Abstract
Objectives
To develop bladder cancer-specific ligands using a combinatorial chemistry approach.
Materials and methods
We performed a high-throughput one-bead one-compound combinatorial chemistry approach to identify ligands that bound to bladder transitional cell carcinoma cells. The whole-cell binding assay allowed successful identification of a few peptides that bound selectively to bladder cancer cells. Single cell suspensions derived from clinical bladder cancer specimens and cell lines were used to determine the binding specificity. Studies with mouse xenografts were performed to determine the in vivo binding and targeting efficiency, specificity, and biodistribution of one of the ligands.
Results
One cyclic peptide named PLZ4 (amino acid sequence: cQDGRMGFc) was identified that could selectively bind to bladder cancer cell lines and all of the 5 primary bladder cancer cells from human patients, but not to normal urothelial cells, cell mixtures from normal bladder specimens, fibroblasts, and blood cells. Comparison of PLZ4 binding to cell lines of different cancer origins showed that it was bladder cancer-specific (P _ 0.05). PLZ4 could bind to tumor cells treated with urine at pH 6.0, but not to noncancerous cells collected from the urine of 4 patients actively being treated with intravesical Bacillus Calmette-Guerin therapy. In vivo and ex vivo imaging studies showed that PLZ4 linked to Cy5.5 fluorescent dye administered via tail vein injection was specifically taken up in mouse xenografts developed from excised fresh human bladder cancer specimens. Several ligands contain the same DGR motif, but only PLZ4 was bladder cancer-specific. We performed alanine walk and rainbow bead coding experiments, and found that the C-terminal GF residues were also important for cell binding and modulated the binding specificity.
Conclusions
PLZ4 has the potential to be used for targeted therapy and imaging detection during diagnosis and follow-up/surveillance of noninvasive and advanced bladder cancer.
Introduction
Bladder cancer is the fourth most common cancer in men and ninth in women [1]. At diagnosis, about 75% of patients are at the noninvasive stages [2]. Noninvasive bladder cancer is ideal for imaging and targeted therapy with cancer-specific ligands because it is easily accessible through intravesical instillation, relatively isolated from the rest of the human body, and has only a few confounding cells. The treatment for noninvasive cancer is usually transurethral resection of bladder tumor (TURBT) followed by intravesical instillation of Bacillus Calmette-Guerin (BCG) or mitomycin C to reduce recurrence. Despite this treatment, 20% to 80% of patients will recur and 25% will have disease progression [3–5]. All these patients require long-term follow- up with urine cytology and cystoscopy. The sensitivity of urine cytology ranges between 29% and 74%, with the overall sensitivity of approximately 35% [6–8]. Cystoscopy is intrusive, uncomfortable, and costly. Because of the long term survival and the need for monitoring over an extended period of time, the cost per case for bladder cancer is the highest among all cancer types, ranging from $96,000 to $187,000 (2001 values) per case [9,10]. Thus, novel alternative diagnostic and monitoring strategies are warranted.
We hypothesized that combinatorial chemistry could be used to develop bladder cancer-specific ligands for imaging and targeted therapy during the diagnosis, treatment, and follow- up of bladder cancer. Screening of a phage display peptide library identified several peptides that have the potential to be used for diagnosis of bladder cancer [11]. However, the in vivo targeting with human primary bladder cancer cells has not yet been determined. We used the one-bead one-compound combinatorial peptide library technology (OBOC) developed by one of us [12,13]. Each bead is ~90 μm in diameter that bears up to 1013 copies of ligands with the same chemical identity (hence the name OBOC). In order to develop cancer-specific ligands, millions of beads (each with a unique ligand sequence) can be screened in parallel to identify those ligands binding to cancer cell surface molecules. “Positive beads” that bear ligands specific for the desired target can be selected using an enzyme-linked colorimetric assay similar to the western blot, or by the evidence of cell attachment on the bead surface [14,15]. Unnatural amino acids, D-amino acids, or even nonpeptide moieties can be incorporated in the library to make the molecules resistant to proteolysis and increase the binding affinity [16]. The ligand leads identified through screening of OBOC libraries can be further optimized to achieve high affinity and specificity [16]. Here, we used OBOC methodology to develop PLZ4 as a bladder-specific ligand that has the potential use for imaging, targeted therapy of bladder cancer, and for capturing of cancer cells in urine for diagnosis and follow-up.
Materials and Methods
Synthesis of the initial and focused OBOC libraries
OBOC libraries were synthesized on solid phase TentaGel S NH2 resin (Rapp Polymere GmbH, Tübingen, Germany). A “split-mix” synthesis method was performed to construct the combinatorial OBOC libraries, each containing random libraries of millions of beads/ligands [12,13,16]. The ligands on the bead surface was synthesized by standard solid-phase peptide synthesis techniques using 9-fluorenylmethoxycarbonyl (Fmoc) chemistry and N-hydroxybenzotriazole (HOBt)/N,N′-diisopropylcarbodiimide (DIC) coupling. The completion of coupling was confirmed with a ninhydrin test. The beads were stored in 70% ethanol at 4°C until use.
Cells
Four human bladder cancer cell lines including 5637 (HTB-9), SCaBER, TCCSUP (HTB-5), and T24 (HTB-4) and other human cell lines were purchased from the American Type Culture Collection (Manassas, VA) and are outlined in detail in Supplement 1, which can be found in the electronic version of this article. The isolation, characterization, and maintenance of normal urothelial cells was performed as previously described [17]. Normal peripheral blood mononuclear cells (PBMC) were prepared by using the Ficoll-Paque gradient method from peripheral blood of healthy donors. Bladder cancer specimens obtained from cystectomy were cut into pieces, digested with collagenase at 37°C for 1 to 2 hours per the manufacturer’s protocol, and filtered through 40μm strainers to make single cell suspensions. Cells (mainly cancer cells) were then isolated with Ficoll-Paque gradient method (800 x g, 30 minutes at 4°C). This protocol was approved by UC Davis IRB before the experiments were started (protocol no. 200614340). Informed consent was obtained from each patient or healthy donor before specimens were collected.
Screening of OBOC library for bladder cancer-specific ligands
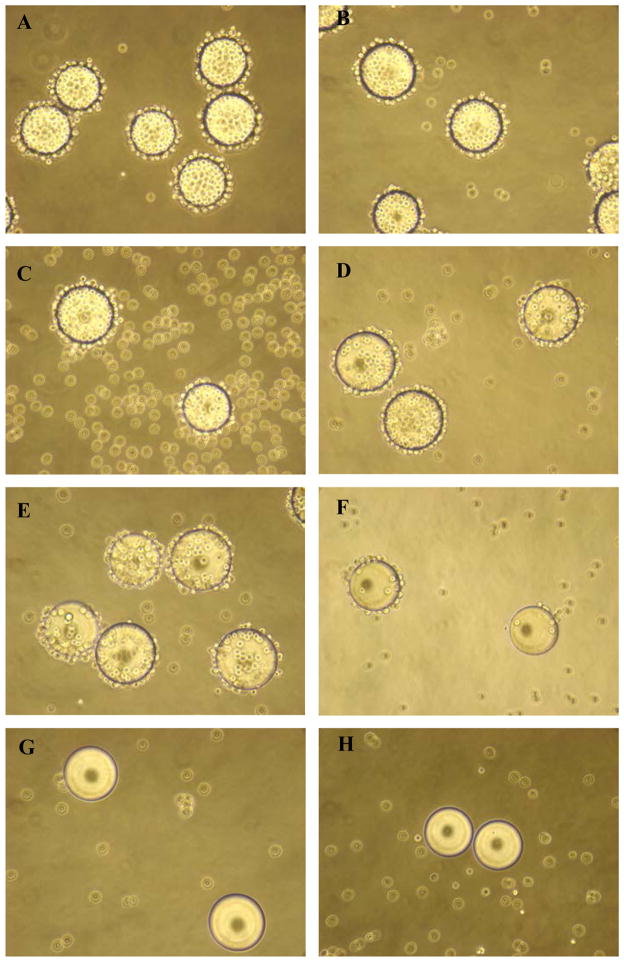
The beads were washed extensively with double-distilled water and phosphate-buffered saline (PBS) before screening. Bladder cancer cells and normal urothelial cells were detached from culture dishes with trypsin/EDTA, washed with their corresponding culture medium, resuspended at 106 cells/ml, and incubated with OBOC beads in Petri dishes in a humidified CO2 incubator at 37°C with shaking (60 rpm). Beads bound by cells appeared as rosettes with a central bead covered by a layer(s) of cells under a microscope (Fig. 1). The positive beads were picked with a pipette under inverted microscope, treated with guanidine-HCl (8M, 20 minutes) to remove cells and proteins on the bead surface, and underwent a second round of screening with the same cells to confirm the binding. Only those beads with cell bindings at both rounds were sent for peptide sequencing as previously described [16].
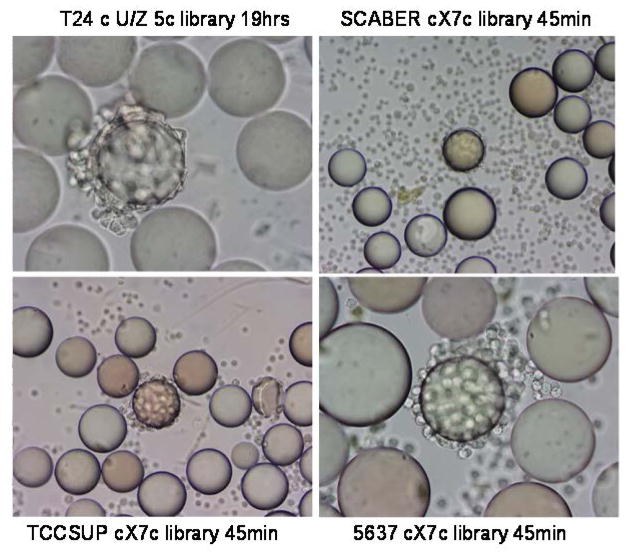
FIGURE 1.
Whole cell binding assay to screen ligands for binding to bladder cancer cells. For the whole cell binding assay, a single cell suspension was incubated with each OBOC library. Beads carrying ligands with affinity for cell surface molecules became covered with cells. In the middle of each panel, one positive bead was covered with bladder cancer cells, suggesting that the peptides on this bead could bind to bladder cancer cells. The cell lines, library types, and incubation time are shown next to each panel. The average bead diameter is approximately 90μm.
Synthesis of peptide and peptide-biotin
The synthetic chemistry of solution phase PLZ4 and PLZ4-biotin for biological testing is similar to that of the library using HOBt/DIC coupling (Supplement 2). Rink amide resin was used as solid support to prepare compounds with carboxyl amide derivatives.
Fluorescence microscopy
Bladder cancer cells and normal urothelial cells (2 ×104 cells per well) were seeded on chamber slides. When the cells grew to confluent of approximately 70%, they were washed with PBS and blocked with 3% BSA-PBS at 4°C for 1 hour, and incubated with the PLZ4-biotin conjugate (1M) for 1 hour at 4°C in TBS buffer. Cells were then washed 3 times with TBS and incubated with FITC-Streptavidin (0.5 μg/ml) (ZYMED, South San Francisco, CA). Cells were washed and examined using an inverted Olympus fluorescence microscope (40x).
Enzyme-linked affinity-sorbent assay (ELASA)
Equal numbers (5 × 103 cells per well) of cells from different cancer origins were inoculated into 96-well plates. Cells were washed 3 times with 0.1 ml cold PBS (pH 7.4)-BSA (1%), and incubated 50 μl PLZ4-biotin solution at 1μM for 1 hour on ice. Cells treated without PLZ4-biotin was used as a control. After washing 6 times with ice-cold PBS-BSA buffer, cells were incubated with 50 μl streptavidin (SA)-HRP, incubated for 1 hour on ice, washed with PBS (pH 7.4) 3 times, and developed per cell-based enzyme- linked immunosorbent assay manufacture’s instructions (R and D Systems, Minneapolis, MN) (Fig. 2).
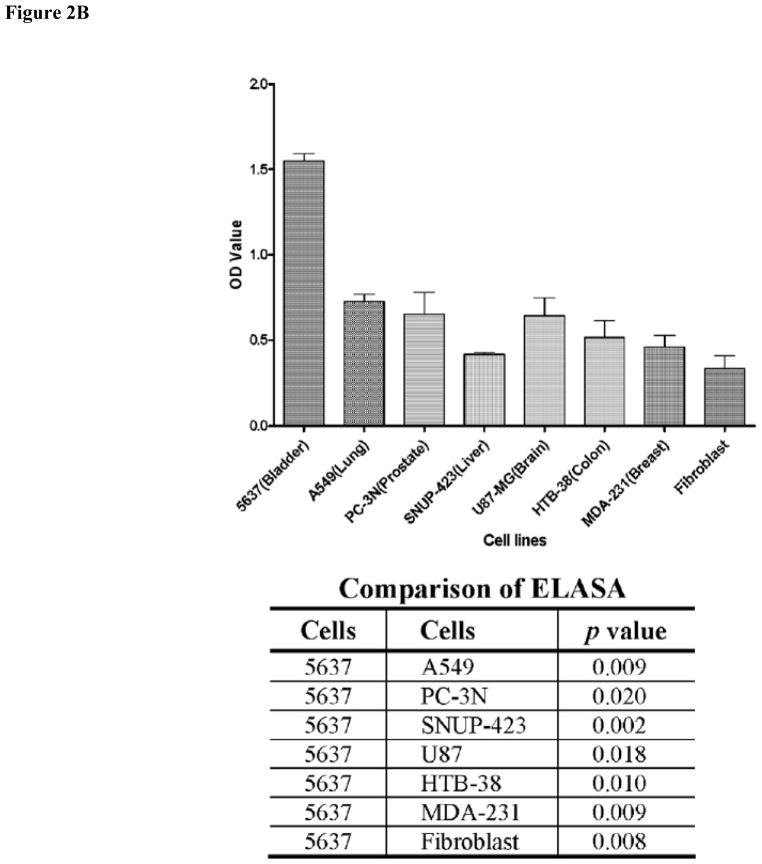
FIGURE 2.
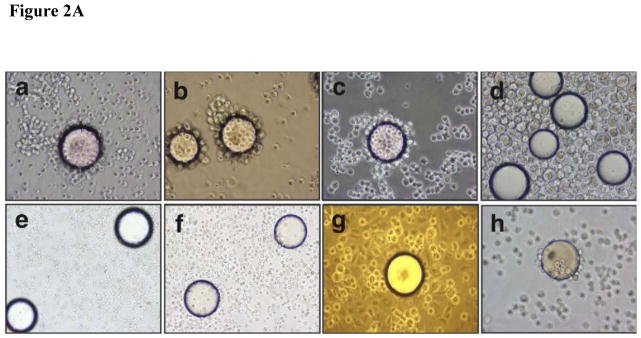
Binding specificity of PLZ4. (A) PLZ4 could bind to bladder tumor cells, but not to normal urothelial cells or other confounding cells. A whole cell binding assay was performed to determine cell binding of PLZ4. To detect any weak binding, cells and beads were incubated for at least 4 hours while rotating in culture. PLZ4 bound to 3 human bladder TCC cell lines: 5637 (a), T24 (b) and TCCSUP (c), but not to normal human urothelial cells (d). The heterogeneity of cell size of normal urothelial cells in Panel d suggests the presence of cells from basal (small cells) to suprabasal layers of urothelium. No binding of PLZ4 was observed with whole blood cells (e), PBMC (f), normal fibroblasts (g). PLZ4-coated beads could bind to 5637 bladder cancer cells after 4 hours of incubation in urine with pH = 6.0 (h). The bead diameter is approximately 90μm. (B) PLZ4 specifically binds to 5637 bladder TCC cell lines. ELASA was performed to determine the binding specificity. The lower panel table showed the statistical analysis (P value) of ELASA results between 5637 and other cell lines (P <0.05).
In vivo and ex vivo mouse imaging
We prepared a PLZ4-Cy5.5 conjugate by incubating PLZ4-biotin conjugates with streptavidin (SA)-Cy5.5 (Rockland Immunochemicals, Gilbertsville, PA). One streptavidin can bind up to 4 molecules of biotin. To ensure at least one PLZ4-biotin molecule was conjugated per molecule of SA-Cy5.5, we mixed PLZ4-biotin with SA-Cy5.5 at molar ratios of 5:1 for 1 h at 4°C. We confirmed the fluorescence labeling by in vitro cell-binding assays. Athymic nude mice were purchased from Harlan Laboratories (Indianapolis, IN). All experiments were performed in compliance with institutional guidelines and according to protocols (no. 12988) approved by the Animal Use and Care Administrative Advisory Committee of University of California Davis. Primary bladder cancer specimens were harvested from cystectomy by a pathologist after informed consent was obtained from patients. This protocol was approved by the UC Davis IRB. Primary cancer tissue was minced and incubated with collagenase at 37°C for 1 hour with rotation. Single cell suspension was obtained by straining through a 40 μm strainer. Some of the primary cells were incubated with OBOC beads to determine cell binding (Fig. 3). We mixed primary bladder cancer cells with Matrigel per manufacturer’s instruction (BD Biosciences, Sparks, MD), and subcutaneously injected into one side of the shoulder of each mouse. When tumor xenografts measured about 0.5–1.0 cm in diameter, we anesthetized mice using intraperitoneal injection of pentobarbital (60 mg/kg), IV injected a dose of PLZ4-Cy5.5, and performed imaging using a Kodak multimodal-imaging system IS2000MM (Kodak, Rochester, NY) equipped with an excitation bandpass filter at 625 nm and an emission filter at 700 nm. Exposure time was 30 seconds per image. Images were analyzed using the imaging station IS2000MM software (Kodak). After in vivo imaging, the mice were euthanized with CO2 overdose. Tumors and other normal organs and tissues were excised and imaged with the Kodak imaging system as described above.
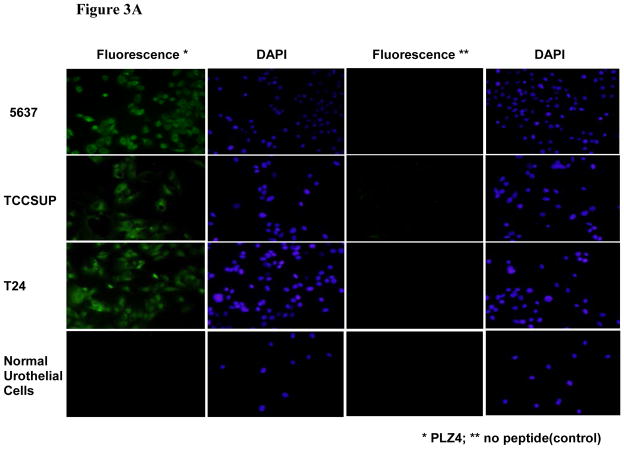
FIGURE 3.
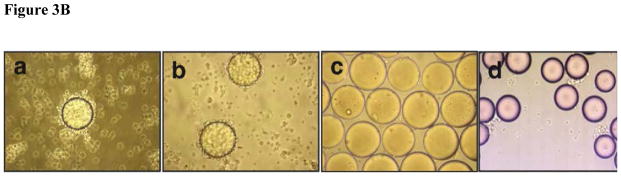
Binding of PLZ4 to human bladder cancer cells. (A) Fluorescence staining of bladder cell lines and normal urothelial cells with PLZ4 conjugated to FITC. Cells were cultured in chamber slides, and stained with PLZ4-biotin-streptavidin-FITC. Bladder cancer cells 5637, TCCSUP, and T24 were stained with green, while normal urothelial cells were not stained (the first column from the left). All cell nuclei were stained purple by DAPI (the second and fourth columns from the left). No cells were stained green when only streptavidin-FITC, but no PLZ4-biotin was added (the third column from the left). Magnification is 40x. (B) Binding of PLZ4 to clinical specimens. Single cell suspensions from freshly resected human bladder cancer specimens were incubated with beads coated with PLZ4 for 2 hours. Significant binding (more than 75% of bead surface covered with cells) was observed for cells from human patients (panels a and b). Cells from normal bladder tissue of the same patient as in panel b did not bind to beads coated with PLZ4 (panel c). Cells in panel c were washed away to decrease overlap of cells with beads. PLZ4 did not bind to cells isolated from urine of a patient who received active treatment with BCG intravesical therapy (panel d). The bead diameter is approximately 90 μm.
Data processing and statistics
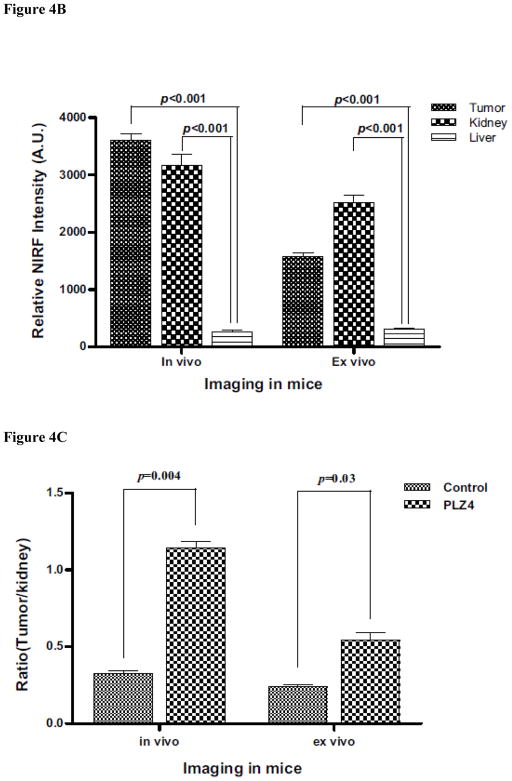
The experiments were repeated in duplicate or triplicate. The mean values and standard deviation were presented for each set of experiments. For determination of tumor contrast, we calculated mean fluorescence intensities of the tumor area and of the normal tissue area by means of the region-of-interest function using Kodak 1D Image Analysis Software (Kodak), then plotted a pseudocolored scale based the semiquantitative information from near-infrared fluorescent (NIRF) images by integrating fluorescence intensities from equal areas within tumor and normal tissue regions (Fig. 4).
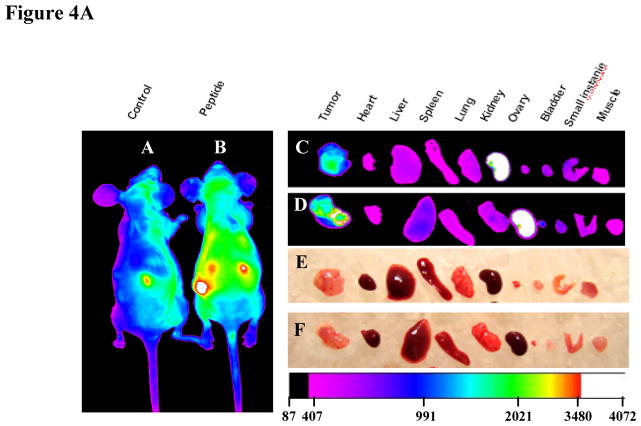
FIGURE 4.
Imaging of tumor xenografts with PLZ4. (A) Imaging. Mouse tumor xenografts were established with the freshly resected bladder cancer specimens from human patients. PLZ4-biotin was linked to SA-Cy5.5 through the strong interaction of biotin and SA. PLZ4-Cy5.5 (7 nmol) was injected through tail vein. (A) Near infrared imaging. (Panel a) In vivo imaging of a control mouse that received SA-Cy5.5 alone. (Panel b) In vivo imaging of a mouse that received PLZ4-Cy5.5. The red arrow points to the tumor xenograft with strong uptake of Cy5.5. (Panel c) Ex vivo imaging of the xenograft and organs from the mouse in panel a that received SA-Cy5.5, showing uptake in kidneys and weak autofluorescence in tumor xenograft. (Panel d) Ex vivo imaging of the xenograft and organs from the mouse in panel b that received PLZ4-Cy5.5, showing fluorescence in tumor xenograft and kidneys. (Panels e and f) Light imaging of panels c and d, respectively. Fluorescence intensity is shown in arbitrary units at the bottom. (B) Quantitative analysis of fluorescence uptake of the mice received PLZ4-Cy5.5. The fluorescence uptake in tumor was compared with kidney and liver (2 normal organs with the highest fluorescence). As in the mice that received SA-Cy5.5, kidneys had high fluorescence uptake. But the fluorescence in tumor xenografts was 15.3 times that of liver (P < 0.001). When analysis was performed with the ex vivo imaging, the absorbance of near-infrared fluorescence was eliminated, and the fluorescence intensity of tumor xenografts is weaker than that of kidneys, but still 5.3 times that of liver (P < 0.001). (C) Comparison of fluorescence uptake in tumor xenografts between mice that received PLZ4-Cy5.5 and mice that received SA-Cy5.5. To compare the fluorescence uptake, the fluorescence in the same amount of tumor was normalized by the fluorescence in kidney of the same mice. The uptake of tumor xenografts from mice that received PLZ4-Cy5.5 was much higher than the uptake of xenografts from the control mice (3.2 times and P = 0.004 for in vivo imaging; 2.4 times and P =0.03 for ex vivo imaging).
Results
Identification of a bladder cancer-specific ligand
We used the whole cell bead binding assay to screen OBOC libraries for peptides that could bind to bladder cancer cell cultures (Fig. 1). Approximately 150,000 library beads (peptides) were screened against each of 4 bladder cancer cell lines (3 transitional cell carcinoma (TCC) lines: T24, TCCSUP and 5637, and 1 squamous cell line SCaBER). From this screening, we identified 28 peptides that could bind to 1 of 4 cell lines (Table 1). After alignment and further analysis of these peptide sequences, several motifs were identified. The NGR/K motif appeared in several peptides. We previously found the DGR motif during the screen of ligands for ovarian cancer cell lines. The RGD motif is a well known ligand for αvβ3 and αvβ5 integrin. Our group has previously shown that peptides containing the Nle-DI motif selectively bind to bronchioloalveolar carcinoma H1650 and lymphoid (Jurkat and RAJI) cells but not other selected human cell lines of lung cancer and fibroblast. We then synthesized these peptide on OBOC beads and determined the binding of these peptides against primary normal urothelial cells in culture [17]. One of these ligands named PLZ4 with the sequence of cQDGRMGFc bound to all 3 bladder TCC cell lines (Fig. 2Aa–c), but did not bind to any of the confounding cells inside bladder, including normal urothelial cells (Fig. 2Ad). PLZ4 did not bind to whole blood cells (Fig. 2Ae), PBMC (Fig. 2Af) or fibroblasts (Fig. 2Ag). This is consistent with the observation that PLZ4 did not bind to 10 out of 12 cell lines with different origins (Supplement 1). The acidic environment in urine may change the 3D structure of ligands and affect ligand binding. Here, we determined whether PLZ4 could bind to 5637 TCC cancer cells in urine. Cells were incubated in urine at pH 6.0 for 4 hours at 37°C before PLZ4- coated beads were added. We used a 4 hour incubation to mimic the in vivo urine retention in patients and to permit potential conformational changes and protease digestion. PLZ4 was still able to bind 5637 cells in urine (Fig. 2Ah). To further quantitate the binding specificity, we performed ELASA to compare the binding of PLZ4 to a variety of cell lines. Compared with fibroblasts and other cancer cell lines, PLZ4 specifically bound to 5637 human bladder cancer cell lines (P < 0.05, Fig. 2B).
Table 1.
Four cell lines, including 3 transitional cell carcinoma (TCC) lines (T24, TCCSUP, and 5637), and 1 squamous cell line SCaBER were screened against 2 cyclic random peptide libraries. These two libraries were 9-mer cX1X2X3X4X5X6X7c and 7-mer c(U/Z)5c in which “c” represents D-cysteine, “X” for 19 natural L-amino acids except cysteine, “U” for 8 unnatural amino acids, and “Z” for 17 L-amino acids except arginine, cysteine, and lysine. Each peptide contained 2 flanking D-cysteine residues at the amino and carboxyl ends. These peptides were cyclized by a disulfide bond in order to more efficiently expose the amino acids in the middle of the ligand for cell binding. Sequence alignment of the peptides identified during this screening is shown in this table.
| Library | Cell lines
|
|||
|---|---|---|---|---|
| T24 | 5637 | TCCSUP | SCaBER | |
| cX7c | 1. cGRLKEKKc 2. cQKGGRKHc 3. cMKKHGKRc |
5. cQDGRMGFc 6. cLHDRGMLc 7. cRGDQFMQc 8. cGMLDGRIc 9. cFGDGRRFc |
15. cARTRARMc 16. cLSKNKNRc 17. cKSFKRIGc 18. cNTEFAHNc 19. cQSHDGKVc 20. cQRRNVVLc 21. cQHPBpaMc 22. cNlePEWHc |
23. cKSVDGRNc 24. cYLMWYAWc 25. cFSEAWAFc 26. cLWQLNGRc 27. cNEGLRSVc 28. cFADDEYSc |
| c(U/Z)5c | 4. cFDDFGc* | 10. cWNleEDVc 11. cDNleNleFEc 12. cYIVDPhgc 13. cNleDIPhgDc 14. cNHypNleQIc |
||
Fluorescence detection of human bladder cancer cells with PLZ4
We determined if fluorescent-labeled PLZ4 could bind to bladder cancer cells. We synthesized PLZ4 conjugated to biotin through hydrophilic linkers (Supplement 2). Biotinylated PLZ4 was then incubated with streptavidin(SA)-FITC to generate PLZ4 -FITC conjugate through the strong binding of biotin and SA. We then determined if PLZ4- FITC could bind to bladder cancer cell lines 5637, TCCSUP, T24, and normal urothelial cells growing on chamber slides. In the control experiment, SA-FITC was added without biotinylated ligand. Compared with control cells, strong fluorescence signals were detected with 5637, TCCSUP, and T24 cells by fluorescence microscopy (Fig. 3A). No significant binding was observed on the normal urothelial cells treated with the same staining conditions.
Beads coated with PLZ4 could bind to bladder tumor cells from patients
The cell surface molecules on established cell lines in culture may not be the same as those molecules on primary bladder cancer cells. Thus we assessed if PLZ4 could bind to bladder cancer cells from patients. We re-synthesized OBOC beads coated with PLZ4 on surface. PLZ4-coated beads could bind to primary bladder cancer cells from patients (Fig. 3Ba and b). PLZ4 could bind to cancer cells from all 5 consecutive freshly resected human bladder cancer specimens. In one patient, both normal bladder tissue and bladder cancer tissue from the same cystectomy specimen were available. Beads coated with PLZ4 could bind to cells from the cancer specimen (Fig. 3B panel b), but not to the cells from the normal specimen of the same bladder (Fig. 3B panel c). PLZ4 did not bind to cells that were isolated from urine specimens collected from 4 consecutive patients who had no evidence of bladder cancer but were actively treated with BCG intravesical therapy (Fig. 3Bd). This suggests that PLZ4 may not bind to inflamed cells commonly seen in patients treated with BCG.
In vivo imaging of nude mice bearing xenografts developed for primary human bladder cancer specimens
To investigate if PLZ4 could be used for in vivo bladder cancer imaging detection and targeted therapy, we used in vivo optical imaging of mice with xenografts developed from primary bladder cancer tissues of human patients who underwent cystectomy. NIRF dye Cy5.5 allows an imaging of deeper tissues because the near infrared light has high penetration, low tissue absorption and scattering rates. When tumor xenografts measured 5 to 10 mm, PLZ4-Cy5.5 conjugate (7 nmol) was injected via the tail vein. Mice were imaged at 0, 1, 2, 4, 8, 16, 24 hours post-inoculation. PLZ4-Cy5.5 uptake by tumors was much higher than that of normal tissue and the tumor area of the control mice receiving SA-Cy5.5 (Fig. 4A panels a and b). The tumor uptake started at 2 hour and reached the greatest difference 4 hours after injection. To further confirm the in vivo uptake of PLZ4-Cy5.5 complex, ex vivo imaging was performed with excised tumors and organs 24 hours after intravenous injection. The fluorescent PLZ4-Cy5.5 complex accumulated primarily in the tumor xenografts and kidney, whereas no significant uptake was observed in other organs, including bladder (Fig. 4A panels c and d). In the control mouse in which only SA-Cy5.5 was injected, strong fluorescent signal (white pseudo-color) was also detected in kidneys, suggesting the accumulation of PLZ4-Cy5.5 complex in kidneys may be secondary to the nonspecific uptake or trapping of SA-Cy5.5 by kidney (Fig. 4Ad). We performed quantitative analysis of Cy5.5 uptake in tumor xenografts and other organs. In mice that were dosed with PLZ4-Cy5.5, the in vivo analysis showed that the fluorescence in tumor xenografts was 15.3 times that in liver, the second highest uptake other than kidney (P < 0.001, Fig. 4B). After eliminating the tissue absorption and deflection, the ex vivo analysis showed that the fluorescence in tumor xenografts was 5.3 times that of liver (P < 0.001, Fig. 4B). Because of the high uptake of fluorescence in kidneys in both groups of mice, we normalized the fluorescence uptake in tumor xenografts with that in kidneys, and found that the uptake of Cy5.5 in tumor xenografts in mice received PLZ4-Cy5.5 is much higher than that in tumor xenografts in mice received SA-Cy5.5 (3.2 times and P = 0.004 for in vivo imaging; 2.4 times and P = 0.03 for ex vivo imaging, Fig. 4C). Histochemical staining was performed and confirmed that the tumor xenografts were bladder TCC (data not shown).
Molecular dissection of PLZ4
PLZ4 and several other ligands contain the same DGR motif (Table 1). However, only PLZ4 was bladder cancer-specific. We hypothesized that, besides DGR, some other amino acids in PLZ4 determined the binding specificity. We performed alanine walk experiments and our patented rainbow bead coding approach to determine which amino acid(s) are critical for cell binding. Alanine has one methyl group side chain, smaller than most other amino acids. In the alanine walk, each amino acid on PLZ4 was sequentially replaced with alanine, one at a time, while all other amino acids remained unchanged. With rainbow bead coding, beads bearing the same ligand are colored with the same dye while beads carrying different ligands are colored with different dyes. After coloring, all of the beads with different colors are mixed together and tested for their binding to the target cells in the same condition. This allows rapid determination of several ligands in one reaction and identification of ligand binding based on the color of the beads. We synthesized these “alanine walk” PLZ4 derivatives on OBOC beads [12,13], colored with different dyes to generate rainbow beads [18], and incubated them with a suspension of 5637 cells to determine the cell binding. Any critical amino acids that were replaced with alanine resulted in diminished binding. The amino acids aspartic acid (D, the X2 position), arginine (R, X4) and phenylalanine (F, X7) were critical for cell binding. Replacement of any of these amino acids with alanine completely abolished the binding of the peptides to 5637 cells (Fig. 5A). The glycine residues (G) at the X3 and X6 positions were important for cell binding because replacement of one of these 2 amino acids significantly decreased, but did not abolish, the binding of PLZ4 to 5637 cells. We could replace the glutamine (Q, X1) and the methionine (M, X5) with many other amino acids without compromising the binding affinity (data not shown). Based on this analysis, the 3 amino acids (D, G, and R) at the N-terminus, together with the 2 amino acids (G and F) at the C-terminus, determine the binding specificity. One interpretation of the data is that the binding pocket may be in a clover shape: 1 for the N-terminal 3 amino acids (DGR), 1 for the C-terminal 2 amino acids (GF), and a middle pocket. To determine the depth of the middle binding pocket, we synthesized PLZ4, but added different numbers of glycine residues to the X 5 position (Supplement 3). Two glycine residues could be added to fill in the central pocket without significant effect on cell binding. Addition of more than 4 glycines completely abolished the binding of PLZ4 to the target cells (Fig. 6).
FIGURE 5.
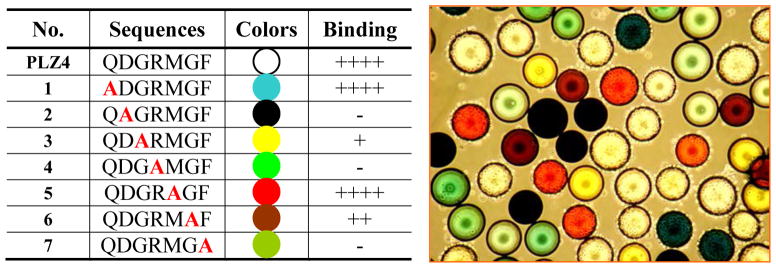
Molecular dissection of PLZ4 with alanine walk. (A) The color of each alanine walk bead and its binding to 5637 bladder TCC cells. Semiquantitative system was used to determine binding activity: ++++ means very strong binding with 75%–100% bead surface covered by cells; +++ means strong binding with 50%–74% bead surface covered by cells; ++ means moderate binding with 25%–49% bead surface covered by cells; +means weak binding with 1% –24% bead surface covered by cells; − means no binding. (B) The binding of 5637 cells to rainbow beads.
FIGURE 6.
The depth of the central pocket of the target molecule. The peptides on beads in each panel have the same backbone sequence (cQDGRKGFc) as PLZ4 (cQDGRMGFc) except the M at the X5 position has been replaced with K (Supplement 3). One (panel c) to 6 (panel i) glycine residues were conjugated to K from panel c to h. The OBOC beads were incubated with 5637 TCC cells. Compared with the parental PLZ4 ligand (panel a), no significant changes in binding to 5637 cells were observed when methionine was replaced by lysine (panel b). This ligand could still strongly bind to 5637 cells when only 1 (panel c) or 2 (panel d) glycine residues were added. There was a progressive decrease in cell binding as the number of glycine residues conjugated to lysine increased: diminished binding with 3 (panel e) or 4 (panel f) glycines, to no binding with 5 (panel g) and 6 (panel h) glycines.
Discussion
Compared with many other cancer types, the better diagnosis and treatment of bladder cancer is an unmet need for which relatively little attention is focused. There is great potential for PLZ4 in the diagnosis and management of bladder cancer. First, PLZ4 has the potential to be used for targeted therapy against noninvasive and advanced bladder cancer. Intravesical instillation of BCG or chemotherapy has been used to reduce the risk of bladder cancer recurrence. However, this therapy is still associated with significant risk of recurrence [3,4]. Intravesical instillation of therapeutic drugs conjugated to targeting ligands like PLZ4 may allow local delivery of high concentration of drugs and kill cancer cells more efficiently. For systemic application, we have already shown that PLZ4 is preferentially taken up at the xenografts developed from primary human bladder cancer specimens (Fig. 4).
PLZ4 may be used for imaging-based detection of bladder cancer. Once bladder cancer has metastasized, the prognosis is poor, and cystectomy is not curative. Current imaging modalities like computed tomography and magnetic resonance imaging are not sensitive and/or specific. While 18F-FDG-PET has been tested in clinic for staging purposes of bladder cancer, it has not been extensively used in clinic because the sensitivity and specificity are not satisfactory [19,20]. We have shown that by linking to NIRF dye Cy5.5, PLZ4 can be used for detection of subcutaneous tumor xenograft similar to metastatic bladder cancer (Fig. 4). Only 7 nmol (equivalent to 20 mg of PLZ4 in a 75-Kg patient) of the cancer-specific ligand was needed for this imaging study. We expect that PLZ4 has the potential to increase the detection specificity and sensitivity of imaging studies. If this targeted approach is successful, it may decrease or supplement the costly cystoscopy procedure during the diagnosis and follow-up of bladder cancer.
PLZ4 may be used for the visualization of tumor localization. When TURBT is used in the treatment of noninvasive bladder cancer, incomplete resection can be found in about one-third of cases regardless of the expertise of the urologists [21]. 5-Aminolevulinic acid (ALA) combined with fluorescence cystoscopy has been used for detecting tumor localization [22]. Nonspecific uptake of ALA by non-cancer urothelial cells, especially in inflamed bladder with BCG treatment, causes high background fluorescence that interferes with the detection of cancer by cystoscopy [23]. PLZ4 can specifically bind to bladder cancer cells, but not to normal urothelial cells, normal cells from the same bladder that contains cancer, or cells in urine specimens from patients who were actively treated with BCG (Figs. 2 and 3). We have also shown that FITC-conjugated PLZ4 could stain bladder cancer cells (Fig. 3), suggesting that this ligand has the potential to be used for fluorescence detection and tumor localization of noninvasive bladder cancer. PLZ4 could bind to cancer cells in urine at pH 6 (Fig. 3Bd), suggesting that the ligand can be used in the acidic urine environment for clinical applications. Because the urinary bladder is relatively isolated from the rest of the human body, we expect minimal side effects from the intravesical instillation of fluorophore-conjugated cancer-specific ligands, although formal clinical testing is warranted.
The binding specificity of PLZ4 is determined by the N-terminal DGR and C-terminal GF amino acids. PLZ4 and several other ligands contain the same DGR motif (Table 1). The DGR motif was the core of the binding motif in a protein that binds to Secreted Frizzled-related proteins [24]. The (D/N)GR motif, also previously identified as a low affinity integrin binding motif, was identified on the capsid of adeno-associated virus, and is critical for the viral capsid integrin α5β1 interaction and cell entry [25–27]. Our preliminary data indicated that PLZ4, like other ligands containing the RGD motif, binds to ανβ3 integrin (data not shown). This integrin heterodimer is expressed in other cancer cell lines including U87 and MDA-MD-231, which we used for this study as controls [28, 29]. However, PLZ4 has higher affinity for bladder cancer cell lines compared with these 2 cell lines (Fig. 2B). Our findings are consistent with the binding specificity of another cancer-specific ligand, LLP2A, identified by one of us (Kit Lam) [16]. LLP2A binds to α4β1 integrin on activated but not on quiescent PBMC. This suggests that conformational changes or other modifications may change the binding specificity. Differential glycosylation or expression level of integrin between normal urothelial and bladder TCC may explain the preferential binding of PLZ4 to bladder cancer cell line over normal urothelial cells [30]. The alanine walk confirmed that besides DGR, amino acids G (X6) and F (X7) are important for cell binding (Fig. 5), and may help determine the binding affinity and specificity. By adding glycine to the X5 position methionine, we found that even within this short peptide, there are 2 domains that determine the binding: the N terminal DGR and the C-terminal GF domains. In summary, we used the OBOC combinatorial library approach to identify the PLZ4 bladder cancer-specific ligand. The potential clinical applications include tumor localization to guide TURBT, imaging detection, and targeted drug delivery for noninvasive and metastatic bladder cancer. These results justify further animal and clinical studies. However, if PLZ4 is validated and approved for clinical applications, the potential cost benefit remains to be determined by additional studies.
Supplementary Material
Acknowledgments
The authors thank Dr. Theresa Koppie and Dr. Chaime Karim for providing bladder cancer cystectomy specimens for this study and Alessia Shahrokh for technical support.
References cited
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Fleming JD, Cooper JS, Jenson DE, et al. AJCC (American Joint Committee on Cancer) Cancer Staging Manuel. 5. Philadelphia: Lippincott-Raven; 1997. [Google Scholar]
- 3.Herr HW, Schwalb DM, Zhang ZF, et al. Intravesical bacillus Calmette-Guerin therapy prevents tumor progression and death from superficial bladder cancer: 10-Year follow-up of a prospective randomized trial. J Clin Oncol. 1995;13:1404–8. doi: 10.1200/JCO.1995.13.6.1404. [DOI] [PubMed] [Google Scholar]
- 4.Herr HW, Badalament RA, Amato DA, et al. Superficial bladder cancer treated with bacillus Calmette-Guerin: A multivariate analysis of factors affecting tumor progression. J Urol. 1989;141:22–9. doi: 10.1016/s0022-5347(17)40575-1. [DOI] [PubMed] [Google Scholar]
- 5.Cookson MS, Herr HW, Zhang ZF, et al. The treated natural history of high risk superficial bladder cancer: 15-Year outcome. J Urol. 1997;158:62–7. doi: 10.1097/00005392-199707000-00017. [DOI] [PubMed] [Google Scholar]
- 6.Eissa S, Kassim S, El-Ahmady O. Detection of bladder tumours: Role of cytology, morphology-based assays, biochemical and molecular markers. Curr Opin Obstet Gynecol. 2003;15:395–403. doi: 10.1097/00001703-200310000-00008. [DOI] [PubMed] [Google Scholar]
- 7.van Rhijn BW, van der Poel HG, van der Kwast TH. Urine markers for bladder cancer surveillance: A systematic review. Eur Urol. 2005;47:736–48. doi: 10.1016/j.eururo.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 8.Lotan Y, Roehrborn CG. Sensitivity and specificity of commonly available bladder tumor markers versus cytology: Results of a comprehensive literature review and meta-analyses. Urology. 2003;61:109–18. doi: 10.1016/s0090-4295(02)02136-2. Discussion 118. [DOI] [PubMed] [Google Scholar]
- 9.Riley GF, Potosky AL, Lubitz JD, et al. Medicare payments from diagnosis to death for elderly cancer patients by stage at diagnosis. Med Care. 1995;33:828–41. doi: 10.1097/00005650-199508000-00007. [DOI] [PubMed] [Google Scholar]
- 10.Botteman MF, Pashos CL, Redaelli A, et al. The health economics of bladder cancer: A comprehensive review of the published literature. Pharmacoeconomics. 2003;21:1315–30. doi: 10.1007/BF03262330. [DOI] [PubMed] [Google Scholar]
- 11.Lee SM, Lee EJ, Hong HY, et al. Targeting bladder tumor cells in vivo and in the urine with a peptide identified by phage display. Mol Cancer Res. 2007;5:11–9. doi: 10.1158/1541-7786.MCR-06-0069. [DOI] [PubMed] [Google Scholar]
- 12.Lam KS, Salmon SE, Hersh EM, et al. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991;354:82–4. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 13.Lam KS, Lebl M, Krchnak V. The “one-bead-one-compound” combinatorial library method. Chem Rev. 1997;97:411–48. doi: 10.1021/cr9600114. [DOI] [PubMed] [Google Scholar]
- 14.Songyang Z, Margolis B, Chaudhuri M, et al. The phosphotyrosine interaction domain of SHC recognizes tyrosine-phosphorylated NPXY motif. J Biol Chem. 1995;270:14863–6. doi: 10.1074/jbc.270.25.14863. [DOI] [PubMed] [Google Scholar]
- 15.Liu R, Marik J, Lam KS. A novel peptide-based encoding system for “one-bead one-compound” peptidomimetic and small molecule combinatorial libraries. J Am Chem Soc. 2002;124:7678–80. doi: 10.1021/ja026421t. [DOI] [PubMed] [Google Scholar]
- 16.Peng L, Liu R, Marik J, et al. Combinatorial chemistry identifies high-affinity peptidomimetics against α4β1 integrin for in vivo tumor imaging. Nat Chem Biol. 2006;2:381–9. doi: 10.1038/nchembio798. [DOI] [PubMed] [Google Scholar]
- 17.Bagai S, Rubio E, Cheng JF, et al. Fibroblast growth factor-10 is a mitogen for urothelial cells. J Biol Chem. 2002;277:23828–37. doi: 10.1074/jbc.M201658200. [DOI] [PubMed] [Google Scholar]
- 18.Luo J, Zhang H, Xiao W, et al. Rainbow beads: A color coding method to facilitate high-throughput screening and optimization of one-bead one-compound combinatorial libraries. J Comb Chem. 2008;10:599–604. doi: 10.1021/cc8000663. [DOI] [PubMed] [Google Scholar]
- 19.Drieskens O, Oyen R, Van Poppel H, et al. FDG-PET for preoperative staging of bladder cancer. Eur J Nucl Med Mol Imaging. 2005;32:1412–7. doi: 10.1007/s00259-005-1886-9. [DOI] [PubMed] [Google Scholar]
- 20.Liu IJ, Lai YH, Espiritu JI, et al. Evaluation of fluorodeoxyglucose positron emission tomography imaging in metastatic transitional cell carcinoma with and without prior chemotherapy. Urol Int. 2006;77:69–75. doi: 10.1159/000092937. [DOI] [PubMed] [Google Scholar]
- 21.Herr HW. Restaging transurethral resection of high risk superficial bladder cancer improves the initial response to bacillus Calmette-Guerin therapy. J Urol. 2005;174:2134–7. doi: 10.1097/01.ju.0000181799.81119.fc. [DOI] [PubMed] [Google Scholar]
- 22.Daniltchenko DI, Riedl CR, Sachs MD, et al. Long-term benefit of 5-aminolevulinic acid fluorescence assisted transurethral resection of superficial bladder cancer: 5-Year results of a prospective randomized study. J Urol. 2005;174:2129–33. doi: 10.1097/01.ju.0000181814.73466.14. Discussion 2133. [DOI] [PubMed] [Google Scholar]
- 23.Grossman HB. Does fluorescence cystoscopy improve the outcome of patients with bladder cancer?. Proceedings of the Society of Urological Oncology Winter Meeting 2005 (Podium presentation); Bethesda, MD. 2005. [Google Scholar]
- 24.Chuman Y, Uren A, Cahill J, et al. Identification of a peptide binding motif for secreted frizzled-related protein-1. Peptides. 2004;25:1831–8. doi: 10.1016/j.peptides.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 25.Koivunen E, Gay DA, Ruoslahti E. Selection of peptides binding to the α5β1 integrin from phage display library. J Biol Chem. 1993;268:20205–10. [PubMed] [Google Scholar]
- 26.Koivunen E, Wang B, Ruoslahti E. Isolation of a highly specific ligand for the α5β1 integrin from a phage display library. J Cell Biol. 1994;124:373–80. doi: 10.1083/jcb.124.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asokan A, Hamra JB, Govindasamy L, et al. Adeno-associated virus type 2 contains an integrin α5β1 binding domain essential for viral cell entry. J Virol. 2006;80:8961–9. doi: 10.1128/JVI.00843-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morini M, Mottolese M, Ferrari N, et al. The α3β1 integrin is associated with mammary carcinoma cell metastasis, invasion, and gelatinase B (MMP-9) activity. Int J Cancer. 2000;87:336–42. [PubMed] [Google Scholar]
- 29.Wang S, Chen KJ, Wu TH, et al. Photothermal effects of supramolecularly assembled gold nanoparticles for the targeted treatment of cancer cells. Angew Chem Int Ed Engl. 2010;49:3777–81. doi: 10.1002/anie.201000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Litynska A, Przybylo M, Ksiazek D, et al. Differences of α3β1 integrin glycans from different human bladder cell lines. Acta Biochim Pol. 2000;47:427–34. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.