Abstract
Persistent adaptive challenges are often met with the evolution of novel physiological traits. Although there are specific examples of single genes providing new physiological functions, studies on the origin of complex organ functions are lacking. One such derived set of complex functions is found in the Lepidopteran bursa copulatrix, an organ within the female reproductive tract that digests nutrients from the male ejaculate or spermatophore. Here, we characterized bursa physiology and the evolutionary mechanisms by which it was equipped with digestive and absorptive functionality. By studying the transcriptome of the bursa and eight other tissues, we revealed a suite of highly expressed and secreted gene products providing the bursa with a combination of stomach-like traits for mechanical and enzymatic digestion of the male spermatophore. By subsequently placing these bursa genes in an evolutionary framework, we found that the vast majority of their novel digestive functions were co-opted by borrowing genes that continue to be expressed in nonreproductive tissues. However, a number of bursa-specific genes have also arisen, some of which represent unique gene families restricted to Lepidoptera and may provide novel bursa-specific functions. This pattern of promiscuous gene borrowing and relatively infrequent evolution of tissue-specific duplicates stands in contrast to studies of the evolution of novelty via single gene co-option. Our results suggest that the evolution of complex organ-level phenotypes may often be enabled (and subsequently constrained) by changes in tissue specificity that allow expression of existing genes in novel contexts, such as reproduction. The extent to which the selective pressures encountered in these novel roles require resolution via duplication and sub/neofunctionalization is likely to be determined by the need for specialized reproductive functionality. Thus, complex physiological phenotypes such as that found in the bursa offer important opportunities for understanding the relative role of pleiotropy and specialization in adaptive evolution.
Keywords: co-option, bursa copulatrix, spermatophore, digestion, evolution, transcriptome
Introduction
Understanding the origin of novel morphological and physiological traits is a central goal in evolutionary biology. Accordingly, there has been focused interest in determining the genetic mechanisms enabling the creation of new traits because it provides direct insight into the adaptive process. Genetic changes underlying the appearance of novel morphological structures have been found to include the co-option and modification of preexisting developmental networks (Moczek et al. 2011; Hallgrimsson et al. 2012; Blank et al. 2014). One of the most striking examples of this process of gene network co-option is the evolutionary origin of the beetle horn, which is used by males to defend breeding females (Arrow and Hincks 1951; Moczek and Emlen 2000; Emlen et al. 2007). Its origin was explained by the recruitment of a network of genes involved in limb patterning (Moczek and Rose 2009). In contrast, other studies have revealed mechanisms by which novel physiological functions have evolved, such as snake venoms and antifreeze proteins, and have emphasized the importance of single gene co-option, gene duplication, and de novo gene formation (Chen et al. 1997; True and Carroll 2002; Casewell et al. 2013).
Although these morphological and physiological examples illustrate the range of mechanisms by which novel functions could arise in a given tissue—including co-option of entire pathways, single gene co-option, gene duplication followed by specialization, and de novo gene formation—the field has yet to determine their relative contributions to the evolutionary origin of complex physiological functions at the entire organ level. Studies of novel functions on an organ-wide scale should yield new insights into the constraints and mechanisms underlying the adaptive process; however, such studies are lacking from the literature. Studies of single-gene novelties suggest that new functions may often be provided by gene duplication, which would allow the protein products to specialize while avoiding pleiotropic consequences in other tissue contexts (Lynch and Force 2000; Long et al. 2003; Casewell et al. 2013). However, the degree to which tissue-specific duplicates provide new physiological functions on an organ-wide scale is not clear. Preexisting genes could also supply many of the molecular functions required by an organ either at its inception or during subsequent modification. If such genes are already expressed in the progenitor tissue giving rise to an organ, they could be simply upregulated to provide the selected functionality. Alternatively, preexisting genes expressed in other tissues could be recruited, either as single genes through cis-regulatory changes or through the adoption of entire expression programs via trans-regulatory changes. All of the above represent plausible mechanisms to supply complex and adaptive physiological functions for a new organ system.
Reproductive systems are widely recognized as dynamic and rapidly evolving, and also exhibit many examples of evolutionary novelties, including unique genital structures, giant sperm, modified sperm storage organs, and copulatory plugs (Pitnick et al. 1999; Dixson and Anderson 2002; Yassin and Orgogozo 2013). This reproductive diversity is thought to be the product of strong, directional selection arising from sexual selection, including male–male competition and male–female reproductive coevolution (Clark et al. 2006). Male–female reproductive interactions in particular are thought to be guided by a mix of cooperative and antagonistic coevolutionary dynamics. These complex coevolutionary interactions remain poorly understood from an empirical standpoint, due in large part to a well-documented male bias in the study of reproductive traits (Ah-King et al. 2014). In order to study the genetic origins of novel physiological functions and also to address the gap in knowledge of female reproductive traits, we focused our efforts on a derived female reproductive organ in Lepidoptera, the bursa copulatrix.
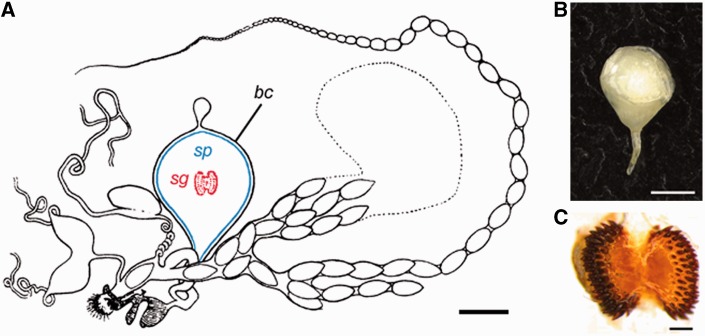
The bursa copulatrix (hereafter bursa) provides unique and specific digestive functionality in the female reproductive tract. During copulation, the bursa receives a complex ejaculate in the form of a nutrient-rich spermatophore produced by the male accessory glands (fig. 1A and B; Karlsson 1995, 1996). Spermatophores may vary in composition depending on the male diet but usually contain protein, carbohydrates, lipids, ions, and minerals (Boggs 1995). Immediately after copulation, the sperm migrate to a storage organ elsewhere in the reproductive tract. The bursa then processes the spermatophore, a nuptial gift, which can represent up to 13% of a male’s body weight (Watanabe and Sato 1993). Stretch receptors in the bursa are able to detect the inflation of the bursa caused by spermatophore transfer. These receptors then trigger nerve responses leading to a decrease in female receptivity and subsequent refusal of courting males until the spermatophore has been sufficiently deflated (Sugawara 1979). In many species, such as Pieris rapae, the male-contributed spermatophore has a tough outer envelope that retards spermatophore digestion for 12–36 h until the signum, a toothed structure within the bursa, breaches the spermatophore envelope, aided by muscular contractions of the bursa (fig. 1C). Subsequent digestion of the spermatophore inner matrix usually takes an additional 24–48 h, after which the female is willing to remate (Suzuki 1979). During digestion, soluble substances and digestion products are imported by the bursa into the hemocoel of the female, with transport being potentially facilitated by pits in the bursa wall (Lai-Fook 1991).
Fig. 1.
Key female reproductive traits in Pieris rapae. (A). Complete reproductive tract (modified from Lorković, 1985) with bursa copulatrix (bc), spermatophore (sp, in blue), and signum (sg, in red). (B) Male spermatophore. (C) The female signum in P. rapae exhibits a “chewing” morphotype. Scale bars: 1 mm (A and B) and 200 µm (C).
The bursa performs complex digestive and absorptive functions but the evolutionary history of these functions, and indeed, of the bursa itself remains poorly understood. Developmental studies have shown that the bursa develops from the same progenitor structure as the remainder of the female reproductive tract (Sendi et al. 1993; Gaikwad et al. 2014). However, the function of the bursa is quite distinct from other reproductive tissues. The forces thought to have shaped bursa function include both cooperative and conflict-based coevolution. The bursa-spermatophore interaction has been traditionally considered a cooperative process because the nutrients donated by the male within the spermatophore are used for both egg production and somatic maintenance, increasing female reproductive output and lifespan (Boggs and Gilbert 1979; Boggs 1981; Boggs and Watt 1981; Greenfield 1982; Drummond 1984). However, the interests of males and females are not entirely aligned, a hallmark of sexual antagonism. Until the spermatophore is almost fully digested by the bursa, females are unable to remate (Sugawara 1979; Vahed 1998; Wiklund 2003; Wedell 2005). This reduction in female remating rate benefits her male mate, because it increases his paternity share (Shapiro 1970). However, it does so at the female’s expense because it reduces the rate with which she can acquire additional spermatophore-based resources. This complex male–female interaction has likely influenced the development of novel functions in the bursa copulatrix, including traits that aid in the rapid digestion of spermatophore nutrients. On one side, novel morphological traits arose, including the bursa itself and the signum. On the other side, the bursa acquired novel physiological traits, including musculature, secretory and digestive function, and finally absorption and transport.
The goal of this study was to characterize the evolutionary acquisition of novel physiological traits in an entire organ system by using the bursa copulatrix of the Cabbage White butterfly (P. rapae) as a model. Female P. rapae are polyandrous, mating on average 2–3 times over the course of their lifespan (Suzuki 1979). The bursa in this species exhibits both mechanical and chemical digestion of the spermatophore (Plakke MS, Deutsch AB, Meslin C, Clark NL, Morehouse NL, in preparation). By using next-generation sequencing, we constructed a comprehensive transcriptome of P. rapae, targeting the female reproductive tract and nonreproductive organs with molecular functions related to those performed in the bursa. To develop a general understanding of the digestive capacity of the bursa copulatrix we identified genes that likely digest, process, and transport male-contributed proteins, carbohydrates, and lipids. Moreover, we placed these genes in an evolutionary context using sequence information from other Lepidopteran and non-Lepidopteran insect species, thereby revealing the importance of genetic co-option—and to a lesser degree, duplication—in the origin of physiological functions in a novel organ system.
Results
Expression Patterns in the Bursa Copulatrix Are Distinct from the Female Reproductive Tract
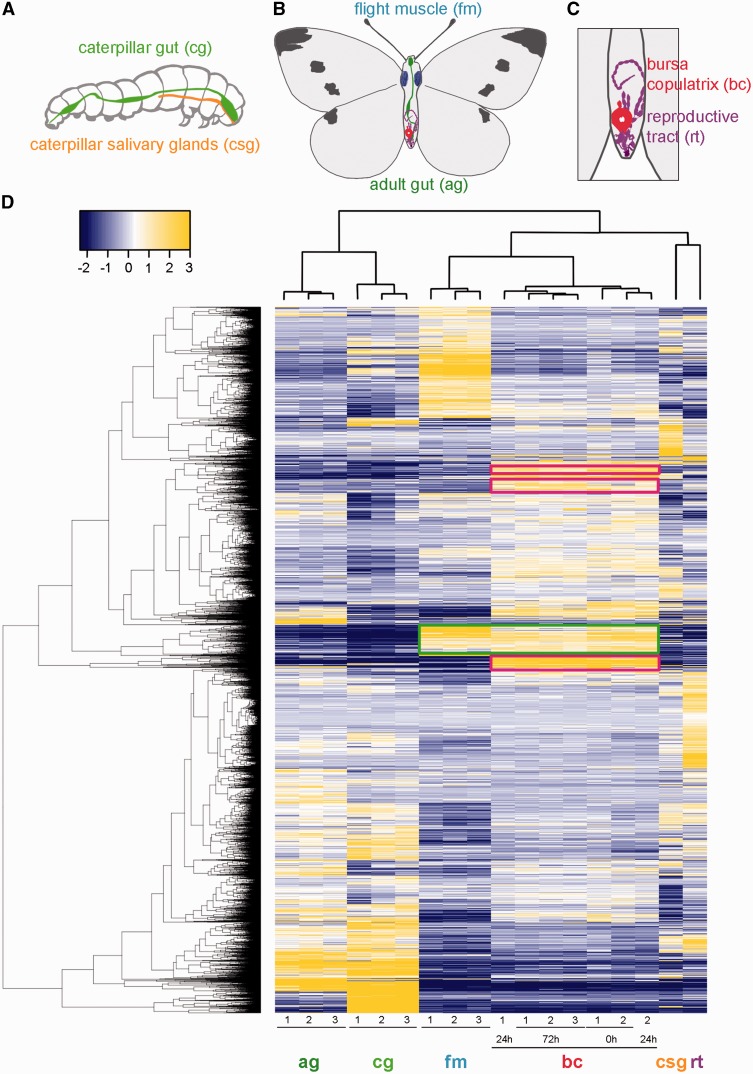
To characterize the potential origins of specific bursa physiological traits, we used transcriptomes of six different P. rapae tissues at two developmental stages: caterpillar gut and caterpillar salivary gland, adult gut, flight muscle, female reproductive tract (without bursa copulatrix), and bursa copulatrix (fig. 2A–C). Additionally, bursae were dissected from virgin females and mated females 24 and 72 h after mating. The 72 h time point is toward the end of the female refractory period, that is, spermatophore digestion is nearing completion in the bursa copulatrix and some females begin to exhibit remating receptivity (Suzuki 1979). Out of the 15,773 protein-coding genes in our de novo transcriptome assembly, 7,105 are differentially expressed between tissues (fig. 2D). Overall, biological replicates within a tissue clustered tightly, indicating that these expression patterns are reproducible and characteristic of these tissues. There are also meaningful similarities between tissues, as observed in the clustering (high correlation) of adult and caterpillar digestive tracts. Bursa transcriptomes clustered together as well, despite the fact that different mating statuses (virgin, 24 and 72 h) were treated as independent samples. There are also groups of tissue-specific genes visible in figure 2. For example, some genes appear to be highly expressed only in the bursa (fig. 2D, magenta boxes). In a quantitative sense, we then classified a gene as bursa-specific when the mean of its expression between bursa replicates represented more than 95% of its total expression across all tissue samples in this study. By using this threshold, we identified 13 bursa-specific genes with expression levels over 10 RPKM (Reads Per Kilobase per Million mapped reads) (supplementary data S1, Supplementary Material online). Based on the developmental origin of the bursa copulatrix, one might expect bursa and female reproductive tract transcriptomes to be similar, but interestingly, the bursa transcriptome is more correlated with the flight muscle transcriptome (fig. 2D, green box) (supplementary data S2, Supplementary Material online), confirming the importance of musculature in this organ. Moreover, genes that are highly expressed in the bursa have very low expression elsewhere in the female reproductive tract. These observations suggest that the bursa has adopted a suite of highly derived functions compared with the remainder of the female reproductive tract.
Fig. 2.
Differentially expressed genes in Pieris rapae. (A) Tissues sampled from P. rapae caterpillar. (B) Samples from P. rapae adult. (C) Detail of female P. rapae reproductive tract and bursa copulatrix. (D) Heatmap of log2-scale expression values for genes significantly differentially expressed among P. rapae tissues. Hierarchical clustering of genes and replicates was based on Euclidean distance. A false discovery rate cutoff (FDR = 0.001) and a minimal 4-fold change were applied to select the genes represented in the heatmap. Cells are colorized according to level of expression (blue = low, yellow = high). Numbers at bottom indicate biological replicates. Each tissue exhibits a unique expression pattern, but there are notable correlations between adult and caterpillar guts (ag, cg) and between the flight muscle and the bursa (fm, bc, green box). Moreover, some genes appear to be highly expressed only in the bursa copulatrix (magenta boxes).
Most Bursa Genes with Digestive Functions Originated from and Remain Coexpressed in Nonreproductive Tissues
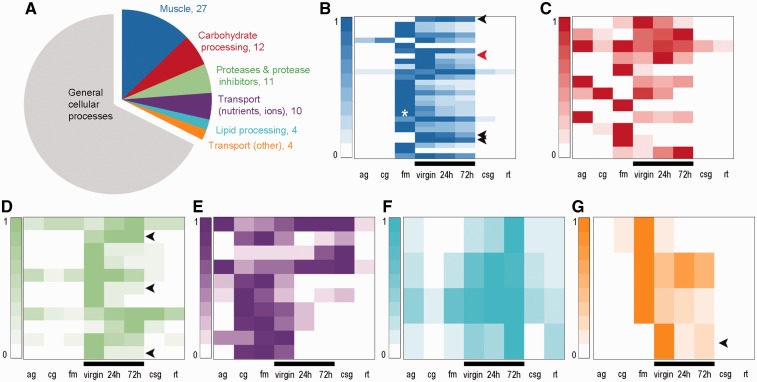
To characterize the origins of the genes expressed in the bursa, we first identified those that were highly expressed and thus likely to be involved in the bursa’s dominant function, processing of the spermatophore. We manually classified the 315 most highly expressed bursa genes into the following molecular function categories relevant to both digestion and spermatophore composition: “muscle,” “carbohydrate processing,” “proteases and protease inhibitors,” “transport (nutrients, ions),” “lipid processing,” and “transport (other)” (fig. 3 and supplementary data S1, Supplementary Material online). Using homologous proteins and domains, we were able to annotate 210 of 315 genes. As expected, most genes were involved in general cell physiological processes such as transcription or translation (supplementary data S1, Supplementary Material online), yet 68 genes (32%) fell into the bursa-relevant categories listed above (fig. 3, colored slices of the pie chart).
Fig. 3.
The bursa copulatrix expresses genes with digestive functions that are coexpressed in other tissues. (A) Functional annotation of 210 of the 315 most expressed genes in the bursa copulatrix and their expression depending on their functional categories. Colored slices of the pie chart are functional categories relevant to the bursa function. (B–G) Each heatmap shows the expression of the genes (rows) belonging to each biological function emphasized in the pie chart. Expression levels for each gene are relative to the maximum expression of that gene in a specific tissue cells are colorized according to each relative level of expression (0 = low relative expression, 1 = high relative expression). (B) Muscle, white star represents Troponin T (comp90911_c0) expression in flight muscle, red arrow represents bursa-specific troponin C (comp90487_c0). (C), Carbohydrate processing; (D) proteases and proteases inhibitors; (E) transport (nutrients, ions); (F) lipid processing; (G) transport (other). ag, Adult gut; cg, caterpillar gut; fm, flight muscle; virgin, virgin bursa copulatrix; 24 h, bursa copulatrix (24 h after mating); 72 h, bursa copulatrix (72 h after mating); csg, caterpillar salivary glands; rt, female reproductive tract. Black bars emphasize bursa samples. Black arrows emphasize genes highly expressed only in the bursa.
The bursa develops from the same progenitor structure as the other organs of the female reproductive tract. Thus, one plausible scenario for the bursa to gain novel physiological functions would be to modify genes already expressed in the reproductive tract itself. However, most of the highly expressed bursa genes exhibit very low expression in the reproductive tract (fig. 3). Furthermore, the vast majority of highly expressed bursa genes are coexpressed in nonreproductive tissues related to their functions, that is, muscle genes in flight muscle (fig. 3B), carbohydrate processing genes in adult/caterpillar guts and flight muscle (fig. 3C). This line of evidence suggests that the bursa adopted most of its physiologically relevant genes from nonreproductive tissues. Although the majority of the genes expressed in the bursa remain coexpressed in other tissues, some genes are highly expressed only in the bursa and may provide bursa-specific functions (fig. 3, black arrows).
Mechanical Digestion
A number of morphological and physiological studies have highlighted the role of muscular contractions in bursa functionality (Sugawara 1979, 1981; Rogers and Wells 1984; Lincango et al. 2013). Consistent with this role, muscle-related genes were the largest functional category (27 genes) among the 315 most highly expressed bursa genes (fig. 3), including homologs of classical muscle-related genes (Troponin T and C, myosin, tropomyosin, myofilin) (supplementary data S1, Supplementary Material online). Interestingly, one particular Troponin C homolog is specifically expressed in the bursa (98.5% of gene expression, comp90487_c0) (fig. 3B, red arrow).
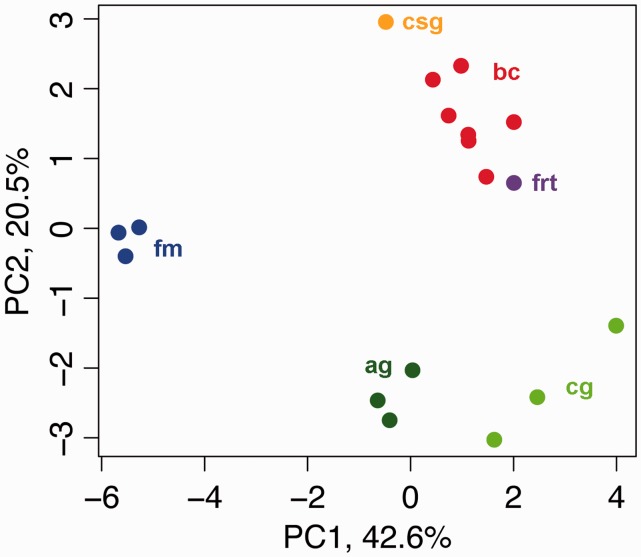
Because several butterfly organ systems contain muscle tissue of varying types, we attempted to determine which organ the bursa musculature most closely resembles. This task is best done using relative gene expression levels, as opposed to absolute levels, because organs contain different proportions of muscle tissue types and associated gene suites. We compared the relative expression levels of 17 characteristic musculature genes between organs (supplementary data S3, Supplementary Material online). A principal component analysis (PCA) of these data explained more than 60% in expression variation between tissues using PC1 and PC2 (42.6% and 20.5%, respectively; fig. 4). Contrary to our observations in transcriptome-wide expression profiles, bursa samples did not cluster with flight muscle, but rather with the female reproductive tract and, more distantly, the caterpillar salivary glands. This result shows that although muscle tissue is apparently less abundant in the general female reproductive tract, the relative expression pattern of muscle-related proteins in the bursa most closely resembles that in the female tract. This observation is consistent with their common developmental origin. Evolutionarily, this finding suggests that muscle-related expression programs—already intact in the ancestral reproductive tract—were simply augmented in the bursa.
Fig. 4.
Expression patterns of musculature genes in the bursa copulatrix are related to those in the female reproductive tract. PCA of expression of the 17 muscle genes that are the most highly expressed in the bursa copulatrix. PC1 and PC2 explain more than 60% of the variation in expression and are sufficient to cluster biological replicates by tissue type. Bursa copulatrix samples are closer to female reproductive tract than flight muscle. bc: bursa copulatrix; frt: female reproductive tract; csg: caterpillar salivary glands; fm: flight muscle; ag: adult gut; cg: caterpillar gut.
Enzymatic Digestion and Nutrient Absorption
There is also evidence of enzyme-based processing based in our detection of 11 genes encoding proteases (aminopeptidase N, endopeptidases) and protease inhibitors (serpin, cysteine proteinase inhibitor) (fig. 3A and D). This number of highly expressed proteolysis proteins is high compared with a nondigestive tissue, such as flight muscle, in which we found only one highly expressed protease (P-value = 0.0034, one-tailed Fisher’s exact test). Secreted proteins are likely to interact more directly with the spermatophore in the bursa lumen, and among the 315 most highly expressed genes, we identified 60 with putative secretion signal peptides (19%), ten of which were involved in proteolysis. The identification of bursa proteases and proteases inhibitors is in agreement with results from a distant Lepidopteran, the European Corn Borer moth (Al-Wathiqui et al. 2014). Furthermore, proteolytic cascades are well known to be involved in reproductive interactions between males and females over a wide range of taxonomic groups (Kelleher et al. 2007, 2011). Because the spermatophore is a complex structure composed of lipids, proteins, and carbohydrates, we explored two additional functional categories potentially important in spermatophore digestion: carbohydrate processing and lipid processing. We identified catalytic enzymes in carbohydrate processing (e.g., alpha-amylase) and three lipid-binding proteins (perilipin and two apolipophorins) and an enhancer of lipid digestion (saposin-like protein). A total of three carbohydrate and lipid processing proteins contained predicted secretion signal peptides. These proteases and proteases inhibitors and lipid processing genes seem to provide crucial bursa functions because their constituent genes are consistently expressed at their highest levels in the bursa compared with other tissues (fig. 3D and F).
The bursa is also thought to absorb spermatophore substrates that are subsequently used for egg production and somatic maintenance (Boggs and Gilbert 1979). We found highly expressed genes supporting this absorptive function in the transport (nutrients, ions) category, including amino acid, peptides, and carbohydrate transporters as well as ferritin, organic cation, and sodium transporters) (fig. 3A and supplementary data S1, Supplementary Material online). In the category transport (other), we identified genes encoding proteins that are involved in molecule diffusion (porins, permeases) (fig. 3G). One particularly notable permease, comp99299_c0, was predominantly and highly expressed in the bursa, indicating a potentially specialized transport protein in the bursa (fig. 3G, black arrow).
Evolutionary Histories of Bursa-Specific Genes Point toward Their Specialization
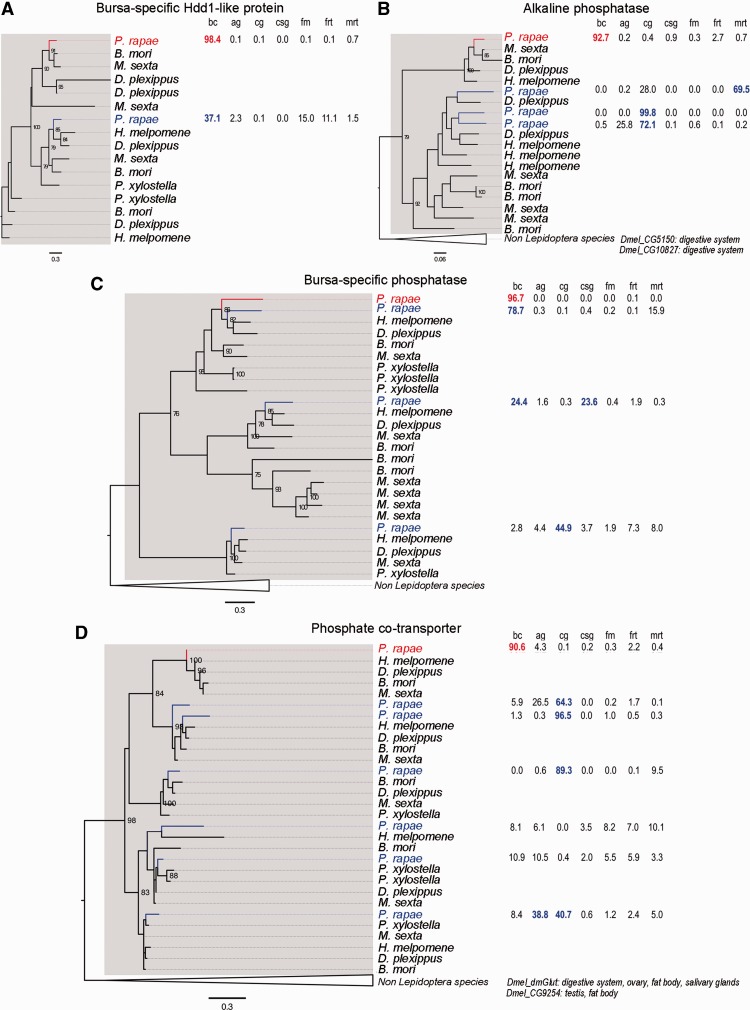
We also asked about the evolutionary origins of bursa-specific genes, as they represent strong candidates for highly specialized functions. To this end, we reconstructed phylogenetic trees for eight of the most highly expressed bursa-specific genes (>50 RPKM) using their homologs from six Lepidoptera and four non-Lepidopteran species. For comparison, we constructed gene phylogenies for 67 bursa genes that also exhibit general expression in other tissues (>50 RPKM). We found two bursa-specific genes with no recognizable homolog outside of Lepidoptera, even when performing PSI-BLAST searches outside of the ten focal species. Moreover, no conserved domains were detected in either gene. These genes are either evolving too rapidly to detect homology outside of Lepidoptera, or alternatively, they potentially arose through de novo gene formation in Lepidoptera. Either explanation, rapid evolution or de novo gene formation, is consistent with functional specialization. We directly addressed the question of rapid evolution in 7 bursa-specific and 64 nonspecific genes by measuring their divergence between P. rapae and the monarch butterfly, Danaus plexippus. Indeed, bursa-specific genes were more rapidly evolving than genes with more general expression, with median amino acid distances of 0.17 and 0.02, respectively (P-value = 0.023, one-sided Wilcoxon rank sum test; supplementary data S1, Supplementary Material online). We found duplication events within Lepidoptera for 2 out of 8 bursa-specific genes (25%) and 10 out of 67 other bursa genes (14.9%; table 1). Although the duplication rate is higher for bursa-specific genes, this difference does not achieve statistical significance (P-value = 0.3769; one-tailed Fisher’s exact test). Finally, we hypothesized that bursa-specific genes would tend to contain secretion signals because secreted proteins would directly interact with the spermatophore and hence may require functional specialization. For this test we considered a set of 13 bursa-specific genes with expression of at least 10 RPKM in the bursa. In agreement with our hypothesis, we found that 54% of bursa-specific genes encode secreted proteins compared with only 14% of nonspecific genes (table 2) (P-value = 0.00093, one-tailed Fisher’s exact test).
Table 1.
Bursa-Specific Genes Tend to Duplicate at a Higher Rate than Other Bursa Genes.
| Bursa-Specific Genes | Other Bursa Genes | |
|---|---|---|
| Duplication in Pierisrapae | 2 | 10 |
| No duplication | 6 | 57 |
Table 2.
Bursa-Specific Genes Tend to Encode Secreted Proteins.
| Bursa-Specific Genes | Other Bursa Genes | |
|---|---|---|
| Secreted | 7 | 602 |
| Not secreted | 6 | 3636 |
Together, the evidence for rapid rates of sequence evolution, formation of new genes, and an association with secreted proteins suggest that bursa-specific genes are highly specialized. Using homology and domain searches, we were able to assign likely functions to four of these bursa-specific genes: a secreted acid phosphatase, an enabler of muscle-contraction (Troponin C homolog), a lipase enzyme, and a transmembrane receptor. The remaining four bursa-specific genes have no recognized domains, although three of these have clear orthologs in other Lepidopteran species. Notably, one of these bursa-specific proteins of unknown function is the most highly expressed transcript in the bursa, and hence represents a strong candidate for a unique bursa-specific function (comp86587_c1, BUMP for “Bursa Maximal expression in Pieris,” supplementary data S4, Supplementary Material online).
Gene Duplicates with Highly Biased Bursa Expression Frequently Stemmed from Digestive Tract Genes
Another outstanding question for highly bursa-enriched genes is to determine their ancestral tissues of origin, and thereby infer their ancestral functional milieu. We studied the expression patterns of the set of bursa-enriched genes, genes for which 90% of their total expression across all tissues sampled in the study was found in the bursa that have paralogs within Lepidoptera (fig. 5 and supplementary data S1, Supplementary Material online). These gene family phylogenies demonstrate that expression patterns diverged greatly during the evolution of the various paralogs. The clearest observation is that 3 out of 4 bursa-enriched genes originated from genes predominantly expressed in the digestive tract or salivary glands (fig. 5B–D, supplementary data S5, Supplementary Material online). Interestingly, these three genes all have molecular functions involving phosphate; specifically they are a secreted alkaline phosphatase, a secreted histidine phosphatase, and a membrane-bound phosphate cotransporter. The next observation is that all four bursa-enriched genes considered here originated from genes not expressed to any large degree in the female reproductive tract. This mirrors the pattern of gene borrowing from nonreproductive tissues detailed in previous sections.
Fig. 5.
Evolutionary histories of highly enriched genes in the bursa. Each section presents a phylogenetic tree and relative expression levels of highly enriched genes in the bursa (in red) and their paralogs in Pieris rapae (in blue). Expression values for P. rapae genes (percentage of total expression) are presented on the right of the tree; bc, bursa copulatrix; cg, caterpillar gut; ag, adult gut; csg, caterpillar salivary glands; fm, flight muscle; frt, female reproductive tract; mrt, male reproductive tract. Bootstrap values are indicated for strongly supported nodes (>75%). Genes from the Lepidoptera clade are emphasized with a gray box. Non-Lepidoptera species are collapsed. Tissues of expression for Drosophila melanogaster orthologs were retrieved from FlyBase and are indicated at the bottom of the tree. (A) Bursa-specific Hdd1-like protein (comp98119_c6). (B) Alkaline phosphatase (comp98536_c0). (C) Bursa-specific phosphatase (comp92490_c0). (D) Phosphate cotransporter (comp100370_c2).
The Digestion of the Spermatophore by the Bursa Copulatrix Is a Transcriptionally Dynamic Process
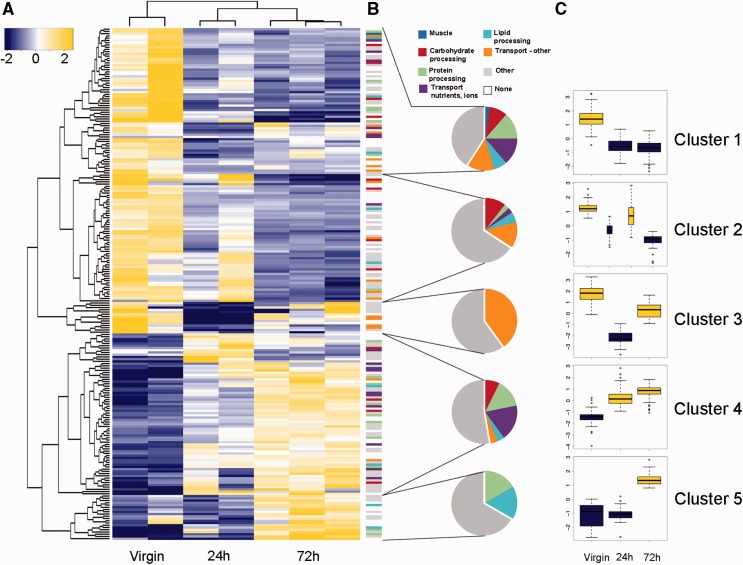
We studied bursa transcription at three different time points (before mating, 24 and 72 h postmating) to ask whether the transcriptional program changes during spermatophore digestion. All replicates for individual time points clustered together showing that the bursa response to mating is reproducible (fig. 6A). In addition, the 24 and 72 h postmating time points were more similar to each other than to the virgin bursa. Yet they were still distinct, demonstrating that the mating cycle persists over many days (fig. 6A and supplementary data S1, Supplementary Material online). There were 228 differentially expressed genes between the three time points, 73 of which were annotated to the previously mentioned nutrient processing and transport categories relevant to bursa functionality. We divided the 228 genes into five clusters according to their expression pattern across mating time points (fig. 6C and supplementary data S1, Supplementary Material online). All enzymatic and transport-related functional categories are represented among the differentially expressed genes, as well as among the five expression pattern clusters (fig. 6B). In contrast, only one muscle gene is differentially expressed (comp90487_c0, troponin C). This suggests that genes related to mechanical functions are transcriptionally static relative to enzymatic and absorptive functions.
Fig. 6.
Spermatophore digestion in the bursa copulatrix is a transcriptionally dynamic process. (A) Heatmap of log2-scale expression values for genes significantly differentially expressed in the bursa copulatrix depending on the timing after mating (virgin, 24 and 72 h). Hierarchical clustering of genes and replicates was based on Euclidean distance. A q-value cutoff for the false discovery rate (FDR = 0.001) and a minimal fold change of 4-fold were applied to select the genes represented in the heatmap. Cells are colorized according to level of expression (blue = low, yellow = high). (B) Functional categories of genes differentially expressed in the bursa for each mating time point (virgin, 24 and 72 h). Each color represents a particular molecular function. Pie charts summarize the proportion of genes belonging to each category. (C) Variation in expression of differentially expressed genes during mating (log2-fold change). Means are represented for each mating time point, except for the 24 h time point for cluster 2. Boxplots are colorized according to level of expression as in A (blue = low, yellow = high).
The specific timing of transcriptional changes provides clues to bursa function. Even though a virgin bursa has never been in contact with a spermatophore, it shows large numbers of highly expressed enzymatic and transport genes, including secreted proteases and protease inhibitors (fig. 6C, clusters 1, 2, and 3). This is in accordance with our measurements in the lab showing that proteolytic activity is maximal in the virgin bursa compared with postmating time points, and at even higher levels than in the entire digestive tract (Plakke et al., in preparation). These observations in the virgin bursa suggest that it is primed to digest a spermatophore before mating begins. One day after mating, the spermatophore is still in the bursa but is typically not yet breached. At this time point we observed major changes in the transcriptional program, with the downregulation of genes (fig. 6C, clusters 1 and 3) and the upregulation of other genes (cluster 4). As in the virgin bursa, all main functional categories are represented in cluster 4, with major representation in the transport (nutrients, ions) category, suggesting that the bursa may prepare for nutrient absorption as soon as 24 h after mating. By 72 h after mating, the outer envelope of the spermatophore is ruptured and nutrients are actively processed and absorbed. Genes from cluster 3, 4, and 5 are upregulated, with all processes represented except for the muscle category. It is interesting to see that even after 72 h, the expression pattern has not yet returned to resemble that of a virgin bursa. If previously mated bursas do eventually return to a virgin-like transcriptional state, our observations would indicate that the duration of one “mating cycle” is at least 72 h in P. rapae. An interesting extension of this work will be to determine how the bursa reacts after completing digestion of the first spermatophore, that is, the first mating cycle. An effective strategy would be to measure gene expression at even later time points after mating and determine if the same genes that were upregulated in the virgin bursa are upregulated again in order to prepare for the digestion of a new spermatophore.
Discussion
In this study, we characterized a unique female reproductive organ, the Lepidopteran bursa copulatrix, with the goals of determining how it carries out its digestive functions and how it became equipped with the genes enabling those functions. The bursa copulatrix develops from the same tissues as the rest of the female reproductive tract (Sendi et al. 1993; Gaikwad et al. 2014); however, its digestive function is highly derived, requiring high expression of enzymes and absorptive proteins not found elsewhere in the reproductive tract. Overall, the bursa copulatrix seems to have developed most of its novel digestive functions through the co-option and continued coexpression of genes found in at least five nonreproductive tissues (fig. 3). These genes included secreted enzymes and membrane-bound transporters enabling digestion and absorption of proteins, carbohydrates, and lipids, all of which are known constituents of the spermatophore (Marshall 1982, 1985). Furthermore, the expression of these genes changed in a repeatable fashion over the mating cycle, which indicates that the bursa is responsive to mating status and/or stage of spermatophore digestion (fig. 6). Finally, muscle-related genes are an exception to the pattern of gene borrowing from disparate organs, in that they are broadly expressed in the female reproductive tract, where they continue to have a similar expression pattern to that in the bursa (fig. 4). Thus, musculature expression programs were likely already intact when the bursa developed its digestive capacity, while the new functions in this organ system were supplied by preexisting genes from a suite of nonreproductive tissues.
Cooption of preexisting genes is not the only means by which the bursa could have been outfitted with novel digestive traits. Many studies have highlighted the roles of gene duplication and de novo gene formation in the origin of novel physiological genes such as those encoding lens crystallins and antifreeze proteins (de Jong and Hendriks 1986; Wistow et al. 1987; True and Carroll 2002). However, these mechanisms seem to contribute to a lesser degree in the origin of organ-wide physiological functions because we found a significantly smaller number of these cases. Perhaps there are fewer organ-specific genes because the process of gene formation requires additional evolutionary steps and could not supply the selected functionalities in a relevant time span. It would then remain a possibility that over longer timescales, more organ-specific genes will accrue and allow for further specialization of function. Alternatively, a low number of organ-specific genes could reflect a lack of need for specialized genes. Representation of these opposing scenarios perhaps depends on resolving the effects of gene pleiotropy and the limiting rate of new gene formation. Pleiotropic effects pit the needs of tissues against each other and provide a driving force for the creation of new genes. However, the rate of new gene formation may constrain this process. Male-reproductive tissues typically contain larger numbers of tissue-specific genes than we observed in the female bursa (13 bursa-specific genes over 10 RPKM compared to 661 in the male-reproductive tract). This disparity could be explained by the relatively recent origin of the bursa’s function, although it has existed for millions of years. Alternatively, there may be less of a need for specialized genes in this organ. We did however reveal a number of bursa-specific gene duplicates that would allow for the evolution of highly specialized bursa functions. For example, a number of these involved the liberation and absorption of phosphate (fig. 5). We did not find direct evidence of de novo gene formation in the bursa, an admittedly rare phenomenon, although there were 2 bursa-specific genes with no detectable homolog outside of Lepidoptera. Further investigation is required to determine if these could be de novo-formed genes.
As a result of these diverse evolutionary mechanisms, the bursa copulatrix exhibits a unique combination of stomach- and gut-like traits within the reproductive tract. Overall, observations from the bursa copulatrix argue that gene co-option coupled with continued coexpression is the dominant mechanism for innovation in organ systems that adopt a wide range of functions, and that only a relatively limited set of specialized duplicate genes is required. This seminal finding opens the door to additional lines of investigation into the origin of novel functionality in tissue and organ systems. Our findings from the bursa suggest a fundamental disparity between the evolution of physiological and morphological traits. Bursa physiology relied upon extensive gene borrowing from several, disparate organs, whereas morphological traits are often described as arising through the adoption of one or relatively few intact developmental pathways (Shubin et al. 1997; Gould 2002).
Multiple studies have shown that genes and traits involved in sexual reproduction evolve rapidly, and this is often hypothesized to result from dynamic interactions between males and females (Swanson et al. 2001; Swanson and Vacquier 2002; Torgerson et al. 2002; Clark et al. 2006, 2009; Panhuis et al. 2006; Meslin et al. 2011, 2012; Yassin and Orgogozo 2013). These interactions are often influenced by selective regimes such as intrasexual selection (e.g., male-male competition), sexual conflict and even cooperative coevolution between the sexes (for review see Swanson and Vacquier [2002]). In our system, the interaction between the spermatophore and the bursa copulatrix sets the stage to study both conflict and cooperation. On the one hand, the nutrients in the spermatophore benefit females because they use them for both egg production and somatic maintenance. However, females are unable to remate until the spermatophore is almost fully digested by the bursa. Both conflict and cooperation are reflected in the morphological and physiological traits of the spermatophore and the bursa. Although males coat their spermatophores with a thick outer envelope that resists spermatophore digestion (Sánchez and Cordero 2014), females equip their bursa with tools for both mechanical and chemical digestion, the signum and nutrient processing enzymes, respectively. The bursa-spermatophore system then offers the possibility to assess the role of sexual conflict and cooperation in the evolution of these traits. Genes with their expression enriched in the bursa copulatrix and with specialized functions are of interest because they are potentially subjected to both evolutionary pressures. For example, proteases are prime candidates to determine the enzymes digesting the spermatophore outer envelope. We may then expect proteases to experience directional selection for increased efficacy during spermatophore digestion. Here, we identified key genes on the female side of reproduction, thereby paving the way to reveal molecular interactions between males and females. Because such studies have suffered from a strong male bias in the past (Córdoba-Aguilar 2010; Ah-King et al. 2014), more work focused on female reproductive adaptations promises to help us to begin to understand the coevolutionary dynamics between the sexes and the selective regimes that define them.
Materials and Methods
Butterfly Stocks
Pieris rapae were sampled in September 2013 from a continuous, inbred lab colony established in October 2012 from 4 females collected in Rochester, Pennsylvania (40°44′45″N, 80°9′45″W). Individuals were raised in insect growth chambers under controlled climatic conditions (24 °C, 60% relative humidity, 16 L:8D photoperiod). Larvae were fed on a diet of kale leaves grown on site, fertilized twice a week with Peter’s Profession General Purpose 20-20-20.
Sample Preparation
Laboratory areas used for dissection were sterilized and sprayed with RNaseZap RNase Decontamination Solution (Life Technologies, Grand Island, NY). Live samples had their abdomen and thorax separated and immediately spread open to expose tissues of interest. One hundred microliters of RNAlater RNA Stabilization Reagent (Qiagen, Valencia, CA) was added to the tissues to immediately preserve the RNA before being submerged in 200 µl of RNAlater and frozen at −20 °C until further dissection. Each sample was then defrosted and tissues carefully dissected out and separated for RNA extraction. Microdissected tissues were stored at −20 °C in 100 µl of RNAlater until RNA extraction.
To obtain a tissue-specific view of gene expression in P. rapae to inform our interpretation of the bursa transcriptome, we dissected and sequenced mRNA from seven tissues from caterpillars and adults: bursa copulatrix, female reproductive tract, male reproductive tract, flight muscle, adult gut, caterpillar gut, and caterpillar mandibular salivary gland. For several tissue types, pooling of tissue from multiple individuals was required to collect sufficient RNA for sequencing. In addition, for tissues of particular interest, biological replicates were performed. These included virgin bursae, both time points of mated bursae (24 and 72 h postmating), flight muscle, adult gut, and caterpillar gut. Pooled tissue samples for biological replicates were always composed of tissues collected from full siblings. For the male reproductive tract, reproductive tissues from three full siblings were pooled. RNA extraction from caterpillar salivary glands required pooling of 80 glands from 40 individuals. Female reproductive tract samples were pooled from five individuals. Both mandibular salivary glands were extracted from the tissues, rinsed in RNAlater, and frozen in 100 µl of RNAlater at −20°C. The gut was then separated and emptied of contents and the remaining carcass rinsed before both tissues were frozen in 100 µl of RNAlater at −20°C. Bursa copulatrix for the two time points after mating (24 and 72 h) were carefully rinsed to remove any male contribution. We also sequenced mRNA from a whole butterfly and a whole caterpillar in order to assemble a more complete transcriptome.
Total RNA extractions were performed with TRIzol (Life Technologies, Grand Island, NY). For the caterpillar salivary glands, total RNA extraction was performed using the Arcturus PicoPure RNA Isolation kit (Life Technologies) and treated with Ambion TURBO DNase. The quality of each RNA sample was validated on a Bioanalyzer RNA 6000 nanochip (Agilent Technologies, Santa Clara, CA), and RNA concentrations were measured with a Qubit 2.0 fluorometer (Life Technologies). Total RNA samples were stored at −80°C until library preparation and sequencing.
RNA Sequencing
All samples were processed at the Genomics Resources Core Facility of Weill Cornell Medical College (New York, NY). Enrichment for mRNAs was performed using polyA selection from total RNA. All libraries were prepared separately using the TruSeq RNA sample prep kit v2, RS-122-2001 (Illumina) and sequenced on an Illumina HiSeq 2500 to generate paired-end reads of 100 bp. The raw sequence reads for P. rapae are available upon request to the authors.
Sequence Data Processing and Trancriptome De Novo Assembly
After trimming off the adaptor sequences, the raw reads were filtered based on both quality and length using the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/, last accessed March 12, 2015; Version 0.0.13). Reads were scanned and trimmed off when a base with a quality score <30 was encountered. Reads with a length <20 were discarded. Cleaned reads from a previous round of sequencing (Axeq Technologies, Seoul, South Korea) of P. rapae tissues were also used for the de novo assembly. The sequencing generated a total of 400,366,479 raw paired-end reads. After removing bases with a quality score lower than 30 (base call accuracy of 99.9%) and sequences shorter than 20 bp, a total of 397,119,041 paired-end reads remained and were used for the de novo assembly of the P. rapae transcriptome using the Trinity pipeline with default parameters (version r2013-02-25) (Haas et al. 2013). Using the high-quality reads, Trinity generated an assembly of 115,574 components. Two subsequent filtration steps were applied to the assembly. The first step removed the components to which no read was mapped by Bowtie (v1.0.0) (Langmead et al. 2009) and led to an assembly of 106,320 components. The second step of filtration removed the components with no open reading frame (ORF) or with an ORF shorter than 51 amino acids. The final de novo transcriptome assembly is composed of 15,773 protein-coding components with a GC content of 39.15% (table 3). Our transcriptome characteristics are consistent with the range of 35–40% GC content commonly observed across Lepidopteran genomes (International Lepidopteran Genome Project, 8/2002) and the 12,669 and 16,866 predicted protein-coding genes for Heliconius melpomene and D. plexippus, respectively (Zhan et al. 2011; Heliconius Genome Consortium 2012). Among our final data set of 15,773 genes, stringent sequence similarity searches against Bombyx mori protein databases using BLASTx showed that 10,068 (63.8% of total contigs) had significant matches with a high E-value threshold of 1 e−20, reflecting the superior quality of our assembly. The final version of the assembly is available upon request to the authors.
Table 3.
Statistics of the Final De Novo Assembly of Pieris rapae Transcriptome.
| Number of components | 15,773 |
| Number of transcripts | 41,378 |
| N50 (nt) | 4,159 |
| Median contig length (nt) | 2,406 |
| GC content | 39.15% |
Abundance Estimation, Differential Expression Analysis, and Functional Annotation
All cleaned reads were then mapped on the assembly using Bowtie (v1.0.0) (Langmead et al. 2009). Transcript abundance was estimated for each sample using RSEM (v1.2.3) (Li and Dewey 2011) and was measured as fragments per kilobase of exon per million fragments mapped (FPKM) values. Differentially expressed transcripts were identified by the EdgeR Bioconductor package (Robinson et al. 2010) by taking into account biological replicates. RNA-seq count was normalized between the different samples and replicates using trimmed mean of M-values normalization method (TMM; Robinson and Oshlack 2010). Annotation was manually refined for genes of specific interest. Component sequences were blasted against B. mori protein sequences from the SilkDB database (Duan et al. 2010) with a cutoff E-value of 1 e−20. The presence/absence of secretion signal peptides and transmembrane domains were determined using SignalP and TMHMM, respectively (Petersen et al. 2011). Tissue-specificity was assessed by computing normalized expression values proportions for each gene (TMM normalized values). A gene was considered tissue-specific when the mean of its expression (TMM normalized FPKM values) between replicates in one tissue represented more than 95% of its total expression across all tissues sampled in this study. A gene was considered tissue-enriched when the mean of its expression (TMM normalized FPKM values) between replicates in one tissue represented more than 90% of its total expression across all tissues sampled in the study.
Principal Component Analysis
A PCA was used to determine similarities between expression patterns of muscle genes from different tissues (bursa, adult, and caterpillar guts, female reproductive tract, caterpillar salivary glands, and flight muscle). TMM-normalized RPKM expression values for 17 highly expressed muscle-related genes were used for each tissue to calculate proportional expression levels. For each gene, we calculated its proportional contribution to total expression of these 17 genes in a given tissue. These proportions were used for the PCA. By normalizing these muscle genes using proportions we were then able to compare only the muscle tissue itself within each organ. Each tissue and replicate was considered independently for the analysis.
Phylogenetic Analyses
Homologous genes searches were performed with BLASTP using translated ORFs from our P. rapae de novo transcriptome assembly as queries against protein databases of the following species: six Lepidoptera: P. rapae translated ORFs (our data), B. mori (http://silkworm.genomics.org.cn/, last accessed March 12, 2015; Bombyx_mori.Bmor1.21.pep.all.fa), D. plexippus (http://monarchbase.umassmed.edu/, last accessed March 12, 2015; Dp_geneset_OGS1_pep.fasta), H. melpomene (http://www.butterflygenome.org/, last accessed March 12, 2015; heliconius_melpomene_v1.1_primaryScaffs_Protein.faa), Manduca sexta (http://agripestbase.org/manduca/, last accessed March 12, 2015; Manduca_sexta_OGS2_20140407_proteins.fa), Plutella xylostella (http://iae.fafu.edu.cn/DBM/, last accessed March 12, 2015; P.xylostella.pep.fasta); two Diptera: Drosophila melanogaster (http://flybase.org/, last accessed March 12, 2015; dmel-all-translation-r5.57.fasta) and Aedes aegypti (Liverpool strain, https://www.vectorbase.org/, last accessed March 12, 2015; Aedes-aegypti-Liverpool_PEPTIDES_AaegL3.1.fa); and two additional insects, Apis mellifera (www.beebase.org/, last accessed March 12, 2015; amel_OGSv3.2_pep.fa) and Tribolium castaneum (http://beetlebase.org/, last accessed March 12, 2015; Tcas20051011-glean_peptide). BLASTP outputs were then parsed to cluster homologous gene sequences together (E-value threshold of 1 e−5). Multiple sequence alignments of clusters were performed using Clustal Omega (Sievers et al. 2011), and phylogenetic trees were reconstructed using PhyML (Guindon et al. 2010). As a first approach, branch support values were computed using aLRT (approximate Likelihood Ratio Test) statistics (Anisimova and Gascuel 2006). For publication purposes, branch support values were estimated using 100 bootstraps (Felsenstein 1985). For two genes with no homologs outside from Lepidoptera, we performed PSI-BLAST searches. PSI-BLAST uses position-specific scoring matrices to score matches between query and databases sequences instead of a predefined scoring matrix such as BLOSUM62 and is then thought to be more sensitive than a regular BLAST and might be able to find distantly related sequences that could be missed in a BLAST search.
Supplementary Material
Supplementary data S1–S5 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by startup funds to NIM and NLC from the University of Pittsburgh, by a Charles E. Kaufman New Investigator Award to NLC from The Pittsburgh Foundation (KA-2014-73920) and by NIH 1S10OD010693-01 for data collected on the Illumina HiSeq 2500 (Cornell University Biotechnology Resource Center [BRC]). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Ah-King M, Barron AB, Herberstein ME. Genital evolution: why are females still understudied? PLoS Biol. 2014;12:e1001851. doi: 10.1371/journal.pbio.1001851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Wathiqui N, Lewis SM, Dopman EB. Using RNA sequencing to characterize female reproductive genes between Z and E Strains of European Corn Borer moth (Ostrinia nubilalis) BMC Genomics. 2014;15:189. doi: 10.1186/1471-2164-15-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- Arrow GJ, Hincks WD. Horned beetles. The Hague: Junk; 1951. [Google Scholar]
- Blank D, Wolf L, Ackermann M, Silander OK. The predictability of molecular evolution during functional innovation. Proc Natl Acad Sci U S A. 2014;111:3044–3049. doi: 10.1073/pnas.1318797111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs CL. Selection pressures affecting male nutrient investment at mating in heliconiine butterflies. Evolution. 1981:931–940. doi: 10.1111/j.1558-5646.1981.tb04959.x. [DOI] [PubMed] [Google Scholar]
- Boggs CL. Male nuptial gifts: phenotypic consequences and evolutionary implications. In: Leather SR, Hardie J, editors. Insect reproduction. Cleveland: CRC Press; 1995. pp. 215–242. [Google Scholar]
- Boggs CL, Gilbert LE. Male contribution to egg production in butterflies: evidence for transfer of nutrients at mating. Science. 1979;206:83–84. doi: 10.1126/science.206.4414.83. [DOI] [PubMed] [Google Scholar]
- Boggs CL, Watt WB. Population structure of pierid butterflies IV. Genetic and physiological investment in offspring by male Colias. Oecologia. 1981;50:320–324. doi: 10.1007/BF00344970. [DOI] [PubMed] [Google Scholar]
- Casewell NR, Wüster W, Vonk FJ, Harrison RA, Fry BG. Complex cocktails: the evolutionary novelty of venoms. Trends Ecol Evol. 2013;28:219–229. doi: 10.1016/j.tree.2012.10.020. [DOI] [PubMed] [Google Scholar]
- Chen L, DeVries AL, Cheng C-HC. Evolution of antifreeze glycoprotein gene from a trypsinogen gene in Antarctic notothenioid fish. Proc Natl Acad Sci U S A. 1997;94:3811–3816. doi: 10.1073/pnas.94.8.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark NL, Aagaard JE, Swanson WJ. Evolution of reproductive proteins from animals and plants. Reproduction. 2006;131:11–22. doi: 10.1530/rep.1.00357. [DOI] [PubMed] [Google Scholar]
- Clark NL, Gasper J, Sekino M, Springer SA, Aquadro CF, Swanson WJ. Coevolution of interacting fertilization proteins. PLoS Genet. 2009;5:e1000570. doi: 10.1371/journal.pgen.1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Córdoba-Aguilar A. The evolution of primary sexual characters in animals. In: Leonard J, Córdoba-Aguilar A, editors. Oxford: Oxford University Press; 2010. [Google Scholar]
- de Jong WW, Hendriks W. The eye lens crystallins: ambiguity as evolutionary strategy. J Mol Evol. 1986;24:121–129. doi: 10.1007/BF02099960. [DOI] [PubMed] [Google Scholar]
- Dixson AL, Anderson MJ. Sexual selection, seminal coagulation and copulatory plug formation in primates. Folia Primatol. 2002;73:63–69. doi: 10.1159/000064784. [DOI] [PubMed] [Google Scholar]
- Drummond B. Multiple mating and sperm competition in the Lepidoptera. In: Smith RL, editor. Sperm competition and the evolution of animal mating systems. New York: Academic Press; 1984. pp. 291–370. [Google Scholar]
- Duan J, Li R, Cheng D, Fan W, Zha X, Cheng T, Wu Y, Wang J, Mita K, Xiang Z, et al. SilkDB v2.0: a platform for silkworm (Bombyx mori) genome biology. Nucleic Acids Res. 2010;38:D453–D456. doi: 10.1093/nar/gkp801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emlen DJ, Lavine LC, Ewen-Campen B. On the origin and evolutionary diversification of beetle horns. Proc Natl Acad Sci U S A. 2007;104:8661–8668. doi: 10.1073/pnas.0701209104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Gaikwad S, Koli Y, Bhawane G. Histomorphology of the Female Reproductive System in Papilio polytes polytes Linnaeus, 1758 (Lepidoptera: Papilionidae) Proc Natl Acad Sci India Section B: Biol Sci. 2014;84:901–908. [Google Scholar]
- Gould SJ. The structure of evolutionary theory. Cambridge (MA): Harvard University Press; 2002. [Google Scholar]
- Greenfield MD. The question of paternal investment in Lepidoptera: male-contributed proteins in Plodia interpunctella. Int J Invertebr Repr. 1982;5:323–330. [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallgrimsson B, Jamniczky HA, Young NM, Rolian C, Schmidt-Ott U, Marcucio RS. The generation of variation and the developmental basis for evolutionary novelty. J Exp Zool B Mol Dev Evol. 2012;318:501–517. doi: 10.1002/jez.b.22448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heliconius Genome Consortium. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487:94–98. doi: 10.1038/nature11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson B. Resource allocation and mating systems in butterflies. Evolution. 1995:955–961. doi: 10.1111/j.1558-5646.1995.tb02330.x. [DOI] [PubMed] [Google Scholar]
- Karlsson B. Male reproductive reserves in relation to mating system in butterflies: a comparative study. Proc R Soc Lond B Biol Sci. 1996;263:187–192. [Google Scholar]
- Kelleher ES, Clark NL, Markow TA. Diversity-enhancing selection acts on a female reproductive protease family in four subspecies of Drosophila mojavensis. Genetics. 2011;187:865–876. doi: 10.1534/genetics.110.124743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher ES, Swanson WJ, Markow TA. Gene duplication and adaptive evolution of digestive proteases in Drosophila arizonae female reproductive tracts. PLoS Genet. 2007;3:e148. doi: 10.1371/journal.pgen.0030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai-Fook J. Absorption of phosphorus from the spermatophore through the cuticle of the bursa copulatrix of the butterfly, Calpodes ethlius. Tissue Cell. 1991;23:247–259. doi: 10.1016/0040-8166(91)90079-9. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincango P, Fernandez G, Baixeras J. Microstructure and diversity of the bursa copulatrix wall in Tortricidae (Lepidoptera) Arthropod Struct Dev. 2013;42:247–256. doi: 10.1016/j.asd.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Long M, Betrán E, Thornton K, Wang W. The origin of new genes: glimpses from the young and old. Nat Rev Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- Lorkovic Z. Enzyme electrophoresis and interspecific hybridization in Pieridae (Lepidoptera) Journal of Research on the Lepidoptera. 1985;24:334–358. [Google Scholar]
- Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall L. Male nutrient investment in the Lepidoptera: what nutrients should males invest? Am Nat. 1982;120:273. [Google Scholar]
- Marshall L. Protein and lipid composition of Colias philodice and C. eurytheme spermatophores and their changes over time (Pieridae) J Res Lepid. 1985;24:21–30. [Google Scholar]
- Meslin C, Brimau F, Nagnan-Le Meillour P, Callebaut I, Pascal G, Monget P. The evolutionary history of the SAL1 gene family in eutherian mammals. BMC Evol Biol. 2011;11:148. doi: 10.1186/1471-2148-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meslin C, Mugnier S, Callebaut I, Laurin M, Pascal G, Poupon A, Goudet G, Monget P. Evolution of genes involved in gamete interaction: evidence for positive selection, duplications and losses in vertebrates. PLoS One. 2012;7:e44548. doi: 10.1371/journal.pone.0044548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moczek AP, Emlen DJ. Male horn dimorphism in the scarab beetle, Onthophagus taurus: do alternative reproductive tactics favour alternative phenotypes? Anim Behav. 2000;59:459–466. doi: 10.1006/anbe.1999.1342. [DOI] [PubMed] [Google Scholar]
- Moczek AP, Rose DJ. Differential recruitment of limb patterning genes during development and diversification of beetle horns. Proc Natl Acad Sci U S A. 2009;106:8992–8997. doi: 10.1073/pnas.0809668106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moczek AP, Sultan S, Foster S, Ledón-Rettig C, Dworkin I, Nijhout HF, Abouheif E, Pfennig DW. The role of developmental plasticity in evolutionary innovation. Proc R Soc B Biol Sci. 2011;278:2705–2713. doi: 10.1098/rspb.2011.0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panhuis TM, Clark NL, Swanson WJ. Rapid evolution of reproductive proteins in abalone and Drosophila. Philos Trans R Soc B Biol Sci. 2006;361:261–268. doi: 10.1098/rstb.2005.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Pitnick S, Markow TA, Spicer GS. Evolution of multiple kinds of female sperm-storage organs in Drosophila. Evolution. 1999;53:1804–1822. doi: 10.1111/j.1558-5646.1999.tb04564.x. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SH, Wells H. The structure and function of the bursa copulatrix of the monarch butterfly (Danaus plexippus) J Morphol. 1984;180:213–221. doi: 10.1002/jmor.1051800305. [DOI] [PubMed] [Google Scholar]
- Sánchez V, Cordero C. Sexual coevolution of spermatophore envelopes and female genital traits in butterflies: evidence of male coercion? PeerJ. 2014;2:e247. doi: 10.7717/peerj.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendi J, Srivastava K, Singh M. Postembryonic study on the development of the female efferent genital system in the lemon-butterfly, Papilio demoleus L.(Lepidoptera) J Res Lepid. 1993;32:162–169. [Google Scholar]
- Shapiro AM. The role of sexual behavior in density-related dispersal of pierid butterflies. Am Nat. 1970;104:367–372. [Google Scholar]
- Shubin N, Tabin C, Carroll S. Fossils, genes and the evolution of animal limbs. Nature. 1997;388:639–648. doi: 10.1038/41710. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara T. Stretch reception in the bursa copulatrix of the butterfly,Pieris rapae crucivora, and its role in behaviour. J Comp Physiol. 1979;130:191–199. [Google Scholar]
- Sugawara T. Fine structure of the stretch receptor in the bursa copulatrix of the butterfly, Pieris rapae crucivora. Cell Tissue Res. 1981;217:23–36. doi: 10.1007/BF00233822. [DOI] [PubMed] [Google Scholar]
- Suzuki Y. Mating frequency in females of the small cabbage white, Pieris rapae crucivora Boisduval (Lepidoptera: Pieridae) Kontyu. 1979;47:335–339. [Google Scholar]
- Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nat Rev Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Yang Z, Wolfner MF, Aquadro CF. Positive Darwinian selection drives the evolution of several female reproductive proteins in mammals. Proc Natl Acad Sci U S A. 2001;98:2509–2514. doi: 10.1073/pnas.051605998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgerson DG, Kulathinal RJ, Singh RS. Mammalian sperm proteins are rapidly evolving: evidence of positive selection in functionally diverse genes. Mol Biol Evol. 2002;19:1973–1980. doi: 10.1093/oxfordjournals.molbev.a004021. [DOI] [PubMed] [Google Scholar]
- True JR, Carroll SB. Gene co-option in physiological and morphological evolution. Annu Rev Cell Dev Biol. 2002;18:53–80. doi: 10.1146/annurev.cellbio.18.020402.140619. [DOI] [PubMed] [Google Scholar]
- Vahed K. The function of nuptial feeding in insects: a review of empirical studies. Biol Rev. 1998;73:43–78. [Google Scholar]
- Watanabe M, Sato K. A spermatophore structured in the bursa copulatrix of the small white Pieris rapae (Lepidoptera, Pieridae) during copulation, and its sugar content. J Res Lepidoptera. 1993;32:26–36. [Google Scholar]
- Wedell N. Female receptivity in butterflies and moths. J Exp Biol. 2005;208:3433–3440. doi: 10.1242/jeb.01774. [DOI] [PubMed] [Google Scholar]
- Wiklund C. Sexual selection and the evolution of butterfly mating systems. In: Boggs CL, Watt WB, Ehrlich PR, editors. Butterflies: ecology and evolution taking flight. Chicago: University of Chicago Press; 2003. p. 90. [Google Scholar]
- Wistow GJ, Mulders JW, de Jong WW. The enzyme lactate dehydrogenase as a structural protein in avian and crocodilian lenses. Nature. 1987;326:622–624. doi: 10.1038/326622a0. [DOI] [PubMed] [Google Scholar]
- Yassin A, Orgogozo V. Coevolution between male and female genitalia in the Drosophila melanogaster species subgroup. PLoS One. 2013;8:e57158. doi: 10.1371/journal.pone.0057158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan S, Merlin C, Boore JL, Reppert SM. The monarch butterfly genome yields insights into long-distance migration. Cell. 2011;147:1171–1185. doi: 10.1016/j.cell.2011.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.