Abstract
Differentiated 3T3-L1 adipocytes are a widely used in vitro model of white adipocytes. In addition to classical white and brown adipocytes that are derived from different cell lineages, beige adipocytes have also been identified, which have characteristics of both white and brown adipocytes. Here we show that 3T3-L1 adipocytes display features of multiple adipocytes lineages. While the gene expression profile and basal bioenergetics of 3T3-L1 adipocytes was typical of white adipocytes, they responded acutely to catecholamines by increasing oxygen consumption in an UCP1-dependent manner, and by increasing the expression of genes enriched in brown but not beige adipocytes. Chronic exposure to catecholamines exacerbated this phenotype. However, a beige adipocyte differentiation procedure did not induce a beige adipocyte phenotype in 3T3-L1 fibroblasts. These multiple lineage features should be considered when interpreting data from experiments utilizing 3T3-L1 adipocytes.
Keywords: 3T3-L1 adipocyte, beige adipocyte, norepinephrine, UCP-1, uncoupling
Introduction
Differentiation of multi-potential 3T3-L1 fibroblasts into lipid laden adipocyte-like cells is one of the most common in vitro models used in the study of adipocyte biology. This model has been critical in advancing our understanding of adipogenesis, lipid metabolism and the actions of hormones. 3T3-L1 cells were isolated and expanded from Swiss 3T3 cells based on their ability to accumulate lipid.1 Differentiation of this preadipocyte cell line into mature adipocytes involves treatment with a number of pro-differentiation agents after growth arrest, including agents such as insulin,1 synthetic glucocorticoids such as dexamethasone2 and the phosphodiesterase inhibitor 1-methyl-3-isobutyl xanthine (IBMX).3 The most common differentiation protocol used throughout the literature employs a combination of all 3 agents together.4 In this protocol, cells undergo cell cycle arrest, before being exposed to differentiation media containing insulin, dexamethasone and IBMX for 2 d. Cells are then exposed to insulin only for a further 2 days, before being place back into normal growth media.4 An early response to differentiation media exposure is up regulation of genes that drive the adipogenic program, including C/EBP and PPAR isoforms.5 Turning on the adipogenic gene expression program increases glucose uptake and triglyceride synthesis and cells first start to show the obvious signs of lipid accumulation ∼4 d after the first exposure to differentiation medium.2,4,6
In recent years, adipocyte biology has been transformed by the recognition that white and brown adipocytes are derived from different cell lineages. Brown adipocytes are thought to originate from a myogenic cell lineage that expresses the myogenic factor 5 (Myf5) protein, while white adipocytes arise from non-myogenic (Myf5 negative) cell lineages.7,8 More recently, a sub-population of white adipocytes has been identified with some characteristics resembling those of brown fat. These cells, termed beige adipocytes, are not derived from a myf-5 positive lineage, but show typical phenotypic characteristics of brown adipocytes in that they increase uncoupling protein 1 (UCP1)-mediated uncoupled respiration in response to acute stimulation with hormones that increase cAMP, such as catecholamines.9 This occurs despite the fact that they have lower basal uncoupled respiration than classical brown adipocytes and have a gene expression signature that is distinct from brown adipocytes.9 Chronic exposure of beige adipocytes to these same neuroendocrine signals that increase cAMP also induces a multilocular appearance, similar to brown adipocytes.9
To our knowledge, no studies have examined the phenotypic characteristics of 3T3-L1 adipocytes and it is unclear whether 3T3-L1 adipocytes assume exclusive white adipocyte characteristics. This is particularly important to establish given that agents that increase cAMP concentrations similar to neuroendocrine signals that promote beige adipocyte formation, such as the phosphodiesterase inhibitor IBMX and synthetic glucocorticoid dexamethasone,10,11 are routinely used to differentiate 3T3-L1 fibroblasts.4 Should 3T3-L1 adipocytes assume some beige cell characteristics, this would have wide ranging implications for the use of this cell model and interpretation of data arising from experiments employing their use. Therefore, in this study we undertook a comprehensive phenotypic investigation of 3T3-L1 adipocytes to better understand their physiology and suitability as an experimental model of white adipocytes.
Results
Differentiation of 3T3-L1 fibroblasts into adipocytes alters cellular bioenergetics and increases uncoupled respiration
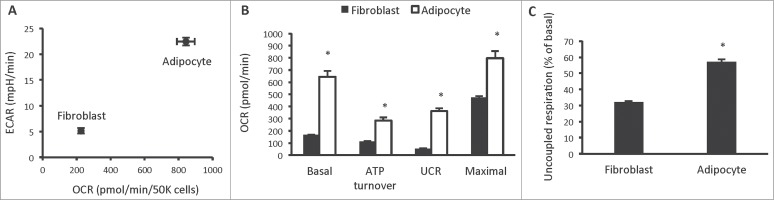
The first phenotypic investigation we undertook was assessment of cellular bioenergetics in 3T3-L1 fibroblasts and adipocytes, using the Seahorse XF analyzer. Differentiation of 3T3-L1 fibroblasts into adipocytes using a commonly used differentiation protocol, increased both aerobic and anaerobic flux, measured by oxygen consumption rate (OCR) and extracellular acidification rate (ECAR), a proxy measure of glycolysis, respectively (Fig. 1A). Mitochondrial function analysis revealed that 3T3-L1 adipocytes increased all parameters of mitochondrial respiration, including basal mitochondrial respiration, respiration due to ATP turnover, uncoupled respiration (UCR) and maximal respiratory capacity (Fig. 1B). In particular, uncoupled respiration was markedly increased in 3T3-L1 adipocytes. In fibroblasts, uncoupled respiration constituted 32% of basal mitochondrial respiration, whereas in adipocytes, it constituted 57% (Fig. 1C). This marked increase in uncoupled respiration is consistent with that observed in primary white adipocyte cell lines.9
Figure 1.
Cellular bioenergetics and mitochondrial function in 3T3-L1 fibroblasts and adipocytes. (A) Cellular bioenergetics measured by cellular oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) in 3T3-L1 fibroblasts and adipocytes. (B) Mitochondrial function represented by basal mitochondrial respiration (basal), respiration due to ATP turnover, uncoupled respiration (UCR) and maximal respiratory capacity in 3T3-L1 fibroblasts and adipocytes. (C) Uncoupled respiration as a percentage of basal mitochondrial respiration in 3T3-L1 fibroblasts and adipocytes. Data are represented as mean ± SEM, n = 10 biological replicates per group. † Denotes significantly different from fibroblasts for both OCR and ECAR. * Denotes significantly different from fibroblasts.
Differentiated 3T3-L1 adipocytes have a gene expression profile most similar to white adipocytes
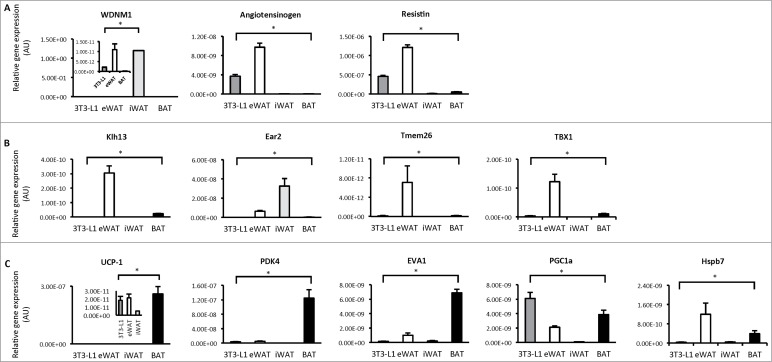
We next profiled the expression of genes that best distinguish white, beige and brown adipocytes9 in differentiated 3T3-L1 adipocytes, epipdidymal white adipose tissue (eWAT), inguinal white adipose tissue (iWAT) and brown adipose tissue (BAT) from C57BL6 mice. There was a high level of diversity in gene expression profiles between these groups, such that all groups were significantly different from each other, even eWAT and iWAT. 3T3-L1 adipocytes expressed white specific genes such as WDNM1, angiotensinogen and resistin (Fig. 2A). Beige specific genes such as klh13, ear2, Tmem and Tbx1 were highest in eWAT and iWAT and were almost undetectable in 3T3-L1 adipocytes. Similarly, the expression of BAT specific genes such as UCP1, PDK4 and eva1 were almost undetected in 3T3-L1 adipocytes and were also low in WAT, but enriched in BAT as expected (Fig. 2C). 3T3-L1 adipocytes had the highest PGC-1α expression of all groups (Fig. 2C). These data show that differentiated 3T3-L1 adipocytes do not show evidence of beige or brown fat characteristics as assessed by expression profiling of genes typically enriched in these adipocyte types.
Figure 2.
Gene expression profiling in 3T3-L1 adipocytes, white and brown adipose tissue. (A) Expression of genes that distinguish white adipocytes in 3T3-L1 adipocytes, epididymal white adipose tissue (eWAT), inguinal white adipose tissue (iWAT) and brown adipose tissue (BAT; subscapular brown fat). (B) Expression of genes that distinguish beige adipocytes in 3T3-L1 adipocytes, WAT and BAT. (C) Expression of genes that distinguish brown adipocytes in 3T3-L1 adipocytes, WAT and BAT. Data are represented as mean ± SEM, n = 6 biological replicates/individual mouse samples per group. * Denotes significantly different from all other groups.
3T3-L1 adipocytes increase oxygen consumption and the expression of brown adipocyte genes in response to norepinephrine
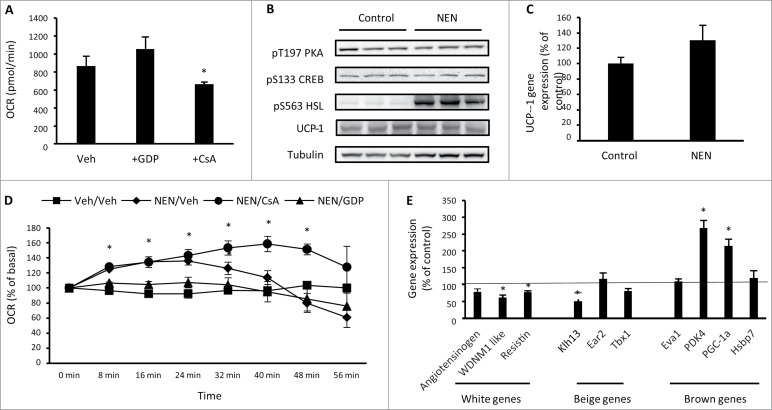
In brown and beige adipocytes, catecholamines are key stimulators of uncoupled respiration through activation of PKA, which rapidly increases UCP1 transcription via phosphorylation of CREB at S133.12 Simultaneously, PKA increases lipolysis through phosphorylation of lipases such as hormone sensitive lipase (HSL) and adipose triglyceride lipase (ATGL) so that fatty acids can serve as a substrate for uncoupled respiration.13 Catecholamines also increase uncoupled respiration in white adipocytes through the generation of fatty acids by lipolysis that opens the mitochondrial transition pore.14 We next assessed these mechanisms in differentiated 3T3-L1 adipocytes and first sought to determine the mechanisms contributing to basal uncoupled respiration in these cells. Treatment of 3T3-L1 adipocytes with the UCP1 inhibitor GDP15 for 24 hr had no effect on basal oxygen consumption, while cyclosporine A (CsA), an inhibitor of mitochondrial transition pore opening,16 reduced basal oxygen consumption by ∼25%, when compared with vehicle treated cells (Fig. 3A). We next assessed whether 3T3-L1 adipocytes respond to an acute exposure of the catecholamine norepinephrine through either of these mechanisms. Exposure to norepinephrine for 20 min had no effect on T197 PKA or S133 CREB phosphorylation, but did increase phosphorylation of HSL at S563 (Fig. 3B). There was no effect of norepinephrine on UCP1 protein (Fig. 3B) or gene expression (Fig. 3C). Oxygen consumption significantly increased in response to norepinephrine that persisted for ∼30 min after exposure when compared with vehicle treated cells (Fig. 3D). The increase in oxygen consumption was not sensitive to oligomycin (data not shown), suggesting that an increase in uncoupled respiration is driving this response. Pre-treatment of cells with GDP was sufficient to prevent the norepinephrine-mediated increased in oxygen consumption, while pre-treatment with CsA sustained the increase in oxygen consumption in response to norepinephrine for longer than vehicle treated cells (Fig. 3D). These data show that mitochondrial transition pore opening contributes to basal oxygen consumption in 3T3-L1 adipocytes, while UCP1 contributes to the increase in oxygen consumption in response to norepinephrine, without changes in UCP1 expression. This is not observed in primary white adipocytes14 and suggests that the acute response of 3T3-L1 adipocytes to catecholamines is more similar to beige and brown adipocytes than white adipocytes. To determine whether acute norepinephrine exposure resulted in gene expression changes that could precede beige or brown phenotypic transformation of 3T3-L1 adipocytes, the expression of white, beige and brown gene markers was assessed, 60 min after norepinephrine exposure. There was a decrease in the WAT enriched genes angiotensinogen and WDNM1 and the beige enriched gene Klh13 (Fig. 3E). In contrast, there was an increase in the BAT enriched genes PDK4 and PGC-1α. This suggests that 3T3-L1 adipocytes could undergo functional lineage transformation toward a brown adipocyte phenotype in response to chronic norepinephrine exposure.
Figure 3.
3T3-L1 adipocytes increase oxygen consumption and the expression of brown adipocyte genes in response to norepinephrine. (A) Basal cellular oxygen consumption rate (OCR) in 3T3-L1 adipocytes treated with 100 μM GDP (UCP1 inhibitor) or 5 μM CsA (inhibitor of mitochondrial transition pore opening) for 24 hrs. (B) Phosphorylation of PKA T197, CREB S133, HSL S563 and total UCP-1 and tubulin in 3T3-L1 adipocytes exposed to vehicle (H2O) or norepinephrine (1 μM) for 20 min. (C) UCP1 gene expression in 3T3-L1 adipocytes following exposure to vehicle (H2O) or norepinephrine (1 μM) for 60 min. (D) OCR immediately following acute exposure to vehicle (H2O) or norepinephrine (1 μM) in 3T3-L1 adipocytes that had been previously treated with vehicle (0.1% DMSO), 100μM GDP or 5 μM CsA for 24 hrs. (E) Expression of genes that discriminate white, beige and brown adipocytes in 3T3-L1 adipocytes treated with norepinephrine (1 μM) for 60 min relative to control (vehicle) treated cells. Data are represented as mean ± SEM, n = 3 −6 biological replicates per group. * Denotes significantly different from vehicle treated cells.
Chronic norepinephrine exposure increases basal oxygen consumption in 3T3-L1 adipocytes
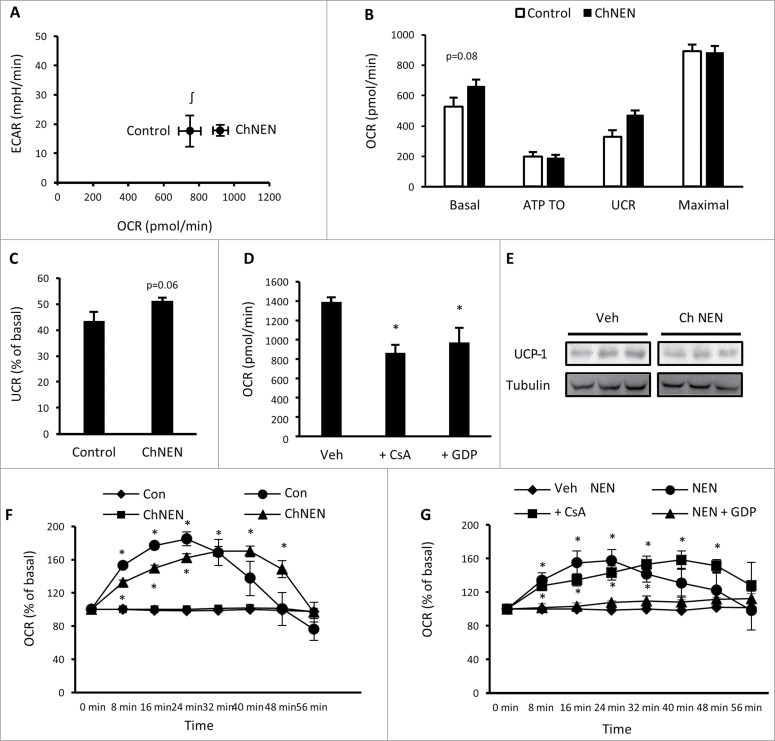
To determine whether prolonged catecholamine exposure can alter the characteristics of 3T3-L1 adipocytes more toward a brown adipocyte phenotype, these cells were continuously treated with norepinephrine or vehicle for the final 3 d of the differentiation protocol, after cells were placed back into growth media. Norepinephrine was removed from media 2 hr prior to experiments. Assessment of basal bioenergetics showed that chronic norepinephrine (ChNEN) increased basal oxygen consumption (Fig. 4A), which was due to an increase in basal uncoupled respiration (Fig. 4B and C). Basal glycolytic rate was unaltered (Fig. 4A). We examined the mechanisms contributing to basal uncoupled respiration in 3T3-L1 adipocytes following ChNEN exposure through treatment with GDP and CsA, and found that both of these compounds reduced basal respiration (Fig. 4D). This contrasts to 3T3-L1 adipocytes that were not exposed to norepinephrine (Fig. 3A) and suggests that UCP1- dependent uncoupled respiration is contributing to the increase in oxygen consumption following ChNEN exposure. However, there was no increase in UCP1 protein (Fig. 4E). We next assessed responses to acute norepinephrine exposure. 3T3-L1 adipocytes that were treated with ChNEN had slower oxygen consumption kinetics than untreated adipocytes (Fig. 4F). Similar to our previous analyses in untreated adipocytes (Fig. 3D), the increase in oxygen consumption in ChNEN treated cells was inhibited by GDP, but not CsA (Fig. 4G), suggesting that UCP1 was still the primary contributor to the increase in oxygen consumption by norepinephrine. Together with our previous gene expression data, these analyses show that chronic exposure to catecholamines induced phenotypic effects similar to brown adipocytes, signified by increased basal oxygen consumption in 3T3-L1 adipocytes through UCP1.
Figure 4.
Chronic norepinephrine exposure in 3T3-L1 adipocytes. (A) Cellular bioenergetics measured by cellular oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) in 3T3-L1 adipocytes treated with vehicle (H2O; Control) or 1 μM norepinephrine for 48 hrs (ChNEN). (B) Mitochondrial function represented by basal mitochondrial respiration (basal), respiration due to ATP turnover, uncoupled respiration (UCR) and maximal respiratory capacity in Control and ChNEN 3T3-L1 L1 adipocytes. (C) Uncoupled respiration as a percentage of basal mitochondrial respiration in in Control and ChNEN 3T3-L1 adipocytes. (D) Basal cellular oxygen consumption rate (OCR) in Control and ChNEN 3T3-L1 adipocytes that were co-treated with vehicle (0.1% DMSO), 100 μM GDP or 5 μM CsA in the final 24 hrs. (E) Total UCP-1 and tubulin protein in Control and ChNEN 3T3-L1 L1 adipocytes. (F) OCR immediately following acute exposure to vehicle (H2O) or norepinephrine (1 μM) in Control and ChNEN 3T3-L1 adipocytes. (G) OCR immediately following acute exposure to vehicle (H2O) or norepinephrine (1 μM) in Control and ChNEN 3T3-L1 adipocytes that were co-treated with vehicle (0.1% DMSO), 100 μM GDP or 5 μM CsA in the final 24 hrs. Data are represented as mean ± SEM, n = 6 biological replicates per group. ∫ Denotes significantly different from Control cells for OCR. * Denotes significantly different from vehicle treated and control cells.
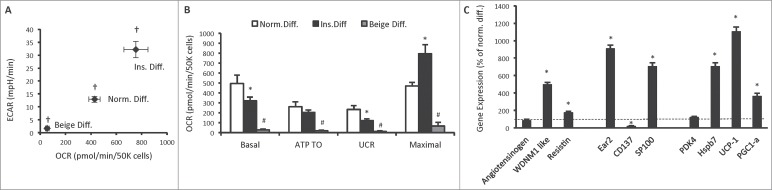
Alternate differentiation protocols alter the bioenergetics and gene expression profile of 3T3-L1 adipocytes
The most common 3T3-L1 adipogenic differentiation protocol in the literature utilises the phosphodiesterase antagonists IBMX and dexamethasone to increase cAMP levels.4 Therefore these compounds have similar effects to catecholamines. We reasoned that exclusion of IBMX and dexamethasone from the adipogenic protocol could alter the phenotypic traits of 3T3-L1 adipocytes. Similarly, a differentiation cocktail containing PPARγ agonists and COX inhibitors have been used to differentiate stromal vascular fraction (SVF) pre-adipocytes into beige adipocytes.17 To determine the effect of these agents on 3T3-L1 adipocyte differentiation, we compared 3T3-L1 adipocytes that were differentiated using the normal differentiation protocol (Norm. diff.) with 3T3-L1 adipocytes that were differentiated using insulin only throughout the differentiation protocol (Ins. diff.) and with 3T3-L1 adipocytes that were differentiated with a beige adipocyte protocol (Beige diff.).17 Adipocytes that underwent Ins. diff. did not accumulate lipid as readily as Norm. diff. and Beige diff. adipocytes (data not shown). Bioenergetically, Ins. diff. adipocytes had lower basal oxygen consumption and glycolytic rates, which were reduced further again with Beige diff. (Fig. 5A). The reduction in basal oxygen consumption with Ins. diff. was primarily due to a reduction in uncoupled respiration (Fig. 5B). Ins. diff. adipocytes had markedly different gene expression profiles of WAT, beige and BAT enriched genes. The WAT enriched genes WDNM1 like and resistin were increased, as were the beige enriched genes Ear2 and SP100 (Fig. 5C). The BAT enriched genes Hspb7, UCP1 and PGC-1α were all increased in Ins. diff. adipocytes. The expression of almost all genes was undetectable in Beige diff. cells (data not shown). These data show that the absence of enhanced cAMP signaling throughout the differentiation procedure, while having divergent effects on the expression of genes used to classify distinct adipocyte lineages, also reduces uncoupled respiration, which is a key feature of all adipocytes, Therefore, these data show that cAMP signaling is essential for adipogenesis and together with chronic norepinephrine data suggests the existence of a cAMP signaling continuum that determines the phenotype of 3T3-L1 adipocytes, while a differentiation protocol that induces beige adipocyte formation in SVF pre-adipocytes appeared to differentiate 3T3-L1 fibroblasts into an unrecognisable cell type.
Figure 5.
Bioenergetics and gene expression in 3T3-L1 adipocytes following differentiation without phosphodiesterase inhibitors. (A) Cellular bioenergetics measured by cellular oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) in 3T3-L1 cells following normal adipogenic differentiation (Norm. diff.), insulin only differentiation (Ins. diff.), or beige adipocyte differentiation (Beige diff.) (B) Mitochondrial function represented by basal mitochondrial respiration (basal), respiration due to ATP turnover, uncoupled respiration (UCR) and maximal respiratory capacity in Norm. diff., Ins. diff. and Beige diff. 3T3-L1 adipocytes. (C) Expression of genes that discriminate white, beige and brown adipocytes in Norm. diff. and Ins. diff. 3T3-L1 adipocytes. Data are represented as mean ± SEM, n = 6 −10 biological replicates per group. † Denotes significantly different from Ins. diff. cells for both OCR and ECAR. * Denotes significantly different from Norm. diff. # Denotes significantly different from Norm. diff and Ins. diff.
Discussion
The study of 3T3-L1 adipocytes has contributed greatly to our understanding of adipogenesis, adipocyte metabolism and hormone action. The realization of distinct adipocyte lineages more recently prompted us to characterize the lineage characteristics of 3T3-L1 adipocytes, using the most common differentiation protocol found in the literature. Our findings show that 3T3-L1 adipocytes display gene expression profiles and basal bioenergetics consistent with white adipocytes. However, upon acute stimulation with the catecholamine norepinephrine, 3T3-L1 adipocytes increased uncoupled respiration through a mechanism similar to beige and brown adipocytes, which is UCP1-dependent. This response occurred, despite limited expression of genes that distinguish beige and brown adipocytes, including relatively low UCP-1 expression when compared with BAT. Furthermore, acute exposure to norepinephrine induced the expression of brown, but not beige, adipocyte enriched genes. Notably, there was no increase in UCP1 expression. Chronic exposure to norepinephrine further transitioned 3T3-L1 adipocytes toward a brown adipocyte phenotype, characterized by increased basal UCP1-dependent oxygen consumption. These findings show that differentiated 3T3-L1 adipocytes display aspects of multiple adipocyte lineages, which should be considered when employing their use in studies of adipocyte metabolism.
The enhanced oxygen consumption in response to catecholamine stimulation in primary white adipocytes is due to PKA/HSL/ATGL-mediated lipolysis that increases intracellular fatty acids, which open the mitochondrial transition pore to increase mitochondrial proton leak, driving uncoupled respiration.14 We observed distinct differences in the 3T3-L1 adipocyte response to norepinephrine stimulation. This included an increase in HSL activation and an increase in oxygen consumption that was UCP1-dependent, but not dependent on opening of the mitochondrial transition pore. The underlying mechanism of this response is also distinct from that seen in brown adipocytes, which includes transcriptional activation of UCP1 via PKA and CREB activation.12 Indeed, we did not observe activation of this signaling and transcriptional response at all in 3T3-L1 adipocytes. Given that HSL phosphorylation was robustly increased in response to norepinephrine, it is possible that the increase in UCP1 mediated oxygen consumption was mediated entirely by lipolysis driven alterations in fatty acid mobilisation. That there was no engagement of mitochondrial transition pore-mediated uncoupled respiration will also require further investigation, but could be due to this mechanism being maximally engaged in the basal state. Indeed, we found that inhibition of the mitochondrial transition pore significantly reduced basal oxygen consumption in 3T3-L1 adipocytes. Furthermore, a modified differentiation protocol deficient in agents that increase cAMP where cells did not accumulate substantial lipid drastically reduced uncoupled respiration, highlighting the important role of lipid availability in increasing uncoupled respiration.
The increase in brown, but not beige, adipocyte gene expression and assumption of basal bioenergetics that are typical of brown adipocytes, characterized by a high basal uncoupled respiration,9 following chronic norepinephrine exposure is an unusual phenotypic transformation. Indeed, white and brown adipocytes are thought to be derived from different cell lineages7,8 and therefore transformation between these cell types should not be possible. However, recent lineage tracing experiments suggest that the notion that white and brown adipocytes originate from distinct cell lineages (Myf5 negative and positive, respectively) is overly simplistic. These studies suggest that white adipocytes that do not express brown adipocyte markers can also express Myf5 and the proportion of these cells in any given white adipose depot is dependent on its location and other functional characteristics, and is also dynamic in nature.18,19 Furthermore, it has been shown that PDGFRα+ stem cells can differentiate into both white and brown adipocytes upon adrenergic stimulation.20 This suggests that multi-potential pre-adipocytes exist and that the traditional uni-potential lineage view of adipogenesis does not account for the emerging complexity in adipocyte phenotypic determination. These findings along with our own in the present study suggest that it is possible that the 3T3-L1 fibroblast pool, which was originally isolated for its potential to accumulate lipid,21 could contain adipocyte precursors with multiple lineage characteristics.
In conclusion, our findings show that phenotypic profiling of 3T3-L1 adipocytes shows that they possess characteristics typical of both white and brown adipocytes. This should be considered and examined when using 3T3-L1 adipocytes as model systems to study adipocyte differentiation and metabolism.
Materials and Methods
Cell culture
Mouse immortalised 3T3-L1 fibroblasts were cultured in 10% CO2 at 37°C in growth media consisting of DMEM (4.5g/L glucose; Invitrogen), 10% heat-inactivated foetal bovine serum (HI-FBS; Thermo Scientific) and antibiotics (100units/mL penicillin and 100µg/mL streptomycin; Life Technologies). Cells were induced to differentiate 2 d after reaching confluence (day 0), by supplementing growth media with 3nM insulin (Humulin R; Eli Lilly), 0.25 µM dexamethasone (Sigma-Aldrich) and 0.5mM 1-methyl-3-isobutyl-xanthine (Sigma-Aldrich). From day 3 until day 7, cells were maintained in growth media supplemented with 3 nM insulin after which the mature adipocytes were maintained in growth media. For chronic norepinephrine treatment, adipocytes were incubated in 1 μM norepinephrine (Sigma-Aldrich) from day 8 to 9. For differentiation experiments without cAMP agonists, cells were maintained in 3nM insulin only until day 7. For beige differentiation experiments, cells were treated as previously described.17 Adipogenesis was monitored by oil-red O staining, as previously described.22 For acute norepinephrine treatments, cells were exposed to 1μM norepinephrine or vehicle (H2O) for 20 min for signaling analyses, or for 60 min for gene expression analyses.
Animals
All experimental procedures were approved by the Deakin University Animal Welfare Committee, which is subject to the Australian Code for the Responsible Conduct of Research. Male C57Bl6 mice (7/8 weeks old) were obtained from the Animal Resource Center (WA) and were housed in a temperature (22°C) and humidity controlled environment with a 12:12-h light/dark cycle, with food and water provided ad libitum. Mice were killed at 0900 and their epididymal and subscapular brown fat pads were rapidly excised and stored at −80°C for later analysis.
Bioenergetics assessment
3T3-L1 fibroblasts were seeded in Seahorse (Seahorse Bioscience) plates at a density of 50,000 cells per well, before being differentiated to adipocytes as outlined above. Prior to all assays, cell media was changed to unbuffered DMEM (DMEM base medium supplemented with 25 mM glucose, 1mM sodium pyruvate, 1mM GlutaMax, pH 7.4) and incubated at 37°C in a non-CO2 incubator for 60 min. Cellular bioenergetics, mitochondrial function assays and parameter calculations were performed as previously described23 using the Seahorse XF24 analyzer. For oxygen consumption responses to norepinephrine, 4 basal measurement periods were performed prior to injection of norepinephrine (1μM final) or vehicle (H2O) and a subsequent 7 measurements were obtained. For studies with GDP and CsA, 3T3-L1 adipocytes were incubated with 100 μM GDP (Sigma-Aldrich), 5 μM CsA (Sigma-Aldrich) or vehicle (0.1% DMSO) for 24 hrs prior to assay. GDP, CsA or vehicle was also added to assay media.
Gene expression
3T3-L1 adipocytes, epididymal, inguinal and brown subscapular fat pads from male C57BL6 mice were homogenized in Trizol and total RNA isolated using RNeasy columns (Qiagen). RNA was reverse transcribed to cDNA using the SuperScript III First-Strand Synthesis System (Life Technologies) in a GeneAmp PCR System 9700 thermal cycler (Applied Biosystems). cDNA was quantitated using Quant-iT™ OliGreen® ssDNA Reagent and Kit (Life Technologies). Gene expression was analyzed by quantitative PCR (qPCR) using Brilliant SYBR master mix (Stratagene) on the MX3005P QPCR system (Stratagene). The PCR conditions were: 95°C for 10 min (1 cycle); 95°C for 30 s and 60°C for 1 min (40 cycles). Primer sequences used in the study are shown in Table 1. Relative gene expression was calculated from mean CTs from triplicate samples that were power transformed out of their logarithmic format, before being divided by sample cDNA content. Primers were designed using Beacon Designer software (PREMIER Biosoft International) and were synthesized by Geneworks.
Table 1.
Real time RT-PCR primer sequences
| Gene | Forward (5'−3') | Reverse (5'−3') |
|---|---|---|
| Angiotensinogen | GCAGGAGAGGAGGAACAG | AGATGGCGAACAGGAAGG |
| WDNM1 | CCTGGGCTCTGTCTAACC | CATCGTTCATCACAAGTTCC |
| Resistin | CTCCCTGTTTCCAAATGC | TGTCCAGTCTATCCTTGC |
| EAR2 | CCTGTAACCCCAGAACTCCA | CAGATGAGCAAAGGTGCAAA |
| CD137 | CGTGCAGAACTCCTGTGATAAC | GTCCACCTATGCTGGAGAAGG |
| Sp100 | TGATGGAGGGAACCCAAACTC | CTTCCTTGAGAATAGCTGGCAC |
| PDK4 | TGTGATGTGGTAGCAGTAGTC | ATGTGGTGAAGGTGTGAAGG |
| HSBP7 | GATGCCTACGAGTTTACAGTGGAC | ATGTGGTTGTTGAAGGTGGTGAC |
| UCP-1 | GATGGTGAACCCGACAACTT | CTGAAACTCCGGCTGAGAAG |
| PGC-1α | CCCTGCCATTGTTAAGACC | TGCTGCTGTTCCTGTTTTC |
Western blotting
20µg of total protein from 3T3-L1 adipocytes was subjected to SDS-PAGE and transferred to PVDF membranes, before being blocked in 1% bovine serum albumin in Tris buffered saline with 0.05% Tween-20 (TBST) for 60 min at room temp. Membranes were exposed to primary antibodies to pT197 PKA, pS133 CREB, pS565 HSL (Cell Signaling Technology), UCP-1 (Abcam) and α-tubulin (Sigma Aldrich), overnight on a rocker at 4°C on separate membranes. Membranes were washed with TBST and exposed to appropriate species HRP conjugated secondary antibodies for 45 min at room temp. Membranes were washed again with TBST before being exposed to enhanced chemiluminescence substrate (Life Technologies). Images were captured using the BioRad Chemidoc (Hercules, USA) and bands were quantified using BioRad Quantity One software.
Statistical analysis
All data are expressed as mean ± SEM and were assessed for normality using SPSS software. Normally distributed data were analyzed by unpaired t-test or one-way ANOVA as appropriate, using Minitab statistical software. Non-normally distributed data were analyzed by Kruskal-Wallis one-way ANOVA non-parametric tests using SPSS software. Statistical significant differences were identified where p < 0.05.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This research was supported by the Deakin University Molecular and Medical Research Strategic Research Center. SLM is supported by a Career Development Fellowship from the NHMRC (1030474).
References
- 1.Green H, Meuth M. An established pre-adipose cell line and its differentiation in culture. Cell 1974; 3:127-33; PMID:4426090; http://dx.doi.org/ 10.1016/0092-8674(74)90116-0. [DOI] [PubMed] [Google Scholar]
- 2.Rubin CS, Hirsch A, Fung C, Rosen OM. Development of hormone receptors and hormonal responsiveness in vitro. Insulin receptors and insulin sensitivity in the preadipocyte and adipocyte forms of 3T3-L1 cells. J Biol Chem 1978; 253:7570-8; PMID:81205. [PubMed] [Google Scholar]
- 3.Russell TR, Ho R. Conversion of 3T3 fibroblasts into adipose cells: triggering of differentiation by prostaglandin F2alpha and 1-methyl-3-isobutyl xanthine. Proc Natl Acad Sci U S A 1976; 73:4516-20; PMID:188043; http://dx.doi.org/ 10.1073/pnas.73.12.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zebisch K, Voigt V, Wabitsch M, Brandsch M. Protocol for effective differentiation of 3T3-L1 cells to adipocytes. Anal Biochem 2012; 425:88-90; PMID:22425542; http://dx.doi.org/ 10.1016/j.ab.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 5.MacDougald OA, Lane MD. Transcriptional regulation of gene expression during adipocyte differentiation. Annu Rev Biochem 1995; 64:345-73; PMID:7574486; http://dx.doi.org/ 10.1146/annurev.bi.64.070195.002021. [DOI] [PubMed] [Google Scholar]
- 6.Mackall JC, Student AK, Polakis SE, Lane MD. Induction of lipogenesis during differentiation in a “preadipocyte” cell line. J Biol Chem 1976; 251:6462-4; PMID:10298. [PubMed] [Google Scholar]
- 7.Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scimè A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al.. PRDM16 controls a brown fat/skeletal muscle switch. Nature 2008; 454:961-7; PMID:18719582; http://dx.doi.org/ 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Timmons JA, Wennmalm K, Larsson O, Walden TB, Lassmann T, Petrovic N, Hamilton DL, Gimeno RE, Wahlestedt C, Baar K, et al.. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc Natl Acad Sci U S A 2007; 104:4401-6; PMID:17360536; http://dx.doi.org/ 10.1073/pnas.0610615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, Khandekar M, Virtanen KA, Nuutila P, Schaart G, et al.. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 2012; 150:366-76; PMID:22796012; http://dx.doi.org/ 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wollenberger A, Irmler R. Effects of adrenaline and methylisobutylxanthine on adenosine 3':5'-monophosphate levels in cultures of beating heart cells of the newborn rat. Recent Adv Stud Cardiac Struct Metab 1976; 12:689-95; PMID:202003. [PubMed] [Google Scholar]
- 11.Costa M, Manen CA, Russell DH. In vivo activation of cAMP-dependent protein kinase by aminophylline and 1-methyl, 3-isobutylxanthine. Biochem Biophys Res Commun 1975; 65:75-81; PMID:167773; http://dx.doi.org/ 10.1016/S0006-291X(75)80063-5. [DOI] [PubMed] [Google Scholar]
- 12.Collins S, Yehuda-Shnaidman E, Wang H. Positive and negative control of Ucp1 gene transcription and the role of β-adrenergic signaling networks. Int J Obes 2010; 34 Suppl 1:S28-33; PMID:20935662; http://dx.doi.org/ 10.1038/ijo.2010.180. [DOI] [PubMed] [Google Scholar]
- 13.Collins S. β-Adrenoceptor Signaling Networks in Adipocytes for Recruiting Stored Fat and Energy Expenditure. Front Endocrinol 2011; 2:102; PMID:22654837; http://dx.doi.org/ 10.3389/fendo.2011.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yehuda-Shnaidman E, Buehrer B, Pi J, Kumar N, Collins S. Acute stimulation of white adipocyte respiration by PKA-induced lipolysis. Diabetes 2010; 59:2474-83; PMID:20682684; http://dx.doi.org/ 10.2337/db10-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jezek P, Borecky J. Mitochondrial uncoupling protein may participate in futile cycling of pyruvate and other monocarboxylates. Am J Physiol 1998; 275:C496-504; PMID:9688604. [DOI] [PubMed] [Google Scholar]
- 16.Broekemeier KM, Pfeiffer DR. Inhibition of the mitochondrial permeability transition by cyclosporin A during long time frame experiments: relationship between pore opening and the activity of mitochondrial phospholipases. Biochemistry 1995; 34:16440-9; PMID:8845372; http://dx.doi.org/ 10.1021/bi00050a027. [DOI] [PubMed] [Google Scholar]
- 17.Aune UL, Ruiz L, Kajimura S. Isolation and differentiation of stromal vascular cells to beige/brite cells. J Vis Exp 2013; PMID:23568137; http://dx.doi.org/ 10.3791/50191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez-Gurmaches J, Hung CM, Sparks CA, Tang Y, Li H, Guertin DA. PTEN loss in the Myf5 lineage redistributes body fat and reveals subsets of white adipocytes that arise from Myf5 precursors. Cell Metab 2012; 16:348-62; PMID:22940198; http://dx.doi.org/ 10.1016/j.cmet.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanchez-Gurmaches J, Guertin DA. Adipocytes arise from multiple lineages that are heterogeneously and dynamically distributed. Nat Commun 2014; 5:4099; PMID:24942009; http://dx.doi.org/ 10.1038/ncomms5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee YH, Petkova AP, Mottillo EP, Granneman JG. In vivo identification of bipotential adipocyte progenitors recruited by beta3-adrenoceptor activation and high-fat feeding. Cell Metab 2012; 15:480-91; PMID:22482730; http://dx.doi.org/ 10.1016/j.cmet.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green H, Kehinde O. Sublines of mouse 3T3 cells that accumulate lipid. Cell 1974; 1:113-6; http://dx.doi.org/ 10.1016/0092-8674(74)90126-3. [DOI] [Google Scholar]
- 22.Konstantopoulos N, Foletta VC, Segal DH, Shields KA, Sanigorski A, Windmill K, Swinton C, Connor T, Wanyonyi S, Dyer TD, et al.. A gene expression signature for insulin resistance. Physiol Genomics 2011; 43:110-20; PMID:21081660; http://dx.doi.org/ 10.1152/physiolgenomics.00115.2010. [DOI] [PubMed] [Google Scholar]
- 23.Martin SD, Morrison S, Konstantopoulos N, McGee SL. Mitochondrial dysfunction has divergent, cell type-dependent effects on insulin action. Mol Metab 2014; 3:408-18; PMID:24944900; http://dx.doi.org/ 10.1016/j.molmet.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]