Abstract
The Gibbs sampler has been used extensively in the statistics literature. It relies on iteratively sampling from a set of compatible conditional distributions and the sampler is known to converge to a unique invariant joint distribution. However, the Gibbs sampler behaves rather differently when the conditional distributions are not compatible. Such applications have seen increasing use in areas such as multiple imputation. In this paper, we demonstrate that what a Gibbs sampler converges to is a function of the order of the sampling scheme. Besides providing the mathematical background of this behavior, we also explain how that happens through a thorough analysis of the examples.
Keywords: Gibbs chain, Gibbs sampler, Potentially incompatible conditional-specified distribution
1. INTRDUCTION
The Gibbs sampler is, if not singularly, one of the most prominent Markov chain Monte Carlo (MCMC)-based methods. Partly because of its conceptual simplicity and elegance in implementation, the Gibbs sampler has been increasingly used across a very broad range of subject areas including bioinformatics and spatial analysis. While its root dates back to earlier work (e.g., [1]), the popularity of the Gibbs sampling is commonly credited to Geman and Geman [2], in which the algorithm was used as a tool for image processing. Its use in statistics, especially Bayesian analysis, has since grown very rapidly [3–5]. For a quick introduction of the algorithm, see Casella and George [6].
One of the recent developments of the Gibbs sampler is in its application to potentially incompatible conditional-specified distributions (PICSD) [7, 8]. When statistical models involve high-dimensional data, it is often easier to specify conditional distributions instead of the entire joint distributions. However, the approach of specifying a conditional distribution has the risk of not forming a compatible joint model. Consider a system of d discrete random variables X = {X1, X2, ⋯, Xd}, whose fully conditional model is specified by F = {f1, f2, ⋯, fd}, where . If the conditional models are individually specified, then there may not exist a joint distribution that will give rise to the specified set of conditional distributions. In such a case, we call F incompatible.
The study of PICSD is closely related to the Gibbs sampler because the latter relies on iteratively drawing samples from F to form a Markov chain. Under mild conditions, the Markov chain converges to the desired joint distribution, if F is compatible. However, if F is not compatible, then the Gibbs sampler could exhibit erratic behavior [9].
In this paper, our goal is to demonstrate the behavior of the Gibbs sampler (or the pseudo Gibbs sampler as it is not a true Gibbs sampler in the traditional sense of presumed compatible conditional distributions) for PICSD. By using several simple examples, we show mathematically that what a Gibbs sampler converges to is a function of the order of the sampling scheme in the Gibbs sampler. Furthermore, we show that if we follow a random order in sampling conditional distributions at each iteration—i.e., using a random-scan Gibbs sampler [10]—then the Gibbs sampling will lead to a mixture of the joint distributions formed by each combination of fixed-order (or more formally, fixed-scan) when d = 2 but the result is not true when d > 2. This result is a refinement of a conjecture put forward in Liu [11].
Two recent developments in the statistical and machine-learning literature underscore the importance of the current work. The first is in the application of the Gibbs sampler to a dependency network, which is a type of generalized graphical model specified by conditional probability distributions [7]. One approach to learning a dependency network is to first specify individual conditional models and then apply a (pseudo) Gibbs sampler to estimate the joint model. Heckerman et al. [7] acknowledged the possibility of incompatible conditional models but argued that when the sample size is large, the degree of incompatibility will not be substantial and the Gibbs sampler is still applicable. Yet another example is the use of the fully conditional specification for multiple imputation of missing data [12, 13]. The method, which is also called multiple imputation by chained equations (MICE), makes use of a Gibbs sampler or other MCMC-based methods that operate on a set of conditionally specified models. For each variable with a missing value, an imputed value is created under an individual conditional-regression model. This kind of procedure was viewed as combining the best features of many currently available multiple imputation approaches [14]. Due to its flexibility over compatible multivariate-imputation models [15] and ability to handle different variable types (continuous, binary, and categorical) the MICE has gained acceptance for its practical treatment of missing data, especially in high-dimensional data sets [16]. Popular as it is, the MICE has the limitation of potentially encountering incompatible conditional-regression models and it has been shown that an incompatible imputation model can lead to biased estimates from imputed data [17]. So far, very little theory has been developed in supporting the use of MICE [18]. A better understanding of the theoretical properties of applying the Gibbs sampler to PICSD could lead to important refinements of these imputation methods in practice.
The article is organized as follows: First, we provide basic background to the Gibbs chain and Gibbs sampler and define the scan order of a Gibbs sampler. In Section 3, we offer several analytic results concerning the stationary distributions of the Gibbs sampler under different scan patterns and a counter-example to a surmise about the Gibbs sampler under a random order of scan pattern. Section 4 describes two simple examples to numerically demonstrate the convergence behavior of a Gibbs sampler as a function of scan order, both by applying matrix algebra to the transition kernel as well as using MCMC-based computation. Finally in Section 5 we provide a brief discussion.
2. GIBBS CHAIN AND GIBBS SAMPLER
Continuing the notation in the previous section, let a = (a1, a2, ⋯, ad) denote a permutation of {1, 2, ⋯, d}, x = {x1, x2, ⋯, xd} denote a realization of X with xk ∈ {1, 2, ⋯, Ck}, where Ck is the number of categories of the kth variable. Thus, xa ≡ (xa1, xa2, ⋯, xad) is a realization of X defined in the order of a. We also denote , the relative complement of xak with respect to xa.
For a specified F, the associated fixed (systemic)-scan Gibbs chain governed by a scan pattern a can be implemented as follows:
Pick an arbitrary starting vector .
- On the tth cycle, successively draw from the full conditional distributions according to scan pattern a = (a1, a2, ⋯, ad) as follows:
The series , ⋯ obtained by a single draw (iteration) is called a realization of Gibbs chain defined by F with scan pattern a; and the series , ⋯ obtained by a single cycle is a realization of the associated Gibbs sampler. For example, let = {X1, X2, X3, X4}, a = (2, 4, 1, 3), and given initial value , the Gibbs chain in cycle 1 performs the following draws and produces the corresponding states:
In this example, the series , ⋯, is the realization of Gibbs sampler defined by F with scan pattern a.
We can also express a Gibbs sampler of random scan order as a Gibbs chain. Let r = {r1, r2, ⋯, rd} be the set of selection probabilities, where rk > 0 is the probability of visiting a conditional fk, and . The random-scan Gibbs sampler can be stated as follows [19]:
Pick an arbitrary starting vector .
- At the sth iteration, s = 1, 2, ⋯,
- Randomly choose k ∈ {1,2, ⋯, d} with probability rk;
Repeat step 2 until a convergence criterion is reached.
Example 1
Consider the following bivariate 2 × 2 joint distribution π and for (X1, X2) defined on the domain {1, 2}, with its corresponding conditional distributions f1 (x1|x2) and f2 (x2|x1) [20, page 242]:
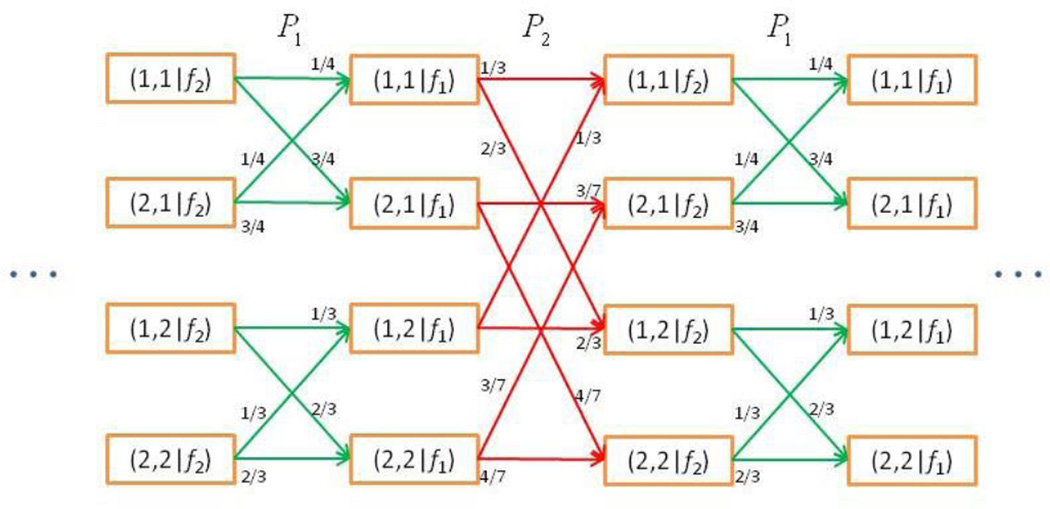
There are 4 possible states, (1, 1), (1, 2), (2, 1), and (2, 2) for the Gibbs chain. The transition from one state to another is governed by the conditional matrices f1 and f2 as shown in Figure 1. As a shorthand, we denote an entry in the matrix as f1 (·,·); e.g. f1 (1,2) = 1/3. In order to keep track of the scan order, we denote the state in the Gibbs chain as (x(s)|fk), if the current state x(s) at iteration s is the result of drawing from the conditional fk. To fix ideas, we use a fixed-scan Gibbs sampler with a = (1,2) and the conditional distributions {f1, f2}. In such case, the realization of this Gibbs sampler is the collection of states {(x(2t)|f2)|t = 1,2, ⋯}.
Figure 1.
Transition probabilities of the Gibbs chain in Example 1.
The local transition probability Pak [21, page 71] for two successive states of Gibbs chain, (x(s−1)|fak−1) and (x(s)|fak), can be defined by
For example, as shown in Figure 1, the local transition probability from (x(2t) = (1,1)|f2) to (x(2t+1) = (1,1)|f1) is f1 (1,1) = 1/4, and (x(2t) = (1,1)|f2) to (x(2t+1) = (1,2)|f1) is 0.
By arranging the state in lexicographical order such that the first index changes the fastest and the last index the slowest (as shown in Figure 1), a transition probability matrix Tak can be constructed according to local transition probability Pak. In Example 1, the transition probability matrices T1 and T2 that correspond respectively to matrices P1, and P2:
The matrices T1 and T2 have two pairs of identical rows and are idempotent but not irreducible.
As the above example illustrates, the local transition probability matrices defined by the given conditional distributions are in general not identical. Thus a Gibbs chain implemented using either fixed or random scan order is not homogeneous. However, if one defines a surrogate transition probability matrix Ta ≡ Ta1Ta2Ta3 ⋯ Tad, then a homogeneous chain with transition matrix Ta can be formed for the scan pattern a = (a1, a2, ⋯, ad) [21]. In other words, for a collection of full conditional distributions F and a scan pattern a the fixed-scan Gibbs sampler is a homogeneous Markov chain with transition matrix Ta. Analogously, a random-scan Gibbs sampler with selection probability r = (r1, r2, ⋯, rd) can be also transferred to a homogeneous Markov chain by defining as the surrogate transition probability matrix [21]. Their corresponding stationary distributions πa and πr can be directly computed (under a mild condition) respectively by evaluating and , where , and 1C is a C-dimensional vector of 1’s [22].
3. SOME ANALYTIC RESULTS
In this section, we offer several general results regarding the behaviors of the fixed-scan and the random-scan Gibbs sampler for discrete variables in which the transition matrices are finite. For these results, it is not necessary to assume compatibility, unless stated otherwise. Besides providing theoretical underpinning to the previous illustrative examples, the results here allow a closer look at the mechanisms through which incompatibility impacts the behaviors of the different Gibbs sampling schemes. Note that these results are special cases that can be derived from more general theories for Markov chains, but for our purpose, focusing on the special case of discrete variables and scan patterns makes it easier to examine the dynamics of convergence. General results regarding convergence of Markov chains can be found elsewhere [5, 24]. All of the proofs of the following results are included in the Appendix.
Theorem 1
If F is positive (fk > 0, ∀ k) then the Gibbs sampler, either fixed-scan with a scan pattern a or random scan with selection probability r > 0, converges to a unique stationary distribution πa and πr respectively.
Note that Theorem 1 does not require F to be compatible. The result assures that when F is positive—a stronger condition than F being non-negative—any scan pattern can have one and only one stationary distribution. Furthermore, the transition for any fixed-scan pattern is governed by the following theorem:
Theorem 2
If F is positive then for each state set, (x1, x2, ⋯, xd|fak), k = 1, ⋯, d, of the Gibbs chain with scan pattern a = (a1, a2, ⋯, ad) has exactly one stationary distribution πak. In particular, and , k = 2, ⋯, d, and πad = πa.
A direct consequence of Theorem 2 is that for any fixed-scan pattern, one of the specified conditional distributions in F can always be derived from its stationary distribution. This is summarized in the following corollary:
Corollary 1
If F is positive then the stationary distribution πa of the Gibbs sampler has fad as one of its conditional distributions for the scan pattern a = (a1, a2, ⋯, ad), i.e., πa (xad|xa1, xa2, ⋯, xad−1) = fad.
When F is compatible, all scan patterns converge to the same joint distribution. The following theorem provides a formal statement.
Theorem 3
Given F is positive. F is compatible if and only if there exists a unique joint distribution π with either πa = π, ∀ a or πr = π, ∀ r. Furthermore, π is the joint distribution characterized by F.
An interesting observation about the random scan is that it forms a mixture of the fixed-scan patterns only for d = 2. We state the Corollary for the case d = 2 and give a counter-example for d = 3.
Corollary 2
If F > 0 and d = 2 then πr, r = (r, 1 − r), is formed by the convex combination of πa2 = (2,1) and πa1 = (1,2); i.e., for all r ∈ [0, 1], πr = (1 − r) πa1 + rπa2.
A three-dimensional counter example to Corollary 2 for the case d = 3 is presented in Table 1. In this example, F = {f1, f2, ⋯, f3} is positive but not compatible. There are a total of six scan patterns and for each scan pattern, the solution to which the individual Gibbs sampler converges (using matrix multiplication) is shown as a row in Table 1. Convergence is defined here as all cell-wise differences between estimates from two consecutive iterations to be less than 0.5 × 10−4. The average of all six fixed-scan Gibbs sampler is provided, as well as a reference. In order to solve for a non-negative linear combination (mixture) of the fixed-scan distributions,
| (1) |
where , we treated equation (1) as a system of linear equations and solved for c = (ci), i = 1, ⋯, 6. As it turned out, our result indicated that there was no solution that satisfied c > 0. This observation led us to believe that the surmise [11] that the stationary distribution for a random-scan Gibbs sampler is a mixture of the stationary distributions for all systematic scan Gibbs samplers is not true in general. It only holds for d = 2.
Table 1.
A three-dimensional counter example for Corollary 2.
| (1,1,1) | (2,1,1) | (1,2,1) | (2,2,1) | (1,1,2) | (2,1,2) | (1,2,2) | (2,2,2) | ||
|---|---|---|---|---|---|---|---|---|---|
| f1 | 0.1 | 0.9 | 0.2 | 0.8 | 0.3 | 0.7 | 0.4 | 0.6 | |
| f2 | 0.5 | 0.6 | 0.5 | 0.4 | 0.7 | 0.8 | 0.3 | 0.2 | |
| f3 | 0.9 | 0.1 | 0.1 | 0.9 | 0.1 | 0.9 | 0.9 | 0.1 | |
| πa1 = (2,3,1) | 0.0199 | 0.1795 | 0.0411 | 0.1646 | 0.1484 | 0.3462 | 0.0401 | 0.0602 | |
| πa2 = (3,1,2) | 0.0305 | 0.2064 | 0.0305 | 0.1376 | 0.1319 | 0.3251 | 0.0565 | 0.0813 | |
| πa3 = (1,2,3) | 0.1462 | 0.0532 | 0.0087 | 0.1970 | 0.0162 | 0.4784 | 0.0784 | 0.0219 | |
| πa4 = (3,2,1) | 0.0228 | 0.2050 | 0.0355 | 0.1421 | 0.1399 | 0.3263 | 0.0513 | 0.0770 | |
| πa5 = (1,3,2) | 0.0775 | 0.1502 | 0.0775 | 0.1002 | 0.0661 | 0.4001 | 0.0283 | 0.1000 | |
| πa6 = (2,1,3) | 0.1464 | 0.0531 | 0.0087 | 0.1972 | 0.0163 | 0.4782 | 0.0782 | 0.0219 | |
| π̄ | 0.0739 | 0.1412 | 0.0337 | 0.1565 | 0.0865 | 0.3924 | 0.0555 | 0.0604 | |
| 0.0728 | 0.1406 | 0.0331 | 0.1532 | 0.0873 | 0.3944 | 0.0575 | 0.0613 |
4. NUMERICL EXAMPLES
We illustrate the mathematical results using two numerical examples. In Example 1, the transition matrices for fixed- and random-scans are respectively Ta1 = T1T2 for a1 = (1,2) and Ta2 = T2T1 for a2 = (2,1) and Tr = (T1 + T2)/2. Table 2 directly compares the joint distributions obtained from the following computations: (i) direct MCMC Gibbs sampler for the only two possible fixed-scan patterns a1 = (1,2) and a2 = (2,1); (ii) direct MCMC Gibbs sampler for random-scan patterns with the following selection probabilities: ; , and , and (iii) matrix multiplication using (Ta)m and (Tr)m with power m. To achieve numerical convergence, we used the first 5,000 cycles as burn-in and the subsequent 1,000,000 cycles for sampling for both (i) and (ii). And the smallest m ∈ {2k|k = 0,1,2,⋯} such that all cell-wise differences between estimates from two consecutive iterations to be less than 0.5 × 10−4 is adopted for (iii).
Table 2.
Joint distributiuons produced by various Gibbs samplers for Example 1.
| (1,1) | (2,1) | (1,2) | (2,2) | ||
|---|---|---|---|---|---|
| π | 0.1 | 0.3 | 0.2 | 0.4 | |
| a1 = (1,2) | 0.1002 | 0.3002 | 0.2000 | 0.3997 | |
| a1 = (1,2) | 0.0998 | 0.3004 | 0.1999 | 0.4000 | |
| 0.1007 | 0.2998 | 0.2000 | 0.3995 | ||
| πa1(m = 4) | 0.1000 | 0.3000 | 0.2000 | 0.4000 | |
| πa2(m = 4) | 0.1000 | 0.3000 | 0.2000 | 0.4000 | |
| πr0(m = 32) | 0.1000 | 0.3000 | 0.2000 | 0.4000 | |
| πr1(m = 32) | 0.1000 | 0.3000 | 0.2000 | 0.4000 | |
| πr2(m = 32) | 0.1000 | 0.3000 | 0.2000 | 0.4000 |
As expected, both the fixed-scan, regardless of scan order, and the random-scan Gibbs samplers numerically converge to the same joint distribution. Table 2 also demonstrates that direct matrix multiplication of the transition probabilities produces rapid convergence even for a small m and different values of r. However, we also observed that if r was heavily imbalanced, it took many more iterations to achieve numerical convergence (not shown). For example, if , it took m > 120 to achieve the same numerical convergence (up to 4 decimal places).
Example 2
(Incompatible conditional distributions). Consider a pair of 2 × 2 conditional distributions f1 (x1|x2) and f2 (x2|x1) defined on the domain {1, 2} as follows [20, page 242]:
These two conditional distributions are not compatible. Table 3 shows the results for the joint distributions derived from the simulated Gibbs samplers and matrix-multiplication of Example 2 for conditions that are identical to those presented in Table 2. Several observations can be made here: (1) The Gibbs samplers that use the fixed-scan pattern a1 and a2 respectively converge to two distinct joint distributions; (2) Each individual fixed-scan Gibbs sampler converges to the corresponding solution computed from the matrix-multiplication method; (3) The random-scan Gibbs sampler converges to the mixture distribution of the individual fixed-scan distributions—i.e., πr = (1 − r) πa1 + rπa2; and (4) Regardless F is compatible or not, a random-scan Gibbs sampler always needs much larger m (using matrix-multiplication) to achieve numerical convergence than a fixed-scan Gibbs sampler does. Such phenomena should result from the idempotent property of the transition probability matrices, and it also implies slower convergence of random-scan Gibbs sampler should be expected in a MCMC simulation.
Table 3.
Joint distributiuons produced by various Gibbs samplers for Example 2.
| (1,1) | (2,1) | (1,2) | (2,1) | ||
|---|---|---|---|---|---|
| a1 = (1,2) | 0.1062 | 0.0680 | 0.2128 | 0.6130 | |
| a1 = (1,2) | 0.0435 | 0.1314 | 0.2753 | 0.5498 | |
| 0.0748 | 0.0990 | 0.2447 | 0.5815 | ||
| πa1(m = 4) | 0.1063 | 0.0681 | 0.2125 | 0.6131 | |
| πa2(m = 4) | 0.0436 | 0.1308 | 0.2752 | 0.5504 | |
| πr0(m = 32) | 0.0749 | 0.0995 | 0.2439 | 0.5817 | |
| πr1(m = 32) | 0.0854 | 0.0890 | 0.2334 | 0.5922 | |
| πr2(m = 32) | 0.0645 | 0.1099 | 0.2543 | 0.5713 |
5. DISUSSION
In this paper, we show that for a given scan pattern, a homogeneous Markov chain is formed by the Gibbs sampling procedure and under mild conditions, the Gibbs sampler converges to a unique stationary distribution. Unlike compatible distributions, different scan patterns lead to different stationary distributions for PICSD. The random-scan Gibbs sampler generally converges to “something in between” but the exact weighted equation only holds for simple cases – i.e., when the dimension is two.
Our findings have several implications for the practical application of the Gibbs sampler, especially when they operate on PICSD. For example, the MICE often relies on a single fixed-scan pattern. This implies that the imputed missing values could change beyond expected statistical bounds when a seemingly innocuous change in the order of the variable is being made. Although in this paper we have not studied the issue of which fixed-scan pattern produces the “best” joint distribution, some recent work has been done in that direction. For example, Chen, Ip, and Wang [8] proposed using an ensemble approach to derive an optimal joint density. The authors also showed that the random-scan procedure generally produces promising joint distributions. It is possible that in some cases the gain from using multiple Gibbs chains, as in the case of random-scan, is marginal. As argued by Heckerman et al. [7], the single-chain fixed-scan (pseudo) Gibbs sampler asymptotically works well when the extent to which the specified conditional distributions are incompatible is minimal. This may be true for models that are applied to one single data set with a large sample size. However, the extent of incompatibility could be much higher when multiple data sets are used and when multiple sets of conditional models are specified. While it is likely that even in more complex applications a brute-force implementation of the (pseudo) Gibbs sampler will still provide some kinds of solutions, the qualities and behaviors of such “solutions” will need to be rigorously evaluated.
Acknowledgements
The work was supported by NIH grants 1R21AG042761-01 and 1U01HL101066-01.
APPENDIX: PROOFS OF ANALYTIC RESULTS
Proof of Theorem 1
We need a lemma to prove Theorem 1 about irreducibility (ability to reach all interesting points of the state-space) and aperiodicity (returning to a given state-space at irregular times).
Lemma 1
If F is positive then Ta and Tr are irreducible and aperiodic w.r.t. any given a and r > 0.
Proof
Let d and be two states for the chain induced by Ta or Tr, and define (T)ij as the (ij)th element in the matrix T. Without loss of generality, we also let a = (1,2, ⋯, d) and , Ta = T1T2T3 ⋯ Td and .
To prove that Ta and Tr are aperiodic, we must have (Ta)ii > 0 and (Ta)ii > 0, ∀ i. By the definition of local transition probability, we have (Tk)ii > 0, ∀ k, if F is positive. Consequently, and .
To prove that Ta and Tr are irreducible is equivalent to prove that x1 and x2 commute, i.e., to show the transition probability P (x1 → x2) > 0 and P (x2 → x1) > 0. Given the scan pattern a we have
And
Similarly, for the random-scan case we have
and
It is well known that if a Markov chain is irreducible and aperiodic, then it converges to a unique stationary distribution [24]. Consequently, we have the uniqueness and existence theorem (Theorem 1) for the Gibbs sampler.
Proof of Theorem 2
We need a lemma to prove Theorem 2.
Lemma 2
If F is positive then the stationary distribution πa of the Gibbs sampler has fad as one of its conditional distribution for the scan pattern a = (a1, a2, ⋯, ad), i.e., πa(xad|xa1, xa2, ⋯, xad−1) = fad.
Proof
Since , t = 1,2,3, ⋯, it follows πa ∝ fad (xad|xa1, xa2, ⋯, xad−1). Consequently, πa (xad|(xad)c) = fad(xad|(xad)c).
Theorem 2 easily follows from Lemma 2.
Proof of Theorem 3
“If” part. Since F is positive and compatible, there exists a positive joint distribution π > 0 characterized by F. Under the positivity assumption of π, it is well known that the Gibbs sampler governed by F determines π [25].
“Only if” part. Let a = (a1, a2, ⋯, ad) be a scan pattern with ad = i, i = 1, ⋯, d. Assuming that there exists a π such that πa = π, ∀ a. From Theorem 1 and Lemma 2, it follows that πa (xad|(xad)c) = fad(xad|(xad)c), ∀ a. Thus, π (xi|(xi)c) = πai(xi|(xi)c) = fi (xi|(xi)c), ∀ i. Hence F is compatible and π is the joint distribution of F.
Assuming that there exists a π such that πr = π, ∀ r. We only need to prove that πa = π, ∀ a. From Theorem 1, we have πaTa = πa. By the definition of random-scan Gibbs sampler, we have πrTk = πr = π, ∀ k, ∀ r. It follows that
Form Theorem 1, π is uniquely determined by Ta. As a result, πr = π = πa, ∀ r, ∀ a.
Proof of Corollary 1
The proof follows directly from Theorem 2 and 3.
Proof of Corollary 2
Since F is positive, πa1, πa2 and π are stationary distributions uniquely determined by a1, a2 and r, respectively. Therefore,
Because Tr = rT1 + (1 − r) T2 is the transition kernel for the random-scan Gibbs chain with selection probability r, we have the uniquely determined πr which equals rπa2 + (1 − r) πa1.■
REFERENCES
- 1.Hastings WK. Monte Carlo Sampling Methods Using Markov Chains and Their Applications. Biometrika. 1970;87:97–109. [Google Scholar]
- 2.Geman S, Geman D. Stochastic Relaxation, Gibbs Distribution, and the Bayes Restoration of Images. IEEE Trans Pattern Anal. 1984;6:721–741. doi: 10.1109/tpami.1984.4767596. [DOI] [PubMed] [Google Scholar]
- 3.Gelfand AE, Smith FM. Sampling-based Approaches to Calculating Marginal Densities. J Am Stat Assoc. 1990;85:398–409. [Google Scholar]
- 4.Smith AFM, Roberts GO. Bayesian Computation via the Gibbs Sampler and Related Markov Chain Monte Carlo Methods. J Roy Stat Soc Ser B. 1993;55:3–23. [Google Scholar]
- 5.Gilks WR, Richardson S, Spiegelhalter D. Markov Chain Monte Carlo in Practice. London: Chapman & Hall; 1996. [Google Scholar]
- 6.Casella G, George E. Explaining the Gibbs Sampler. Am Stat. 1992;46:167–174. [Google Scholar]
- 7.Heckerman D, Chickering DM, Meek C, Rounthwaite R, Kadie C. Dependence Networks for Inference, Collaborative Filtering, and Data Visualization. J Mach Learn Res. 2000;1:49–57. [Google Scholar]
- 8.Chen SH, Ip EH, Wang Y. Gibbs Ensembles for Nearly Compatible and Incompatible Conditional Models. Comput Stat Data Anal. 2011;55:1760–1769. doi: 10.1016/j.csda.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hobert JP, Casella G. Functional Compatibility, Markov Chains and Gibbs Sampling with Improper Posteriors. J Comput Graph Stat. 1998;7:42–60. [Google Scholar]
- 10.Liu JS, Wong HW, Kong A. Correlation Structure and Convergence Rate of the Gibbs Sampler with Various Scans. J Roy Stat Soc Ser B. 1995;57:157–169. [Google Scholar]
- 11.Liu JS. Discussion on Statistical Inference and Monte Carlo Algorithms, by G. Casella. Test. 1996;5:305–310. [Google Scholar]
- 12.Van Buuren S, Boshuizen HC, Knook DL. Multiple Imputation of Missing Blood Pressure Covariates in Survival Analysis. Stat Med. 1999;18:681–694. doi: 10.1002/(sici)1097-0258(19990330)18:6<681::aid-sim71>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 13.van Buuren S, Brand JPL, Groothuis-Oudshoorn CGM, Rubin DB. Fully Conditional Specifcation in Multivariate Imputation. J Stat Comput Sim. 2006;76:1049–1064. [Google Scholar]
- 14.Rubin DB. Nested Multiple Imputation of NMES via Partially Incompatible MCMC. Stat Neerl. 2003;57:3–18. [Google Scholar]
- 15.Schafer JL. Analysis of Incomplete Multivariate Data. London: Chapman & Hall; 1997. [Google Scholar]
- 16.Rässler S, Rubin DB, Zell ER. Incomplete Data in Epidemiology and Medical Statistics. In: Rao CR, Miller JP, Rao DC, editors. Handbook of Statistics 27: Epidemiology and Medical Statistics. The Netherlands: Elsevier; 2008. pp. 569–601. [Google Scholar]
- 17.Drechsler J, Rässler S. Does Convergence Really Matter? In: Shalabh, Heumann C, editors. Recent Advances in Linear Models and Related Areas. Heidelberg: Physica-Verlag; 2008. pp. 341–355. [Google Scholar]
- 18.White IR, Royston P, Wood AM. Multiple Imputation Using Chained Equations: Issues and Guidance for Practice. Stat Med. 2011;30:377–399. doi: 10.1002/sim.4067. [DOI] [PubMed] [Google Scholar]
- 19.Levine R, Casella G. Optimizing Random Scan Gibbs Samplers. J Multivar Anal. 2006;97:2071–2100. [Google Scholar]
- 20.Arnold BC, Castillo E, Sarabia JM. Exact and Near Compatibility of Discrete Conditional Distributions. Comput Stat Data Anal. 2002;40:231–252. [Google Scholar]
- 21.Madras N. Lectures on Monte Carlo Methods, Providence. Rhode Island: American Mathematical Association; 2002. [Google Scholar]
- 22.Seneta E. Non-negative Matrices and Markov Chains. 2nd ed. New York: Springer; 1981. [Google Scholar]
- 23.Tierney L. Markov Chains for Exploring Posterior Distributions. Ann Stat. 1994;22:1701–1728. [Google Scholar]
- 24.Norris JR. Markov Chain. Cambridge, UK: Cambridge University Press; 1998. [Google Scholar]
- 25.Besag JE. Discussion of Markov Chains for Exploring Posterior Distributions. Ann Stat. 1994;22:1734–1741. [Google Scholar]