Abstract
For there were very short of excellent strains inoculated to ferment tea products, the lactic acid bacteria from pickled tea were isolated, characterized and identified, and the acid production capacity of part better strains was determined. There are only 22 strains isolated from pickled tea, and 2 of them were yeast, and 8 strains selected from the other 20 strains all were identified as Lactobacillus plantarum. A1, L2 and L5 of L. plantarum with a high acid production capacity were screened out and could obviously shorten the fermentation time of pickled tea by the verification, which suggests that they have a potential use of inoculating to ferment tea products. It was the first report on screening lactic acid bacteria with high yielding-acid capacity from pickled tea, which will bring benefits to fermenting tea products by artificial inoculation.
Electronic supplementary material
The online version of this article (doi:10.1007/s13197-015-1803-6) contains supplementary material, which is available to authorized users.
Keywords: Pickled tea, High yielding acid, Screening, Lactobacillus plantarum
Introduction
Tea as a natural beverage with a long history is widely consumed in the world. As a traditional tea, there are mainly three kinds of microbial fermented tea: dark tea, Kombucha and pickled tea (Mo et al. 2008; Zhang et al. 2013; Jayabalan et al. 2014). Dark tea like Pu-erh tea, Fuzhuan brick tea and Laoqing brick tea is produced only in China (Mo et al. 2008; Zhang et al. 2013), Kombucha is produced and drunk widely in the world (Greenwalt et al. 2000; Jayabalan et al. 2014), and pickled tea such as Acid tea (Tamang 2011), Miang tea (Reichart et al. 1988; Lee 1997) and Goishi-cha (Kato et al. 1994; Nanba et al. 1998) is fermented and chewed or drunk in many Asian countries. Dark tea and pickled tea both are fermented in solid state with tea leaves as substrates (Reichart et al. 1988; Lee 1997; Zhang et al. 2013), but Kombucha is fermented by tea liquid added sugar (Greenwalt et al. 2000; Jayabalan et al. 2014).
At present, it is well known that microbial fermented tea has many unique functions such as losing weight, and lowering blood pressure in that microbial fermentation gifted it with special qualities and special active substances (Mo et al. 2008; Hirota et al. 2011; Zhang et al. 2013; Jayabalan et al. 2014). There are many reports on microbial fermented tea, but almost concentrated on the fermentation technology, health function and chemical components as well as isolation and identification of microbial strains (Okada et al. 1986; Tanasupawat and Komagata 1995; Mo et al. 2005; Jayabalan et al. 2007; Sukontasing et al. 2007; Tanasupawat et al. 2007; Abe et al. 2008; Jayabalan et al. 2008; Mo et al. 2008; Klayraung and Okonogi 2009; Klayraung; and Okonogi 2009; Jayabalan et al. 2011). In recent years, microbial fermented tea has been paid more and more attention in the world (Mo et al. 2008; Zhang et al. 2013; Jayabalan et al. 2014). However, most of microbial fermented tea products were fermented in a natural way, which results in that their fermentation process, product quality and product safety are not easily controlled and guaranteed. For this reason, it is an urgent need for artificial inoculation fermentation, which also benefits to implementing a mechanized and standardized processing of microbial fermented tea (Mo et al. 2008). However, there are few studies on artificial inoculation of fermented tea products, and the inoculated strains only were isolated directly from samples, without being screened (Jeng et al. 2007; Abe et al. 2008; Chen et al. 2008; Chen et al. 2010; Wang et al. 2014). Furthermore, there is little research on screening predominant strains from microbial fermented tea, and there is a lack of better strains to be artificially inoculated to ferment tea products (Jeng et al. 2007; Abe et al. 2008; Chen et al. 2008; Chen et al. 2010; Wang et al. 2014).
In order to ensure product quality and safety, microbial fermentation of tea products would be artificially inoculated in the future, and the most important naturally is to have some excellent strains which could be used for artificial inoculation fermentation (Mo et al. 2008; Wang et al. 2014). To this end, microbial strains were isolated and identified from a special kind of pickled tea, and then screened out three high-yielding-acid strains of lactic acid bacteria. The verification test confirmed the screened strains possess a great potential in application to ferment tea products. It was the first report on screening lactic acid bacteria with high yielding-acid capacity from pickled tea, which will bring benefits to ferment dark tea, Kombucha and pickled tea by artificial inoculation.
Materials and methods
Bacterial medium and reagents
MRS agar was used for cultivation, isolation and count of lactic acid bacteria (Man et al. 1960). MRS with 30 % glycerol was used for strains preservation, with 20 g/L calcium carbonate for further strain selection. All the reagents used in this study were purchased from Tianyuan Reagent Co. Ltd., Wuhan City, China, and mostly were of analytical grade.
Samples of pickled tea
After being cleaned and drained, the fresh leaves, Camellia sinensis buds with three or four leaves plucked in summer and autumn at the tea plantation of Huazhong Agricultural University, were blanching-deactivated about 90 s and cooled immediately. The fixed leaves were spread out on bamboo plaque with a fan blowing. When its weight loss was about 30 % by continuous fan blowing, the leaves were filled into sterilized glass bottles. The boiled and cooled saline with 40 g/L salt was added into the bottles according to a solid-to-liquid weight ratio of 1:5 and the leaves were submerged into the saline. The bottles were completely sealed and placed at room temperature to ferment for 7 days in a natural way. The fermented broth of pickled tea was collected into sterilized tubes for strains analysis in 24 h. Lactococcus lactis subsp. lactis (Lister) (ATCC 11454) from State Key laboratory of Agricultural Microbiology, Huazhong Agricultural University, was as a control.
Isolation, purification and screening methods of the strains
Serial dilutions of fermented broth samples were prepared and plated to MRS agar. MRS agar plates were incubated at 30 °C for 48 h-72 h. Single colonies with different shapes and colors were selected from the MRS agar plates and transferred into tubes containing MRS broth that were incubated at 37 °C for 24 h. Appropriate dilutions of the MRS broths cultured with the selected colonies were plated to MRS agar to form single colony which was picked to microscopic evaluation. The previous two steps for purification were repeated continuously until the Gram-stain color and size of each cell observed by microscope were same, and only the single colony was as a selected one. Then, the isolated strains were saved for further analysis. Cell morphology of strains was observed and pictured by microscope (Nikon E600, Japan).
Strains were incubated at 30 °C for 24 h, and then plated to MRS agar with 20 g/L calcium carbonate at 30 °C for 48 h. Strains with larger soluble calcium ring size were selected and numbered for further research.
Autoploid of 16S rDNA gene sequence analysis
Genomic DNA of the selected strains was extracted from 1.5 ml colony MRS broths cultured at 37 °C about 8 h. The primers of 27 F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’-GGTTACTTGTTACGACTT-3’) were devised and synthesized according to previously reports (Eden et al. 1991; Weisburg et al. 1991). PCR (polymerase chain reaction) was performed by Takara Easy Taq polymerase (Takara, Shiga, Japan) on a Bio-rad S1000 Thermal Cycler PCR instrument (Bio-Rad, USA). Amplifications were carried out in a final volume of 25 μl, containing 2 μl of template DNA, 2.5 μl of 10 × Eazy Taq PCR Buffer, 2 μl of dNTP Mixture, 0.2 μl of TaKaRa Easy Taq polymerase (5 U, Takara, Shiga, Japan), 0.3 μl of primers, and 17.7 μl of sterilized double-distilled water. Reactions were run for 36 cycles of denaturation at 94 °C for 60 s, annealing at 52 °C for 60 s, extension at 72 °C for 90 s and a final extension at 72 °C for 10 min.
Products of PCR were sequenced by BGI (BGI, Wuhan) and the obtained gene sequences were done by BLAST (http://www.ncbi.nlm.nih.gov) to find the similar sequences. The obtained sequences and their similar sequences were analyzed by the Clustal W (Eden et al. 1991; Thompson et al. 1994). Phylogenetic tree was constructed by MEGA version 4.1 based on the Neighbor-joining Method (Kumar et al. 2008). Confidence values of branches of the phylogenetic tree were determined by bootstrap analysis (Felsenstein 1985) based on 1000 resamples.
Physiological and biochemical characteristics of the strains
The physiological and biochemical characteristics of isolates selected from pickled tea, including Gram stain, catalase test, arginin hydrolysis test, nitrate reduction test, and H2S production test, were done. Each test had a corresponding control. Each isolate was propagated in MRS broth before use. Fermentation of carbohydrates (including aescin glycosides, cellobiose, maltose, mannitol, salicin, sorbitol, sucrose, raffinose, inulin, and lactose) was determined by bacterial micro tube for biochemical identification produced by Qingdao Hope Bio-Technology Co. Ltd.. Reagent blank and strain ATCC 11454 were as a control.
Acid production capacity of the strains
The four selected strains were cultured at 30 °C for 16 h-18 h and then inoculated with 4 % (v/v) inoculum to the fermentation broth containing 40 g blanched tea leaves and 200 ml sterile brine with 40 g/L NaCl in a flask. The fermentation was cultured in a static way at 30 °C for 36 h. pH value and total acid of the fermented broths were measured every 12 h. Total acid was determined by the method of NaOH titration, and pH value measured directly by pH meter (FE 20 type, China).
Verification of the selected strains
According to the previous method of determining the acid production capacity, the selected strains were inoculated to ferment C. sinensis leaves about 60 h. As a control, the pickled tea was fermented in a natural way about 60 h and 7 d, respectively.
Evaluation of the sensory quality
In the verification, the wet samples of pickled tea were evaluated directly according to the scoring criteria of evaluating the sensory quality of pickled tea (Suppl. 1) for pickled tea mainly was eaten as a snack or dish (Reichart et al. 1988). 10 sensory assessors, all major in tea science at Huazhong Agricultural University and professionally trained in sensory evaluation, evaluated the sensory quality of the wet fermented leaf samples.
Statistical analysis
All experiments were repeated three times. One-way Analyses of Variance were performed using SPSS software (PASW Statistics 18.0) and the multiple range tests were used to compare difference among mean values. The significance was defined at p < 0.05 (significant difference) and p < 0.01 (highly significant difference).
Results
Bacterial isolates from pickled tea
The broths of pickled tea were serially diluted and plated on MRS agar and those plates were cultured at 30 °C. From the initial separation of plates, it was known that pickled tea had a much pure microbial environment in that the number of colonies on the plates was relatively little and the shape and size of the colonies were much similar. Through isolation and purification, there were about 22 strains isolated from the MRS agar plates and 2 of them were yeast determined by colony morphology and cell morphology. The rest 20 bacteria were divided into two groups according to their characteristics and numbered with A1 to A10, and L1 to L10, respectively.
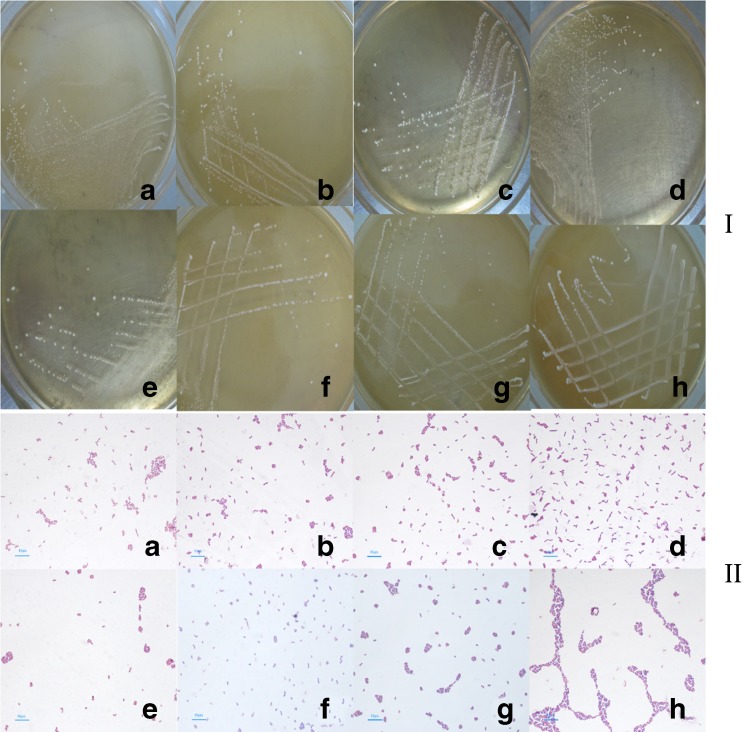
Then, the 8 isolated strains, A1, A2, A3, A10, L2, L4, L5, and L6 were further screened out by MRS agar plate with calcium carbonate for they had a larger dissolved calcium ring on the plates. The colony morphology of the 8 strains on MRS agar and the Gram stain results were shown in Fig. 1, and the colony characteristics were shown in Suppl. 2. The 8 strains basically were short rod and single or in pairs, and all of them were Gram-positive. Most of the colonies in about 2 mm-3.5 mm of diameter were round, smooth, convex surface, neat edge, shiny, soft, off-white or milky, and opaque (Fig. 1, Suppl. 2). These characteristics showed that the 8 strains were similar to lactic acid bacteria.
Fig. 1.
The colony morphology of the 8 strains isolated from pickled tea in MRS agar plates (I) and their Gram stain results (II). Both in (I) and (II), A to H is A1, A2, A3, A10, L2, L4, L5, and L6, respectively
Physiological and biochemical characteristics of the 8 isolates
The physiological and biochemical characteristics of the 8 strains isolated from pickled tea were further analyzed (Fig. 1, Suppl. 3 and 4). Biochemical characteristics of the 8 strains isolated from pickled tea showed that only the results of MR were positive and all the others were negative (Suppl. 3), and the 8 isolates had the same reaction at tests of catalase, NH3 from Arginine, H2S from Cysteine, MR and VP, which were equal to those of Lactococcus lactis subsp. lactis (Lister) (ATCC 11454). The sugar fermentation of the 8 strains all showed positive, except that the melitose fermentation of A2 was negative (Suppl. 4). The results of aescin glycosides, maltose, mannitol, sorbitol, and raffinose fermented by the 8 strains, except the result of raffinose fermented by A2, were different to those fermented by Lactococcus lactis subsp. lactis (Lister) (ATCC 11454), but the results of other sugar fermented by the 8 strains were same to those fermented by ATCC 11454. These results showed that the 8 strains are very likely belong to Lactobacillus but different to Lactococcus lactis subsp. lactis (Lister) (ATCC 11454).
Identification of the isolates by 16S rDNA gene sequences
To further identify the 8 isolates, the genomic DNA of the 8 strains was extracted respectively and the 16S rDNA gene sequences were amplified by PCR. From Fig. 2, there was an obvious rough band about 1,500 bp for all the amplified fragments of A1, A3, A10, L2, L4, and L6, respectively (Fig. 2). However, although the amplified contents were much low and there was no obvious DNA band about L5 and A2 in Fig. 2, their amplified fragments were still purified and sequenced.
Fig. 2.

The results of bacterial 16S rDNA PCR amplification for the 8 strains isolated from Yancha. The concentration of agarose gel was 10 g kg−1. 1 to 3 is L2; 4 to 6, and 25 to 27 are L4; 7 to 9 is L5; 10 to 12 is L6; 13 to 15 is A1; 16 to 18 is A2; 19 to 21 is A3; 22 to 24 is A10. M is the DNA molecular mass standards (DL2000), and CK is the blank control
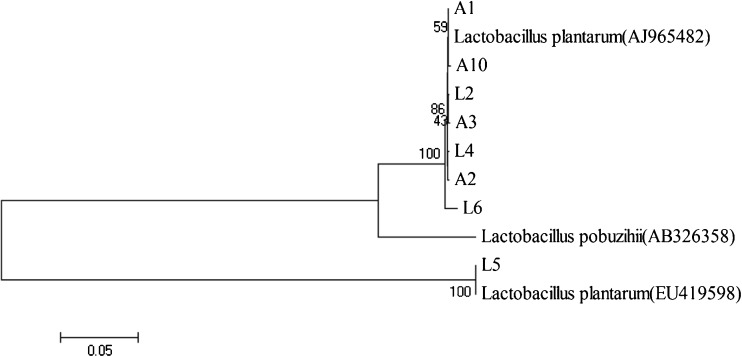
The amplified fragments from the genomic DNA of the 8 isolates were all sequenced, and the sequences were accessed in GenBank (Their GenBank accession numbers: L2, KC684521; L4, KC684522; L5, KC684523; L6, KC684524; A1, KC684525; A2, KC684526; A3, KC684527; A10, KC684528). And the 8 sequences were assembled and compared with those available in GenBank. It was interesting that the similarities of the 16S rDNA gene sequences amplified from the 8 strains were more than 99 % between each other, which means that the 8 isolates may be due to the same strain. The results of sequences aligned block by Clustal W showed 5 of them were almost the same strain (Fig. 3). Comparison of the 16S rDNA gene sequences of the 8 strains with those in the DNA Data Bank in the NCBI revealed their homologies of greater than 99 % to Lactobacillus plantarun. The results indicated that the 8 strains were much related to Lactobacillus plantarum subsp. (DQ239696), Lactobacillus sp. KLDS plantarum (EU600909), Lactobacillus plantarum JDM1 (CP001617) (Fig. 3). Considering the results of their physiological and biochemical characteristics and the 16S rDNA sequence analysis, the 8 strains were all identified as Lactobacillus plantarum.
Fig. 3.
Phylogenetic tree based on 16S rDNA sequences showing the relationship between the isolated strains from pickled tea and the related bacterial species. Phylogenetic tree was constructed using the Neighbor-joining Method. The most similar sequence in GenBank database were as the reference sequences and NCBI numbers were in brackets. Numbers on the branches said the confidence coefficient with 1000 computed result. Plotting scale was the genetic distance
The acid production capacity of strains isolated from pickled tea
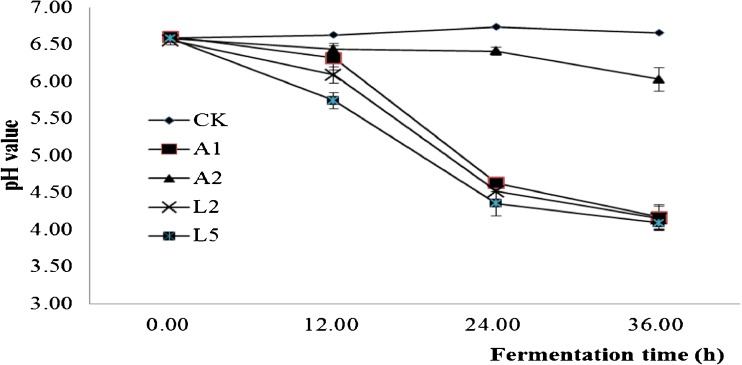
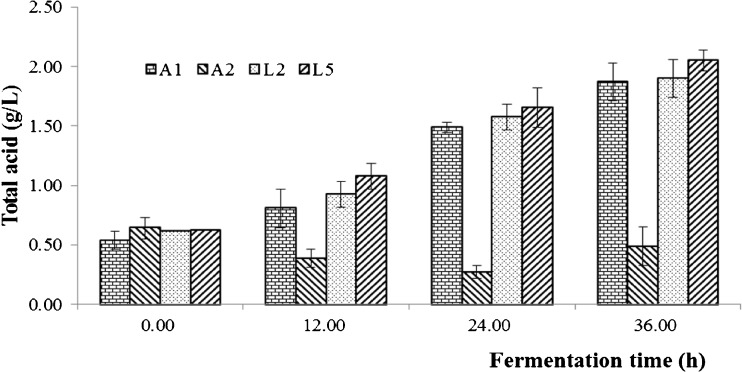
Acid production of microbe plays an essential role in the quality of pickled teas, and rapid acid production is conducive to the growth of beneficial microorganisms in a natural fermentation. The acid production capacity of A1, A2, L2 and L5 selected from the 8 strains were analyzed by directly fermenting C. sinensis leaves (Fig. 4 and Fig. 5). In 36 h, the pH values of the broths fermented by the 4 strains decreased linearly except of A2, and their acidity also increased linearly except of A2. In general, A2 produced little acid and its pH value decreased very slowly. The significance of difference analysis showed that the acid production of A2 was highly significant (ρ < 0.01) lower than those of A1, L2 and L5, but the differences of acid production among A1, L2 and L5 were not significant at the 0.05 level. In a word, A1, L2 and L5 all have a high acid production capacity, and their acidity reached more than 1.5 g/L in 24 h when they fermented C. sinensis leaves directly (Fig. 5).
Fig. 4.
The pH changes of the fermentation broths fermented by A1, A2, L2, and L5. CK, uninoculated
Fig. 5.
The acidity changes of the fermentation broths fermented by A1, A2, L2, and L5. CK, uninoculated
The sensory qualities of the pickled tea fermented by inoculating the selected strains
To further validate the screened strains, A1, L2 and L5 were inoculated to ferment pickled tea about 60 h, and the pickled tea was fermented in a natural way at different time as a control. From Table 1, the pickled tea (CK1) fermented only 60 h had a dark green of its color, an obviously grass odor, hard texture and much bitter taste; in a word, the sensory quality of CK1 was much similar to that of unfermented tea leaves, which meant that the quality of pickled tea was not formed during the short 60-h fermentation. However, the qualities of pickled tea inoculated the A1, L2 and L5 were closed to, even better than that of the pickled tea (CK2) fermented in a natural way about 7 d. In a 60-h fermentation, the pickled tea inoculated the selected strains can possess its normal quality characteristics, which suggests that the selected strains can speed up the fermentation process of pickled tea, and shorten the fermentation time.
Table 1.
The sensory qualities of pickled tea fermented by the selected strains
| Strains | Color and appearance | Aroma | Texture | Taste | Scores |
|---|---|---|---|---|---|
| L2 | Yellow-green, lightly dark | Great Lactic acid fermentation incense | Much Crisp | Slight bitter and astringent, sour | 91.00 ± 1.76Aa |
| L5 | Yellow-green | Lactic acid fermentation incense | crisp | Slight bitter and astringent, sour | 84.15 ± 1.05Bb |
| A1 | Yellow-green | Lactic acid fermentation incense | crisp | Slight bitter and astringent, sour | 82.07 ± 1.57Bb |
| CKl | Dark green | Obvious grass odor, only weak Lactic acid fermentation incense | Hard | Much bitter and astringent, weak sour | 45.60 ± 2.94Dd |
| CK2 | Dark yellow-green | Lactic acid fermentation incense | crisp | Slight bitter and astringent, sour | 83.72 ± 1.26Bb |
The samples were evaluated directly in a wet form when the fermentation was over. The fermentation time of L2, L5 and A1 was 60 h. The pickled tea was fermented about 60 h (CK1) and 7 d (CK2) in a natural way as the control, respectively
Discussions
Pickled tea as a traditional tea with a long history is produced and eaten or drunk in a lot of Asian countries (Reichart et al. 1988; Nanba et al. 1999). There were some studies about pickled tea, but there is little research on screening the lactic acid bacteria with high yielding-acid capacity from microbial fermented teas (Reichart et al. 1988; Kato et al. 1994; Nanba et al. 1998; Nanba et al. 1999). The strains of pickled tea fermented by soaking method were isolated, purified and identified, and the acid production capacity of better strains was determined and verified. There are only 22 strains isolated from pickled tea, and 2 of them were yeast, and 8 strains selected from the other 20 strains all were identified as L. plantarum (Fig. 3). It showed that L. plantarum is the dominant species of pickled tea, which was same to Miang tea fermentation (Okada et al. 1986). Yet, the microbial species of pickled tea were simpler than those of Miang tea. It is known that there were different lactic acid bacteria in Miang tea, such as L. plantarum, L. pentosus, L. vaccinostercus, L. sp., Enterococcus casseliflavus, and Enterococcus sp., L. thailandensis sp. nov., L. camelliae sp. nov., and Pediococcus siamensis sp. nov. sp. et al. (Okada et al. 1986; Tanasupawat and Komagata 1995; Sukontasing et al. 2007; Tanasupawat et al. 2007; Klayraung and Okonogi 2009; Klayraung; and Okonogi 2009). The reason for this difference may be that the salt was added into pickled tea fermentation, and Miang tea is a non-salt fermented product (Okada et al. 1986). The strains of pickled tea also were different to those of Kombucha which included lactic acid bacteria, yeasts and acetic acid bacteria (Blanc 1996), and to those of the pile-fermentation of Pu-erh tea in which the main microbe was Aspergillus niger (LeBar 1967; Zhou et al. 2004).
L. plantarum is a safe and probiotic microorganism, and has been widely used in food industry (Li et al. 2012). It was known that A1, L2 and L5 produced more than 1.5 g/L acidity in 24 h when they fermented C. sinensis leaves directly (Fig. 5), which means the selected strains have a high yielding-acid capacity (Fig. 5). The selected strains could tolerate the bactericidal or bacteriostatic environment with a high concentration of tea ingredients (Tanasupawat et al. 2007), and would play an important role in controlling the whole fermentation environment and ecosystem by secreting high acidity quickly (Tanasupawat et al. 2007), which would promote to keep product safety. The verification of the screened strains showed that the sensory qualities of pickled tea inoculated the selected strains were formed during a 60-h pure fermentation, but the fermentation time of pickled tea in a natural way generally was 7 d. It confirmed that when A1, L2 and L5 were artificially inoculated to ferment pickled tea, the fermentation time was shortened and the quality was guaranteed (Table 1, Figs. 4 and 5). These selected strains can secret high acid quickly in a short time, which will reduce the chance of harmful microbe contamination and improve food safety (Jayabalan et al. 2007). These strains also have the obvious potential to be used to ferment Pu-erh tea, Kombucha, and lactobacillus liquid tea drinks, and will play an important role in producing microbial fermented tea products in future (Jayabalan et al. 2007; Mo et al. 2008; Zhang et al. 2013; Jayabalan et al. 2014).
Conclusion
This study isolated and characterized the species of lactic acid bacteria from pickled tea and screened out A1, L2 and L5 of L. plantarum with a high acid production capacity. L. plantarum is a safe and probiotic microorganism, and could be artificially inoculated to ferment tea products. This also is the first report on screening lactic acid bacteria with high yielding-acid capacity from pickled tea, which will bring benefits to ferment dark tea, Kombucha and pickled tea by artificial inoculation.
Electronic supplementary material
(DOC 36 kb)
(DOC 38 kb)
(DOC 35 kb)
(DOC 40 kb)
Acknowledgment
The authors are very grateful for the financial support from Huazhong Agricultural University Scientific & Technological Self-innovation Foundation (Grant No. 2009SC013) and the Fundamental Research Funds for the Central Universities, China (Grant No. 2009PY023).
Footnotes
Highlights
--The microbial species of pickled tea were much simple.
--Lactobacillus plantarum is the dominant specie in pickled tea fermentation.
--Three strains with a high acid production capacity were screened out.
--The selected strains can shorten the fermentation time of pickled tea.
Ping Xiao and Youyi Huang contributed equally to this work.
References
- Abe M, Takaoka N, Idemoto Y, Takagi C, Imai T, Nakasaki K. Characteristic fungi observed in the fermentation process for Puer tea. Int J Food Microbiol. 2008;124(2):199–203. doi: 10.1016/j.ijfoodmicro.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Blanc PJ. Characterization of the tea fungus metabolites. Biotechnol Lett. 1996;18(2):139–142. doi: 10.1007/BF00128667. [DOI] [Google Scholar]
- Chen K, Zhang X, Zhu H, Yang C, Zhang Y. The effects of Aspergillus on the post-fermentative process of Pu-er tea. Acta Botanica Yunnanica. 2008;30(5):624–628. [Google Scholar]
- Chen Y, Liu B, Chang Y. Bioactivities and sensory evaluation of Pu-erh teas made from three tea leaves in an improved pile fermentation process. J Biosci Bioeng. 2010;109(6):557–563. doi: 10.1016/j.jbiosc.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Eden PA, Schmidt TM, Blakemore RP, Pace NR. Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction-amplified 16S rRNA-specific DNA. Int J Syst Bacteriol. 1991;41(2):324–325. doi: 10.1099/00207713-41-2-324. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- Greenwalt CJ, Steinkraus KH, Ledford RA. Kombucha, the fermented tea: Microbiology, composition, and claimed health effects. J Food Prot. 2000;63(7):976–981. doi: 10.4315/0362-028x-63.7.976. [DOI] [PubMed] [Google Scholar]
- Hirota R, Ngatu NR, Miyamura M, Nakamura H, Suganuma N. Goishi tea consumption inhibits airway hyperresponsiveness in BALB/c mice. BMC Immunol. 2011;12(25):1–8. doi: 10.1186/1471-2172-12-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayabalan R, Marimuthu S, Swaminathan K. Changes in content of organic acids and tea polyphenols during kombucha tea fermentation. Food Chem. 2007;102(1):392–398. doi: 10.1016/j.foodchem.2006.05.032. [DOI] [Google Scholar]
- Jayabalan R, Subathradevi P, Marimuthu S, Sathishkumar M, Swaminathan K. Changes in free-radical scavenging ability of kombucha tea during fermentation. Food Chem. 2008;109(1):227–234. doi: 10.1016/j.foodchem.2007.12.037. [DOI] [PubMed] [Google Scholar]
- Jayabalan R, Chen P-N, Hsieh Y-S, Prabhakaran K, Pitchai P, Marimuthu S, Thangaraj P, Swaminathan K, Yun SE. Effect of solvent fractions of kombucha tea on viability and invasiveness of cancer cells-Characterization of dimethyl 2-(2-hydroxy-2-methoxypropylidine) malonate and vitexin. Indian J Biotechnol. 2011;10(1):75–82. [Google Scholar]
- Jayabalan R, Malbasa RV, Loncar ES, Vitas JS, Sathishkumar M. A review on Kombucha tea microbiology, composition, fermentation, beneficial effects, toxicity, and tea fungus. Compr Rev Food Sci Food Saf. 2014;13(4):538–550. doi: 10.1111/1541-4337.12073. [DOI] [PubMed] [Google Scholar]
- Jeng KC, Chen CS, Fang YP, Hou RCW, Chen YS. Effect of microbial fermentation on content of statin, GABA, and polyphenols in Pu-erh tea. J Agric Food Chem. 2007;55:8787–8792. doi: 10.1021/jf071629p. [DOI] [PubMed] [Google Scholar]
- Kato M, Tamura A, Omori M, Nanba A, Miyagawa K, Nishimura O, Kameda W. Changes of flavor during manufacturing process of Japanese fermented tea (Goishi-cha) and its characteristic. Japan Soc Home Econ. 1994;45(6):527–532. [Google Scholar]
- Klayraung S, Okonogi S. Antibacterial and antioxidant activities of acid and bile resistant strains of lactobacillus fermentum isolated from Miang. Braz J Microbiol. 2009;40(4):757–766. doi: 10.1590/S1517-83822009000400005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9(4):299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBar FM. Miang: fermented tea in North Thailand. Cross-Cultural Res. 1967;2(2):105–121. doi: 10.1177/106939716700200206. [DOI] [Google Scholar]
- Lee CH. Lactic acid fermented foods and their benefits in. Asia Food Control. 1997;8(5–6):259–269. doi: 10.1016/S0956-7135(97)00015-7. [DOI] [Google Scholar]
- Li SY, Zhao YJ, Zhang L, Zhang X, Huang L, Li D, Niu CH, Yang ZN, Wang Q. Antioxidant activity of Lactobacillus plantarum strains isolated from traditional Chinese fermented foods. Food Chem. 2012;135(3):1914–1919. doi: 10.1016/j.foodchem.2012.06.048. [DOI] [PubMed] [Google Scholar]
- Man JCD, Rogosa M, Sharpe ME. A culture medium for the cultivation of lactobacillus. J Appl Microbiol. 1960;23(1):130–135. [Google Scholar]
- Mo H, Xu X, Yan M, Zhu Y. Microbiological analysis and antibacterial effects of the indigenous fermented Puer tea. Agro Food Ind Hi-Tech. 2005;16(6):16–18. [Google Scholar]
- Mo H, Zhu Y, Chen Z. Microbial fermented tea - a potential source of natural food preservatives. Trends Food Sci Technol. 2008;19(3):124–130. doi: 10.1016/j.tifs.2007.10.001. [DOI] [Google Scholar]
- Nanba A, Miyagawa K, Omori M, Kato M, Tamura A, Saito H. Non-salted pickled tea (Sour tea) in south-east Yunnan in China. Japan Soc Home Econ. 1998;49(8):907–915. [Google Scholar]
- Nanba A, Nyein MM, Win SY, Miyagawa K. Post-heated and fermented edible teas and their dried forms used for drinking in Myanmar. J Home Econ Japan. 1999;50(6):639–646. [Google Scholar]
- Okada S, Daengsubha W, Uchimura T, Ohara N, Ki AIK. Flora of lactic acid bacteria in miang produced in northern Thailand. J Gen Appl Microbiol. 1986;32(1):57–65. doi: 10.2323/jgam.32.57. [DOI] [Google Scholar]
- Reichart PA, Philipsen HP, Mohr U, Geerlings H, Srisuwan S. Miang chewing in northern Thai villagers. Trop Geogr Med. 1988;40(1):39–44. [PubMed] [Google Scholar]
- Sukontasing S, Tanasupawat S, Moonmangmee S, Lee J-S, Suzuki K-I. Enterococcus camelliae sp nov., isolated from fermented tea leaves in Thailand. Int J Syst Evol Microbiol. 2007;57:2151–2154. doi: 10.1099/ijs.0.65109-0. [DOI] [PubMed] [Google Scholar]
- Tamang, JP (2011) Prospects of Asian fermented foods in global markets. The 12th ASEAN FOOD CONFERENCE, BITEC Bangna, Bangkok, Thailand (June 16–18), 67–76
- Tanasupawat S, Komagata K. Lactic acid bacteria in fermented foods in Thailand. World J Microbiol Biotechnol. 1995;11(3):253–256. doi: 10.1007/BF00367094. [DOI] [PubMed] [Google Scholar]
- Tanasupawat S, Pakdeeto A, Thawai C, Yukphan P, Okada S. Identification of lactic acid bacteria from fermented tea leaves (miang) in Thailand and proposals of Lactobacillus thailandensis sp. nov., Lactobacillus camelliae sp. nov., and Pediococcus siamensis sp. nov. J Gen Appl Microbiol. 2007;53(1):7–15. doi: 10.2323/jgam.53.7. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ji B, Wu W, Wang R, Yang Z, Zhang D, Tian W. Hepatoprotective effects of kombucha tea: identification of functional strains and quantification of functional components. J Sci Food Agric. 2014;94(2):265–272. doi: 10.1002/jsfa.6245. [DOI] [PubMed] [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173(2):697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhang ZZ, Zhou YB, Ling TJ, Wan XC. Chinese dark teas: Postfermentation, chemistry and biological activities. Food Res Int. 2013;53(2):600–607. doi: 10.1016/j.foodres.2013.01.016. [DOI] [Google Scholar]
- Zhou H, Li J, Zhao L, Han S, Yang X, Yang W, Wu X. Study on main microbes on quality formation of Yunnan Puer tea during pilefermentation process. J Tea Sci. 2004;24(3):212–218. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 36 kb)
(DOC 38 kb)
(DOC 35 kb)
(DOC 40 kb)