Primary human fibroblasts were cultured in either insulin-rich defined medium or in a conventional medium to study the significance of proliferation/growth factor signaling for reprogramming. A direct inverse correlation of high cell proliferation and reprogramming efficiency was seen in human somatic cells grown in the defined medium. These results have important implications for utility of these cells for translational studies in humans.
Keywords: Reprogramming, Insulin signaling, Cell proliferation, Human pluripotency
Abstract
The impact of somatic cell proliferation rate on induction of pluripotent stem cells remains controversial. Herein, we report that rapid proliferation of human somatic fibroblasts is detrimental to reprogramming efficiency when reprogrammed using a lentiviral vector expressing OCT4, SOX2, KLF4, and cMYC in insulin-rich defined medium. Human fibroblasts grown in this medium showed higher proliferation, enhanced expression of insulin signaling and cell cycle genes, and a switch from glycolytic to oxidative phosphorylation metabolism, but they displayed poor reprogramming efficiency compared with cells grown in normal medium. Thus, in contrast to previous studies, our work reveals an inverse correlation between the proliferation rate of somatic cells and reprogramming efficiency, and also suggests that upregulation of proteins in the growth factor signaling pathway limits the ability to induce pluripotency in human somatic fibroblasts.
Significance
The efficiency with which human cells can be reprogrammed is of interest to stem cell biology. In this study, human fibroblasts cultured in media containing different concentrations of growth factors such as insulin and insulin-like growth factor-1 exhibited variable abilities to proliferate, with consequences on pluripotency. This occurred in part because of changes in the expression of proteins involved in the growth factor signaling pathway, glycolysis, and oxidative phosphorylation. These findings have implications for efficient reprogramming of human cells.
Introduction
Breakthrough discoveries from the Yamanaka laboratory [1, 2] and the establishment of human induced pluripotent stem cells (hiPSCs) have opened new avenues for generating patient-specific stem cell derivatives that can be used for in vitro modeling of human disease, drug development, and cell replacement therapy. However, current methodologies of induced pluripotent stem cell (iPSC) generation continue to face technical challenges, in part because of relatively poor reprogramming efficiencies. As efforts to make iPSCs more useful in human transplantation studies continue, many groups have contributed to significant progress in this field, including the use of reduced numbers of reprogramming factors, and adopting nonintegrating methods of their delivery, cell permeable proteins, and stand-alone small molecules or direct reprogramming [3–7]. Despite these efforts, reprogramming efficiency and its relationship with cell proliferation continues to remain poorly understood in the iPSC field. For example, vitamin C has been suggested to promote reprogramming by limiting cell senescence and indirectly promoting proliferation [8], and mitochondrial regression has been reported to be associated with a pluripotent state [9]. Other studies suggest that a high proliferation rate of human somatic fibroblasts is essential for efficient reprogramming by decreasing apoptosis rates and limiting reprogramming barriers, including senescence [10, 11]. In contrast, Xu et al. [12] reported that the slow proliferation of mouse somatic cells is beneficial for reprogramming. Consistently, several small molecule inhibitors of cell proliferation have also been reported to enhance somatic cell reprogramming [13–15].
To directly address the significance of proliferation for reprogramming, we cultured primary human fibroblasts in either defined insulin-rich AmnioMAX (Ax) medium or in normal, conventional Dulbecco’s modified Eagle’s medium (N) (both from Thermo Fisher Scientific Inc., Waltham, MA, http://www.thermofisher.com). AmnioMAX, a well-defined medium, has been used for culturing human amniotic fluid cells and fibroblasts [16, 17]. Our study demonstrates a direct inverse correlation of high cell proliferation and reprogramming efficiency for human somatic cells in Ax medium. These results have important implications for utility of these cells for translational studies in humans.
Results and Discussion
AmnioMAX Medium Accelerates the Growth Kinetics of Somatic Fibroblasts
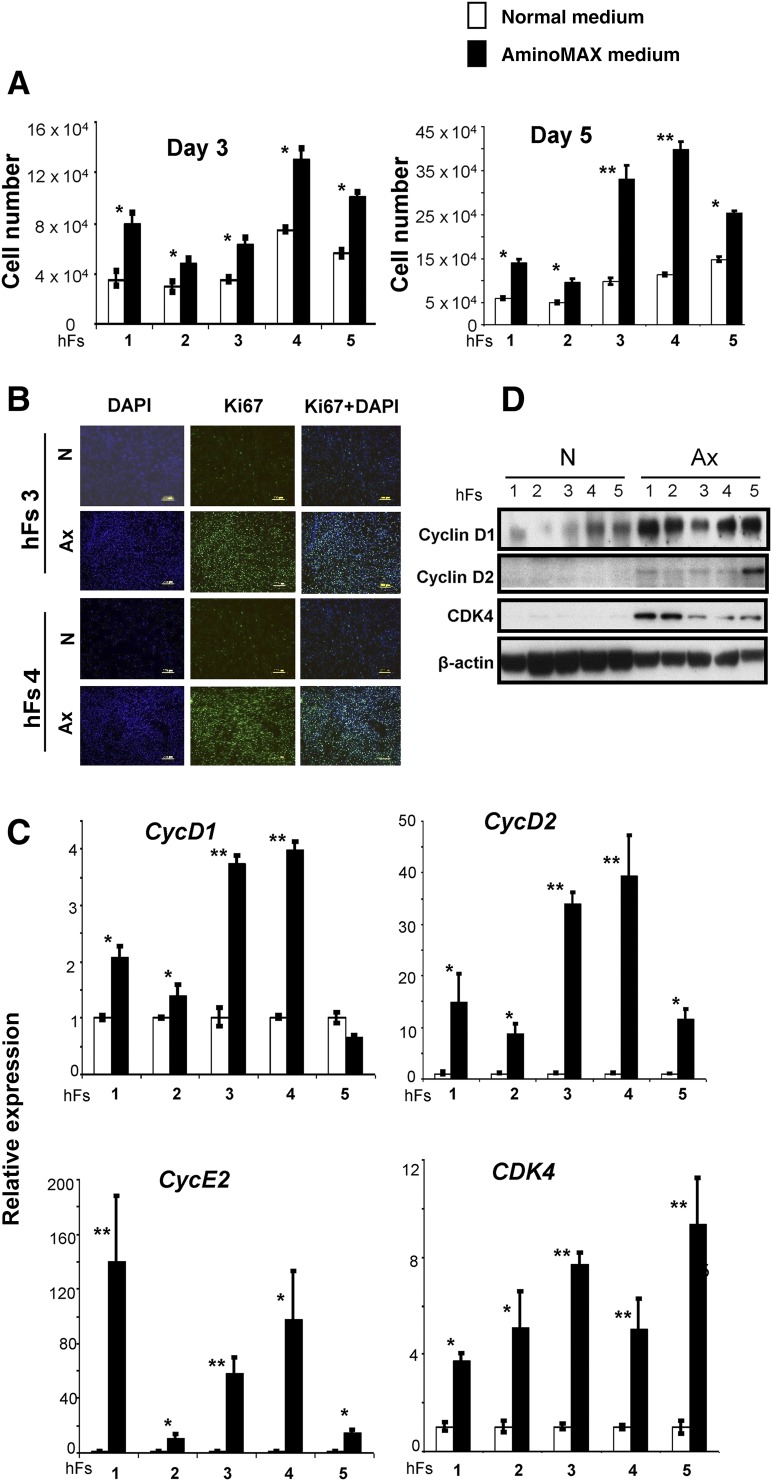
Human fibroblasts (hFs) obtained from healthy individuals—AG16104 (hFs 1), AG16086 (hFs 2), 120111 (hFs 3), 120116 (hFs 4), and AG16102 (hFs 5)—were either grown in conventional normal growth medium (N) or Ax medium. Strikingly, fibroblasts grew faster when grown in Ax medium compared with cells grown in N medium (supplemental online Fig. 1A). To quantify proliferation, we seeded an equal number of fibroblasts (2 × 104) on day 0 and counted the fibroblasts at days 3 and 5. We observed a 1.5- to 2-fold increase at day 3 and a 2- to 3-fold increase in number of cells at day 5, when cultured in Ax medium compared with N medium (Fig. 1A). An increased rate of proliferation in Ax medium was confirmed by Ki67/4′,6-diamidino-2-phenylindole immunostaining in hFs 3 and hFs 4 (Fig. 1B). Further quantification by flow cytometry analysis of hFs 3, hFs 4, and hFs 5 at day 5 revealed an average 2- to 3-fold increase in Ki67-positive cells grown in Ax medium (supplemental online Fig. 1B, 1C). We also observed an increase in expression of cell cycle genes CycD1 (2- to 5-fold), CycD2 (2- to 10-fold), CycE2 (20- to 40-fold), and CDK4 (2- to 6-fold) in human fibroblasts grown in Ax medium compared with fibroblasts cultured in N medium (Fig. 1C). Detection of increased CycD1, CycD2, and CDK4 proteins by Western immunoblotting confirmed enhanced cell cycle progression in fibroblasts cultured in Ax medium (Fig. 1D). Attempts to culture human fibroblasts in mTeSR human medium were not successful and the cells failed to grow in contrast to robust growth when cultured in N or Ax medium (supplemental online Fig. 1D). Together, these data suggest that growth of human fibroblasts in Ax medium leads to a greater rate of proliferation and the enhanced expression of cell cycle proteins.
Figure 1.
The growth medium determines growth kinetics of human somatic fibroblasts. (A): The cell numbers of 5 primary hFs—AG16104 (hFs 1), AG16086 (hFs 2), 120111 (hFs 3), 120116 (hFs 4), and AG16102 (hFs 5)—were measured by a hemocytometer on day 3 and day 5 after excluding dead cells by trypan blue staining. (White boxes represent cell numbers in N and black boxes indicate cell numbers in defined Ax.) The cells were seeded at equal densities (30,000 cells per well) on day 0 and counted on day 3 and day 5 in triplicate wells. The x-axis denotes the hFs. (B): hFs 3 and hFs 4 were seeded at equal densities (30,000 cells per well) and cultured in N or Ax medium. Cells were harvested on day 3 and subjected to Ki67 immunostaining to identify cycling cells. DAPI was used to stain nuclei. Scale bars = 200 μm. (C): The relative expression of cell cycle genes CycD1, CycD2, CDK4, and CycE2 in hFs grown in N (white bars) or Ax medium (black bars). Expression was normalized to the β-actin gene and is shown relative to the average N medium level. The x-axis denotes the number of hFs. (D): Expression of CycD1, CycD2, and CDK4 proteins was demonstrated by Western immunoblot analysis. β-actin was used as an internal control. All experiments were performed three times. Data are shown as mean ± SD. Statistical significance was determined by Student’s t test. ∗, p < .05; ∗∗, p < .01 for Ax versus N. Abbreviations: Ax, AminoMAX medium; DAPI, 4,6-diamidino-2-phenylindole; hF, human fibroblast; N, Dulbecco’s Modified Eagle’s medium.
Enhanced Growth Factor (Insulin) Signaling Contributes to Higher Proliferation of Fibroblasts Cultured in Ax Medium
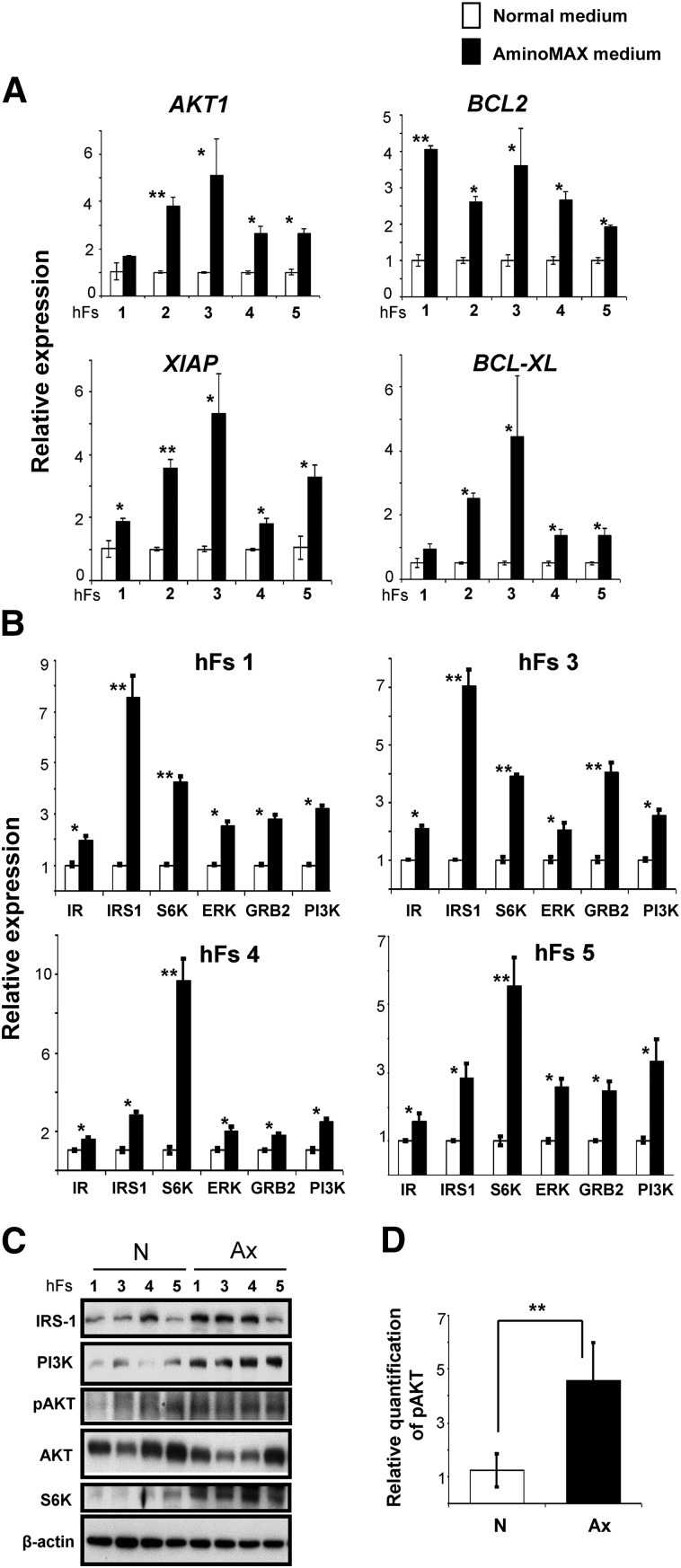
The more rapid proliferation of somatic fibroblasts in defined Ax medium led us to investigate the expression of genes associated with cell survival and growth factor (insulin/insulin-like growth factor-1 [IGF-1]) signaling pathways. We observed a 2- to 6-fold upregulation of genes involved in blocking apoptosis, including AKT1, BCL2, XIAP, and BCL-XL in human fibroblasts grown in Ax medium (Fig. 2A). Furthermore, growth factor (insulin/IGF-1) signaling pathway-related genes, including IR, IRS1, S6K, ERK, GRB2, and PI3K, were also significantly upregulated (2- to 3-fold) in fibroblasts grown in Ax medium (Fig. 2B). Western immunoblot analysis confirmed an increased phosphorylation of AKT (approximately 2.5-fold) and higher protein expression of phosphatidylinositol 3-kinase (PI3K), IRS-1, and S6K (approximately 5-fold) in human fibroblasts grown in defined Ax medium (Fig. 2C, 2D; supplemental online 2A). However, we did not observe any significant change in the protein levels of GRB2 or ERK (supplemental online Fig. 2B), suggesting Ax medium has a more prominent effect on the PI-3 kinase pathway than the MAP kinase pathway.
Figure 2.
Fibroblasts cultured in Ax growth medium exhibit altered expression of genes in cell survival and growth factor (insulin/insulin-like growth factor-1 [IGF-1]) signaling pathways. (A): The relative expression of AKT1, BCL2, XIAP, and BCL-XL genes by real-time polymerase chain reaction in human fibroblasts (hFs 1, 2, 3, 4, and 5) cultured in N (white bars) or Ax (black bars) medium. The x-axis denotes the hFs. (B): Gene expression analysis of proteins in the growth factor (insulin/IGF-1) signaling pathway, including IR, IRS1, S6K, ERK, GRB2, and PI3K, in four hFs (hFs 1, hFs 3, hFs 4, and hFs 5) grown in N or Ax medium. In both (A) and (B), expression was normalized to the β-actin gene and is shown relative to the average N medium level. The x-axis denotes the number of hFs. (C): Western blot analysis of IRS1, PI3K, S6K, pAKT, and AKT proteins in hFs 1, hFs 3, hFs 4, and hFs 5 cultured in N or Ax medium. β-Actin was used as an internal control. (D): Graph representing the relative quantity of phosphorylation of AKT normalized to total AKT band density by ImageJ software (US National Institutes of Health, Bethesda, MD, http://imagej.nih.gov/ij). All experiments were performed three times, represented as mean ± SD. Statistical significance was determined by Student’s t test. ∗, p < .05; ∗∗, p < .01 for Ax versus N. Abbreviations: Ax, AminoMAX medium; D, day; ERK, extracellular signal-regulated kinase; GRB2, growth factor receptor-bound protein; IR, insulin receptor; IRS1, insulin receptor substrate-1; N, Dulbecco’s modified Eagle’s medium; PI3K, phosphatidylinositol 3-kinase; S6K, S6 kinase.
Our findings are consistent with previous studies [18] reporting increased expression of cell survival genes (AKT1, BCL2, XIAP, and BCL-XL) during increased proliferation. Consistent with the observation that insulin is a potent inducer of cell proliferation during development [19], we detected an upregulation of phosphorylated AKT and of proteins in the insulin/IGF-1 signaling pathway, including PI3K, IRS1, and S6K, in fibroblasts grown in defined Ax medium.
Our focus on examining the insulin signaling pathway and cell proliferation in the context of reprogramming of human somatic fibroblasts gains significance given several opposing reports in this field. Ruiz et al. [11] reported that high proliferation of somatic cells is beneficial to reprogramming by providing evidence that expression of CycD1, CycD2, and CycE2, but not CDK1, CDK2, and CDK4, increased the reprogramming efficiency of human keratinocytes by more than 2-fold. On the contrary, Xu et al. [12] reported that low proliferation of somatic cells is helpful to induce pluripotency. They demonstrated that removing cMyc from among the four reprogramming factors led to a 10-fold increase in reprogramming efficiency of mouse fibroblasts, whereas forced expression of cMyc led to hyperproliferation and correlated negatively with overall reprogramming efficiency. In contrast to these two studies, our observations implicate increased cell cycle markers and an upregulation of proteins in the insulin/IGF-1 signaling pathway in the reprogramming process.
High Proliferation Rate and Upregulation of Insulin Signaling in Somatic Fibroblasts Correlates With Lower Reprogramming Efficiency
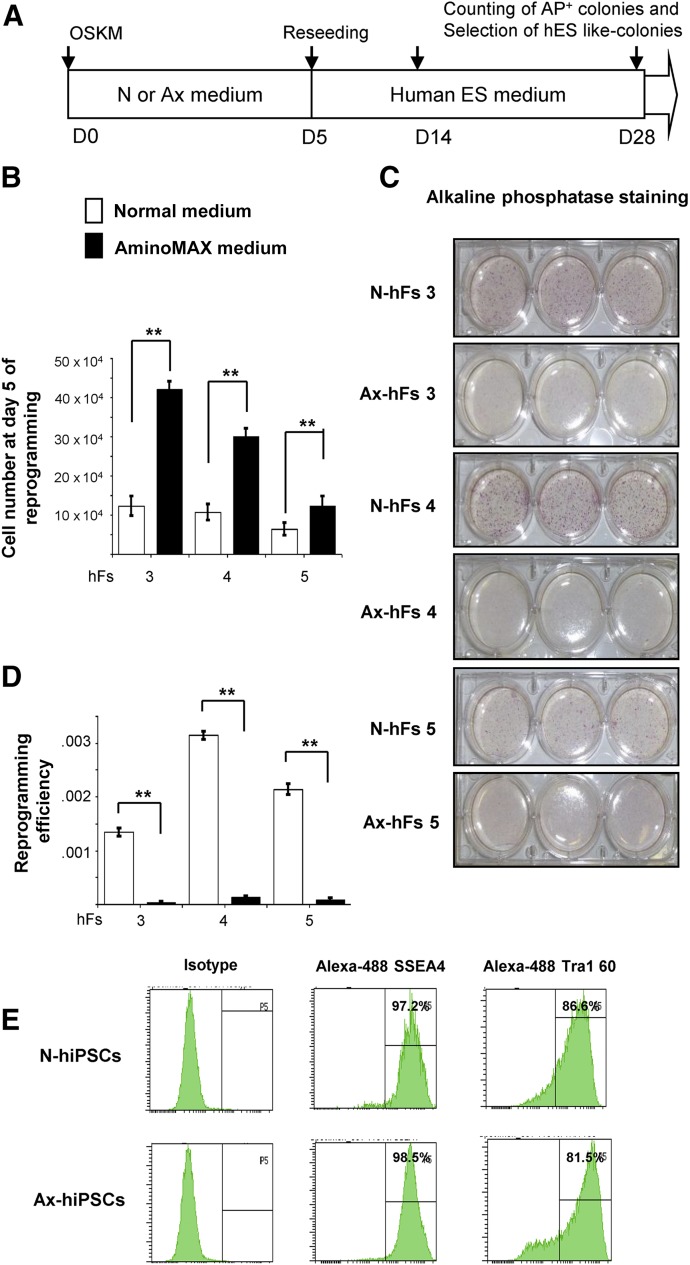
Next, to examine whether cell cycle progression and increased growth determine somatic cell reprogramming, we subjected fibroblasts individually cultured in N or Ax medium to reprogramming into iPSCs, using a protocol reported previously [20] (Fig. 3A). As expected, even after human lentiviral transduction, we observed a 2- to 3-fold increase in viable cell numbers of both transduced and nontransduced fibroblasts in Ax medium (Fig. 3B). However, surprisingly, this increase in cell numbers was associated with a significant reduction in reprogramming efficiency of cells cultured in Ax medium compared with those cultured in N medium, as demonstrated by alkaline phosphatase staining. A similar outcome in three independent samples (hFs 3, hFs 4, and hFs 5) confirmed a uniform effect (Fig. 3C, 3D; supplemental online Fig. 2C). Real-time polymerase chain reaction analysis showed no difference in the expression levels of OCT4 or NANOG between N-hiPSCs and Ax-hiPSCs, and there was no detection of OCT4 and NANOG in their respective parental fibroblasts (supplemental online Fig 2D). Furthermore, we observed a similar level of expression of SSEA4 (>90%) and TRA1 60 (>80%) pluripotent surface markers by flow cytometry (Fig. 3E) and OCT4 expression by immunohistochemistry (supplemental online Fig. 2E) in hiPSCs derived from the fibroblasts cultured in either medium. The hiPSCs from both groups were able to form embryoid bodies as well as develop teratomas that included cells from the three lineages, as shown by immunostaining (supplemental online Fig. 2F, 2G). These results indicate that a higher proliferation and an upregulation in expression of proteins in the growth factor (insulin/IGF-1) signaling pathway does not impact pluripotency of the derived hiPSCs that are successfully reprogrammed, but does influence the frequency of cells that undergo reprogramming. Consistent with our results, Xu et al. [12] reported that low proliferation of mouse fibroblasts is beneficial for reprogramming. Although these authors did not explain the precise mechanism, their data reveal that different small molecules that are antiproliferative agents (e.g., amphidicolin, cisplatin, aloisine A, CDK9 inhibitor II) enhanced the reprogramming efficiency of mouse somatic fibroblasts. One possible explanation for the altered reprogramming is that higher proliferation rates affect some epigenetic markers and/or influence the heterochromatin stage of the cells to eventually limit cellular reprogramming.
Figure 3.
Cellular state of fibroblast growth affects cellular reprogramming. (A): Scheme of reprogramming of human somatic fibroblasts into hiPSCs using a cocktail of a Cre-excisable STEMCCA lentivirus vector expressing OSKM. (B): Human fibroblasts 120111 (hFs 3), 120116 (hFs 4), and AG16102 (hFs 5) were subjected to reprogramming by STEMCCA lentiviral vector. On day 5, at the time of further splitting during reprogramming process, cells were counted growing in N or Ax medium. (C): On day 5 of reprogramming, hFs 3, hFs 4, and hFs 5, grown in N or Ax medium, were seeded at a density of 3 × 104 fibroblasts in triplicate in 6-well plates. AP staining was performed using a Stemgent kit (Stemgent Inc., Cambridge, MA, https://www.stemgent.com/products/227) between days 21 and 28 after iPSC colonies were visualized in reprogramming culture. (D): Reprogramming efficiencies from hFs 3, hFs 4, and hFs 5 grown in N or Ax medium were determined by dividing the number of AP-positive colonies by the number of fibroblasts that were initially seeded and transduced (5 × 104 cells). (E): hiPSCs from both the groups were subjected to flow cytometry analysis to evaluate the surface pluripotency markers SSEA4 and Tra-1 60, using Alexa Fluor 488 antibodies. All experiments were performed three times, represented as mean ± SD. Statistical significance by Student’s t test. ∗, p < .05; ∗∗, p < .01 for Ax versus N. Abbreviations: AP, alkaline phosphatase; Ax, AminoMAX medium; ES, embryonic stem cell; hF, human fibroblast; hiPSC, human induced pluripotent stem cell; N, Dulbecco’s modified Eagle’s medium; OSKM, OCT4, SOX2, KLF4, and cMYC.
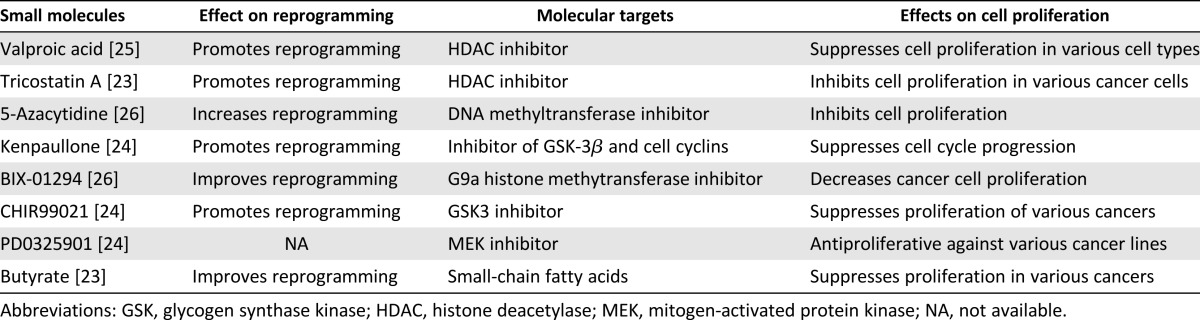
Vitamin C, a small molecule, was reported to improve somatic cell reprogramming by enhancing cell proliferation. In contrast, valproic acid has been shown to increase reprogramming efficiency and to induce pluripotency in human amniotic-fluid cells alone, without ectopic expression of reprogramming factors [15, 21, 22]. Indeed, several small molecules, such as kenpaullone, trichostatin A, 5-azacytidine, and CHIR99021, have all been identified as antiproliferative agents in various cell types [23–26] that also promote reprogramming of fibroblasts [13, 15] (Table 1; supplemental online data).
Table 1.
Small molecules that suppress cellular proliferation increase reprogramming efficiency
To further validate the role of IGF-1 and insulin signaling, we reprogrammed hFs 3, 4, and 5 cultured in N medium with or without supplementation with IGF-1 (100 nM) or insulin (43 ng/ml). We used the same concentration of insulin as that present in Ax medium. Interestingly, we observed a significant decrease in reprogramming efficiency in the presence of either IGF-1 or insulin (supplemental online Fig. 3A–3C). This supplementation experiment further supported a potential role for insulin or IGF-1 signaling in reprogramming of human fibroblasts.
Activation of Metabolic Switch From Glycolysis to Oxidative Phosphorylation Leads to Significant Decrease in Reprogramming Efficiency of Somatic Fibroblasts
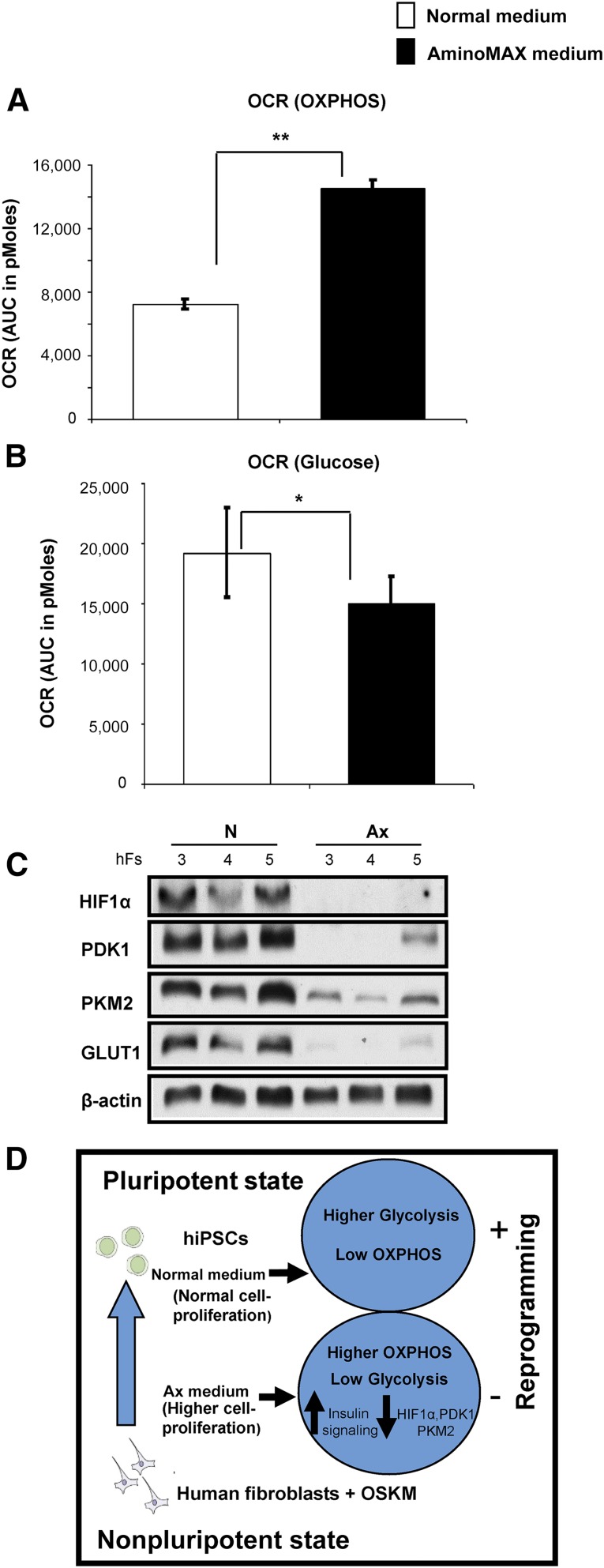
Previous reports indicating that cell-fate conversion is associated with a transition between oxidative phosphorylation and glycolytic metabolism [9], coupled with the observation that insulin/IGF-1 is known to regulate mitochondrial function [27, 28], prompted us to explore whether a similar switch appears in the phenotype of human fibroblasts that show altered insulin/IGF-1 signaling. To this end, we undertook metabolic profiling by investigating cellular metabolism in the context of reprogramming, using the Seahorse Bioflux Analyzer (Seahorse Bioscience, Billerica, MA ,http://www.seahorsebio.com). This analysis revealed that human fibroblasts cultured in defined Ax medium exhibit increased basal respiration, as shown by a 2-fold higher oxygen consumption rate (OCR) compared with fibroblasts grown in conventional N medium (Fig. 4A; supplemental online Fig. 4A). Interestingly, fibroblasts cultured in N medium displayed increased glycolytic capacity compared with fibroblasts grown in Ax medium. Thus, in response to glucose stimulation, fibroblasts grown in N medium showed a higher OCR and extracellular acidification rate than fibroblasts grown in Ax medium (Fig. 4B; supplemental online Fig. 4B). Consistent with a role for altered glycolysis and hypoxia in the regulation of reprogramming [29], we observed a significantly reduced protein expression of HIF1α (93%), PDK1 (77.1%), PKM2 (91.6%), and GLUT1 (95.9%) in total cell extracts of fibroblasts grown in Ax medium (Fig. 4C; supplemental online Fig. 4C).
Figure 4.
Metabolic shift and the expression of genes regulating reprogramming in fibroblasts grown in N or Ax medium. (A): OCR during basal respiration, displayed as AUC, of human hFs 3 and hFs 4 grown in N or Ax medium (B): OCR in the presence of glucose as sole energy substrate, displayed as AUC, of human somatic fibroblasts hFs 3 and hFs 4 grown in N or Ax medium. (C): Western blot analysis of HIF1α, PDK1, PKM2, and GLUT1 in hFs 3, hFs 4, and hFs 5 cultured in N or Ax medium. β-Actin was used as an internal control. (D): A proposed model for the effects of high cell proliferation and insulin signaling on reprogramming of human fibroblasts. In fibroblasts with normal proliferation, cells maintain higher glycolysis and low OXPHOS. Cells cultured in Ax medium, with higher cell proliferation, exhibit increased growth factor (insulin/IGF-1) signaling and a higher OXPHOS by downregulating the expression of HIF1a, PDK1, PKM2, and GLUT1 proteins, leading to a significant decrease in the efficiency of induction of pluripotency (hiPSCs). All experiments were performed three times, represented as mean ± SD. Statistical significance was determined by Student’s t test. ∗, p < .05; ∗∗, p < .01 for Ax versus N. Abbreviations: AUC, area under the curve; GLUT1, glucose transporter-1; Ax, AminoMAX medium; HIF1α, hypoxia inducible factor-1 α; hiPSC, human induced pluripotent stem cell; N, Dulbecco’s modified Eagle’s medium; OCR, oxygen consumption rate; OSKM, OCT4, SOX2, KLF4, and cMYC; OXPHOS, oxidative phosphorylation; PDK1, phosphoinositide dependent kinase-1; PKM2, pyruvate kinase M2 isoform.
HIF1α signaling enhances reprogramming efficiency via metabolic switch toward glycolysis by upregulating expression of PDK1. Therefore, activation of HIF1α regulates Oct4 expression and augments the induction of human stemness signature in various tumor cell lines [29, 30]. Consistent with this notion, our findings demonstrate that reduction in HIF1α protein in highly proliferating somatic fibroblasts grown in Ax medium promotes refractoriness to reprogramming. Previous reports implicated an upregulation of PDK1 by small molecules in an increase in reprogramming [3]. Similarly, PKM2 may be involved in positive regulation of OCT4 and GLUT1 in glycolysis [31]. In our study, we noted that key regulators of glycolysis (e.g., PDK1, PKM2, and GLUT1) are all decreased in human fibroblasts that are rapidly proliferating when cultured in Ax medium and, consequently, exhibit a significant loss of reprogramming efficiency (Fig. 4D).
To further validate the role of PDK1 in reprogramming, a central regulator of glycolysis, we knocked down PDK1 in hFs 3, hFs 4, and hFs 5 using scrambled or PDK1-specific siRNAs (supplemental online Fig. 5A). Knocked-down PDK1 human fibroblasts showed significantly reduced reprogramming efficiency as compared with fibroblasts cultured in scrambled control small interfering RNA (supplemental online Fig. 5B, 5C). This loss-of-function study further validated our findings in regard to a potential role of PDK1 and glycolysis in reprogramming of human fibroblasts.
Conclusion
We report that stimulation of cell proliferation limits human somatic cell reprogramming via upregulation of proteins in the insulin/IGF-1 signaling pathway and by promoting a metabolic switch from glycolysis to oxidative phosphorylation. These data provide a previously unidentified perspective on the roles of cell proliferation and growth factor signaling in induction of pluripotency and have implications for studies aimed at reprogramming of cells derived from humans with pathological states associated with impaired metabolism and/or cell proliferation, such as diabetes or cancer.
Supplementary Material
Acknowledgments
We thank Dr. G. Mostoslavsky (Boston University) for the kind gift of lentiviral plasmids and R. Martinez for technical assistance. We thank Dr. C.R. Kahn (Joslin Diabetes Center) for allowing access to the Seahorse instrument. The Joslin DRC iPS Core is supported by NIH Grant 5 P30 DK036836-27. A.K.K.T. is supported by a Juvenile Diabetes Research Foundation Postdoctoral Fellowship. A.K. was supported by Deutsche Forschungsgemeinschaft projects KL2399/1-1 and KL2399/3-1. R.N.K. is supported by NIH Grant R01 DK67536 and a grant from AstraZeneca.
Author Contributions
M.K.G.: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; A.K.K.T. and T.N.R.: collection and assembly of data, data analysis and interpretation, final approval of manuscript; S.B., J.S., T.T., J.H., D.F.D.J., and R.W.: collection and assembly of data, final approval of manuscript; A.K.: collection and assembly of data of metabolic study, data analysis and interpretation, final approval of manuscript; A.J.W.: provision of suggestions, manuscript editing, final approval of manuscript; R.N.K.: conception and design, manuscript writing, financial support, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
A.J.W. is on the scientific advisory board of FATE Therapeutics. R.N.K. has compensated research funding from AstraZeneca. The other authors indicated no potential conflicts of interest.
References
- 1.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Daley GQ. Stem cells: Roadmap to the clinic. J Clin Invest. 2010;120:8–10. doi: 10.1172/JCI41801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu S, Li W, Zhou H, et al. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell. 2010;7:651–655. doi: 10.1016/j.stem.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hou P, Li Y, Zhang X, et al. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–654. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
- 5.Kim JB, Greber B, Arauzo-Bravo MJ, et al. Direct reprogramming of human neural stem cells by OCT4. Nature. 2009;461:649–643. doi: 10.1038/nature08436. [DOI] [PubMed] [Google Scholar]
- 6.Park IH, Zhao R, West JA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 7.Hanna J, Markoulaki S, Schorderet P, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esteban MA, Wang T, Qin B, et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6:71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Folmes CD, Nelson TJ, Martinez-Fernandez A, et al. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Son MJ, Son MY, Seol B, et al. Nicotinamide overcomes pluripotency deficits and reprogramming barriers. Stem Cells. 2013;31:1121–1135. doi: 10.1002/stem.1368. [DOI] [PubMed] [Google Scholar]
- 11.Ruiz S, Panopoulos AD, Herrerias A, et al. A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Curr Biol. 2011;21:45–52. doi: 10.1016/j.cub.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y, Wei X, Wang M, et al. Proliferation rate of somatic cells affects reprogramming efficiency. J Biol Chem. 2013;288:9767–9778. doi: 10.1074/jbc.M112.403881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W, Zhou H, Abujarour R, et al. Generation of human-induced pluripotent stem cells in the absence of exogenous Sox2. Stem Cells. 2009;27:2992–3000. doi: 10.1002/stem.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyssiotis CA, Foreman RK, Staerk J, et al. Reprogramming of murine fibroblasts to induced pluripotent stem cells with chemical complementation of Klf4. Proc Natl Acad Sci USA. 2009;106:8912–8917. doi: 10.1073/pnas.0903860106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huangfu D, Maehr R, Guo W, et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye L, Chang JC, Lin C, et al. Induced pluripotent stem cells offer new approach to therapy in thalassemia and sickle cell anemia and option in prenatal diagnosis in genetic diseases. Proc Natl Acad Sci USA. 2009;106:9826–9830. doi: 10.1073/pnas.0904689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Staunstrup NH, Madsen J, Primo MN, et al. Development of transgenic cloned pig models of skin inflammation by DNA transposon-directed ectopic expression of human beta1 and alpha2 integrin. PloS One. 2012;7:e36658. doi: 10.1371/journal.pone.0036658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu X, Zhang X, Dhakal IB, et al. Induction of cell proliferation and survival genes by estradiol-repressed microRNAs in breast cancer cells. BMC Cancer. 2012;12:29. doi: 10.1186/1471-2407-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strassburger K, Tiebe M, Pinna F, et al. Insulin/IGF signaling drives cell proliferation in part via Yorkie/YAP. Dev Biol. 2012;367:187–196. doi: 10.1016/j.ydbio.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 20.Teo AK, Windmueller R, Johansson BB, et al. Derivation of human induced pluripotent stem cells from patients with maturity onset diabetes of the young. J Biol Chem. 2013;288:5353–5356. doi: 10.1074/jbc.C112.428979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moschidou D, Mukherjee S, Blundell MP, et al. Valproic acid confers functional pluripotency to human amniotic fluid stem cells in a transgene-free approach. Mol Ther. 2012;20:1953–1967. doi: 10.1038/mt.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moschidou D, Mukherjee S, Blundell MP, et al. Human mid-trimester amniotic fluid stem cells cultured under embryonic stem cell conditions with valproic acid acquire pluripotent characteristics. Stem Cells Dev. 2013;22:444–458. doi: 10.1089/scd.2012.0267. [DOI] [PubMed] [Google Scholar]
- 23.Medina V, Edmonds B, Young GP, et al. Induction of caspase-3 protease activity and apoptosis by butyrate and trichostatin A (inhibitors of histone deacetylase): Dependence on protein synthesis and synergy with a mitochondrial/cytochrome c-dependent pathway. Cancer Res. 1997;57:3697–3707. [PubMed] [Google Scholar]
- 24.Tighe A, Ray-Sinha A, Staples OD, et al. GSK-3 inhibitors induce chromosome instability. BMC Cell Biol. 2007;8:34. doi: 10.1186/1471-2121-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia Q, Sung J, Chowdhury W, et al. Chronic administration of valproic acid inhibits prostate cancer cell growth in vitro and in vivo. Cancer Res. 2006;66:7237–7244. doi: 10.1158/0008-5472.CAN-05-0487. [DOI] [PubMed] [Google Scholar]
- 26.Weller EM, Poot M, Hoehn H. Induction of replicative senescence by 5-azacytidine: fundamental cell kinetic differences between human diploid fibroblasts and NIH-3T3 cells. Cell Prolif. 1993;26:45–54. doi: 10.1111/j.1365-2184.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 27.Liu S, Okada T, Assmann A, et al. Insulin signaling regulates mitochondrial function in pancreatic beta-cells. PloS One. 2009;4:e7983. doi: 10.1371/journal.pone.0007983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zarse K, Schmeisser S, Groth M, et al. Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell Metab. 2012;15:451–465. doi: 10.1016/j.cmet.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshida Y, Takahashi K, Okita K, et al. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell. 2009;5:237–241. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Prigione A, Rohwer N, Hoffman S, et al. HIF1α modulates reprogramming through early glycolytic shift and upregulation of PDK1-3 and PKM2. Stem Cells. 2014;32:364–376. doi: 10.1002/stem.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J, Kim HK, Han YM, et al. Pyruvate kinase isozyme type M2 (PKM2) interacts and cooperates with Oct-4 in regulating transcription. Int J Biochem Cell Biol. 2008;40:1043–1054. doi: 10.1016/j.biocel.2007.11.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.