Abstract
A novel intracellular β-galactosidases produced by Lactobacillus plantarum HF571129, isolated from an Indian traditional fermented milk product curd was purified and characterized. The β-galactosidases is a hetrodimer with a molecular weight of 60 kDa (larger subunit) and 42 kDa (smaller subunit), as estimated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE). The enzyme was purified 7.23 fold by ultrasonication, ultrafiltration and gel filtration chromatography with an overall recovery of 30.41 %. The optimum temperature for hydrolysis of its preferred substrates, o-nitrophenyl- β-D-galactopyranoside (ONPG) and lactose, are 50 °C (both), and optimum pH for these reactions is 6.5 and 7.5, respectively. The β-galactosidases showed higher affinity for ONPG (Km, 6.644 mM) as compared to lactose (Km, 23.28 mM). Galactose, the end product of lactose hydrolysis was found to be inhibited (47 %). The enzyme activity was drastically altered by the metal ion chelators EDTA, representing that this enzyme is a metalloenzyme. The enzyme was activated to a larger extent by Mg2+ (73 % at 1 mM), while inhibited at higher concentrations of Na+ (54 % at 100 mM), K+ (16 % at 100 mM) and urea (16 % at 100 mM). The thermal stability study indicated an inactivation energy of Ed = 171.37 kJ mol−1. Thermodynamic parameters such as ∆H, ∆S and ∆G, were determined as a function of temperature. About 88 % of lactose was hydrolyzed at room temperature within 1 h. The study suggested that this enzyme showed its obvious superiority in the industrial lactose conversion process.
Keywords: β-galactosidases, Lactobacillus plantarum HF571129, Purification, Thermodynamic parameters, Inactivation kinetics
Introduction
Lactose intolerance occurs in 70 % of the world’s adult population, and Eastern Asia has the highest number of lactose malabsorbers with more than 90 % of its population (Vrese et al. 2001). To avoid lactose intolerance problems, milk and milk related products are enzymatically hydrolyzed using β-galactosidase. Lactose hydrolysis causes several changes with potential value for the manufactured and marketing of dairy products (Hettwer and Wang 1990).
Lactic acid bacteria (LAB), which comprise an assorted group of Lactococci, Streptococci and Lactobacilli, have been studied intensively with respect to their enzymes for various different reasons including their generally recognized as safe (GRAS) status. L. plantarum survives even at low pH of the stomach and duodenum, resisting the effects of bile acids in the upper small intestine when ingested, and temporarily colonizing the gastrointestinal tract by binding to the intestinal and colonic mucosa.
Beta galactosidases (EC 3.2.1.23) are present in a wide variety of organisms including plants, animals and microorganisms, and are known to catalyze both hydrolytic and transglycosylation reactions. Microbial β-galactosidases have a prominent position in terms of their role in the production of various industrially relevant products like biosensor, lactose hydrolyzed milk, the production of galacto-oligosaccharides for use in probiotic foodstuffs, etc. The enzyme can be obtained from microbial cells such as bacteria, fungi or yeast with variable properties depending on the species (Richmond et al. 1981; Hubber et al. 1994; Hung et al. 2001). The major β-galactosidase enzymes of commercial interest are isolated mainly from the yeast Kluyveromyces lactis, Kluyveromyces fragilis, Kluyveromyces marxianus, Candida kefyr, Saccharomyces cerevisae and the fungi Aspergillus niger, Aspergillus oryzae (Neri et al. 2008).
β-galactosidase is mainly intracellular enzyme in bacteria and yeasts, but in case of fungi, is an extracellular enzyme. β-galactosidases from LAB are secreted within the cell. Cell disruption is the first step in the isolation of intracellular enzyme and a wide range of disruption methods can be used in the laboratory, both chemical (detergents, alkali, or enzymes) and mechanical (ultrasonication, homogenizer). Deactivation studies help understand the relation between structure and function of a particular enzyme (Sadana 1995). Enzymes are deactivated by many ways to an inactive state, which forms a major constraint in many biotechnological processes (Joly 1965). It is also important to analyze the estimated thermodynamic parameters, since these aids in understanding the probable denaturation mechanism, which is very important in enzymatic processes (Ustok et al. 2010).
Few studies have been performed on the thermophilic β-galactosidases in certain microorganisms, including yeast, bacteria and fungi (Di Lauro et al. 2008). The optimum temperature of most commercially available β-galactosidases is less than 58 °C. Thus, identification of thermostable β-galactosidases could greatly benefit the dairy industry. In the past, many studies have reported on the properties of Lactobacillus plantarum, including its molecular mass, pH and temperature optimum, stability, inhibitor effects and metal ion effects. However, there are no studies to till date regarding the thermal deactivation kinetics and thermodynamic parameters from Lactobacillus plantarum.
Our study aims at purifying, partially characterizing β-galactosidases enzyme from a newly isolated Lactobacillus plantarum HF571129 and to study its optimal enzymatic conditions (temperature and pH), effect of metal ions, end product, its stability, and its kinetics for different substrates. Also, using the purified enzyme lactose hydrolysis was performed which will be useful for the lactose free product development.
Materials and methods
Materials
Ortho Nitro phenol (ONP), Ortho Nitro Phenyl Galacto-pyranoside (ONPG), X Gal, IPTG, Lactose, MRS and all other chemicals were purchased from Hi Media, Mumbai, India. Sephadex G-100 is from Sigma−Aldrich (St. Louis, MO, USA). All other reagents used were of analytical grade unless otherwise stated.
Isolation of Lactobacillus sp.
The various food samples such as idli batter, milk, curd and rice wine was aseptically collected in cold condition from Vellore district, Tamil Nadu and microbiologically processed within 2 h of collection by following standard microbiological procedure. De Mann Rogosa Sharpe (MRS) agar and broth medium pH 6.5 ± 0.2 was used for the enumeration and culturing lactic acid bacteria. The samples were serially diluted using saline and pour plated on MRS agar plates. For 24 h, the plates were incubated at 37 °C and a few isolates with the colony morphology similar to that of Lactobacillus were selected. The selected isolates were further subculture in MRS medium to get pure culture. After purification, the selected strains were numbered VITES01–VITES31, and stored at −20 °C in MRS broth with 20 % glycerol. The stock cultures were propagated twice in MRS broth for 18 h before each experiment.
β-galactosidase screening
X-gal substrate: X-gal (5-Bromo-4-chloro-3-indoxyl-β-D-galactopyranoside) is a substrate, which has been used to screen β-galactosidase positive organisms. Single colony of isolated bacteria were grown on MRS agar plates containing 60 μL X-gal and 10 μL of IPTG (Iso-propyl β-D-1-thiogalactopyranoside) solution as an inducer. For 72 h, the plates were incubated at 37 °C. Colonies producing β-galactosidase were green (Vinderola and Reinheimer 2003) in colour.
Biochemical identification of the HF571129 isolate
A bacterium designated as strain HF571129, isolated from the curd sample, showed the most intense β-galactosidase activity and then was selected for further work. Pure culture of strain HF571129 was tested for Gram staining, catalase, nitrate reductase test and sugar fermentation test (Akolkar et al. 2006). Characteristics of the isolate were compared with data from Bergey’s Manual of Determinative Bacteriology (Bergey et al. 1923).
Molecular characterization
The strain was further identified by 16S rRNA gene sequencing for molecular level identification. The nucleic acids of the isolate were extracted using a DNA purification kit (Amnion Biosciences Pvt Ltd, India), according to the manufacturer’s instructions. Two primers namely Forward (5′- CWG RCC TAN CAC ATG SAA GTC- 3) and Reverse (5′ - GRC GGW GTG TAC NAG GC- 3′) were used for the DNA amplification isolated from HF571129 isolate for 16S rRNA sequencing. The obtained sequences were subjected to BLAST search at http://www.ncbinlm-nih.gov/ in the NCBI database. Phylogenetic tree was constructed in MEGA 4.0.2 based on partial 16S rRNA gene sequences using neighbor-joining (NJ) method.
β-galactosidase production from Lactobacillus plantarum HF571129
L. plantarum HF571129 (1 % v/v) was inoculated in MRS broth and incubated for 18 h at 25 °C under stationary conditions. Cell pellets were harvested and suspended in sterile saline so that the final OD 1.0 when measured at 600 nm. This sterile cell suspension was used as inoculum for the production of β-galactosidase enzyme which was carried out in a medium containing 10 g/L lactose, 20 g/L yeast extract, 10 g/L protease peptone, 50 mg/L magnesium sulphate and 25 mg/L manganese sulphate, 4 g/L tri-ammonium citrate, 2.5 g/L potassium acetate and 4 g/L dipotassium hydrogen phosphate, at 25 °C for 24 h under stationary conditions.
Purification of β-galactosidase
L. plantarum was found to produce β-galactosidase within the cells (intracellular enzyme); the cells were further ultrasonicated for enzyme extraction. Cell pellets were harvested from the fermented broth by centrifuging at 5000 rpm for 15 min at 15 °C. The pellet was washed thrice by resuspending the cells in double distilled water, followed by centrifugation (Remi C-30BL, India) at 8000 rpm at 4 °C for 10 min. The cell pellet obtained after washing was again suspended in 100 mM phosphate buffer pH 7.2 and this cell suspension (25 mL) was subjected to ultrasonication treatment using the Sonics-Ultra cell, UK, with a ½″ diameter tapped bio horn that delivers ultrasonic sound at constant frequency of 20 kHz. The ultrasonication process was carried out at constant acoustic power, constant duty cycle of 20 W and 50 % respectively. The samples were kept in an ice bath to avoid denaturation of the product during the lysis process. The total lysis process was carried out for 12 min. Subsequently, the cell suspension was centrifuged for 15 min at 8000 rpm, at 4 °C and the resulting cell free supernatant was used for further purification. The intracellular enzyme was subjected for ultrafiltration (Amicon Ultra-15, Millipore, India) with a 50 kDa molecular weight cut-off membrane and assayed for both protein content and β galactosidase activity. Gel filtration were carried out with a self-packed Sephadex G-100 column (1.6-cm in diameter; 25-cm gel bed height; 1-mL sample volume) equilibrated with sterile column buffer 0.01 M Tris–HCl and 0.01 M NaCl (pH7.5). Retentate (1 mL) collected from ultrafiltration was loaded on the prepared column with a flow rate of 0.5 mL/min. The fractions were eluted (3 mL), and protein content was analyzed at 280 nm. Further, the fractions containing protein content were checked for enzyme activity at 420 nm.
β-galactosidase activity assay
ONPG substrate: β-galactosidase activity was measured by a modified method of Dickson (Dickson and Markin 1980) involving the hydrolysis of substrate o-nitrophenyl- β-galactopyranoside (ONPG) to o-nitrophenol (ONP). The cell-free supernatant was appropriately diluted to 1 mL using 100 mM phosphate buffer (pH 7.0) for the assay. Two mL ONPG solution (2 mg/mL), prepared in the same buffer was added to the enzyme solution. The reaction mixture was incubated at 50 °C in a water bath for 10 min and the reaction was then stopped by adding 1 mL (100 g/L) Na2CO3 solution. The yellow colour of ONP obtained as a result of ONPG hydrolysis was measured at 420 nm using ONP as the standard. The amount of ONP released per min by the cell-free supernatant was directly proportional to the quantity of enzyme released (Choonia and Lele 2011).
Lactose substrate: Activity towards lactose was estimated in 50 μL of the reaction mixture containing 50 mM lactose prepared in 50 mM acetate buffer, pH 6.5, and 10 μL of appropriately diluted enzyme. The reaction was carried out at 30 °C for 10 min. The reaction was stopped by heating the reaction mixture at 99 °C for 5 min. Glucose released was estimated by the GOD—POD method, using a commercially available kit (Span Diagnostics Ltd., Surat, India). Ten microlitres of the reaction mixture were added to 1000 μL of glucose reagent. Colour was developed for 10 min at 37 °C; absorbance was recorded at 505 nm. Glucose concentration was calculated by absorbance of test solution/absorbance of standard glucose solution (1 mg/mL) to give a concentration in mg/mL, which is converted to concentration in mM. One unit of enzyme is defined as 1 μmol of glucose released per min at 30 °C
Determination of molecular weight and estimation of protein
Molecular weight of purified β-galactosidase was determined by 12 % SDS PAGE. The enzyme samples were mixed with sample preparation buffer (1 M Tris–HCl buffer pH 6.8 containing 4 % SDS, 1 % bromophenol blue and 20 mL of glycerol) and 5 % 2-mercaptoethanol and heated at 95 °C for 5 min. The samples were loaded onto a polyacrylamide gel consisting of a 5 % stacking gel and a 12 % separating gel. After the gel was run, the protein bands were stained with 0.25 % Coomassie Brilliant Blue G-250 in 40 % methanol and 10 % acetic acid and destained with 10 % acetic acid and 40 % methanol. The molecular weight was determined with standard known marker such as Myosin (205 kDa), β-Galactosidase (116 kDa), BSA (66 kDa), Carbonic anhydrase (29 kDa), the Soybean tripsin inhibitor (20.1 kDa), Lysozyme (14.4 kDa) and Aprotinin (6.5 kDa), obtained from Sigma Aldrich. Protein estimation was done by Lowry’s method with bovine serum albumin as a standard (Lowry et al. 1951).
Study of optimum Temperature and pH for β-galactosidase
The activity of the β-galactosidase at different temperatures was determined. Therefore, the ONPG and lactose assay was performed using different incubation temperatures: 20, 25, 30, 37, 40, 45, 50, 55, and 60 °C. For each temperature, a blank sample was tested. For the determination of the pH optimum 3 different buffers were prepared [Acetic acid buffer, pH 4.5, 5.0, 5.5, 6.0 Sodium phosphate buffer, pH 6.5, 7.0, 7.5 Glycine buffer, pH 8.0, 8.5, 9.0]. In total 10 different pH values were tested to find the optimum pH for the β-galactosidase. All enzyme assays were performed in triplicate and reported as mean values ± SD.
Study of β-galactosidase stability as function of temperature and pH
For the determination of temperature stability, the purified enzyme was incubated in 50 mM sodium phosphate buffer (pH 6.5) at different temperatures (4 °C, 20 °C, 30 °C, 37 °C and 40 °C). The activity was measured using the ONPG standard assay before and after storage. For the determination of the pH stability the enzyme was stored at 37 °C in the same buffers as used for the pH optimum: Acetic acid buffer (pH 4.5, 5.0, 5.5, 6.0), Sodium phosphate buffer (pH 6.5, 7.0, 7.5) and Glycine buffer (pH 8.0, 8.5, 9.0). Activity measurements using the ONPG standard assay were done for 8 h. All enzyme assays were performed in triplicate and reported as mean values ± SD.
Effect of metal ions and reagents on β-galactosidase activity
The purified enzyme samples were incubated with 1 mM, 10 mM and 100 mM of various metal ions such as Na+, K+, Urea, EDTA, Mg2+ and Mn2+ for 10 min. Enzyme activity without metal ions was used as a control. β-galactosidase activity was measured using ONPG as substrate.
Effect of different carbohydrates on β-galactosidase activity
The purified enzyme samples were incubated with 10 mM of various carbohydrates such as Glucose, Galactose, Maltose, Lactose, Sucrose, Inositol and Fructose for 10 min. Enzyme activity without carbohydrate was used as a control. β-galactosidase activity was measured using ONPG as substrate.
Thermal deactivation kinetics and estimation of the deactivation energy
To investigate the thermal deactivation kinetics of the β-galactosidase, the purified enzyme was incubated at different temperatures (35, 37, 40, 45, 50, and 55 °C) in the absence of substrate. Aliquots were withdrawn at periodic intervals and cooled in an ice bath prior to the assay. The residual activity was expressed as a percentage of the initial activity. From a semi-natural logarithmic plot of the residual activity versus time, the deactivation rate constants (Kd) were calculated, and the half-lives estimated using Eq. 1. The half-life t1/2 is defined as the time taken for the residual activity to reach 50 %.
| 1 |
The temperature dependence of Kd was analyzed using the Arrhenius plot (Shuler and Kargi 2005). The deactivation energy was calculated from the Arrhenius equation as
| 2 |
The value for the deactivation energy (Ed) was estimated from the slope of the plot of ln (Kd) against 1/T
Estimation of the thermodynamic parameters
The enthalpy of inactivation (∆H) for each temperature was calculated according to Eq. (3)
| 3 |
The values for the Gibb’s free energy (∆G) of inactivation at different temperatures were calculated from the first-order constant of inactivation process by using Eq. (4)
| 4 |
From Eqs. (3) and (4) the entropy of inactivation (∆S) of β-galactosidase was calculated from Eq. (5)
| 5 |
Determination of kinetic parameters
A Lineweaver–Burk (1/V versus 1/S) plot (Shuler and Kargi 2005) was used in order to determine the kinetic constants (Vmax and Km), where different concentrations of ONPG (1–22 mM) at pH 6.5 and lactose (1–600 mM) at pH 6.5 were used as the substrate.
| 6 |
With a slope of Km/Vm and intercept of 1/Vm (Eq. (6)), Km and Vm values were determined.
Lactose hydrolysis
Milk lactose was prepared according to the methodology given by (Dwevedi et al. 2009). Defatted milk powder (10 %) was dissolved in double distilled water, to avoid turbidity of the solution for uninterrupted spectroscopic analysis. The reaction was carried out by adding 50 U enzymes in 5 mL of prepared milk at 37 ± 2 °C. Aliquots of 20 μL were withdrawn at regular time intervals and released glucose content was estimated using GOD–POD method. Percentage of unhydrolyzed lactose was calculated as,
A plot was generated with log % unhydrolyzed lactose versus time, and the rate constant of lactose hydrolysis was determined from the slope of the plot using the formula:
where ‘x’ is unhydrolyzed lactose. Therefore, the time required for 50 % lactose hydrolysis could be calculated as
Results and discussion
Screening for β-galactosidase secreting bacterial strains
Thirty one lactic acid bacterial strains were isolated from idli batter, milk, rice wine, and curd and screened for production of β-galactosidase by β-galactosidase activity assay (X Gal). Only one strain from curd showed positive for screening and named as HF571129, was selected for further studies (Fig. 1a and b).
Fig. 1.
a Pure culture of L.plantarum HF571129, b Green coloured colonies on X-gal plate
Biochemical characterization of the strain HF571129
Identification of bacterial isolate HF571129 was carried out according to a great variety of morphological, cultural, physiological and biochemical features. The isolate HF571129 had a rod shape (pairs or chains) and was Gram-positive, negative for catalase and nitrate reductase -negative. The organism was able to grow over a wide range of pH from 4.0 to 9.0 and temperature range from 15 to 45 °C. It did not produce CO2 from glucose. Biochemical identifications showed that the strain HF571129 cells could grow on glucose, lactose, raffinose, dextrose, fructose, galactose, and cellobiose (Table 1). However, they could not grow on Xylose. Biochemical and physiological studies indicated that the strain belongs to the Lactobacillus genus.
Table 1.
Morphological and biochemical characteristics of L. plantarum HF571129
| Features | Specifications | Outcomes |
|---|---|---|
| Appearance | Cell shape | Rods |
| Cell arrangement | pairs or chains | |
| Gram staining | + | |
| Biochemical reactions | Catalase | − |
| Nitrate reductase | − | |
| Gas production from glucose | − | |
| Growth at Temperature (°C) | 15 37 45 |
+ ++ + |
| Growth at pH | 4 7 9 |
W- ++ W+ |
| Growth at NaCl (%) | 2 4 6 |
+ + − |
| Carbohydrate utilization | Lactose | + |
| Xylose | − | |
| Mannose | + | |
| Fructose | + | |
| Dextrose | + | |
| Galactose | + | |
| Raffinose | + | |
| Cellobiose | + |
+, positive; −, negative; W+, weakly positive; W-, weakly negative; ++, more positive
Molecular characterization
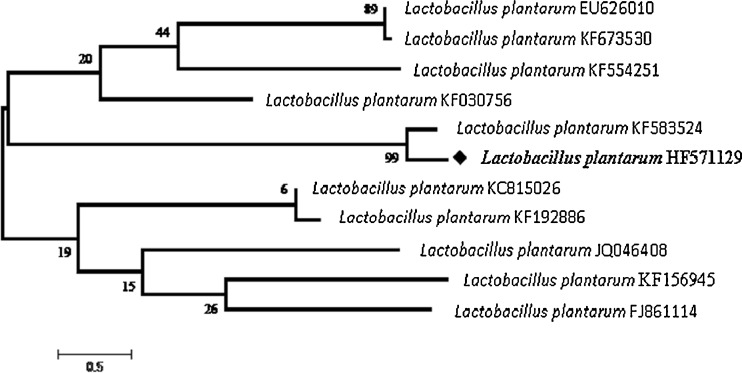
Molecular techniques were used to prove and further confirm the identification of the isolate HF571129 to its species level. 16S rRNA phylogeny studies were primarily carried out. Sequencing results of 16S rRNA were exported to the database and checked for homology alignment. Based on the alignment results HF571129 was found as Lactobacillus plantarum. The 16S rRNA sequence (1494 nucleotides) of HF571129 strain has been submitted to EMBL database, and the accession number was HF571129. The 16S rRNA analysis of the isolate showed 99 % similarity with L.plantarum. Figure 2 shows the phylogenetic tree based on 16S rRNA sequences of strains.
Fig. 2.
Phylogenetic relationships based on 16S r RNA sequences of curd isolates. The neighbor-joining method cladogram showing a phylogenetic relationship based on the 16 s rRNA gene sequence analysis. The numbers at branching point refer to bootstrap values based on 1000 re-samplings. The branch lengths in the cladogram are not to scale
Purification of the β-Galactosidase from the bacterium HF571129
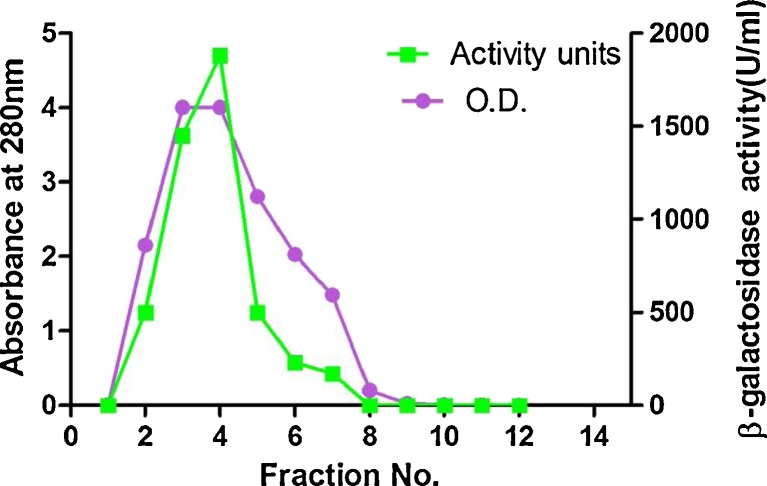
The intracellular β-galactosidase from the bacterium HF571129 was produced and purified according to the procedure described in section “Materials and methods”. Purification of β-galactosidase from the culture medium of L.plantarum was achieved in two steps which include Ultrafiltration and Sephadex G-100 gel filtration chromatography. The enzyme eluted in the form of a single peak (Fig. 3) and most of the maximum activity was confined between the 3rd and 5th fractions. Summary of the purification of β-galactosidase from L.plantarum is presented in Table 2. At the final step of purification, the purified β-galactosidase yielded 7.23-fold, specific activity of 143.7 IU/mg protein and removed 88.8 % of the original protein content in the medium. Similarly, the specific activity and yield of β-galactosidase from L. reuteri L103 was found to be 158 units/mg & 7 % respectively (Nguyen et al. 2006). Whereas there are reports with showed very less enzyme activity of 22.7 U/mL from L. casei subsp. rhamnosus (Gheytanchi et al. 2010).
Fig. 3.
Enzyme elution pattern and enzyme activity in gel permeation column for purified β-galactosidase
Table 2.
Summary of purification of the β-galactosidase from L. plantarum HF571129
| Purification steps | Volume (ml) | Total enzyme activity(IU) | Total protein (mg) | Specific activity(IUmg−1) | Purification factor | Yield |
|---|---|---|---|---|---|---|
| Crude extract (after sonication) | 30 | 3360 | 63 | 53.33 | 1 | 100 |
| Ultrafiltration | 10 | 930.5 | 16.2 | 57.438 | 2.695 | 83.08 |
| Sephadex G100 | 1 | 34.07 | 0.237 | 143.755 | 7.234 | 30.41 |
Molecular mass determination
The molecular weight of the purified enzyme (Sephadex G100, fractions) was determined by 12 % (w/v) SDS polyacrlymide gel under denaturing condition and showed two isoforms of approximately 60 kDa and 42 kDa with CBB staining solution (Fig. 4, Lane 2) The molecular mass of β-galactosidase was calculated to be 102 kDa by SDS-PAGE. Similar results were obtained by L.plantarum WCFS1 with molecular weight of 107 kDa (Iqbal et al. 2010). β-galactosidase from Lactobacilli often shows a dimeric structure, like the one from L. pentosus KUB-ST10-1 with the molecular mass of 105 kDa (Maischberger et al. 2010). β-galactosidases from L. plantarum (Kleerebezem et al. 2003) are heterodimer which is encoded by two genes, lacL and lacM. These two genes belong to LacLM type has been classified as members of glycoside family GH2 in the CAZy (Carbohydrate Active Enzymes) databank. Similarly in the present research, after SDS-PAGE analysis, two bands were obtained at 60 kDa and 42 kDa which is more evident that obtained β-galactosidase is a heterodimer with 102 kDa.
Fig. 4.
SDS-PAGE of purified β -galactosidase from Lactobacillus plantarum. The enzyme was electrophoresed through 12 % acrylamide gel and stained with Commassie Brilliant Blue G-250. Lane 1, crude extract; Lane 2, ultra filtration; Lane 3, gel permeation fraction 4; Lane 4, gel permeation fraction 5; Lane 5, protein molecular mass marker (20 kDa to 205 kDa)
Temperature and pH optimum of β-galactosidase from L. plantarum HF571129
The effect of temperature on HF571129 β-galactosidase was studied by assaying the β galactosidase activity at different temperatures. The enzyme was active in a wide range of temperatures ranging from 20 to 60 °C, with optimum value around 50 °C for both ONPG, as well as lactose, (Fig. 5a). Purified β-galactosidase activity was found to be very active at 50 °C, beyond which there was a rapid decline in activity This optimum was similar to that of β-galactosidase enzymes from the L.acidophillus R22 (Nguyen et al. 2007) and L.amylophilus GV6 (Mahalakshmi et al. 2013). The activity of β-galactosidase was investigated using both ONPG & lactose as substrate over a wide pH range of 4.5–9.0 (Fig. 5b). Results indicated that this enzyme presents an optimal activity at pH 6.5 (ONPG) and pH 7.5 (Lactose). The activity with lactose shows a narrow optimum (pH 6.5–7.5), whereas, for ONPG, it shows a broader optimum (pH 5.5–7.0). This optimum pH was in accordance with several earlier reports for other mesophilic β-galactosidase such as L.acidophillus at pH 6 (Pan et al. 2010), L.plantarum WCFS1 at pH 7.0 & 7.5 for lactose and ONPG (Iqbal et al. 2010).
Fig. 5.
a Temperature and pH optimum (b) of β-galactosidase from L. plantarum for ONPG and lactose as the substrate. c pH stability and d temperature stability of β-galactosidase from L. plantarum HF571129. Bars indicate the standard deviation from triplicate determinations
Effect of pH and temperature on enzyme stability
A pH stability study is an essential part of any enzyme characterization before it can be exploited commercially The purified β galactosidase was incubated at 37 °C for various pH values, and its stability was found to be maximum at pH 8 (Fig. 5c). The β-galactosidase was not stable in acetic acid buffer at any pH value that was tested, and was not stable at the highest tested value (pH 9.0) in glycerine buffer. The β-galactosidase was also found to be fairly stable at pH (Sodium phosphate buffer) 6.5 and 7.5: 60.82 and 68.65 % of the activity was still present after 2 h of incubation, respectively. The activity decreased rapidly at pH 4.5 and pH 5.5, with relative activities of 16.5 and 24.25 %, respectively
At the lower temperatures (4 °C and 20 °C) the enzyme was quite stable and did not lose much of its activity even after 5 h (Fig. 5d). Activity was lost rapidly after 4 h when stored at 40 °C. At 30 °C, a constant level of activity was reached even after 4 h incubation. At higher temperature stability was lost to 51 % at 50 °C after 2 h of incubation. The β-galactosidase could, therefore, be assumed as stable enzyme. β-galactosidase activity at higher temperatures was lost due to unfolding and loss of the active site (Haider and Husain 2007). There are only few reports on thermal stability of β-galactosidase enzyme from L. pentosus KUB-ST10-1 (Maischberger et al. 2010).
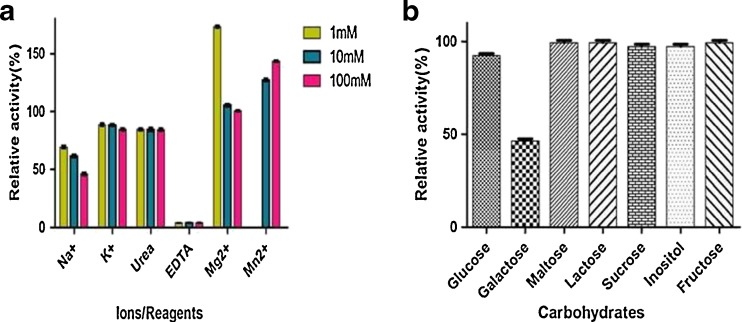
Effect of metal ions and reagents on β-galactosidase activity
Maximum activation of β-galactosidase was observed by the addition of MgSO4 (1 mM). β-galactosidase activity was enhanced to 173 %. Similar effect was observed by the addition of 100 mM MnSO4 (143 %). Na+ ions reduced the activity to 69.3 % when added in a concentration of 1 mM (Fig. 6a), K+ by 88.5 % (1 mM and10 mM). Only EDTA reduced the activity at all tested concentrations to 3.9 % (100, 10 and 1 mM). This study is consistent with other β-galactosidase from other microbial sources. The enzyme activity was strongly activated on divalent ions such as Mg2+ and Mn2+, whereas, for monovalent ions such as K+ and Na+, the enzyme activity was reduced. EDTA strongly inactivates the enzyme even at very low concentration (Iqbal et al. 2010). However, the chelating agent EDTA inactivated the enzyme, suggesting that the β-galactosidase of strain HF571129 is a metalloenzyme, Mg2+ or Mn2+ instead of K+ could be essential for the activity of this metalloenzyme. The β-galactosidase activity from Bacillus sp MTCC 3088 was inhibited by most of the metal ions with the exception of Mg2+ (Chakraborti et al. 2000).
Fig. 6.
Effect of metal ions (a) on β-galactosidase from L. plantarum HF571129. Effect of different carbohydrates (b) on β-galactosidase from L. plantarum HF571129
Effect of different carbohydrates on β-Galactosidase activity
Glucose, Lactose, Maltose, Sucrose, Inositol, and Fructose showed a negligible inhibitory effect on ONPG hydrolysis activity (Fig. 6b), whereas with respect to galactose; it was found to significantly inhibit the activity to 47 %. End product inhibition (galactose) has been reported previously for the β-galactosidase from Bacillus sp MTCC 3088 (Chakraborti et al. 2000) where the enzyme activity was strongly inhibited (68 %) in the presence of galactose. This reveals the product inhibition was very less when compared to the earlier reports
Determination of kinetic parameters
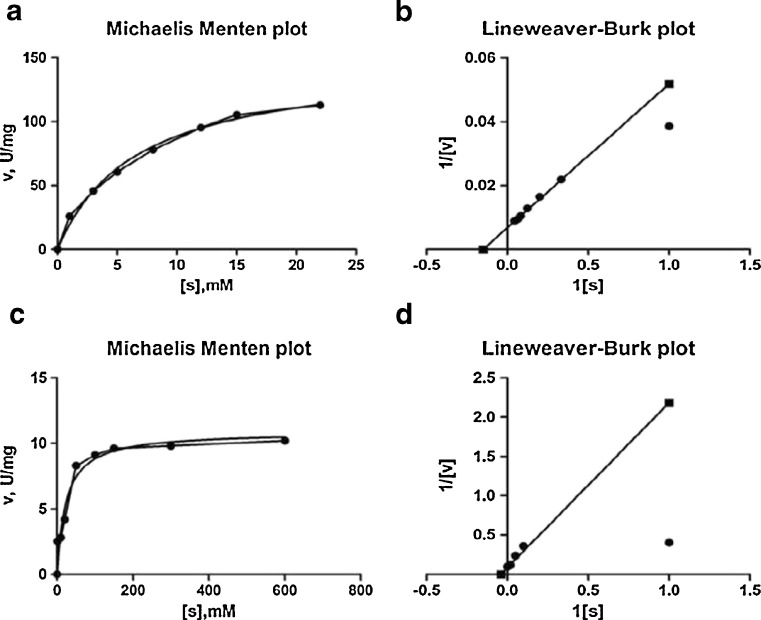
Kinetic parameters like maximum reaction velocity (Vmax) and kinetic constant (Km) were determined for purified β-galactosidase with respect to its artificial and natural substrates ONPG and lactose, respectively, by Lineweaver–Burk plots. The kinetic constant (Km) measured for ONPG was 6.644 mM and Vmax was found to be 147.5 μmol min−1 mg−1 (Fig. 7a and b). The Km and Vmax values of purified β-galactosidase activity towards lactose were 23.28 mM and 10.88 μmol min−1 mg−1, respectively (Fig. 7c and d). The values of Km indicate that the β-galactosidase had a relatively lower affinity for lactose than for the ONPG. The turn over number (Kcat) and second order rate constant (Kcat/Km) were 60.24 s−1 and 9.06 respectively, for ONPG, while 64.02 s−1 and 2.75 respectively for lactose (Table 3).
Fig. 7.
a Michaelis Menten plot and b Lineweaver- Burk plot of β-galactosidase from L. plantarum HF571129 for ONPG. c Michaelis Menten plot and d Lineweaver- Burk plot of β-galactosidase from L. plantarum HF571129 for lactose
Table 3.
Kinetic parameters for β galactosidase from L. Plantarum HF571129 for the hydrolysis of ONPG and lactose
| Substrate | Vmax (μmol min-1 mg-1) | Km (mM) | kcat (s−1) | kcat / Km (mM-1min-1) |
|---|---|---|---|---|
| ONPG | 147.5 | 6.644 | 60.24 | 9.06 |
| Lactose | 10.88 | 23.28 | 64.02 | 2.75 |
The Km value of 23.28 mM determined for lactose is analogous with the reported values for other β-galactosidases such as 29 mM for L. plantarum WCFS1 (Iqbal et al. 2010), 31 mM for L. reuteri L461 (Nguyen et al. 2006) and 14 mM for L. crispatus (Kim and Rajagopal 2000). The Km value of 6.64 mM estimated for ONPG from L. plantarum β-galactosidase compares furthermore very favourably with the reported values for other β-galactosidases 2.3 mM for K. marxianus ATCC 16045; 3.3 mM for K. marxianus CCT 7082 (Braga et al. 2013) and 6.34 mM for Bacillus Sp. MTCC 3088 (Chakraborti et al. 2000).
The Km value of β-galactosidases from L. plantarum HF571129 is lower than that of L. plantarum WCFS1 (Iqbal et al. 2010), L. reuteri L461 (Nguyen et al. 2006); this means that the β-galactosidase from L. plantarum HF571129 possesses considerably more lactose hydrolysing activity compared with the activity of β-galactosidase from many other Lactobacillus strains. These features make L. plantarum strain HF571129 an attractive β-galactosidase producer for potential applications in the dairy industry.
Thermal deactivation kinetics and estimation of the deactivation energy
The thermal inactivation studies are crucial towards understanding the relationship between structure and function of enzymes. The extent of deactivation is measured by the deactivation rate. The deactivation rate is proportional to the active enzyme concentration, and Kd (deactivation rate constant) is the proportional constant. Figure 8a and b shows linear inactivation curves in Arrhenius plots for heat induced inactivation of β-galactosidase at 35–55 °C temperature range. First-order inactivation kinetic model fitted these curves based on which the deactivation rate constant was calculated. Inactivation rate constants (Kd) of β-galactosidase, presented in (Tables 4 and 5) were calculated from the slope of the semilogarithmic plot of residual activity versus time. Likewise, half-life values estimated using these constants and Eq. (1) are presented in (Table 5). The half-life of β-galactosidase at 35 °C was 1.35, 10.71, 19.24 and 45.67 times higher than the half-life values at temperatures of 40, 45, 50 and 55 °C, respectively. This issue reveals the thermal stability of this enzyme at 35 °C and its easy inactivation at higher temperatures. Comparison of half-life of the enzymes at 35 °C revealed that the purified β-galactosidase was more stable than crude one. With increasing temperature, the t1/2 and D-value decreased, and the first order thermal deactivation rate constants (Kd) increased. From the results, it is clear that the enzyme is less thermostable at higher temperatures as a higher rate constant means the enzyme is less thermostable.
Fig. 8.
a Thermal inactivation of β -galactosidase at 35 °C, 40 °C, 45 °C, 50 °C, 55 °C; b Arrhenius plot of the inactivation rate constants for β -galactosidase. c Rate of Lactose unhydrolyzed, d Lactose hydrolysis by β -galactosidase enzyme in milk at 37 °C. Bars indicate the standard deviation from triplicate determinations
Table 4.
Kinetic parameters for the thermal inactivation of the crude β-galactosidases from L.plantarum HF571129
| Temperature (K) | Kd (min−1) | R2 % | t1/2 (min) | ∆G(kJ/mol) | ∆H(kJ/mol) | ∆S(kJ/mol.K) | D(min) |
|---|---|---|---|---|---|---|---|
| 308 | 0.00813 | 82.58 | 82.25 | 87.88 | 136.70 | 0.158 | 283.2 |
| 313 | 0.02667 | 98.77 | 26.65 | 86.26 | 136.66 | 0.160 | 86.33 |
| 318 | 0.0873 | 92.5 | 7.96 | 84.54 | 136.62 | 0.163 | 26.37 |
| 323 | 0.1161 | 94.89 | 5.97 | 85.14 | 136.57 | 0.159 | 19.83 |
| 328 | 0.2873 | 85.17 | 2.41 | 84.03 | 136.53 | 0.159 | 8.014 |
Table 5.
Kinetic parameters for the thermal inactivation of the purified β-galactosidases from L.plantarum HF571129
| Temperature (K) | Kd (min−1) | R2 % | t1/2 (min) | ∆G(kJ/mol) | ∆H(kJ/mol) | ∆S(kJ/mol.K) | D(min) |
|---|---|---|---|---|---|---|---|
| 308 | 0.0056 | 86.27 | 123.77 | 77.041 | 171.37 | 0.306 | 411.17 |
| 313 | 0.0076 | 98.88 | 91.20 | 89.529 | 171.33 | 0.261 | 302.97 |
| 318 | 0.0600 | 87.05 | 11.55 | 85.535 | 171.29 | 0.269 | 38.37 |
| 323 | 0.1077 | 94.39 | 6.43 | 85.349 | 171.25 | 0.265 | 21.379 |
| 328 | 0.2557 | 85.97 | 2.71 | 84.353 | 171.20 | 0.264 | 9.005 |
Inactivation energies (Ed) of crude and purified β-galactosidase were found to be 33.28, 41.57 kcal mol−1, respectively. These values are in the range (40–70 kcal mol−1) estimated for many microbial enzymes (Shuler and Kargi 2005).
Estimation of the thermodynamic parameters
Thermodynamic parameters viz. enthalpy of activation (∆H), the entropy of activation (∆S) and Gibbs free energy of activation (∆G) measured during the enzymatic substrate hydrolysis process provide a relative idea about the functionality of the enzyme toward the specific substrate (Maisuria et al. 2010). With the increase in temperature, a meagre decrease in the value of ∆G takes place for both crude and purified enzymes. Braga et al. reported similar findings; with ∆G values of 94.3–109.75 kJ/mol for the β-galactosidases produced by Kluyveromyces marxianus ATCC 16045 cultures (Braga et al. 2013). The plots of residual activity versus incubation time for the enzyme were linear, with R2>86 %, in the temperature range 35–55 °C indicating that the inactivation could be expressed as first order kinetics.
Determination of lactose hydrolysis
In order to assess whether β-galactosidases is suitable for removal of lactose from milk, the lactose hydrolysis was studied. The ability of the enzyme towards continuous lactose hydrolysis was examined by incubating the enzyme with milk (defatted) at 37 ± 2 °C. Rate of lactose hydrolysis and t1/2 was determined from a plot of log % residual activity versus time (Fig. 8c and d). Rate of lactose hydrolysis was found to be 0.0111 min−1 whereas t1/2 was calculated to be 62.4 min for milk. The hydrolysis of lactose increased up to 50 min of reaction time, at which point approximately 88 % of the lactose had been converted
Conclusion
From the present study, it is evident that potential L. plantarum HF571129 possessing β-galactosidase property could be isolated from the curd samples. Current results obtained, revealed us to understand the enzyme stability and its potential characteristic properties of β-galactosidase from L. plantarum HF571129. Half-life time of purified β-galactosidases was 123.77 min at 35 °C. Moreover, the enzyme was found to be metalloenzyme and enhanced with metal ions (Mg2+ or Mn2+). With these promising results obtained, it may also be considered as an alternative to the commercial strain, A. niger. The stability of the β-galactosidases appears to be an attractive feature for its use in industrial applications. Some salient characteristic of the β-galactosidases from L. plantarum HF571129 are; higher stability for larger duration values at high temperature, enhanced activity in the presence of Mg2+ or Mn2+ metal ions, very less percentage of end product inhibition, lactose hydrolysis in the short period of time. Therefore, the β-galactosidase produced by L. plantarum HF571129 should be a novel one. Conclusively, the work furnished here is novel β-galactosidases from the new strain L. plantarum which revealed interesting properties, as well as considerable economic values for applications in the processes of lactose hydrolysis.
Acknowledgments
The authors would like to thank VIT University, Vellore, India for supporting and performing the study. ES is obliged to the management of VIT for partial completion of doctoral works.
Conflict of interest
The authors declare that they have no conflict of interest.
Abbreviation
- ∆H
Enthalpy
- ∆G
Gibbs free energy
- ∆S
Entropy
- R
Universal gas constant
- Kd
Deactivation rate constants
- Vmax
Maximum reaction velocity
- Km
Michaelis–Menten constants
- Kcat
Turn over number
- Kcat/Km
Second order rate constant
- T
Absolute temperature
- κ
Boltzmann’s constant
Half-life time
References
- Akolkar SK, Sajgure AD, Lele SS. β -Galactosidase from Lactobacillus acidophilus isolated from fermented ragi. Indian J Biotechnol. 2006;5:184–188. [Google Scholar]
- Bergey DH, Harrison FC, Breed RS, Hammer BW, Huntoon FM, editors. Bergey’s manual of determinative bacteriology. 1. Baltimore: The Williams & Wilkins Co; 1923. pp. 1–442. [Google Scholar]
- Braga ARC, Manera AP, Ores JC, Sala L. Kinetics and thermal properties of crude and purified β-Galactosidase with potential for the production of Galacto oligosaccharides. Food Technol Biotechnol. 2013;51:45–52. [Google Scholar]
- Chakraborti S, Sani RK, Banerjee UC, Sobti RC. Purification and characterization of a novel β -galactosidase from Bacillus sp MTCC 3088. J Ind Microbiol Biotechnol. 2000;24:58–63. doi: 10.1038/sj.jim.2900770. [DOI] [Google Scholar]
- Choonia HS, Lele SS. β -Galactosidase release kinetics during ultrasonic disruption of Lactobacillus acidophilus isolated from fermented Eleusine coracana. Food Bio Prod Process. 2011;89:288–293. doi: 10.1016/j.fbp.2010.08.009. [DOI] [Google Scholar]
- Di Lauro B, Strazzulli A, Perugino G, La Cara F, Bedini E, Corsaro MM, Rossi M, Moracci M. Isolation and characterization of a new family 42 β-galactosidase from the thermoacidophilic bacterium Alicyclobacillus acidocaldarius: identification of the active site residues. Biochim Biophys Acta. 2008;1784:292–301. doi: 10.1016/j.bbapap.2007.10.013. [DOI] [PubMed] [Google Scholar]
- Dickson RC, Markin JS. Physiological studies of β-galactosidase induction in Kluveromyces lactis. J Bacteriol. 1980;142:777–785. doi: 10.1128/jb.142.3.777-785.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwevedi A, Singh AK, Singh DP, Srivastava ON, Kayastha AM. Lactose nano-probe optimized using response surface methodology. Biosens Bioelectron. 2009;25:784–790. doi: 10.1016/j.bios.2009.08.029. [DOI] [PubMed] [Google Scholar]
- Gheytanchi E, Heshmati F, Shargh BK, Nowroozi J. Study on β -galactosidase enzyme produced by isolated lactobacilli from milk and cheese. Afr J Microbiol Res. 2010;4:454–458. [Google Scholar]
- Haider T, Husain Q. Preparation of lactose free milk by using salt fractionated almond (Amygadalus communis) β-galactosidase. J Sci Food Agric. 2007;87:1278–1283. doi: 10.1002/jsfa.2840. [DOI] [Google Scholar]
- Hettwer DJ, Wang HY. Protein release from chemically permeabilized Escherichia coli. In: Asenjo JA, Hong J, editors. Separation, recovery and purification in biotechnology—recent advances and mathematical modeling, ACS Symposium series 314. Washington, DC: American Chemical Society; 1990. pp. 2–8. [Google Scholar]
- Hubber RE, Gupta MN, Khare SK. The active site and mechanism of the β-galactosidase from Escherichia coli. Int J Biochem. 1994;26:309–318. doi: 10.1016/0020-711X(94)90051-5. [DOI] [PubMed] [Google Scholar]
- Hung MN, Xia Z, Hu NT, Lee BH. Molecular and biochemical analysis of two β-galactosidases from Bifidobacterium infantis HL96. Appl Environ Microbiol. 2001;67:4256–4263. doi: 10.1128/AEM.67.9.4256-4263.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal S, Nguyen T, Nguyen TT, Maischberger T. β -Galactosidase from Lactobacillus plantarum WCFS1: biochemical characterization and formation of prebiotic galacto-oligosaccharides. Carbohydr Res. 2010;345:1408–1416. doi: 10.1016/j.carres.2010.03.028. [DOI] [PubMed] [Google Scholar]
- Joly M. Physico-chemical approach to the denaturation of proteins. New York: Academic; 1965. [Google Scholar]
- Kim JW, Rajagopal SN. Isolation and characterization of β-galactosidase from Lactobacillus crispatus. Folia Microbiol. 2000;45:29–34. doi: 10.1007/BF02817446. [DOI] [PubMed] [Google Scholar]
- Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc Natl Acad Sci U S A. 2003;100:1990–1995. doi: 10.1073/pnas.0337704100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Mahalakshmi S, Kiran KK, Hameeda B, Gopal R. Fermentative production of lactase from Lactobacillus amylophilus GV6. J Sci Ind Res. 2013;72:548–552. [Google Scholar]
- Maischberger T, Leitner E, Nitisinprasert S, Juajun O. β Galactosidase from Lactobacillus pentosus: purification, characterization and formation of galacto oligosaccharides. Biotechnol J. 2010;5:838–847. doi: 10.1002/biot.201000126. [DOI] [PubMed] [Google Scholar]
- Maisuria VB, Patel VA, Nerurkar AS. Biochemical and thermal stabilization parameters of polygalacturonase from Erwinia carotovora subsp. carotovora BR1. J Microbiol Biotechnol. 2010;20:1077–1085. doi: 10.4014/jmb.0908.08008. [DOI] [PubMed] [Google Scholar]
- Neri DFM, Balcao VM, Carneiro MG, Carvalino LB, Teixeira JA. Immobilization of β-galactosidase from Kluyveromyces lactis onto a polysiloxane-polyvinyl alcohol magnetic (mPOS-PVA) composite for lactose hydrolysis. Catal Commun. 2008;9:2334–2339. doi: 10.1016/j.catcom.2008.05.022. [DOI] [Google Scholar]
- Nguyen TH, Splechtna B, Steinböck M, Kneifel W, Lettner HP, Kulbe KD, Haltrich D. Homodimeric β-galactosidase from Lactobacillus delbrueckii subsp. bulgaricus DSM 20081: expression in Lactobacillus plantarum and Biochemical Characterization. J Agric Food Chem. 2006;54:4989–4998. doi: 10.1021/jf053126u. [DOI] [PubMed] [Google Scholar]
- Nguyen TH, Splechtna B, Krasteva S, Kneifel W, Kulbe KD, Divne C, Haltrich D. Characterization and molecular cloning of a heterodimeric beta-galactosidase from the probiotic strain Lactobacillus acidophilus R22. FEMS Microbiol Lett. 2007;269:136–144. doi: 10.1111/j.1574-6968.2006.00614.x. [DOI] [PubMed] [Google Scholar]
- Pan Q, Zhu J, Liu L, Cong Y, Hu F, Li J. Functional identification of a putative beta-galactosidase gene in the special lac gene cluster of Lactobacillus acidophilus. Curr Microbiol. 2010;60:172–178. doi: 10.1007/s00284-009-9521-9. [DOI] [PubMed] [Google Scholar]
- Richmond ML, Gray JI, Stine CM. Beta-galactosidase: review research related to technological application, nutritional concerns, and immobilization. J Dairy Sci. 1981;64:1759–1771. doi: 10.3168/jds.S0022-0302(81)82764-6. [DOI] [Google Scholar]
- Sadana A. Biocatalysis: fundamentals of deactivation kinetics. Englewood Cliffs: Prentice-Hall; 1995. [Google Scholar]
- Shuler ML, Kargi F (eds) (2005) Bioprocess Engineering-Basic Concepts. Prentice Hall of India Pvt. Ltd
- Ustok FI, Tari C, Harsa S. Biochemical and thermal properties of β-galactosidase enzymes produced by artisanal yoghurt cultures. Food Chem. 2010;119:1114–1120. doi: 10.1016/j.foodchem.2009.08.022. [DOI] [Google Scholar]
- Vinderola CG, Reinheimer JA. Lactic acid starter and probiotic bacteria, a comparative “in vitro” study of probiotic characteristics and biological barrier resistance. J Food Res Int. 2003;36:895–904. doi: 10.1016/S0963-9969(03)00098-X. [DOI] [Google Scholar]
- Vrese M, Stegelmann A, Richter B, Fenselau S, Laue C, Schrezenmeir J. Probiotics—compensation for lactase insufficiency. Am J Clin Nutr. 2001;73:421–429. doi: 10.1093/ajcn/73.2.421s. [DOI] [PubMed] [Google Scholar]