Abstract
Hadrontherapy is an advanced form of radiotherapy that uses beams of charged particles (such as protons and carbon ions). Compared with conventional radiotherapy, the main advantages of carbon ion therapy are the precise absorbed dose localization, along with an increased relative biological effectiveness (RBE). This high ballistic accuracy of particle beams deposits the maximal dose to the tumor, while damage to the surrounding healthy tissue is limited. Currently, hadrontherapy is being used for the treatment of specific types of cancer. Previous in vitro studies have shown that, under certain circumstances, exposure to charged particles may inhibit cell motility and migration. In the present study, we investigated the expression of four motility-related genes in prostate (PC3) and colon (Caco-2) cancer cell lines after exposure to different radiation types. Cells were irradiated with various absorbed doses (0, 0.5 and 2 Gy) of accelerated 13C-ions at the GANIL facility (Caen, France) or with X-rays. Clonogenic assays were performed to determine the RBE. RT-qPCR analysis showed dose- and time-dependent changes in the expression of CCDC88A, FN1, MYH9 and ROCK1 in both cell lines. However, whereas in PC3 cells the response to carbon ion irradiation was enhanced compared with X-irradiation, the effect was the opposite in Caco-2 cells, indicating cell-type–specific responses to the different radiation types.
Keywords: carbon ion irradiation, colony survival assay, motility genes, PC3 prostate adenocarcinoma, Caco-2 colon adenocarcinoma, gene expression
INTRODUCTION
In cancer radiation treatment, one of the main goals is to efficiently target the tumor, thereby sparing the surrounding, healthy tissue. In this respect, radiation therapy has made many advances since its initial use [1]. Of these advances, the use of hadrontherapy, which uses accelerated charged particle beams (such as protons and carbon ions), was proposed by Wilson in 1946 [2]. Charged particle beams offer the ballistic advantage of having an inverted depth–dose profile and a sharp dose fall-off after the Bragg peak [3], which results in more specific energy deposition to the tumor. Besides this more precise dose localization, high-linear energy transfer (LET) carbon ion therapy also offers biological advantages [4]. Hadrontherapy with carbon ions is more effective than conventional radiotherapy in inducing DNA damage, cell cycle arrest and cell death in tumor cells [5–7], which accounts for the highly lethal effects. Because the fate of irradiated cells is believed to be controlled by a network of signaling pathways, we recently investigated the effects of a different radiation types on genome-wide gene expression in human prostate adenocarcinoma (PC3) cells [8]. We performed a microarray study on PC3 cells after carbon ion irradiation and X-irradiation and observed a downregulation in several motility-related genes at 8 h after exposure [8] that was more pronounced after carbon ion irradiation compared with X-irradiation. This was in line with previous studies showing that exposure to different radiation types induces changes in the motility phenotype of cancer [9–20]. Studies investigating the effect of sublethal doses of photon irradiation of cancer cells indicated an increase in the migration and invasion potential of the cells [9, 10]. Additionally, in vitro studies comparing the effect of hadron beams with photon irradiation have demonstrated that particle beams decrease the migration potential of cancer cells, while photon irradiation induces only a slight decrease in (or even increases) the cells' capacity to migrate [16–20]. Because changes in the cellular phenotype originate from changes in gene expression, many studies have explored how a range of radiation types can change gene expression patterns [8, 17, 21–27]. These studies have generally indicated that charged particle beams have a more pronounced effect on gene expression with regard to both the number of differentially expressed genes, and also the magnitude of the changes [23–26]. Furthermore, pathway analysis has demonstrated that, besides typical radiation response pathways such as cell cycle regulation and DNA repair, genes involved in invasion and angiogenesis are differentially expressed, features which could eventually lead to an enhanced aggressive phenotype for surviving cancer cells. Other studies have focused more specifically on the radiation response of selected genes [17, 21, 24]. Meador et al. validated the radiation response of some genes, which had been found after microarray analysis of human lymphoblastoid and colon cancer cell lines to be differentially expressed [21]. They showed that the decreased expression of histone genes was dose- and time-dependent. Furthermore, expression of CDKN1A, a gene involved in cell cycle processes, showed a transient increase in expression after exposure to low-dose, low-LET γ-rays, which was prolonged after high-LET iron ion irradiation. Girdhani et al. focused on angiogenic genes in human non-small-cell lung carcinoma cells [24]. They reported that high-energy proton irradiation was able to induce a dose-dependent suppression of pro-angiogenic signaling genes. In co-culture experiments, they found that endothelial cell invasion was inhibited in cultures with irradiated cells, suggesting that proton irradiation modulates paracrine signaling to suppress angiogenesis [24]. Therefore, proton irradiation might have important biological consequences that should be considered in therapy. Akino et al. studied gene expression changes in ANLN and GADD45A (two genes involved in cell migration) after carbon ion and photon irradiation [17]. They also observed differential effects between carbon ion irradiation and X-irradiation. Whereas carbon ion irradiation induced downregulation of ANLN expression 12 h after irradiation and did not change GADD45A expression, X-irradiation did not alter ANLN expression and induced downregulation of GADD45A expression. Furthermore, these induced changes were also time-dependent. Boyden chamber assays confirmed a decreased migration and invasion potential after irradiation, which were dose- and radiation-type–dependent.
Other previous data has also indicated differences in radiation response depending on the cell type. Fujita et al. irradiated four pancreatic cell lines with carbon ions or X-rays to study changes in the cell phenotype. They observed that whereas carbon ion irradiation was able to reduce the migration potential in three out of four cell lines, a more aggressive phenotype was induced in one cell line [19]. Additionally, cell survival and gene expression are known to vary widely when different cancer cell types are exposed to ionizing radiation [21, 28, 29].
Although several studies have compared gene expression changes after exposure to a variety of radiation types, many questions still remain. Given the importance of cell motility in cancer progression, the present study specifically focused on the impact of carbon ion irradiation and X-irradiation on the expression levels of motility-related genes (CCDC88A, FN1, MYH9 and ROCK1.) Expression of these genes was evaluated both in the prostate adenocarcinoma cell line PC3 and in the colorectal adenocarcinoma Caco-2 cell line using RT-qPCR analysis at different time-points after radiation exposure. Our results demonstrated that different radiation types have a time-, dose- and cell-type–dependent response in the expression of these four genes.
MATERIALS AND METHODS
Cell culture
PC3 cells and Caco-2 cells were obtained from the American Type Culture Collection (ATCC, Molsheim Cedex, France). PC3 cells were cultured in Kaighn's Modification of Ham's F-12 Medium (F-12 K) (ATCC) supplemented with 10% fetal bovine serum (FBS) (GIBCO, Life Technologies, Ghent, Belgium). Caco-2 cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) (GIBCO) supplemented with 10% FBS and 1% non-essential amino acids (GIBCO). Cell cultures were maintained in a humidified incubator (37°C; 5% CO2). For each irradiation experiment, the same passage number (P30) of cells was used. Cell cultures were not synchronized in cell distribution prior to the irradiation experiments. Cell cultures were irradiated between 70% and 80% confluence. Cell cultures were regularly tested for mycoplasma contamination (DSMZ, Braunschweig, Germany).
X-irradiation
X-irradiation experiments were performed at the irradiation facility available at SCK•CEN (Mol, Belgium). Medium was replaced prior to irradiation in a horizontal position. Cells were exposed to different doses of X-rays (0, 0.1, 0.25, 0.5, 1, 2, 3 and 5 Gy for the colony survival assays and 0, 0.5 and 2 Gy for the gene expression analysis) using a Pantak HF420 RX machine (250 kV, 15 mA, 1.2 mm Al equivalent, 1 mm Cu-filtered X-rays and a calculated dose rate of 0.25 Gy/min).
Carbon ion irradiation
Cells were transported by car in a transportable incubator at 37°C to the Grand Accélérateur National d'Ions Lourds (GANIL) (Caen, France). For the gene expression analysis, cells were plated in 175-cm2 tissue culture flasks (Falcon; VWR) at SCK•CEN (Mol, Belgium) for transport and replated at GANIL for irradiation experiments. For the colony survival assays, 105 cells were plated in 12.5-cm2 tissue culture flasks (Falcon; VWR), which were used during irradiation. In the course of transportation, all culture flasks were completely filled with medium. After arrival, medium was changed and cells were placed overnight in a humidified incubator. The flasks were completely filled with medium to allow irradiation in a vertical position. The cells were irradiated with a 13C beam with an initial energy of 75 MeV/u (LET = 33.7 keV/µm). The requested doses were 0, 0.5, 1, 1.5, 2 and 3 Gy for the colony survival assays and 0, 0.5 and 2 Gy for the gene expression assay.
Colony survival assay
For the clonogenic assays, cells were seeded 2 d before irradiation at a concentration of 105 cells/T12.5 flask. After irradiation, cells were trypsinized, counted and replated in appropriate concentrations in six-well plates in culture medium with 1% penicillin–streptavidin (GIBCO). For each condition, triplicates were used. The exact number of cells seeded, used for calculating plating efficiency (PE), was determined with a separate sample that was fixed immediately after cells were allowed to adhere. After 11 d, the six-well plates were washed with phosphate buffered saline (PBS) (GIBCO) and fixed and stained with a 6% glutaraldehyde, 0.5% crystal violet solution (both Sigma–Aldrich, Bornem, Belgium) for at least 20 min. The samples were washed once with 40% EtOH and once with milliQ. Finally, colonies with more than 50 cells were counted. The surviving fraction (SF) was calculated based on the following formulae:
Using GraphPad Prism, the clonogenic survival curve was fitted to an LQ model given by the formula:
in which α and β are radiation sensitivity parameters and D is the dose. Relative biological effectiveness (RBE) at 10% survival is calculated by dividing the dose X-rays at SF10 by the dose carbon ions at SF10.
RNA extraction
For gene expression analysis, cells were plated at a concentration of 3.5 × 105 cells in 12.5-cm2 tissue culture flasks (Falcon; VWR). For each condition, four separate replicates were used. After irradiation, cells were further incubated for 2 h, 8 h or 24 h. Control samples were treated under similar conditions, including transportation and positioning identical to, and simultaneous with, that of treated samples. For RNA collection, medium was removed, and cells were rinsed with PBS and finally collected in 350 µl RLT lysis buffer (Qiagen, Venlo, The Netherlands) supplemented with β-mercaptoethanol (Sigma–Aldrich). Total RNA was isolated according to the manufacturer's instructions using the AllPrep DNA/RNA/protein mini kit (Qiagen). The quantity of RNA was measured with the NanoDrop Spectrophotometer (NanoDrop products, Wilmington, USA). RNA was stored at −80°C until further processing.
cDNA synthesis
cDNA was synthesized with the GoScript™ Reverse Transcription System (Promega, Leiden, The Netherlands) on a Gene Amp PCR System 2700 (Applied Biosystems, Foster City, CA, USA). We used 0.4 µg RNA in 20 µl reactions as per the manufacturer's instructions. cDNA samples were stored at −20°C until further reverse transcriptase PCR analysis.
RT-qPCR
Primers for target gene expression analysis (Table 1) were purchased as pre-made assays (TaqMan Gene Expression Assay) (Applied Biosystems). Assays were performed according to the manufacturer's instructions. Briefly, 2 µl cDNA was added to 1 µl Taqman Gene Expression Primer, 10 µl TaqMan® Fast Advanced Master Mix and 7 µl RNase free water. Assays were run on a 7500 Fast Real Time PCR system (Applied Biosystems). First, the efficiency of the primers was tested using a five-fold dilution series of an independent control sample. Expression ratios (R) were calculated using the method as described by Pfaffl [30]. Finally, data were normalized by a log2 transformation and data were represented as: Average log2(R) ± SD.
Table 1.
List of Applied Biosystems assays used
| Gene symbol | Gene name | Assay ID | Ref seq | Exon boundary | Measured efficiency |
|---|---|---|---|---|---|
| FN1 | Fibronectin 1 | Hs01549967_m1 | NM_002026.2; | 3–4 | 1.98 |
| MYH9 | Myosin; heavy chain 9; non-muscle | Hs01066369_m1 | NM_002473.4 | 23–24 | 1.96 |
| CCDC88A | Coiled-coil domain containing 88A | Hs01559766_m1 | NM_001135597.1 | 18–19 | 1.98 |
| ROCK1 | Rho-associated; coiled-coil containing protein kinase 1 | Hs01127714_mH | NM_005406.2 | 4–5 | 1.99 |
| B2M | Beta-2-microglobulin | Hs00984230_m1 | NM_004048.2 | 3–4 | 2.05 |
Statistical analysis of RT-qPCR
Statistics were performed with GraphPad Prism 5.00 (GraphPad Software, Inc., La Jolla, USA). The statistical significance of differences between log2(R) of control and each experimental condition was determined using one-tailed Mann–Whitney tests, based on the observed downregulation in our previous results [8]. P-values ≤ 0.05 were considered statistically significant.
RESULTS
Survival curves for PC3 and Caco-2 cells after carbon ion irradiation and X-irradiation
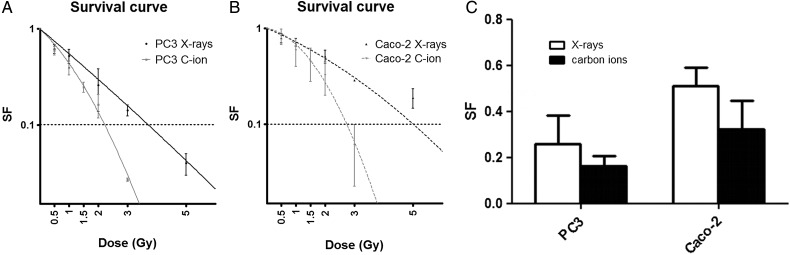
Colony survival was assessed 11 d after irradiation according to the protocol of Franken [31]. Figure 1 represents the colony survival curves of PC3 (Fig. 1A) and Caco-2 cells (Fig. 1B) exposed to different doses of carbon ion irradiation and X-irradiation. As expected, the surviving fractions for PC3 and Caco-2 cells irradiated with X-rays and carbon ions decreased with increasing doses. Doses used for carbon ion irradiations ranged from 0.5 to 3 Gy, and results were fitted according to an LQ model. Average α and β radio sensitivity parameters for these models for carbon ion irradiation were 0.67 ± 0.10 and 0.17 ± 0.04, respectively, for the PC3 cell line and 0.13 ± 0.16 and 0.26 ± 0.07, respectively, for the Caco-2 cells. For the X-ray experiments, doses ranged from 0.5 to 5 Gy. In this case, parameters α and β were 0.59 ± 0.12 and 0.01 ± 0.05, respectively, for the PC3 cell line and 0.30 ± 0.07 and 0.03 ± 0.03, respectively, for the Caco-2 cells. The RBE of carbon ions at 10% survival was calculated to be 1.67 for PC3 cells and 1.83 for Caco-2 cells. In view of our subsequent gene expression analysis, we also compared survival induced by 2 Gy irradiation. Survival fractions at 2 Gy irradiation are shown in Fig. 1C. The percentage cell survival was 2.2 times less for PC3 cells after carbon ion irradiation. For Caco-2 cells, survival at an equal dose of 2 Gy was reduced by a factor of 1.78.
Fig. 1.
Colony survival assay of PC3 and Caco-2 cells exposed to carbon ion or X-irradiation. (A–B) Survival fraction of PC3 (A) and Caco-2 (B) cells calculated using conventional clonogenic assays. The linear quadratic model was applied to experimental data. (C) Surviving fractions of clonogenic cells exposed to 2 Gy carbon ion or X-irradiation.
Dose- and time-dependency of expression of motility genes in PC3 cells after exposure to carbon ion irradiation and X-irradiation
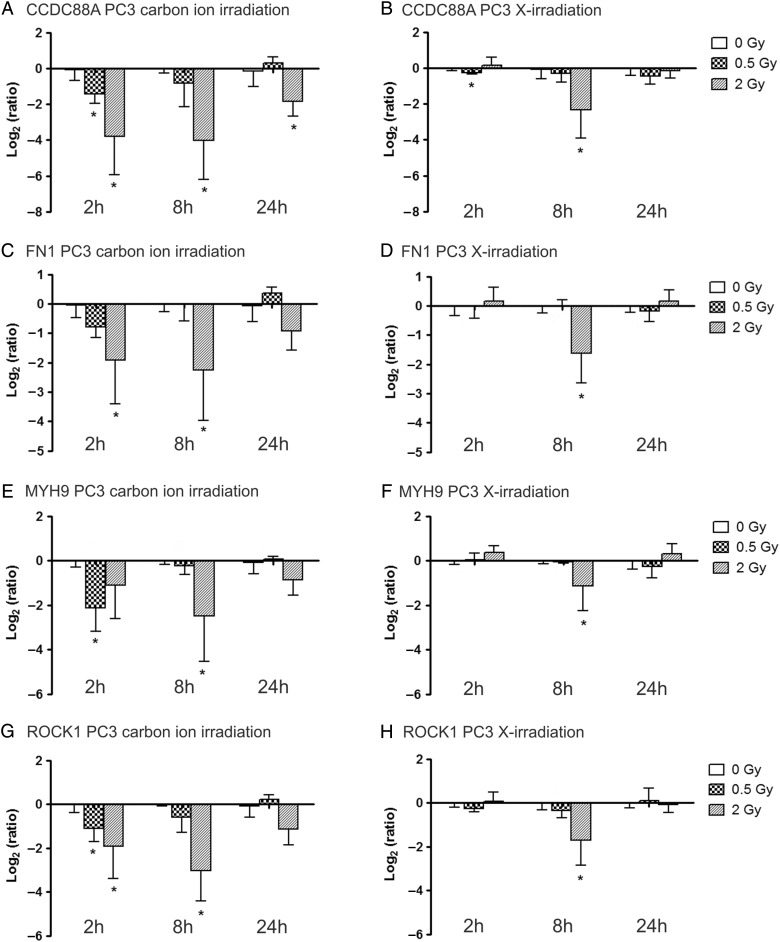
We previously performed a microarray analysis to evaluate gene expression changes in PC3 cells 8 h after exposure to carbon ion irradiation or X-irradiation [8]. Our results showed a high response of motility-related genes after both radiation types. Most of these genes were downregulated, and both the number of responsive genes and the magnitude of the change was found to be increased after carbon ion irradiation compared with X-irradiation. Based on this study, we decided to further investigate the gene expression profiles of the motility genes with the highest fold change, including CCDC88A, FN1, MYH9 and ROCK1, at a range of time-points (2 h, 8 h and 24 h) after exposure to different radiation types using RT-qPCR. Average log2 ratios of these genes in PC3 cells are presented in Fig. 2. Overall, the expression profiles showed that, for most conditions, radiation exposure resulted in a dose-dependent downregulation of these genes in PC3 cells, which was most pronounced at 8 h after irradiation for both beam types. In fact, for X-rays, we could only observe significant changes at 8 h post irradiation with a dose of 2 Gy. At this time-point, CCDC88A expression was reduced 1.7 times more after 2 Gy carbon ion irradiation (Fig. 2A) compared with X-irradiation (Fig. 2B) (respective log2 ratios −4.00 and −2.34). Exposure to 0.5 Gy of either beam type did not induce a significant downregulation in CCDC88A after 8 h, although a downregulating trend was observed. While no significant downregulation of this gene was observed 2 h and 24 h after 2 Gy X-irradiation, downregulation after carbon ion irradiation was more persistent over time (respective log2 ratios for 2 h and 24 h after irradiation were −3.79 and −1.83). Furthermore, 0.5 Gy carbon ion irradiation was also able to significantly downregulate CCDC88A expression 2 h after exposure.
Fig. 2.
Relative gene expression changes of four motility genes in PC3 cells at 2 h, 8 h and 24 h after carbon ion (left column) and X-irradiation (right column). Log2(ratio) of the expression of CCDC88A after carbon ion (A) and X-irradiation (B), FN1 expression after carbon ion (C) and X-irradiation (D), MYH9 expression after carbon ion (E) and X-irradiation (F) and ROCK1 expression after carbon ion (G) and X-irradiation (H) is presented. *marks significantly altered gene expression compared to CTRL samples (P-value ≤ 0.05) based on one-tailed Mann Whitney tests.
Similar patterns were observed for the expression of the other three genes. At the 8 h time-point, the reduction in gene expression after 2 Gy was ∼1.5 to 2 times stronger after carbon ion irradiation compared with after X-irradiation (FN1 Fig. 2C–D; MYH9 Fig. 2E–F; ROCK1 Fig. 2G–H). A downregulating trend was observed 24 h after 2 Gy carbon ion irradiation, which was not present after X-irradiation. While for MYH9, FN1 and ROCK1 expression no significant changes were found 2 h after X-ray exposure, significant downregulation was observed after carbon ion irradiation. Interestingly, this downregulation was found to be significant for 0.5 Gy for MYH9 expression levels (log2 ratio −2.12) but not for exposure to 2 Gy (log2 ratio −1.10). At this early time-point, expression of FN1 was found to be significant after 2 Gy, while ROCK1 expression was changed significantly after both 0.5 and 2 Gy carbon ion irradiation.
Dose- and time-dependency of expression of motility genes in Caco-2 cells after exposure to carbon ion irradiation and X-irradiation
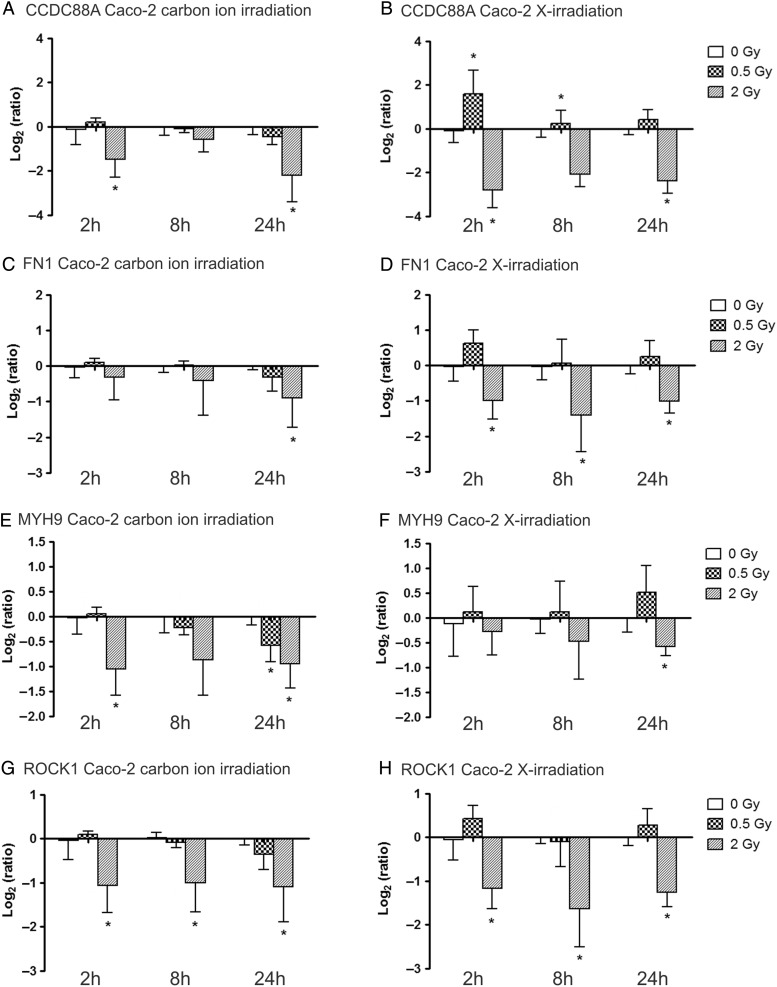
Average log2 (R) of gene expression profiles of CCDC88A, FN1, MYH9 and ROCK1 in Caco-2 cells are presented in Fig. 3. As for PC3 cells, clear dose-dependent changes in gene expression can be observed in irradiated Caco-2 cells for both beam types. CCDC88A expression levels (Fig. 3A) were found to be significantly downregulated 2 h and 24 h after 2 Gy carbon ion exposure (respective log2 ratios −1.48 and −2.20). At the 8 h time-point, a downregulating trend was observed. The lower dose of 0.5 Gy X-irradiation, however, induced upregulation in CCDC88A expression levels, which was found to be significant 2 h and 8 h after exposure (respective log2 ratios 1.58 and 0.25). Exposure to 2 Gy X-rays (Fig. 3B) resulted in more pronounced downregulation in CCDC88A expression when compared with carbon ion exposure. Log2 ratios were significantly different from control samples 2 h and 24 h after irradiation (respectively, −2.79 and −2.39), while a similar trend was observed 8 h after irradiation.
Fig. 3.
Relative gene expression changes of four motility genes in Caco-2 cells at 2 h, 8 h and 24 h after carbon ion (left column) and X-irradiation (right column). Log2(ratio) of the expression of CCDC88A after carbon ion (A) and X-irradiation (B), FN1 expression after carbon ion (C) and X-irradiation (D), MYH9 expression after carbon ion (E) and X-irradiation (F) and ROCK1 expression after carbon ion (G) and X-irradiation (H) is presented. *marks significantly altered gene expression compared to CTRL samples (P-value ≤ 0.05) based on one-tailed Mann Whitney tests.
Carbon ion irradiation induced a downregulating trend in FN1 expression levels (Fig. 3C) that was found to be significant 24 h after exposure (log2 ratio −0.90). Compared with X-irradiation, FN1 gene expression was less altered after exposure to carbon ion irradiation. FN1 expression levels were persistently downregulated after 2 Gy X-irradiation (Fig. 3D) (respective log2 ratios after 2 h, 8 h and 24 h were −0.99, −1.41 and −1.01). Exposure to 0.5 Gy X-rays induced a very subtle upregulation, which was not found to be statistically significant at either time-point.
This was not the case for MYH9 expression (Fig. 3F), where subtle changes with high variability were observed after X-ray exposure. However, after 24 h, downregulation was found to be significant for 2 Gy (log2 ratio −0.59). Exposure to 2 Gy carbon ion irradiation (Fig. 3E) was associated with small but significant downregulation of MYH9 at the 2 h and 24 h time-points (respective log2 ratios −1.06 and −0.95). This was also the case for exposure to 0.5 Gy analyzed 24 h after irradiation (log2 ratio −0.58). At the 8 h time-point, no significant changes were found, but a dose-dependent downregulating trend was observed.
Finally, ROCK1 expression (Fig. 3G–H) was persistently downregulated after 2 Gy irradiation of both beam types at the three different time-points (respective log2 ratios after carbon ion exposure −1.06, −1.00 and −1.10; respective log2 ratios after X-irradiation −1.17, −1.63 and −1.26). Changes induced by 0.5 Gy of both beam types were not found to be significant.
Comparing the irradiation responses of PC3 and Caco-2 cells, it is clear that changes in the expression of motility genes CCDC88A, FN1, MYH9 and ROCK1 are cell-type–dependent in their response to different beam types. In PC3 cells, the strongest response was observed after carbon ion radiation (Fig. 2), whereas Caco-2 cells in general responded more to X-irradiation (Fig. 3). Dose-dependent changes were observed in both cell lines at the different time-points. However, for PC3 cells, expression patterns were generally downregulated for all doses, whereas for Caco-2 cells, 0.5 Gy X-rays induced an upregulating trend that was found to be significant for CCDC88A expression levels (Fig. 3B). Also, time-dependent changes differed between the cell lines. Whereas X-irradiation of PC3 cells induced a fluctuating gene expression profile with the strongest response after 8 h, gene expression levels in Caco-2 cells was more persistently downregulated for up to 24 h for both beam types. Furthermore, expression levels induced by a particular beam type showed a similar pattern for all four genes in PC3 cells, while the response in Caco-2 cells was more variable (in particular the response of FN1 expression (Fig. 3C) after carbon ion exposure and MYH9 expression (Fig. 3F) after X-irradiation).
DISCUSSION
Radiation therapy plays an important role in the management of cancer, with almost half of all cancer patients receiving radiation therapy at some point during their treatment. Recently, a more advanced form of radiotherapy, using accelerated particle beams instead of photons, has become an interesting approach for patient treatment. In order to better understand the fate of an irradiated cancer cell, many studies have been conducted in order to elucidate the network of gene and signaling pathways triggered by different types of ionizing radiation. We previously investigated differential gene expression in PC3 cells 8 h after carbon ion irradiation and X-irradiation, and demonstrated a large number of differentially expressed genes and pathways, among which was a gene signature of motility-related genes, which was in general downregulated after radiation exposure [8]. In order to further explore the observed changes, a more detailed analysis of the alteration in the expression of four motility-related genes (CCDC88A, FN1, MYH9 and ROCK1) was performed after exposure to carbon ions and X-rays in PC3 cells at a range of time-points (2 h, 8 h and 24 h). In addition, Caco-2 colon cancer cells were included in the experimental set-up in order to verify whether the observed changes were cell type-dependent. As a first step, the radio sensitivity of the cancer cells lines and the RBE of the carbon ion beam were determined using the colony survival assay after exposure to both radiation types.
Radio sensitivity of PC3 and Caco-2 cell lines
The LQ model was applied to the colony survival data to examine the radio sensitivity of PC3 and Caco-2 cells exposed to carbon ion irradiation and X-irradiation. These data clearly showed that PC3 cells were more radio sensitive to both beam types compared with Caco-2 cells. The high α values of the parameters in the LQ model of the PC3 cells indicate the higher intrinsic radio sensitivity of this cell line compared with Caco-2 cells. Furthermore, elevated β values after carbon ion irradiation in both cell lines indicates a longer time would be required to achieve complete repair. Interestingly, the RBE is slightly higher for Caco-2 cells compared with PC3 cells, indicating that particle irradiation is relatively more effective on radio resistant cells. To our knowledge, colony survival assays of PC3 and Caco-2 cells exposed to carbon ion irradiation have not previously been performed. However, Friedrich et al. performed a systematic analysis of RBE values of ion beam experiments with several cancer cell lines [32]. In that study, the calculated RBE values at 10% survival were plotted against the LET of the ion beams used, and the results demonstrated that experiments with LET < 50 keV/µm yielded RBE values of around 2, which is consistent with our data. Balcer–Kubiczek et al., who performed irradiation of PC3 cells with iron ions (1 GeV/nucleon) or X-rays, found similar survival rates for PC3 cells after 5 Gy X-rays (± 5%) [33]. Survival after 2.5 Gy iron ion irradiation (± 3%) was slightly lower compared with the carbon ion irradiated samples (± 6%), which also showed a slightly higher RBE value (calculated at 10% survival = 2.4). However, these differences could be explained by the type and LET value of the particle beam.
Radiation-induced expression patterns in genes coding for actin-binding proteins
We (and others) have previously shown that both photons and particle beams have an impact on motility gene signaling in cancer cells [8, 17, 19, 23, 24, 34]. Our results have demonstrated that this downregulation is stronger after carbon ion exposure (LET = 33.7 keV/µm) compared with after X-irradiation [8]. In view of the importance of cell motility in cancer progression, the present study further investigated these observations.
Three genes coding for actin-binding proteins (CCDC88A, MYH9 and ROCK1) were selected for gene expression analysis in two cancer cell lines at different time-points after exposure to X-rays or carbon ions. In PC3 cells, downregulation in gene expression was most prominent 8 h after irradiation, and this was observed for both beam types. The magnitude of the changes was dose-dependent and stronger after carbon ion irradiation compared with X-irradiation. Furthermore, carbon ion irradiation induced significant downregulation at a very early time-point after exposure (2 h). This downregulating trend, although no longer significant, was still visible 24 h after irradiation. In contrast, X-irradiation induced no significant changes in gene expression at these time-points. These results clearly demonstrate that in PC3 prostate cancer cells, the magnitude of downregulation is dependent on radiation type, as well as on the time-point analyzed. Interestingly, in Caco-2 cells, CCDC88A and ROCK1 gene expression changes were more responsive to X-rays compared with carbon ions. Also, Girdhani et al. observed radiation type–dependent changes in gene expression [24]. They focused on how different radiation types could modulate critical processes in tumor progression, such as angiogenesis, invasion and proliferation. Microarray analysis of A549 carcinoma cells exposed to proton irradiation or X-irradiation showed changes in expression patterns of a number of angiogenesis-regulating genes. RT-PCR analysis showed VEGF, IL-8, IL-6 and HIF1-A expression to be downregulated 6 h after proton irradiation, whereas X-irradiation induced a dose-dependent upregulation in these genes.
It is well known that remodeling of the actin–myosin skeleton, by the activation of the serine/threonine kinase Akt, plays an important role in migration, invasion and metastasis [35]. Previous studies demonstrated that CCDC88A, MYH9 and ROCK1 are involved in the progression of various tumor types [35–40]. Although, to our knowledge, the responsiveness of these genes to different radiation types has not been discussed before, radiation-induced gene expression changes related to the Akt pathway have been observed in other studies. Akino et al. investigated radiation-induced changes in the metastatic potential of human A459 lung carcinoma cells [17]. They irradiated their cells with equitoxic doses of carbon ions and X-rays, after which microarray analysis indicated several differentially expressed genes, amongst which was anillin (ANLN). Similar to the genes that we focused on, ANLN codes for an actin-binding protein and is regulated by the PI3 K/Akt pathway. They also demonstrated that radiation decreased the migration potential of the cells, and this could be explained by the observed genetic changes. Decreased expression of our three actin-binding proteins after irradiation could be an indication that in our PC3 cell line too, the motility potential of these cells is changed by radiation exposure. In contrast, radiation-induced expression profiles in Caco-2 cells were very different. This shows that radiation-induced gene expression of these genes is cell-type–dependent and could be an indication that cell motility processes might be differently affected in Caco-2 cells and PC3 cells after irradiation. Furthermore, it should also be noted that irradiated samples taken for gene expression analysis contain a mixture of both doomed and surviving cells. Furthermore, the beam types used have distinct physical characteristics, which induce damage in different ways and might therefore trigger different signaling mechanisms. Since repair kinetics can differ greatly between cell lines and depend on the induced damage (i.e. the type of radiation used), this could explain differences between both cell lines and radiation-type–induced changes.
As mentioned previously, in addition to cell-type–dependent changes, we also observed time-dependent changes in our three motility genes. For carbon ion irradiation, gene expression for CCDC88A, MYH9 and ROCK1 in PC3 cells was significantly reduced 2 h and 8 h after exposure to 2 Gy. For the same dose of X-irradiation, downregulation was only significant 8 h after exposure. In contrast, in Caco-2 cells, ROCK1 expression was persistently downregulated after both carbon ion irradiation and X-irradiation. For CCDC88A expression, downregulation after 2 Gy was found to be significant after 2 h and 24 h; however, exposure to 0.5 Gy induced a significant upregulation of the gene 2 h and 8 h after exposure. Finally, MYH9 expression showed a clear dose-dependent downregulation, which was again significant 2 h and 24 h after carbon ion irradiation, whereas X-rays induced very small and variable changes in gene expression. Time-dependent changes were also found in the studies of Akino et al. [17] and Meador et al. [21]. Furthermore, Akino et al. also observed that time-dependency of gene expression differed between the two radiation types.
Seeing the vast differences in gene expression changes of these three motility genes concerning time-, cell type- and radiation-type–dependency it remains to be clarified whether these genes are involved in radiation-induced changes in the migration potential of both cancer cell lines (and, if so, to what extent). Although we are the first to compare the responsiveness of these three motility genes after exposure to different radiation types, two previous studies have already investigated the involvement of ROCK1 in photon radiation–induced cellular invasion [19, 34]. Zhai et al. observed a dose-dependent increase in invasive potential in three photon irradiated gliobastoma cell lines in vitro. They found that by inhibiting ROCK1, this increase in invasion was stopped; however, blocking ROCK1 without radiation exposure had no effect. In addition, Fujita et al. irradiated pancreatic cancer cells with carbon ions or X-rays and observed a switch from a mesenchymal mode of motility to a protease-independent mechanism of invasion [19]. They concluded that ROCK signaling is involved in this invasive phenotype and, furthermore, that inhibition of ROCK is needed to block invasiveness of the cells, whereas carbon ion radiation is only capable of decreasing the migration potential.
Radiation-induced changes in FN1 gene expression
FN1 codes for a glycoprotein involved in integrin signaling [41, 42], and thereby plays a role in cell adhesion and migration. This gene has been demonstrated to be involved in tumor metastasis [43–45]. With respect to radiation treatment, this gene has previously been identified to be a possible biomarker for radiation resistance in head and neck cancers [46]. We previously showed that high FN1 expression levels are correlated with poor prognosis in prostate cancer patients [8]. We observed that, in PC3 cells, changes in FN1 gene expression showed a similar pattern to the actin-binding gene expression profile. In contrast, in Caco-2 cells the expression of FN1 was completely different.
So far, few articles have reported on FN1 gene expression changes induced by radiation. Two papers investigated the impact of radiation on FN1, although no firm conclusions can be drawn from their studies [47, 48]. More research will be needed to further investigate how FN1 gene expression is influenced by ionizing radiation.
Relation between RBE and gene expression
RBE assessment by the colony survival assay has long since been a gold standard in determining the radiation sensitivity of cell lines and tissues. Although there is no doubt that the RBE value is of huge importance in the clinical world, many difficulties are encountered when analyzing the data obtained in radiobiological experiments. Even though RBE values (based on 10% survival) can be similar for different cell types, gene expression responses vary over time and can differ widely between cell lines. Therefore, in the context of gene expression analysis, we suggest that it can be useful to compare equal doses rather than equitoxic doses.
CONCLUSIONS
In the present study, two different cancer cells lines were exposed to different radiation types. Clonogenic survival assays demonstrated that PC3 prostate cancer cells showed a higher sensitivity to both X-rays and carbon ions compared with Caco-2 colon cancer cells. Moreover, clear dose-, time- and radiation-type–dependent changes in motility gene expression were observed for each cell line. The RBE (at 10% survival) was higher for Caco-2 cells, whereas gene expression changes were more pronounced in PC3 cells. These results indicate that gene expression changes induced by different radiation types are highly cell-type–dependent. Further research is needed to better understand how different radiation types influence motility gene expression and whether this will affect the behavior of a particular cancer cell type.
FUNDING
This work is partly supported by the Federal Public Service in the context of the feasibility study ‘Application of hadrontherapy in Belgium’, which is part of action 30 of the Belgian cancer plan (CO-90-2088-01). Annelies Suetens is a recipient of a SCK•CEN-UCL PhD grant. This work is currently supported by the European Space Agency / Belgian Science Policy (C4000109861). Carbon ion beam time was provided by Grand Accélérateur National d’Ions Lourds (P911-H). Funding to pay the Open Access publication charges for this article was provided by SCK•CEN.
ACKNOWLEDGEMENTS
We would like to thank the iPAC committee of GANIL for the beam time granted (P911-H) and the staff of the LARIA, CIRIL (GANIL, Caen, France) for allowing us access to and use of their facility. We also thank Bart Marlein and Ludo Melis for their continued assistance during the X-irradiations at SCK•CEN. We are grateful to Vanesa Bol, Stefaan Vynckier and Pierre Scalliet (UCL) for their guidance in dosimetry and feedback on the experimental design.
REFERENCES
- 1.Thariat J, Hannoun-Levi JM, Sun Myint A, et al. Past, present, and future of radiotherapy for the benefit of patients. Nat Rev Clin Oncol. 2013;10:52–60. doi: 10.1038/nrclinonc.2012.203. [DOI] [PubMed] [Google Scholar]
- 2.Wilson RR. Radiological use of fast protons. Radiology. 1946;47:487–91. doi: 10.1148/47.5.487. [DOI] [PubMed] [Google Scholar]
- 3.Karger CP, Jakel O. Current status and new developments in ion therapy. Strahlenther Onkol. 2007;183:295–300. doi: 10.1007/s00066-007-1645-x. [DOI] [PubMed] [Google Scholar]
- 4.Kramer M, Weyrather WK, Scholz M. The increased biological effectiveness of heavy charged particles: from radiobiology to treatment planning. Technol Cancer Res Treat. 2003;2:427–36. doi: 10.1177/153303460300200507. [DOI] [PubMed] [Google Scholar]
- 5.Iwadate Y, Mizoe J, Osaka Y, et al. High linear energy transfer carbon radiation effectively kills cultured glioma cells with either mutant or wild-type p53. Int J Radiat Oncol Biol Phys. 2001;50:803–8. doi: 10.1016/s0360-3016(01)01514-0. [DOI] [PubMed] [Google Scholar]
- 6.Wada S, Kobayashi Y, Funayama T, et al. Detection of DNA damage in individual cells induced by heavy-ion irradiation with an non-denaturing comet assay. J Radiat Res (Tokyo) 2002;43(Suppl):S153–6. doi: 10.1269/jrr.43.s153. [DOI] [PubMed] [Google Scholar]
- 7.Hamada N. Recent insights into the biological action of heavy-ion radiation. J Radiat Res (Tokyo) 2009;50:1–9. doi: 10.1269/jrr.08070. [DOI] [PubMed] [Google Scholar]
- 8.Suetens A, Moreels M, Quintens R, et al. Carbon ion irradiation of the human prostate cancer cell line PC3: a whole genome microarray study. Int J Oncol. 2014;44:1056–72. doi: 10.3892/ijo.2014.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wild-Bode C, Weller M, Rimner A, et al. Sublethal irradiation promotes migration and invasiveness of glioma cells: implications for radiotherapy of human glioblastoma. Cancer Res. 2001;61:2744–50. [PubMed] [Google Scholar]
- 10.Zhou YC, Liu JY, Li J, et al. Ionizing radiation promotes migration and invasion of cancer cells through transforming growth factor-beta–mediated epithelial–mesenchymal transition. Int J Radiat Oncol Biol Phys. 2011;81:1530–7. doi: 10.1016/j.ijrobp.2011.06.1956. [DOI] [PubMed] [Google Scholar]
- 11.Yao H, Zeng ZZ, Fay KS, et al. Role of alpha(5)beta(1) integrin up-regulation in radiation-induced invasion by human pancreatic cancer cells. Transl Oncol. 2011;4:282–92. doi: 10.1593/tlo.11133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cordes N, Blaese MA, Meineke V, et al. Ionizing radiation induces up-regulation of functional beta1-integrin in human lung tumour cell lines in vitro. Int J Rad Biol. 2002;78:347–57. doi: 10.1080/09553000110117340. [DOI] [PubMed] [Google Scholar]
- 13.De Bacco F, Luraghi P, Medico E, et al. Induction of MET by ionizing radiation and its role in radioresistance and invasive growth of cancer. JNCI. 2011;103:645–61. doi: 10.1093/jnci/djr093. [DOI] [PubMed] [Google Scholar]
- 14.Fujita M, Otsuka Y, Yamada S, et al. X-ray irradiation and Rho-kinase inhibitor additively induce invasiveness of the cells of the pancreatic cancer line, MIAPaCa-2, which exhibits mesenchymal and amoeboid motility. Cancer Sci. 2011;102:792–8. doi: 10.1111/j.1349-7006.2011.01852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mikheeva SA, Mikheev AM, Petit A, et al. TWIST1 promotes invasion through mesenchymal change in human glioblastoma. Mol Cancer. 2010;9:194. doi: 10.1186/1476-4598-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goetze K, Scholz M, Taucher-Scholz G, et al. The impact of conventional and heavy ion irradiation on tumor cell migration in vitro. Int J Rad Biol. 2007;83:889–96. doi: 10.1080/09553000701753826. [DOI] [PubMed] [Google Scholar]
- 17.Akino Y, Teshima T, Kihara A, et al. Carbon-ion beam irradiation effectively suppresses migration and invasion of human non-small-cell lung cancer cells. Int J Radiat Oncol Biol Phys. 2009;75:475–81. doi: 10.1016/j.ijrobp.2008.12.090. [DOI] [PubMed] [Google Scholar]
- 18.Ogata T, Teshima T, Kagawa K, et al. Particle irradiation suppresses metastatic potential of cancer cells. Cancer Res. 2005;65:113–20. [PubMed] [Google Scholar]
- 19.Fujita M, Otsuka Y, Imadome K, et al. Carbon-ion radiation enhances migration ability and invasiveness of the pancreatic cancer cell, PANC-1, in vitro. Cancer Sci. 2012;103:677–83. doi: 10.1111/j.1349-7006.2011.02190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goetze K, Scholz M, Taucher-Scholz G, et al. Tumor cell migration is not influenced by p21 in colon carcinoma cell lines after irradiation with X-ray or 12C heavy ions. Radiat Environ Biophys. 2010;49:427–35. doi: 10.1007/s00411-010-0297-x. [DOI] [PubMed] [Google Scholar]
- 21.Meador JA, Ghandhi SA, Amundson SA. p53-independent downregulation of histone gene expression in human cell lines by high- and low-let radiation. Radiat Res. 2011;175:689–99. doi: 10.1667/rr2539.1. [DOI] [PubMed] [Google Scholar]
- 22.Ding LH, Shingyoji M, Chen F, et al. Gene expression changes in normal human skin fibroblasts induced by HZE-particle radiation. Radiat Res. 2005;164:523–6. doi: 10.1667/rr3350.1. [DOI] [PubMed] [Google Scholar]
- 23.Matsumoto Y, Iwakawa M, Furusawa Y, et al. Gene expression analysis in human malignant melanoma cell lines exposed to carbon beams. Int J Radiat Biol. 2008;84:299–314. doi: 10.1080/09553000801953334. [DOI] [PubMed] [Google Scholar]
- 24.Girdhani S, Lamont C, Hahnfeldt P, et al. Proton irradiation suppresses angiogenic genes and impairs cell invasion and tumor growth. Rad Res. 2012;178:33–45. doi: 10.1667/rr2724.1. [DOI] [PubMed] [Google Scholar]
- 25.Fushimi K, Uzawa K, Ishigami T, et al. Susceptible genes and molecular pathways related to heavy ion irradiation in oral squamous cell carcinoma cells. Radiother Oncol. 2008;89:237–44. doi: 10.1016/j.radonc.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 26.Higo M, Uzawa K, Kawata T, et al. Enhancement of SPHK1 in vitro by carbon ion irradiation in oral squamous cell carcinoma. Int J Radiat Oncol Biol Phys. 2006;65:867–75. doi: 10.1016/j.ijrobp.2006.02.048. [DOI] [PubMed] [Google Scholar]
- 27.Sanzari JK, Nuth M, Kennedy AR. Induction of cytokine gene expression in human thyroid epithelial cells irradiated with HZE particles (iron ions) Radiat Res. 2009;172:437–43. doi: 10.1667/RR1363.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murray-Zmijewski F, Slee EA, Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev Mol Cell Biol. 2008;9:702–12. doi: 10.1038/nrm2451. [DOI] [PubMed] [Google Scholar]
- 29.Amundson SA, Do KT, Vinikoor LC, et al. Integrating global gene expression and radiation survival parameters across the 60 cell lines of the National Cancer Institute Anticancer Drug Screen. Cancer Res. 2008;68:415–24. doi: 10.1158/0008-5472.CAN-07-2120. [DOI] [PubMed] [Google Scholar]
- 30.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franken NA, Rodermond HM, Stap J, et al. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1:2315–9. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 32.Friedrich T, Scholz U, Elsasser T, et al. Systematic analysis of RBE and related quantities using a database of cell survival experiments with ion beam irradiation. J Radiat Res. 2013;54:494–514. doi: 10.1093/jrr/rrs114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balcer-Kubiczek EK, Zhang XF, Harrison GH, et al. Delayed expression of hpS2 and prolonged expression of CIP1/WAF1/SDI1 in human tumour cells irradiated with X-rays, fission neutrons or 1 GeV/nucleon Fe ions. Int J Radiat Biol. 1999;75:529–41. doi: 10.1080/095530099140177. [DOI] [PubMed] [Google Scholar]
- 34.Zhai GG, Malhotra R, Delaney M, et al. Radiation enhances the invasive potential of primary glioblastoma cells via activation of the Rho signaling pathway. J Neurooncol. 2006;76:227–37. doi: 10.1007/s11060-005-6499-4. [DOI] [PubMed] [Google Scholar]
- 35.Liu C, Xue H, Lu Y, et al. Stem cell gene Girdin: a potential early liver metastasis predictor of colorectal cancer. Mol Biol Rep. 2012;39:8717–22. doi: 10.1007/s11033-012-1731-8. [DOI] [PubMed] [Google Scholar]
- 36.Narumiya S, Tanji M, Ishizaki T. Rho signaling, ROCK and mDia1, in transformation, metastasis and invasion. Cancer Metastasis Rev. 2009;28:65–76. doi: 10.1007/s10555-008-9170-7. [DOI] [PubMed] [Google Scholar]
- 37.Lin SL, Chiang A, Chang D, et al. Loss of mir-146a function in hormone-refractory prostate cancer. RNA. 2008;14:417–24. doi: 10.1261/rna.874808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandquist JC, Swenson KI, Demali KA, et al. Rho kinase differentially regulates phosphorylation of nonmuscle myosin II isoforms A and B during cell rounding and migration. J Biol Chem. 2006;281:35873–83. doi: 10.1074/jbc.M605343200. [DOI] [PubMed] [Google Scholar]
- 39.Derycke L, Stove C, Vercoutter-Edouart AS, et al. The role of non-muscle myosin IIA in aggregation and invasion of human MCF-7 breast cancer cells. Int J Dev Biol. 2011;55:835–40. doi: 10.1387/ijdb.113336ld. [DOI] [PubMed] [Google Scholar]
- 40.Garcia-Marcos M, Jung BH, Ear J, et al. Expression of GIV/Girdin, a metastasis-related protein, predicts patient survival in colon cancer. FASEB J. 2011;25:590–9. doi: 10.1096/fj.10-167304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leiss M, Beckmann K, Giros A, et al. The role of integrin binding sites in fibronectin matrix assembly in vivo. Curr Opin Cell Biol. 2008;20:502–7. doi: 10.1016/j.ceb.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 42.Oppenheimer SB. Cellular basis of cancer metastasis: a review of fundamentals and new advances. Acta Histochem. 2006;108:327–34. doi: 10.1016/j.acthis.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 43.Ding J, Li D, Wang X, et al. Fibronectin promotes invasiveness and focal adhesion kinase tyrosine phosphorylation of human colon cancer cell. Hepatogastroenterology. 2008;55:2072–6. [PubMed] [Google Scholar]
- 44.Qian P, Zuo Z, Wu Z, et al. Pivotal role of reduced let-7 g expression in breast cancer invasion and metastasis. Cancer Res. 2011;71:6463–74. doi: 10.1158/0008-5472.CAN-11-1322. [DOI] [PubMed] [Google Scholar]
- 45.Hébrant A, Dom G, Dewaele M, et al. mRNA expression in papillary and anaplastic thyroid carcinoma: molecular anatomy of a killing switch. PLoS One. 2012;7:e37807. doi: 10.1371/journal.pone.0037807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amundson SA, Smilenov LB. Integration of biological knowledge and gene expression data for biomarker selection: FN1 as a potential predictor of radiation resistance in head and neck cancer. Cancer Biol Ther. 2010;10:1252–5. doi: 10.4161/cbt.10.12.13731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang HJ, Kim N, Seong KM, et al. Investigation of radiation-induced transcriptome profile of radioresistant non-small cell lung cancer A549 cells using RNA-seq. PLoS One. 2013;8:e59319. doi: 10.1371/journal.pone.0059319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hei TK, Zhao YL, Roy D, et al. Molecular alterations in tumorigenic human bronchial and breast epithelial cells induced by high LET radiation. Adv Space Res. 2001;27:411–9. doi: 10.1016/s0273-1177(01)00009-6. [DOI] [PubMed] [Google Scholar]