Abstract
Mechanical strain regulates the development, organization and function of multicellular tissues, but mechanisms linking mechanical strain and cell-cell junction proteins to cellular responses are poorly understood. We showed that mechanical strain applied to quiescent epithelial cells induced rapid cell cycle re-entry, mediated by independent nuclear accumulation and transcriptional activity of first Yap1 and then β-catenin. Inhibition of Yap1- and β-catenin-mediated transcription blocked cell cycle re-entry and progression through G1 into S phase, respectively. Maintenance of quiescence, Yap1 nuclear exclusion and β-catenin transcriptional responses to mechanical strain required E-cadherin extracellular engagement. Our results indicate that activation of Yap1 and β-catenin is a master regulator of mechanical strain-induced cell proliferation, and cadherins are signaling centers required for cellular responses to externally applied force.
Cellular responses to mechanical force are important during development and disease, and involve reinforcing cell-cell and cell-extracellular matrix (ECM) adhesions, increased cytoskeletal stiffness, and regulation of cell fate (1–4). Increased ECM stiffness leads to cytoskeleton reorganization and cell cycle progression by activating the Hippo pathway transcription factors Yap/Taz (5) downstream of actin remodeling factors (6), indicating that Yap is a mechanotransducer. However, less is known about signaling from cadherin-mediated cell-cell junctions following applied force.
Classical cadherins couple neighboring cells through trans interactions between opposed extracellular domains and force-dependent linkage of the cytoplasmic domain to the actin cytoskeleton through β-catenin and α-catenin (7–11), resulting in constitutive tension on E-cadherin at the plasma membrane (10). The cadherin-catenin complex is thought to regulate growth signaling by sequestering the transcription factors β-catenin and Yap1(12–16) in the cytoplasm. However, it is unclear whether cadherin-mediated adhesion is required for the activation of β-catenin and Yap1 in response to mechanical force.
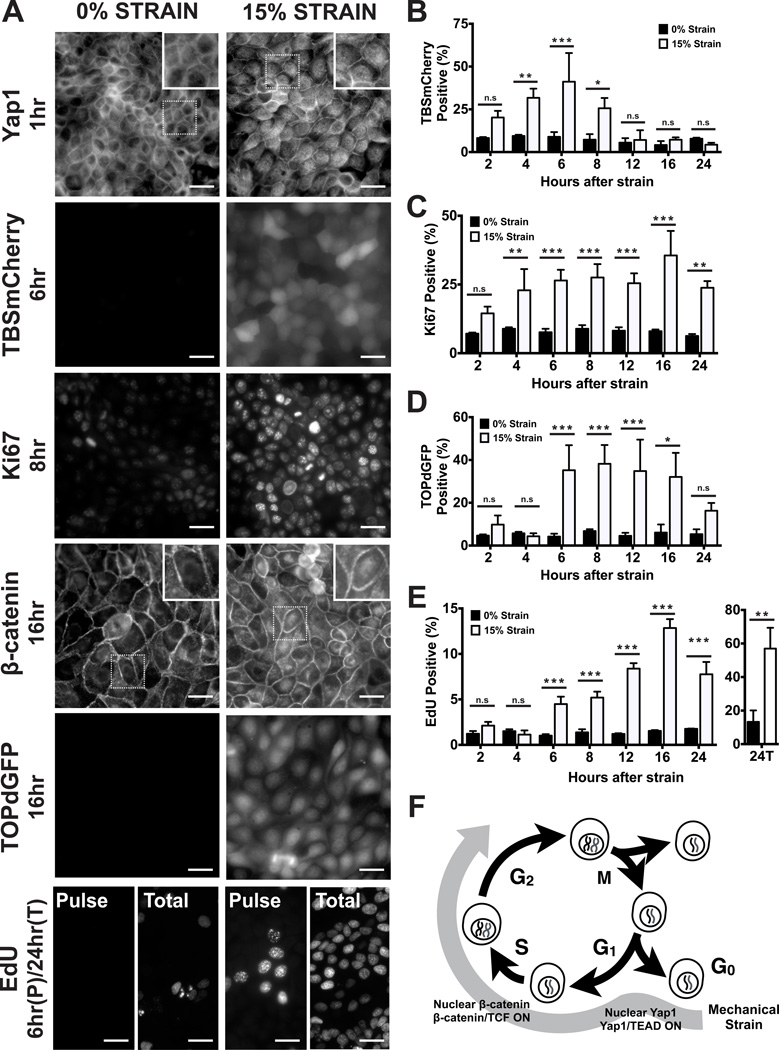
To model mechanical force in multicellular tissues, dense monolayers of quiescent kidney epithelial (MDCK) cells were formed on compliant silicone substrates in an integrated strain array (ISA) (Fig. S1; see also (17)). The ISA was used to apply and maintain different levels of static biaxial stretch for different times (2–24 hours). Cells were then processed for imaging, data acquisition, and analysis (see Supplementary Information). Mechanical strain induced rapid cell cycle re-entry (Ki67 positive, Fig. 1A,C; Fig. S2A,C), and subsequent DNA replication and progression through S phase (EdU positive; Figure 1A,E; Fig. S2B,D; see also (6)) into G2 (Geminin positive; Fig. S3). The majority of cells had entered S phase after 24 hours of strain application (Fig. 1E ‘24C’), and higher levels of strain resulted in higher levels of cell cycle re-entry (Fig. S2).
Figure 1.
Mechanical strain induces cell cycle re-entry and sequential activation of Yap1 and β-catenin. (A) Distribution of Yap1 1hr after no strain or 15% strain, TBSmCherry after 6hrs, Ki67 after 8hrs, β-catenin after 16hrs, TOPdGFP after 16hrs, and EdU incorporation after a 1hr pulse (‘Pulse’) before fixation at 6hrs, or total EdU incorporation over 24hrs (‘Total’). Insets show higher magnification of the region outlined by a dotted line. (B–E) Levels of TBSmCherry (B), Ki67 (C), TOPdGFP (D) and EdU (E) in MDCK monolayers 2–24hrs after mechanical strain; for methods used for quantification, see Supplementary Information. (F) Summary of cell cycle responses to mechanical strain. Scale bars: 25 µm. All quantifications were from at least 3 independent experiments with 2 replicate monolayers per experiment (Table S1). Quantifications were mean +/− SEM; unpaired t-test p values < 0.05 (*), <0.01(**) and <0.001 (***).
We examined whether the cadherin-associated transcriptional activators Yap1 and β-catenin responded to mechanical strain. In the absence of mechanical strain, Yap1 localized in the cytoplasm and cell cortex (Fig. 1A; S4A; see also (13)). β-Catenin localized at cell-cell contacts (Figure 1A; S5A), as expected due to cadherin binding (7) and proteasome-mediated degradation of excess cytoplasmic β-catenin (18, 19). Upon mechanical strain, Yap1 and β-catenin re-localized to the nucleus, but on different time scales. Nuclear Yap1 was detected within 1 hour of strain application, peaked at 6 hours, and then declined rapidly to background levels (Figure 1A; Fig. S4A,B). In contrast, nuclear β-catenin was not observed until 6 hours following strain and remained over 24 hours (Fig. 1A; Fig. S5A,B).
We next determined if nuclear localization of Yap1 and β-catenin corresponded to their transcriptional activities. Analysis of the TBSmCherry reporter for Yap1 transcriptional activity (Fig S4C; (13)) revealed that, like Yap1 nuclear accumulation, activation following strain was rapid and peaked at 6 hours (Fig. 1A,B; Fig. S4D,E), then decreased prior to the majority of cells entering S phase (EdU positive, Fig. 1E; Fig. S2B,D). In contrast, β-catenin transcriptional activity measured with the TOPdGFP reporter (20) increased rapidly 6 hours after strain application, at the same time that nuclear β-catenin was detected (Fig. 1A,D; Fig. S5). β-Catenin transcriptional activity then remained high (Fig. 1D; Fig. S5D,E) as cells proceeded through S phase (Fig. 1E; Fig S2B,D). Thus, mechanical strain induced both Yap1- and β-catenin-mediated transcriptional activities, but at different times after strain application and transiently in the case of Yap1 (Figure 1F).
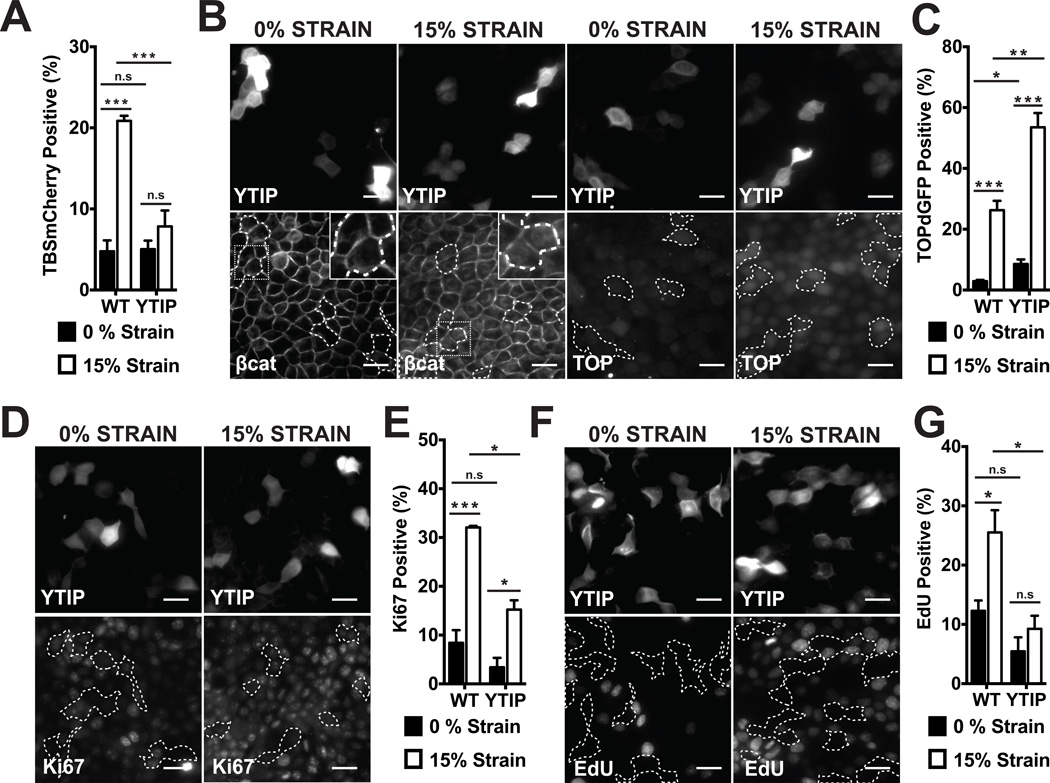
Although Yap1 activation preceded β-catenin activation by several hours following strain, we tested whether their activation was coupled. Expression of the YAP1-TEAD inhibitory peptide (YTIP) disrupts interactions between Yap1 and TEA domain (TEAD) transcription factors and prevents transcription of Yap1/TEAD targeted genes (Fig. S6C; (21)). When mechanical strain was applied to MDCK cells transiently expressing GFP- or RFP-tagged YTIP, YTIP-positive cells did not have increased Yap1 activity (Fig. 2A), Ki67 staining (Fig. 2D,E; Fig. S6E,F), nor EdU incorporation (Fig. 2F,G), in contrast to their un-transfected neighbors. Similar results were obtained with Verteporfin (Fig. S8), a small molecule inhibitor of Yap1 binding to TEAD transcription factors (22). However, inhibition of Yap1 activity with YTIP or Verteporfin did not block increased nuclear β-catenin levels (Fig. 2B; Fig. S8) or β-catenin transcriptional activity following mechanical strain (Figure 2B,C; Fig. S6G,H; Fig. S8). In the presence of Verteporfin, Yap1 was still detected in the nucleus after strain application even though it could not bind to TEAD transcription factors (Fig. S8). Yap1 has been reported to co-regulate β-catenin transcriptional activity (23), but our results showed that in response to mechanical strain nuclear Yap1 levels peaked before nuclear β-catenin was detected, and then decreased while nuclear β-catenin and TOPdGFP levels remained high (Figs. S4A, S5).
Figure 2.
Yap1-mediated transcription is required for cell cycle re-entry after mechanical strain, but not for β-catenin nuclear accumulation or transcriptional activity. (A) % TBSmCherry-positive cells in MDCK monolayers 4hrs after no strain or 15% strain in control (WT) and YTIP-GFP expressing cells; (B) β-catenin and TOPdGFP distributions in YTIP-RFP expressing cells and TOPdGFP quantification (C) after 8 hours; (D) Ki67 distribution and quantification (E) after 8hrs; EdU incorporation (F) and quantification (G) after 24hrs in WT or YTIP-expressing cells after no strain or 15% strain. YTIP-expressing cells outlined with white dashed lines; insets show higher magnification of the region outlined by a dotted line. Scale bars: 25 µm. All quantifications (see Supplementary Information) were from at least 3 independent experiments with 2 replicate monolayers per experiment (Table S1). Quantifications were mean +/− SEM; unpaired t-test p values < 0.05 (*), p values < 0.01(**), p values <0.001 (***).
Thus transient Yap1 activation was required for strain-induced cell cycle re-entry, but neither Yap1/TEAD-mediated gene transcription nor cell cycle re-entry was required for β-catenin nuclear accumulation or transcriptional activity. Additionally, β-catenin transcriptional activity was not sufficient for strain-induced cell cycle re-entry or progression in the absence of Yap1 activation.
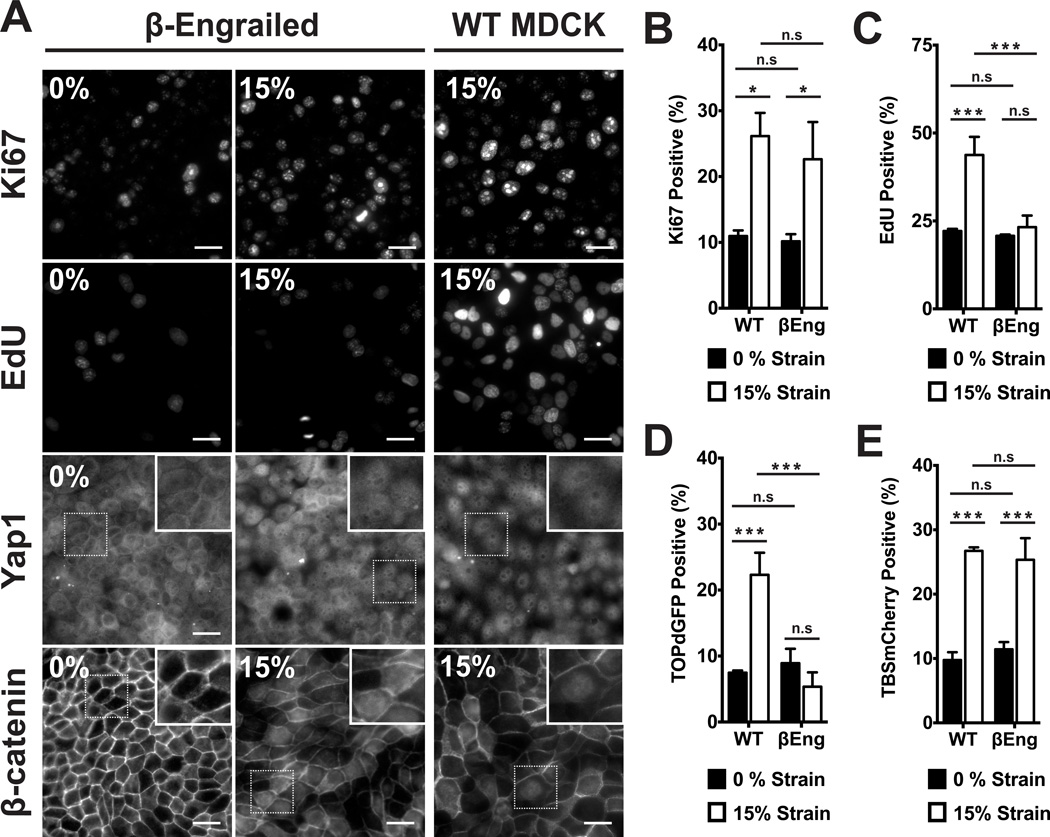
We next determined if β-catenin transcriptional activity was required for strain-induced cell cycle re-entry and progression. The β-catenin-Engrailed chimera (βEng) selectively inhibits β-catenin-mediated transcription without affecting cadherin-mediated cell-cell adhesion (24) nor density dependent inhibition of proliferation (Fig. S7). Application of mechanical strain in βEngexpressing cells induced nuclear accumulation of β-catenin (Fig. 3A), but did not result in β-catenin transcriptional activity (Fig. 3D) nor progression of cells into S phase (Fig. 3A,C). However, cell cycle re-entry (Fig. 3A,B) and Yap1 nuclear localization and transcriptional activity (Fig. 3A,E) were still induced by mechanical strain of βEng cells, similar to normal MDCK cells. Similar results were obtained with iCRT3 (Fig. S8), a small molecule inhibitor of β-catenin binding to TCF (25). Thus β-catenin transcriptional activity was not required for Yap1 nuclear accumulation and transcriptional activity or cell cycle re-entry following strain, but was required for cell cycle progression into S phase.
Figure 3.
β-catenin transcriptional activity was required for cell cycle progression through G1 into S phase after mechanical strain. (A) Distributions of Yap1 4hrs after no strain or 15% strain, Ki67 and β-catenin after 8hrs, and EdU incorporation after 24hrs in control (WT) and βEng-expressing MDCK monolayers. Insets show higher magnification of the region outlined by a dotted line. Quantification of Ki67 (B) and TOPdGFP (D) 8hrs after no strain or 15% strain, EdU (C) after 24hrs, and TBSmCherry (E) after 4hrs in control (WT) and βEng MDCK cells. Scale bars: 25 µm. All quantifications (see Supplementary Information) were from at least 3 independent experiments with 2 replicate monolayers per experiment (Table S1). Quantifications were mean +/− SEM; unpaired t-test p values < 0.05 (*), p values < 0.01(**), p values <0.001 (***).
These results are consistent with a model in which mechanical strain in a quiescent epithelial cell monolayer causes the transient nuclear localization and transcriptional activation of Yap1, which is required for cell cycle re-entry. Independently, strain also induces the nuclear localization and transcriptional activation of β-catenin, which is required for progression into S phase. In mammalian tissues, Yap1/TEAD-targeted genes promote proliferation (CTGF, FGF, Ki67), anti-apoptosis (Birc5, AREG), and adhesion (Dsc3) (26–28), while β-catenin/TCF/LEF-targeted genes include additional cell cycle regulators (c-Myc, Cyclin D1, AuroraA, cdc25) (29). Activation of Yap1 and β-catenin gene targets, therefore, is congruent with our model of cell cycle re-entry following mechanical strain. Mechanisms of Yap1 and β-catenin nuclear localization are complex and involve many pathways (12, 15, 16, 30–32) and many cell surface receptors regulate responses to mechanical strain including integrin-based adhesions to the ECM (33, 34). However, a specific role for E-cadherin extracellular domain binding between cells has not been tested.
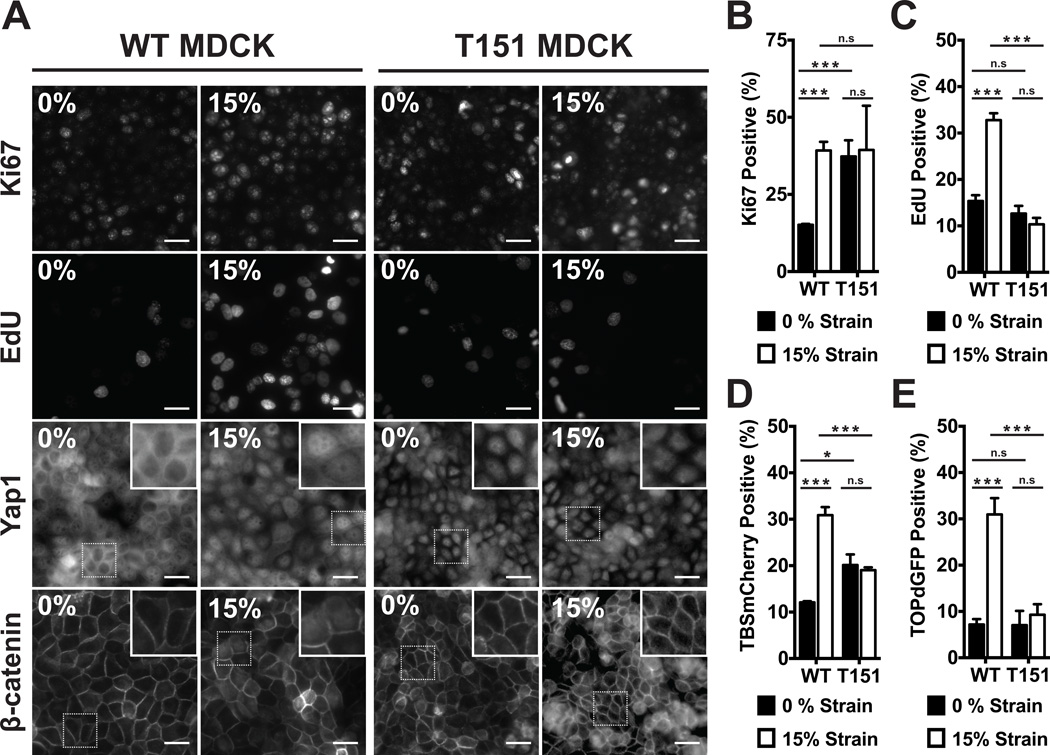
Rather than simply removing E-cadherin from MDCK cells, which would result in the loss of binding sites for Yap1 and β-catenin cytoplasmic sequestration, we used MDCK cells stably expressing a mutant E-cadherin (T151) under control of a doxycycline-repressible promoter (35). T151 comprises a truncated, nonfunctional extracellular domain, but a normal plasma membranete-thered cytoplasmic domain that binds catenins. Importantly, expression of T151 causes the down-regulation of endogenous E-cadherin (Fig. S9A), resulting in the complete loss of E-cadherin mediated cell-cell adhesion, but does not prevent the formation of tight junctions and desmosomes (35), growth to confluence, nor contact inhibition (Fig. S9B–D). Unlike normal MDCK cells at high cell densities, T151 monolayers without externally applied strain were Ki67-positive (Fig. 4A, B) and had nuclear Yap1 (Fig. 4A) and increased TBSmCherry signal (Fig. 4D), consistent with cells being in G1. While T151 monolayers appeared ‘primed’ for cell cycle progression, levels of nuclear β-catenin (Fig. 4A), TOPdGFP (Fig. 4E), and EdU incorporation were all low (Fig. 4C), indicating inhibition of G1 to S phase transitions. Application of mechanical strain to T151 monolayers did not increase the level of EdU incorporation (Fig. 4A,C), nuclear β-catenin (Fig. 4A), or β-catenin transcriptional activity (Fig. 4E), indicating cells had not progressed into S phase. Thus, in multicellular monolayers, coupling between E-cadherin extracellular domains is required to block cell cycle entry and sequester Yap1 in the cytoplasm, and for strain-induced nuclear accumulation and transcriptional activity of β-catenin and subsequent cell cycle progression into S phase.
Figure 4.
E-cadherin extracellular domain interactions were required for quiescence at high density, Yap1 nuclear exclusion, and β-catenin activation following strain. (A) Distribution of Yap1 after 4hrs, Ki67 and β-catenin after 8hrs, and EdU incorporation after 24hrs in control (WT) and T151 MDCK monolayers after no strain or 15% strain. Insets show higher magnification of the region outlined by a dotted line. Quantification of Ki67 (B) and TOPdGFP (E) after 8hrs, EdU (C) after 24hrs, and TBSmCherry (D) after 4hrs in control (WT) and T151 MDCK cell monolayers after no strain or 15% strain. Scale bars: 25 µm. All quantifications (see Supplementary Information) were from least 3 independent experiments with 2 replicate monolayers per experiment (Table S1). Quantifications were mean +/− SEM; unpaired t-test p values < 0.05 (*), p values <0.001 (***).
Mechanical strain in epithelial monolayers results in cell cycle re-entry and progression through S phase by the nuclear accumulation and transcriptional activity of first Yap1 and then β-catenin. Activation of Yap1 is required for cell cycle re-entry, whereas β-catenin is required for progression from G1 to S phase. Specific inhibition of Yap1 and β-catenin transcription using two independent methods blocked cell cycle re-entry and progression, respectively, indicating that other transcription factors were not sufficient for these critical responses to mechanical strain. Thus, activation of Yap1 and β-catenin may be a master regulator for cell cycle re-entry and progression through S phase following mechanical strain, and an underlying mechanism for regulation of homeostasis in adult tissues. Finally, extracellular E-cadherin engagement and β-catenin represent critical regulators of quiescence and strain-induced proliferation in multicellular assemblies. Thus, cell-cell junctions are not only mechanically responsive structural scaffolds, but also signaling centers that coordinate transcriptional responses to externally applied force.
Supplementary Material
Acknowledgements
This work was supported by a National Science Foundation (NSF) Pre-doctoral Fellowship to BWB-P (DGE-114747) and grants from the NSF (EFRI-1136790) to BLP and WJN, and The National Institutes of Health to BLP (EB006745) and WJN (GM 35527). Data reported in this paper are further detailed in the supplementary Materials
Footnotes
Supplementary Materials
Materials and Methods
Figs. S1 to S9
Table S1
References
- 1.Chen CS, Tan J, Tien J. Mechanotransduction at cell-matrix and cell-cell contacts. Annu. Rev. Biomed. Eng. 2004;6:275–302. doi: 10.1146/annurev.bioeng.6.040803.140040. [DOI] [PubMed] [Google Scholar]
- 2.Streichan SJ, Hoerner CR, Schneidt T, Holzer D, Hufnagel L. Spatial constraints control cell proliferation in tissues. Proceedings of the National Academy of Sciences. 2014;111:5586–5591. doi: 10.1073/pnas.1323016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilbert PM, et al. Substrate Elasticity Regulates Skeletal Muscle Stem Cell Self-Renewal in Culture. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 5.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 6.Aragona M, et al. A Mechanical Checkpoint Controls Multicellular Growth through YAP/TAZ Regulation by Actin-Processing Factors. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 7.Pokutta S, Weis WI. Structure and mechanism of Cadherins and catenins in cell-cell contacts. Annu Rev Cell Dev Bi. 2007;23:237–261. doi: 10.1146/annurev.cellbio.22.010305.104241. [DOI] [PubMed] [Google Scholar]
- 8.le Duc Q, et al. Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II-dependent manner. J Cell Biol. 2010;189:1107–1115. doi: 10.1083/jcb.201001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Z, et al. Mechanical tugging force regulates the size of cell-cell junctions. P Natl Acad Sci Usa. 2010;107:9944–9949. doi: 10.1073/pnas.0914547107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borghi N, et al. E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proceedings of the National Academy of Sciences. 2012;109:12568–12573. doi: 10.1073/pnas.1204390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buckley CD, et al. Cell adhesion. The minimal cadherin-catenin complex binds to actin filaments under force. Science. 2014;346:1254211–1254211. doi: 10.1126/science.1254211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlegelmilch K, et al. Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell. 2011;144:782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silvis MR, et al. α-catenin is a tumor suppressor that controls cell accumulation by regulating the localization and activity of the transcriptional coactivator Yap1. Sci Signal. 2011;4:ra33. doi: 10.1126/scisignal.2001823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Azzolin L, et al. YAP/TAZ Incorporation in the β-Catenin Destruction Complex Orchestrates the Wnt Response. Cell. 2014;158:157–170. doi: 10.1016/j.cell.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 16.Kim N-G, Koh E, Chen X, Gumbiner BM. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proceedings of the National Academy of Sciences. 2011;108:11930–11935. doi: 10.1073/pnas.1103345108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simmons CS, et al. Integrated strain array for cellular mechanobiology studies. J Micromech Microeng. 2011;21:54016–54025. doi: 10.1088/0960-1317/21/5/054016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. β-catenin is a target for the ubiquitin–proteasome pathway. Embo J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikeda S, et al. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3 beta and beta-catenin and promotes GSK-3 beta-dependent phosphorylation of beta-catenin. Embo J. 1998;17:1371–1384. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maher MT, Flozak AS, Stocker AM, Chenn A, Gottardi CJ. Activity of the beta-catenin phosphodestruction complex at cell-cell contacts is enhanced by cadherin-based adhesion. J Cell Biol. 2009;186:219–228. doi: 10.1083/jcb.200811108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Gise A, et al. YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proceedings of the National Academy of Sciences. 2012;109:2394–2399. doi: 10.1073/pnas.1116136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu-Chittenden Y, et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Gene Dev. 2012;26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenbluh J, et al. β-Catenin-Driven Cancers Require a YAP1 Transcriptional Complex for Survival and Tumorigenesis. Cell. 2012;151:1457–1473. doi: 10.1016/j.cell.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montross W, Ji H, McCrea P. A beta-catenin/engrailed chimera selectively suppresses Wnt signaling. J Cell Sci. 2000;113:1759–1770. doi: 10.1242/jcs.113.10.1759. [DOI] [PubMed] [Google Scholar]
- 25.Gonsalves FC, et al. An RNAi-based chemical genetic screen identifies three small-molecule inhibitors of the Wnt/wingless signaling pathway. P Natl Acad Sci Usa. 2011;108:5954–5963. doi: 10.1073/pnas.1017496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor Suppressor LATS1 Is a Negative Regulator of Oncogene YAP. J Biol Chem. 2007;283:5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- 27.Dong J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu L, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proceedings of the National Academy of Sciences. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torre C, et al. The transforming growth factor-α and cyclin D1 genes are direct targets of β-catenin signaling in hepatocyte proliferation. Journal of Hepatology. 2011;55:86–95. doi: 10.1016/j.jhep.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 30.Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF -TRCP. Gene Dev. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitehead J, et al. Mechanical factors activate beta-catenin-dependent oncogene expression in APC mouse colon. HFSP J. 2008;2:286–294. doi: 10.2976/1.2955566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Desprat N, Supatto W, Pouille P-A, Beaurepaire E, Farge E. Tissue Deformation Modulates Twist Expression to Determine Anterior Midgut Differentiation in Drosophila Embryos. Dev Cell. 2008;15:470–477. doi: 10.1016/j.devcel.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 33.Alenghat FJ, Ingber DE. Mechanotransduction: all signals point to cytoskeleton, matrix, and integrins. Sci STKE. 2002;2002 doi: 10.1126/stke.2002.119.pe6. pe6-pe6. [DOI] [PubMed] [Google Scholar]
- 34.Vogel V. Mechanotransduction involving multimodular proteins: Converting force into biochemical signals. Annual review of biophysics and biomolecular structure. 2006;35:459–488. doi: 10.1146/annurev.biophys.35.040405.102013. [DOI] [PubMed] [Google Scholar]
- 35.Troxell ML, et al. Inhibiting cadherin function by dominant mutant E-cadherin expression increases the extent of tight junction assembly. J Cell Sci. 2000;113:985–996. doi: 10.1242/jcs.113.6.985. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.