesis. Similarly, comparison of the Ct values combining both the genders also suggests that there is no statistical difference between different dose groups at 0 h and 24 h post-irradiation (Supplementary Table 2). We are aware that small sample size could be a critical factor in this outcome. The order of abundance (1/Ct) of the genes covered in this study, was 18S>MT-ATP6>B2M>GAPDH>ACTB>CDKN1A in all experimental groups comprising both gender and post-irradiation incubation period groups (Fig. 1). In order to determine the effect of radiation upon expression level, the Ct values of the control (or sham-exposed) samples were compared with exposed samples (Supplementary Table 3). In this case also, no significant dose effect was observed. However, different experimental groups individually exhibited intragroup variations. The standard deviation (STDev) of the average Ct values was used to represent the ‘range of variability’ of gene expression level. In males, the order of variability was 18S>ACTB>B2M>GAPDH>CDKN1A>MT-ATP6 at 0 h and ACTB>B2M>18S>GAPDH>MT-ATP6>CDKN1A at 24 h post-irradiation (Fig. 2, top panels), while in the case of females, the orders were CDKN1A>ACTB>18S>MT-ATP6>B2M>GAPDH at 0 h and ACTB>18S>B2M>CDKN1A>MT-ATP6>GAPDH at 24 h post-irradiation (Fig. 2, bottom panels). However, comparison of the variability of each gene across the dose range suggested that there were no significant differences in any of the groups. For instance, in the case of 18S, there was no significant difference in the level of expression between 18S and any other gene under comparison (Supplementary Table 4). From these results, the most abundantly expressed gene comes out to be 18S followed by MT-ATP6 in all the groups (Fig. 1). The minimum average Ct range was found for the MT-ATP6 gene in males and the GAPDH gene in females (Fig. 2).
Fig. 1.
Ct values representing the expression levels of six housekeeping genes in human whole blood samples in the sham-exposed control and groups exposed to 0.5 Gy, 1 Gy, 2 Gy and 4 Gy doses of 60Co γ-radiation at 0 (left panels) and 24 h (right panels) post-irradiation in male (top panels) and female (bottom panels) blood samples. The bars represent the statistical means of the Ct values for different individuals within a group, and the SD represents the range of variation within a group. (Differences of P < 0.05 were considered statistically significant.)
Fig. 2.
Gene variability as determined by comparison of Ct values. Variability of gene expression was estimated by comparing the standard deviations (StdDev) of the Ct values. The average StdDev represents the variation in gene expression level in the different experimental groups. (Differences of P < 0.05 were considered statistically significant.)
Gene expression stability was further evaluated by ΔCt and standard deviation (StdDev) methods by comparing all possible gene combinations [11, 18, 21]. The advantage of this approach was that it bypassed the need to accurately quantify input RNA, and instead employed ΔCt comparisons between the genes. This study involved six genes, making 30 possible gene combinations (Table 2). The increased level of the average StdDev of Ct values across the samples is indicative of the high variability and, therefore, low stability of gene expression, and vice versa. In this test, the genes that scored the highest for the requirements of being suitable endogenous controls were GAPDH and MT-ATP6 (Table 2). The least value of the average standard deviation was observed when the GAPDH and MT-ATP-6 genes were compared against the other five genes (0.755 and 0.781, respectively). CDKN1A and B2M demonstrated an intermediate level of variation (0.823 and 0.923, respectively), whereas 18S and ACTB demonstrated higher levels of variability (Table 2). The variability ranking of all of the endogenous genes covered in this study, therefore, emerged as GAPDH>MT-ATP6>CDKN1A>B2M>18S>ACTB. This result showed that expression of the GAPDH gene, followed by the MT-ATP6 gene, was the most stable in terms of expression across all the parameters in γ-ray-exposed HPBL samples. This also demonstrated that ionizing radiation had the least effect on these two genes, whereas the ACTB gene showed the maximum variation.
Table 2.
Pair-wise comparison of six housekeeping genes
| Sample | Average ΔCt | StdDev | Average StdDev |
|---|---|---|---|
| 18S vs ACTB | 0.885 | 0.918 | 1.597 |
| 18 s vs B2M | 1.605 | 1.599 | |
| 18S vs GAPDH | 2.086 | 1.880 | |
| 18S vs MT-ATP6 | 1.851 | 1.831 | |
| 18S vs CDKN1A | 1.572 | 1.756 | |
| ACTB vs 18S | 1.129 | 1.088 | 1.756 |
| ACTB vs B2M | 1.812 | 1.740 | |
| ACTB vs GAPDH | 2.090 | 2.046 | |
| ACTB vs MT-ATP6 | 2.090 | 1.99 | |
| ACTB vs CDKN1A | 1.775 | 1.912 | |
| B2M vs 18S | 0.622 | 0.625 | 0.923 |
| B2M vs ACTB | 0.551 | 0.574 | |
| B2M vs GAPDH | 1.299 | 1.175 | |
| B2M vs MT-ATP6 | 1.153 | 1.145 | |
| B2M vs CDKN1A | 0.979 | 1.098 | |
| GAPDH vs 18S | 0.479 | 0.531 | 0.755 |
| GAPDH vs ACTB | 0.424 | 0.488 | |
| GAPDH vs B2M | 0.769 | 0.850 | |
| GAPDH vs MT-ATP6 | 0.887 | 0.973 | |
| GAPDH vs CDKN1A | 0.753 | 0.934 | |
| MT-ATP6 vs 18S | 0.539 | 0.545 | 0.781 |
| MT-ATP6 vs ACTB | 0.478 | 0.501 | |
| MT-ATP6 vs B2M | 0.867 | 0.873 | |
| MT-ATP6 vs GAPDH | 1.126 | 1.026 | |
| MT-ATP6 vs CDKN1A | 0.848 | 0.959 | |
| CDKN1A vs 18S | 0.636 | 0.569 | 0.823 |
| CDKN1A vs ACTB | 0.563 | 0.522 | |
| CDKN1A vs B2M | 1.021 | 0.910 | |
| CDKN1A vs GAPDH | 1.327 | 1.070 | |
| CDKN1A vs CDKN1A | 1.177 | 1.042 |
Average ΔCt values represent mean difference between the genes across 30 samples. Standard deviation (StdDev) represents variation in Ct values across the samples.
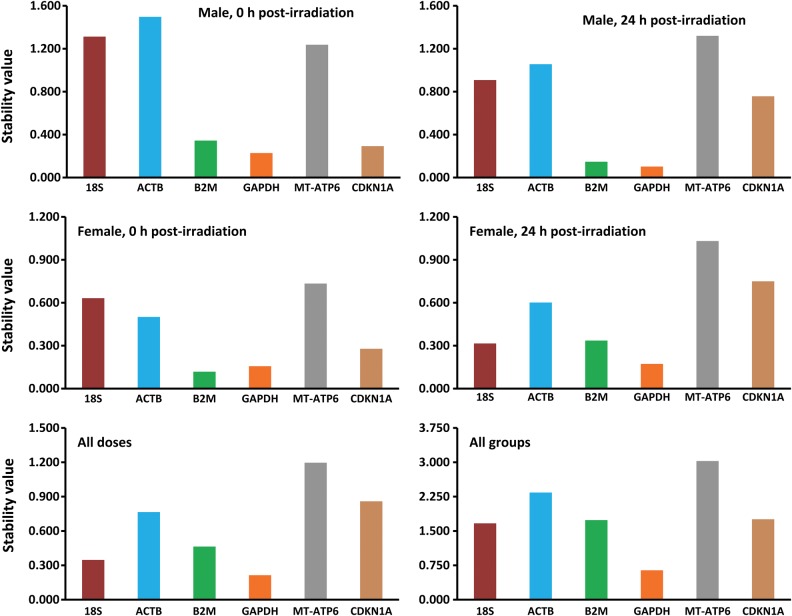
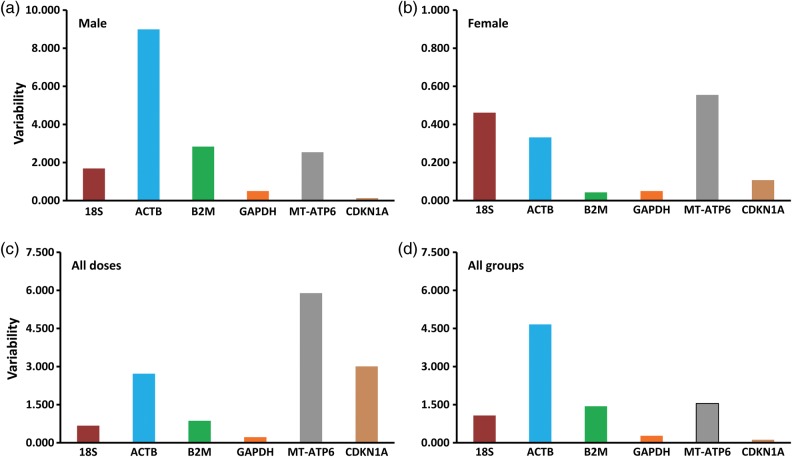
Since all the genes selected for this study have different functions, the possibility of coregulation or coordinate expression can be ruled out. The NormFinder algorithm, being rooted in a mathematical gene expression model, employs a solid statistical framework for estimating the variation between sample subgroups within a sample set [18, 26, 32]. In the earlier Ct approach, we could only estimate the overall gene expression variation, without taking into account the systematic intergroup variation, which is critical in correct interpretation of results [25, 37]. NormFinder can discriminate between different groups based on a given group identifier (e.g. 0 Gy, 1 Gy, 2 Gy and 4 Gy samples) and combines both intra- and inter-group variations into a stability value for each gene [14]. The gene with the lowest stability value signifies the most stable gene within the groups under investigation. Besides, it also suggests the best combinations of two genes within a group. The NormFinder algorithm ranks the six genes from irradiated HPBLs based on their expression stability, as shown in Fig. 3. Overall, the GAPDH gene was the most stably expressed gene with the lowest stability value, closely followed by the B2M and 18S genes (Table 3). The intragroup variations were also estimated by the NormFinder for each of the experimental groups (Fig. 4). In males, the CDKN1A and GAPDH genes were the least variable, whereas in females, the least variable were the B2M and GAPDH genes (Fig. 4, top panels). In the different dosage groups, GAPDH showed least variation, followed by 18S and B2M (Fig. 4, bottom left). When all experimental groups were combined, CDKN1A and GAPDH showed the least variation, followed by 18S (Fig. 4, bottom right). The best combination of two genes was also predicted by the NormFinder program for each experimental group (Table 3), with the best combination represented by the lowest stability values. The GAPDH and 18S genes, by far, appear to be the best combination of two genes to serve as the endogenous control under the experimental conditions employed in our study. The variability observed in the case of the β-actin and CDKN1A genes can be explained by earlier findings that showed the effect of radiation on the expression levels of these genes [22, 28].
Fig. 3.
The gene stability values of six housekeeping genes, as predicted by the NormFinder algorithm for a number of experimental groups comprising both genders (top and middle panels), two post-irradiation periods (top and middle panels), and the different irradiation groups together (bottom left panel), and both genders as well as irradiation groups together (bottom right panel). The lowest stability value indicates the most stable gene and vice versa.
Table 3.
Best combination of two genes predicted by NormFinder
| Male | Female | Different dosage | All doses and genders | |
|---|---|---|---|---|
| Gene Combination | GAPDH MT-ATP6 | B2M 18S | GAPDH 18S | GAPDH 18S |
| Stability Value | 0.63 | 0.41 | 0.2 | 0.8 |
Fig. 4.
Intragroup variations for six housekeeping genes from different study groups, namely: (a) male, (b) female, (c) all doses and (d) all groups combined. The variability value of each gene represents the level of variation of a gene across the different groups.
CONCLUSION
The results we obtained using a number of different approaches are essentially similar, suggesting that GAPDH is the most stable and abundant endogenous control gene, closely followed by the 18S gene. Therefore, from this study, we proposed that gene expression analysis involving qPCR of human whole blood exposed to ionizing radiation, such as 60Co γ-rays, should employ either the GAPDH gene alone or in combination with the 18S gene as the endogenous control for the most accurate and reliable interpretation of results. We do not rule out use of these endogenous controls in other gene expression studies involving interventions other than radiation.
SUPPLEMENTARY DATA
Supplementary data is available at the Journal of Radiation Research online.
FUNDING
The authors thank the funding agencies Board of Research in Nuclear Sciences (BRNS), (Department of Atomic Energy, India), Science and Engineering Research Board (SERB) (Department of Science and Technology, India) and International Atomic Energy Agency (IAEA) (Vienna, Austria) (Grant # 17047) for their shared financial support of this investigation.
Supplementary Material
REFERENCES
- 1.Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–22. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 2.Derveaux S, Vandesompele J, Hellemans J. How to do successful gene expression analysis using real-time PCR. Methods. 2010;50:227–30. doi: 10.1016/j.ymeth.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Dheda K, Huggett JF, Chang JS, et al. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal Biochem. 2005;344:141–3. doi: 10.1016/j.ab.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 4.Hellemans J, Vandesompele J. Selection of reliable reference genes for RT-qPCR analysis. Methods Mol Biol. 2014;1160:19–26. doi: 10.1007/978-1-4939-0733-5_3. [DOI] [PubMed] [Google Scholar]
- 5.Huggett JF, Foy CA, Benes V, et al. The digital MIQE guidelines: minimum information for publication of quantitative digital PCR experiments. Clin Chem. 2013;59:892–902. doi: 10.1373/clinchem.2013.206375. [DOI] [PubMed] [Google Scholar]
- 6.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 7.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfaffl MW. The ongoing evolution of qPCR. Methods. 2010;50:215–6. doi: 10.1016/j.ymeth.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Schmittgen TD, Zakrajsek BA. Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods. 2000;46:69–81. doi: 10.1016/s0165-022x(00)00129-9. [DOI] [PubMed] [Google Scholar]
- 10.Stamova BS, Apperson M, Walker WL, et al. Identification and validation of suitable endogenous reference genes for gene expression studies in human peripheral blood. BMC Med Genomics. 2009;2:49. doi: 10.1186/1755-8794-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barber RD, Harmer DW, Coleman RA, et al. GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol Genomics. 2005;21:389–95. doi: 10.1152/physiolgenomics.00025.2005. [DOI] [PubMed] [Google Scholar]
- 13.Derks NM, Muller M, Gaszner B, et al. Housekeeping genes revisited: different expressions depending on gender, brain area and stressor. Neuroscience. 2008;156:305–9. doi: 10.1016/j.neuroscience.2008.07.047. [DOI] [PubMed] [Google Scholar]
- 14.Jung M, Ramankulov A, Roigas J, et al. In search of suitable reference genes for gene expression studies of human renal cell carcinoma by real-time PCR. BMC Mol Biol. 2007;8:47. doi: 10.1186/1471-2199-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee PD, Sladek R, Greenwood CM, et al. Control genes and variability: absence of ubiquitous reference transcripts in diverse mammalian expression studies. Genome Res. 2002;12:292–7. doi: 10.1101/gr.217802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pachot A, Blond JL, Mougin B, et al. Peptidylpropyl isomerase B (PPIB): a suitable reference gene for mRNA quantification in peripheral whole blood. J Biotechnol. 2004;114:121–4. doi: 10.1016/j.jbiotec.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Radonic A, Thulke S, Mackay IM, et al. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun. 2004;313:856–62. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- 18.Silver N, Best S, Jiang J, et al. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol. 2006;7:33. doi: 10.1186/1471-2199-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki T, Higgins P, Crawford D. Control selection for RNA quantitation. Biotechniques. 2000;29:332–7. doi: 10.2144/00292rv02. [DOI] [PubMed] [Google Scholar]
- 20.Thellin O, Zorzi W, Lakaye B, et al. Housekeeping genes as internal standards: use and limits. J Biotechnol. 1999;75:291–5. doi: 10.1016/s0168-1656(99)00163-7. [DOI] [PubMed] [Google Scholar]
- 21.Chi C, Tian R, Liu H, et al. Follow-up study of abnormal biological indicators and gene expression in the peripheral blood of three accidentally exposed persons. J Radiat Res. 2013;54:840–51. doi: 10.1093/jrr/rrt022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amundson SA, Grace MB, McLeland CB, et al. Human in vivo radiation-induced biomarkers: gene expression changes in radiotherapy patients. Cancer Res. 2004;64:6368–71. doi: 10.1158/0008-5472.CAN-04-1883. [DOI] [PubMed] [Google Scholar]
- 23.Budworth H, Snijders AM, Marchetti F, et al. DNA repair and cell cycle biomarkers of radiation exposure and inflammation stress in human blood. PLoS One. 2012;7:e48619. doi: 10.1371/journal.pone.0048619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grace MB, McLeland CB, Gagliardi SJ, et al. Development and assessment of a quantitative reverse transcription-PCR assay for simultaneous measurement of four amplicons. Clin Chem. 2003;49:1467–75. doi: 10.1373/49.9.1467. [DOI] [PubMed] [Google Scholar]
- 25.Brkljacić J, Tanić N, Milutinović DV, et al. Validation of endogenous controls for gene expression studies in peripheral lymphocytes from war veterans with and without PTSD. BMC Mol Biol. 2010;11:26. doi: 10.1186/1471-2199-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Yan Y, Xu H, et al. 2011. Selection of reference genes for gene expression studies in ultraviolet B-irradiated human skin fibroblasts using quantitative real-time PCR. BMC Mol Biol. 2011;12:8. doi: 10.1186/1471-2199-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reuther S, Reiter M, Raabe A, et al. Effect of irradiation on the expression of DNA repair genes studied in human fibroblasts by real-time qPCR using three methods of reference gene validation. Radiat Environ Biophys. 2013;52:463–9. doi: 10.1007/s00411-013-0482-9. [DOI] [PubMed] [Google Scholar]
- 28.Miller AC, Luo L, Chin WK, et al. Proto-oncogene expression: a predictive assay for radiation biodosimetry applications. Radiat Prot Dosimetry 2002;(1–4):295–302. [DOI] [PubMed]
- 29.Sharungbam GD, Schwager C, Chiblak S, et al. Identification of stable endogenous control genes for transcriptional profiling of photon, proton and carbon-ion irradiated cells. Radiat Oncol. 2012;7:70. doi: 10.1186/1748-717X-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forrester HB, Sprung CN. Intragenic controls utilizing radiation-induced alternative transcript regions improves gene expression biodosimetry. Radiat Res. 2014;181:314–23. doi: 10.1667/RR13501.1. [DOI] [PubMed] [Google Scholar]
- 31.Haberhausen G, Pinsl J, Kuhn CC, et al. Comparative study of different standardization concepts in quantitative competitive reverse transcription-PCR assays. J Clin Microbiol. 1998;36:628–33. doi: 10.1128/jcm.36.3.628-633.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–50. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 33.Turtoi A, Brown I, Oskamp D, et al. Early gene expression in human lymphocytes after gamma-irradiation—a genetic pattern with potential for biodosimetry. Int J Radiat Biol. 2008;84:375–87. doi: 10.1080/09553000802029886. [DOI] [PubMed] [Google Scholar]
- 34.Turtoi A, Sharan RN, Srivastava A, et al. Proteomic and genomic modulations induced by γ-irradiation of human blood lymphocytes. Int J Radiat Biol. 2010;86:888–904. doi: 10.3109/09553002.2010.486016. [DOI] [PubMed] [Google Scholar]
- 35.Sharan RN, Turtoi A, Srivastava A, et al. Proteomic and genomic approach to understanding γ-radiation-induced early cellular response: biotechnology in radiation counter-measures. In: Hasnian SE, Jha B Rashmi, Sharan RN, editors. Biotechnology for Sustainable Development: Achievements and Challenges. New Delhi: McGraw Hill Education; 2010. pp. 193–208. [Google Scholar]
- 36.Glare EM, Divjak M, Bailey MJ, et al. Beta-actin and GAPDH housekeeping gene expression in asthmatic airways is variable and not suitable for normalising mRNA levels. Thorax. 2002;57:765–70. doi: 10.1136/thorax.57.9.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.