Abstract
Eukaryotic regulatory small RNAs (sRNAs) that induce RNA interference (RNAi) are involved in a plethora of biological processes, including host immunity and pathogen virulence. In plants, diverse classes of sRNAs contribute to the regulation of host innate immunity. These immune-regulatory sRNAs operate through distinct RNAi pathways that trigger transcriptional or post-transcriptional gene silencing. Similarly, many pathogen-derived sRNAs also regulate pathogen virulence. Remarkably, the influence of regulatory sRNAs is not limited to the individual organism in which they are generated. It can sometimes extend to interacting species from even different kingdoms. There they trigger gene silencing in the interacting organism, a phenomenon called cross-kingdom RNAi. This is exhibited in advanced pathogens and parasites that produce sRNAs to suppress host immunity. Conversely, in host-induced gene silencing (HIGS), diverse plants are engineered to trigger RNAi against pathogens and pests to confer host resistance. Cross-kingdom RNAi opens up a vastly unexplored area of research on mobile sRNAs in the battlefield between hosts and pathogens.
Introduction
Eukaryotic non-coding small RNAs (sRNAs) are generated by endoribonucleases DICER or DICER-like (DCL) and are loaded into Argonaute (AGO) proteins to induce silencing of genes with complementary sequences. This mechanism is referred to as RNA interference (RNAi). In plants, sRNAs are divided into two subgroups, small interfering RNAs (siRNAs) and microRNAs (miRNAs), based on their precursor structures and biogenesis pathways. Both miRNAs and siRNAs play a pivotal role in regulating and fine-tuning gene expression in diverse cellular processes such as development and growth, genome integrity, epigenetic inheritance, and cellular stress responses, including host immunity [1-4]. Similarly, sRNAs from eukaryotic plant pathogens, pests, and symbionts also play an important regulatory role in developmental processes and pathogenicity [3,5,6]. Remarkably, some sRNAs are mobile signals in plants that transmit gene silencing from cell to cell, or systemically over a long distance [7-10]. Recent attention has been focused on mobile sRNAs that mediate cross-kingdom RNAi in host-pathogen interactions [3,11,12].
Cross-kingdom RNAi is the phenomenon in which gene silencing is induced between unrelated species from different kingdoms, such as a plant host and its interacting microorganism or pest. It requires the translocation of a gene-silencing trigger from a donor into an interacting recipient. Indeed, interaction with other organisms by way of cross-kingdom RNAi has been observed in plant and animal systems [3,11,12]. Cross-kingdom RNAi can occur from the host to the pest/pathogen/parasite/symbiont or vice versa. The most prominent example of cross-kingdom RNAi from a plant to its interacting microorganism is host-induced gene silencing (HIGS), a phenomenon in which a plant-produced RNAi signal triggers silencing of a pathogen gene [13,14]. Conversely, sRNAs produced by pathogens and parasites can also translocate into host cells and trigger gene silencing of host genes [5,11,12,15,16]. Advanced pathogens hijack the host RNAi pathways and suppress host immunity genes to facilitate infection [3,12]. This review focuses on the roles of plant- and pathogen-derived sRNAs in host immunity, and pathogen virulence, respectively.
Plant endogenous sRNAs and sRNA pathway components regulate host innate immunity
In plants, diverse classes of endogenous sRNAs, including miRNAs and siRNAs, are involved in regulating and fine-tuning defense responses against pathogens and pests [3,17-20]. miRNAs and siRNAs are involved in the activation of pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) [21-24] and effector-triggered immunity (ETI) [25-30]. Normally, these sRNAs are either induced or down-regulated upon pathogen attack in order to suppress expression or to release suppression of their targets [20].
Many proteins of sRNA pathways are involved in immune response by manipulating sRNA biogenesis and function, such as DCLs that generate sRNAs, AGOs that execute sRNA-directed target gene suppression, and RNA-dependent RNA polymerase (RDRs) that produce double-stranded sRNA precursors. The Arabidopsis genome encodes 4 DCL proteins. DCL1 is the key protein in miRNA production, and several miRNAs it produces are associated with PTI and ETI against bacterial and fungal pathogens. Consistent with this observation, the Arabidopsis mutants dcl1-9 and dcl1-7 showed enhanced susceptibility toward bacterial [31] and fungal [16] infection. These findings emphasize the notion that miRNAs participate in regulation of immune response. DCL4 is mainly involved in siRNA production and is important in antiviral, antibacterial, and antifungal defense [25,32].
There are 10 AGO proteins in Arabidopsis [33]. Only AGO2 is highly induced by bacterial infection [24], and the ago2-1 mutant is more susceptible to both virulent and avirulent strains of Pseudomonas syringae pv tomato DC3000 (Pst) [24]. Deep sequencing analysis of AGO2-associated sRNAs after immunoprecipitation identified several miRNA*s. miRNA393* is one of the most abundant sRNAs loaded into AGO2, resulting in the suppression of MEMB12 upon infection of Pst (AvrRpt2), and promoting secretion of pathogenesis-related (PR) proteins. Interestingly, the complementary strand of miR393*, miR393, functions through AGO1 to induce antibacterial immune response [34]. This study has demonstrated that miRNA*s, formerly considered non-functional byproducts of miRNAs, can be functional in inducing gene silencing [35]. Similar phenomena have also been observed in animal systems [36,37]. AGO1 generally plays a positive role in plant immunity. The ago1-25 and ago1-27 mutants are hindered in PAMP-perception and in antibacterial immunity [22]. However, ago1 mutants showed enhanced disease resistance against certain fungal pathogens [16,38], indicating a sophisticated role of plant AGO1 protein in plant-fungal interactions, which is discussed in greater detail below. The Arabidopsis genome encodes six RDRs, of which RDR6 is involved in secondary siRNA production. The rdr6 mutant exhibits enhanced susceptibility to fungal pathogens [38] and an avirulent bacterial Pst strain carrying the AvrRpt2 effector gene [26], while the rdr6 mutant exhibits enhanced basal resistance toward a virulent strain of Pst [39,40]. Moreover, mutation in a RDR6 interacting protein SGS3 also enhances susceptibility to Verticillium dahliae [38], suggesting that the sRNA pathway is generally required for antifungal resistance in plants.
Furthermore, heterochromatic siRNAs (hcsiRNAs) direct DNA methylation and/or histone modifications to induce silencing of transposons, repeats and genes at the transcriptional level. This hcsiRNA-mediated so-called RNA-directed DNA methylation (RdDM) pathway also regulates immune responses [41,42]. RdDM mutants display altered disease phenotypes to bacteria or fungal pathogen infection. For instance, the triple mutant of the non-CG loci methyltransferases, drm1-2/drm2-2/cmt3-11 (ddc), the mutant in which the largest subunit of Pol IV being mutated nrpd1a, a chromatin remodeling protein mutant drd-1, and the dcl2/dcl3/dcl4 triple mutant, all show enhanced resistance to Pst [41]. In addition, expression of many of the RdDM pathway genes are down-regulated upon treatment with the bacterial PAMP trigger flg22, supporting the notion that RdDM transcriptionally controls the expression of antibacterial defense genes [42]. Consistent with this, most of these RdDM pathway genes are also repressed during bacterial infection, which leads to demethylation and activation of several defense genes [42]. Furthermore, ROS1, which encodes for a 5-methylcytosine DNA glycosylase that initiates active DNA demethylation, is repressed upon flg22 treatment, and ros1 mutant exhibits enhanced susceptibility to Pst [42], suggesting that active DNA demethylation is part of the regulatory circuit for gene activation in response to pathogen attacks. The ros1/dml2/dml3 (rdd) triple mutant, and the RdDM pathway mutants ago4 and nrpe1 are more susceptible to Fusarium oxysporum [43], and microarray analysis indicates that a much larger group of genes was differentially expressed in the RdDM mutants nrpd1 and nrpe1 than in the rdd mutant. Obviously, RdDM or DNA demethylation re-arranges the transcriptional status of immunity genes. The ago4, drd1, rdr2, drm1 drm2, and nrpd2 mutants showed enhanced susceptibility toward necrotrophic fungal pathogens Botrytis cinerea and Plectospherella cucumerina, which is contrary to the increased resistance against bacterial pathogens [44]. Chromatin immunoprecipitation revealed that the RdDM pathway epigenetically controls salicylic acid–dependent defense responsive genes, which are activated in nrpd2 and other RdDM mutants [41,42], leading to enhanced resistance to bacterial pathogens. However, these mutants compromise jasmonic acid-dependent defense responses and enhance susceptibility to necrotrophs. Furthermore, trans-generational systemic acquired resistance was observed in Arabidopsis, which was dependent on RdDM-mediated hypomethylation at non-CG sites [45].
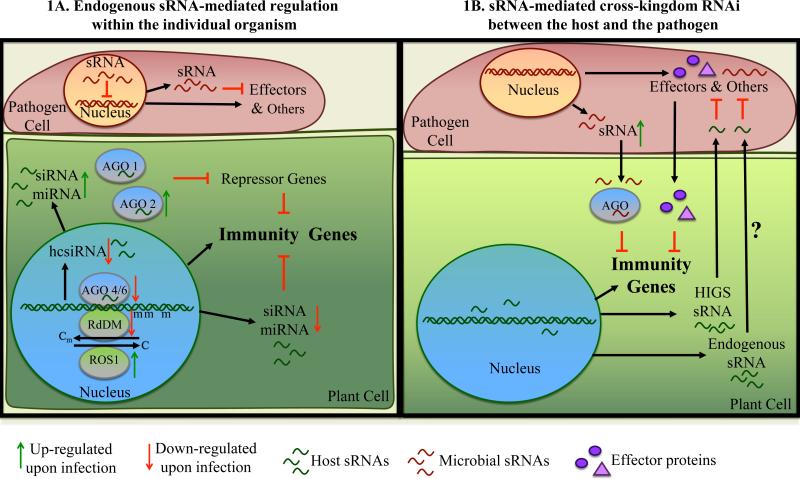
Thus, plant endogenous sRNAs and sRNA pathway components are critical factors in regulating and fine-tuning host immune responses (Figure 1A) against an array of pathogens and pests, including bacteria, fungi, oomycetes, nematodes, and insect pests.
Figure 1.
sRNA-mediated regulation in plant-pathogen interactions
A) Plant and pathogen sRNAs regulate host immunity and pathogen virulence, respectively, within the individual organism via post-transcriptional and transcriptional gene silencing. In plants, miRNAs and siRNAs acting through AGO1 or AGO2 mediate post-transcriptional gene silencing; and transcriptional gene silencing is mediated by hcsiRNAs acting mostly through AGO4 and AGO6 to induce DNA cytosine methylation or histone modifications. In general, AGO2 is up-regulated, and AGO4/AGO6 are down-regulated by pathogen infection.
B) Plant and pathogen sRNAs trigger cross-kingdom RNAi in plant-pathogen interactions. Plant HIGS siRNAs and likely plant endogenous sRNAs target effector genes and other essential genes in pathogens, whereas pathogen sRNAs translate to host cells, and target host immune-responsive genes through the host AGO-RISC.
Microbial sRNAs regulate pathogen virulence
Although plant sRNAs involved in host defense have been extensively studied, relatively little is known about the function of pathogen-derived sRNAs. Recent studies have demonstrated that microbial sRNAs play a regulatory role in pathogen virulence. Both prokaryotic and eukaryotic organisms produce a plethora of non-coding sRNAs; however, the structure of bacterial non-coding sRNAs differs significantly from that of eukaryotic sRNAs.
Bacterial regulatory non-coding sRNAs are heterogeneous in length (50—300 nt), and mostly regulate the translation efficiency and stability of target mRNAs through short and imperfect base pairing. In phytopathogenic bacteria such as Xanthomonas, Agrobacterium, and Pectobacterium, non-coding sRNAs are responsive to stress and may regulate pathogenic development [46-50]. For instance, the non-coding RNAs sX12 (67 nt) and sX13 (105 nt) are important for Xanthomonas campestris pv. vesicatoria pathogenicity, and deletion of these loci attenuates the virulence of the pathogen. HrpX, a key regulator of the type III secretion system (T3SS), induces sX12. Moreover, deletion of sX13 led to reduced accumulation of HrpF, HrcN, and HrcJ, suggesting that both of these non-coding RNAs contribute to virulence by regulating expression of or being regulated by the T3SS components [49,51]. Bacterial non-coding RNAs generally operate through conserved RNA-binding protein complexes, such as Hfq, CsrA/RsmA or CRISPR-Cas to execute gene regulation. It is very likely that such RNA-directed silencing complexes contribute to pathogenesis. Supporting evidence includes the findings that the hfq and rsmA deletion mutants exhibited attenuated virulence [52,53].
Diverse classes of sRNAs contribute to the pathogenesis of various eukaryotic pathogens as well [5,6]. In the rice blast fungus Magnaporthe oryzae, many sRNAs that are derived from tRNAs are enriched in appressoria, an infection-specific organ at the interaction interface [54]. A class of predominantly 24-nt sRNAs that mapped to long terminal repeat (LTR) retrotransposons is up-regulated during invasive growth [55]. The aggressive fungal pathogen B. cinerea can infect more than 200 plant species, including almost all vegetables and fruits. It produces a class of 21 to 22 nt sRNAs that mostly map to LTR retrotransposons. Some of these 21 to 22 nt B. cinerea sRNAs (Bc-sRNAs) are induced during the early stage of the infection process and target host genes from both Arabidopsis and tomato by hijacking host sRNA machinery [16] (Figure 1B). Microbial sRNA-induced host gene silencing is a naturally occurring cross-kingdom RNAi event utilized by aggressive pathogens as a novel virulence strategy. This study has added sRNAs to the list of pathogen effectors, molecules that are secreted and delivered into host cells to suppress host immunity.
A comparative study revealed that the three notorious oomycete Phytophthora plant pathogens, P. infestans, P. sojae, and P. ramorum, produce miRNAs as well as two distinct classes of siRNAs of 21 and 25 nt in length [56]. Several 21 nt siRNAs are generated in antisense orientation to LTR retrotransposon loci. Many RXLR and CRN effector genes, crucial factors of Phytophthora infection and host adaptation, are located in close vicinity (< 2 kb) to LTR retrotransposons. RXLR and CRN effectors are specifically induced during the infection process. Interestingly, siRNAs were found to co-regulate retrotransposons and the surrounding CRN effectors [57,58], which impacts the interaction between the host and the pathogen [59]. How the production of effector gene-related siRNAs is regulated is currently unclear.
In summary, non-coding RNAs of bacterial plant pathogens and diverse classes of sRNAs in fungal and oomycete plant pathogens play pivotal roles in pathogen development, pathogenesis, and host specificity/adaptation (Figure 1A). It is important to understand the evolutionary processes and circumstances that are shaping the repertoires and expression levels of sRNAs in diverse phytopathogens as part of their virulence. Of particular interest is sRNAs that are capable of inducing silencing of host genes, as discussed below.
Cross-kingdom RNAi and sRNA trafficking
Cross-kingdom RNAi has been observed in both animal and plant systems. Plants transfer RNAi signals into interacting organisms, such as filamentous fungi, oomycetes, nematodes, parasitic plants, and pests [14,60,61], to suppress their growth in a process referred to as HIGS, the most prominent example of cross-kingdom RNAi in plants. Similar observations have been made in humans [62]. RNAi signaling in the opposite direction has also been reported. Recent discoveries showed that advanced pathogens and parasites use cross-kingdom RNAi to suppress host immunity for infection, as previously reviewed [3,12]. Such microbial sRNA-induced host gene silencing is not only present in plants as the case of Botrytis-host interaction, but it has also been observed in animal systems [15,63,64]. In addition, sRNAs produced by diverse parasites were observed in the body fluids of infected individuals [15,65-67], suggesting a common strategy of pathogens and parasites to secrete RNAi signals during infection to manipulate host cell immunity.
In plants, B. cinerea delivers some of its sRNAs into the host cells during early infection, and the predicted host targets of these Bc-sRNAs are highly enriched with signaling and regulatory genes. Three Bc-sRNAs are confirmed to suppress Arabidopsis and tomato immunity genes in vivo. Transgenic Arabidopsis lines ectopically expressing Bc-siRNAs show elevated susceptibility toward B. cinerea infection. Those Bc-siRNAs that target host genes share common features to plant miRNAs that are 21-22 nt in length with a 5’ first nucleotide uracil, which are preferentially loaded into Arabidopsis AGO1 for host gene suppression (Figure 1B). Consistent with this result, the ago1-27 mutant, but not ago2-1 or ago4-2, exhibit enhanced resistance toward B. cinerea. Interestingly, ago1-27 also showed enhanced resistance against another fungal pathogen, Verticillium dahliae [38], even though most of the other Arabidopsis sRNA pathway mutants showed enhanced susceptibility. Whether pathogenic Verticillium spp. has evolved similar strategies to hijack host AGO1 to suppress host immunity, has yet to be determined. Furthermore, it is worthwhile to investigate how widespread this strategy is to use sRNAs as effector molecules to suppress host immunity or manipulate host physiology and whether it is present in symbiotic relationships.
Strikingly, recent studies in animal systems indicate that such manipulation strategies via cross-kingdom RNAi have also evolved in animal parasites [15]. The gastrointestinal nematode Heligmosomoides polygyrus and the filarial nematode Litomosoides sigmodontis secrete miRNAs via extracellular vesicles [15], which can be internalized by host cells to suppress host genes efficiently. It is currently unclear whether these miRNAs function through host AGO proteins. Remarkably, the strongest suppressive effect of these parasites’ miRNAs was found on host gene Dusp1, which is targeted by three nematode miRNAs [15]. Dusp1 is an important regulator of MAPK pathways in animal systems. The plant pathogen B. cinerea has evolved to manipulate host MAPKs in Arabidopsis and a MAPKKK in tomato using mobile Bc-sRNAs. MAPK cascades are essential regulatory pathways in plant and animal immune signaling, and it is obvious that members and regulators of MAPK pathways are favorably targeted and manipulated by pathogen and parasite effectors including sRNAs (Figure 1B). Secreted sRNAs of diverse parasites have been identified in various hosts, including miRNAs of Schistosoma japonicum in plasma of infected rabbit [65], tRNA-derived sRNAs of Trypanosoma cruzi in infected susceptible mammalian cells [66], and miRNAs of Onchocerca ochengi and O. volvulus in nodule fluid of cattle, and plasma and serum of infected humans [67], respectively. These findings point to a common strategy of manipulating host immunity through secreted sRNAs.
Clearly, sRNA-guided cross-kingdom RNAi has evolved as an advanced virulence strategy adapted by pathogens and parasites of both plant and animal hosts. Furthermore, cross-kingdom gene silencing has been also found to act in an opposite direction in the case of HIGS (Figure 1B). Scientists have engineered diverse plant species, from model plants to commercial crops, to express exogenous artificial RNAi signals that target mRNAs of parasitic nematodes, herbivores, and fungal and oomycete pathogens for gene suppression, with the ultimate goal to create pest- and pathogen- resistant crops [13,68-70]. HIGS has yielded astonishing effects in enhancing plant resistance against a variety of pathogens and pests [60,61,71], demonstrating that it is an efficient strategy for crop protection. HIGS is also functional against parasitic plants, such as Orobanche and Cuscuta spp. [72]. Incredibly bidirectional transfer of thousands of mRNAs has been documented between the parasitic plant, dodder (Cuscuta pentagona), and two hosts, Arabidopsis and tomato [73-75]. Furthermore, RNA translocation between host and parasite is highly selective, as the profiles of the transferred parasite mRNAs and the total mRNAs within the parasitic plants were rather different. Although not examined, we speculate that sRNAs are also likely to be exchanged between the parasitic plant and its host.
In humans, resistance to the malaria pathogen Plasmodium falciparum is seen in people with sickle-cell anemia disease [76]. A recent study indicates that infected sickle cell erythrocytes overproduce certain miRNAs, which translocate into the Plasmodium cells and bind to P. falciparum PKA-R mRNA and block its translation [62], even though P. falciparum lacks the canonical RNAi pathway. This study demonstrates that animals have also evolved the cross-kingdom gene silencing strategy to suppress virulence of parasites. Examples of pathogen-induced host gene silencing, the cross-kingdom RNAi in the opposite direction, have been recently documented. However, what is the exact form of the mobile silencing signals and how these signals translocate from a host into the pathogen and parasite or vice versa, is still poorly understood. The uptake of external RNAi signals that induces silencing in nematodes and insects (called environmental RNAi) has been well characterized, and nematode specific transporters were identified [69], although no obvious homologues of these transporters are found in other systems. Silencing of the cotton bollworm (Helicoverpa armigera) mono-oxygenase gene CYP6AE14 by HIGS led to impaired tolerance to gossypol in larvae. HIGS against the cotton bollworm was still effective in an Arabidopsis dcl2dcl3dcl4 mutant background that was unable to process the long dsRNA precursors of the CYP6AE14 RNAs into mature siRNAs [77], which suggests that insects are likely to take up long dsRNA precursors. This is supported by findings of Zhang et al. [78], where long double-stranded plastid RNA was sufficient to induce gene silencing in pests.
Secretory pathways in plants, such as the exocytosis and the unconventional secretion pathway [79,80], as well as cellular uptake pathway of environmental substances, such as endocytosis [81,82], have been extensively studied. Secretion and uptake of proteins and other micro- and macromolecules is a hallmark of plant-microbe interactions and play key roles in plant defense against pathogens and parasite [83,84] and in pathogenesis and effector-triggered suppression of host plant immunity [83,85] However, secretion of RNAs, a known feature of cell-to-cell communications in animal systems [10,86], has hardly been documented in plants. We propose that plant export “channels” for RNAi triggers are not only sufficient for movement of artificial transgenic HIGS sRNAs; but it is most likely that some host endogenous sRNAs are also transported into pathogen cells for gene regulation (Figure 1B).
Conclusions
Plant host endogenous sRNAs and pathogen-derived sRNAs play pivotal roles in regulating cellular stress responses and plant immunity. Diverse classes of immune-regulatory sRNAs that are differentially regulated upon pathogen attack have been identified. Some of them are capable of translocation into interacting organisms to induce cross-kingdom RNAi. Hosts produce RNAi triggers, to silence pathogen/parasite genes, while advanced eukaryotic pathogens/parasites secrete sRNAs that mimic host sRNAs to suppress host immunity. Although cross-kingdom RNAi has been shown in many examples as HIGS, the mobile RNAi triggers and the mechanisms and pathway(s) of RNA transport are not known. We expect that well-designed genetic, biochemical, and cell biology assays will shed light on the transport mechanisms of cross-kingdom RNAi signals.
Research highlights.
Small RNAs regulate plant immune responses and pathogen virulence.
Small RNAs can move between interacting organisms and induce cross-kingdom RNAi.
Advanced plant pathogens use cross-kingdom RNAi to suppress host immunity genes.
Host induced gene silencing allows crops produce small RNAs silencing pathogen genes.
Acknowledgments
We regret that we would not be able to include and cite many related interesting studies due to the limited space. This research was supported by an NIH grant (R01 GM093008), an NSF Career Award (MCB-0642843), an NSF Award (IOS- 1257576), California Department Food & Agriculture Award CDFA-SCB12057, Citrus Research Board Award (5100-131), and an AES-CE Award (PPA-7517H) to HJ. We thank Marschal Bellinger for generating the figure, Yifan Lii for critical reading of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Seo JK, Wu J, Lii Y, Li Y, Jin H. Contribution of small RNA pathway components in plant immunity. Mol Plant Microbe Interact. 2013;26:617–625. doi: 10.1094/MPMI-10-12-0255-IA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Staiger D, Korneli C, Lummer M, Navarro L. Emerging role for RNA-based regulation in plant immunity. New Phytol. 2013;197:394–404. doi: 10.1111/nph.12022. [DOI] [PubMed] [Google Scholar]
- 3.Weiberg A, Wang M, Bellinger M, Jin H. Small RNAs: a new paradigm in plant-microbe interactions. Annu Rev Phytopathol. 2014;52:495–516. doi: 10.1146/annurev-phyto-102313-045933. [DOI] [PubMed] [Google Scholar]
- 4.Yang L, Huang H. Roles of small RNAs in plant disease resistance. J Integr Plant Biol. 2014;56:962–970. doi: 10.1111/jipb.12200. [DOI] [PubMed] [Google Scholar]
- 5.Chacko N, Lin X. Non-coding RNAs in the development and pathogenesis of eukaryotic microbes. Appl Microbiol Biotechnol. 2013;97:7989–7997. doi: 10.1007/s00253-013-5160-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang M, Weiberg A, Jin H. Pathogen small RNAs: a new class of effectors for pathogen attacks. Mol Plant Pathol. 2015;16:219–223. doi: 10.1111/mpp.12233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brosnan CA, Voinnet O. Cell-to-cell and long-distance siRNA movement in plants: mechanisms and biological implications. Curr Opin Plant Biol. 2011;14:580–587. doi: 10.1016/j.pbi.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 8.Melnyk CW, Molnar A, Bassett A, Baulcombe DC. Mobile 24 nt small RNAs direct transcriptional gene silencing in the root meristems of Arabidopsis thaliana. Curr Biol. 2011;21:1678–1683. doi: 10.1016/j.cub.2011.08.065. [DOI] [PubMed] [Google Scholar]
- 9.Molnar A, Melnyk C, Baulcombe DC. Silencing signals in plants: a long journey for small RNAs. Genome Biol. 2011;12:215. doi: 10.1186/gb-2010-11-12-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarkies P, Miska EA. Small RNAs break out: the molecular cell biology of mobile small RNAs. Nat Rev Mol Cell Biol. 2014;15:525–535. doi: 10.1038/nrm3840. [DOI] [PubMed] [Google Scholar]
- 11.Knip M, Constantin ME, Thordal-Christensen H. Trans-kingdom cross-talk: small RNAs on the move. PLoS Genet. 2014;10:e1004602. doi: 10.1371/journal.pgen.1004602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiberg A, Bellinger M, Jin H. Conversations between kingdoms: small RNAs. Curr Opin Biotechnol. 2015;32C:207–215. doi: 10.1016/j.copbio.2014.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koch A, Kogel KH. New wind in the sails: improving the agronomic value of crop plants through RNAi-mediated gene silencing. Plant Biotechnol J. 2014;12:821–831. doi: 10.1111/pbi.12226. [DOI] [PubMed] [Google Scholar]
- 14.Nunes CC, Dean RA. Host-induced gene silencing: a tool for understanding fungal host interaction and for developing novel disease control strategies. Mol Plant Pathol. 2012;13:519–529. doi: 10.1111/j.1364-3703.2011.00766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15••.Buck AH, Coakley G, Simbari F, McSorley HJ, Quintana JF, Le Bihan T, Kumar S, Abreu-Goodger C, Lear M, Harcus Y, et al. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat Commun. 2014;5:5488. doi: 10.1038/ncomms6488. [This exciting article shows that a helminth parasite secretes host-mimicking miRNAs through exosomes. Host cells internalize parasite vesicular-emitted miRNAs, which fatally for the host suppress immunity genes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16••.Weiberg A, Wang M, Lin FM, Zhao H, Zhang Z, Kaloshian I, Huang HD, Jin H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science. 2013;342:118–123. doi: 10.1126/science.1239705. [This study showed for the first time that a plant pathogenic fungus produces siRNAs that mimic plant endogenous siRNAs. Pathogen siRNAs were shown to hijack the plant RNAi pathway through an Argonaute protein to supress host immunity genes, as this is part of the fungal virulence strategy.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin H. Endogenous small RNAs and antibacterial immunity in plants. FEBS Lett. 2008;582:2679–2684. doi: 10.1016/j.febslet.2008.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katiyar-Agarwal S, Jin H. Role of small RNAs in host-microbe interactions. Annu Rev Phytopathol. 2010;48:225–246. doi: 10.1146/annurev-phyto-073009-114457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Padmanabhan C, Zhang X, Jin H. Host small RNAs are big contributors to plant innate immunity. Curr Opin Plant Biol. 2009;12:465–472. doi: 10.1016/j.pbi.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 20.Ruiz-Ferrer V, Voinnet O. Roles of plant small RNAs in biotic stress responses. Annu Rev Plant Biol. 2009;60:485–510. doi: 10.1146/annurev.arplant.043008.092111. [DOI] [PubMed] [Google Scholar]
- 21.Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS One. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Zhang Q, Zhang J, Wu L, Qi Y, Zhou JM. Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol. 2010;152:2222–2231. doi: 10.1104/pp.109.151803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, Gao S, Zhou X, Chellappan P, Chen Z, Zhou X, Zhang X, Fromuth N, Coutino G, Coffey M, et al. Bacteria-responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol Biol. 2011;75:93–105. doi: 10.1007/s11103-010-9710-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24••.Zhang X, Zhao H, Gao S, Wang WC, Katiyar-Agarwal S, Huang HD, Raikhel N, Jin H. Arabidopsis Argonaute 2 regulates innate immunity via miRNA393( *)-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol Cell. 2011;42:356–366. doi: 10.1016/j.molcel.2011.04.010. [This article proved for the first time that the passenger strand of a functional microRNA, miR393*, plays a crucial role in antibacterial plant immunity. miR393* functions through AGO2 in Arabidopsis, and with miR393/AGO1 represents a dual regulation system of two distinct signaling pathways in plant immunity.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katiyar-Agarwal S, Gao S, Vivian-Smith A, Jin H. A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev. 2007;21:3123–3134. doi: 10.1101/gad.1595107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A, Jr., Zhu JK, Staskawicz BJ, Jin H. A pathogen-inducible endogenous siRNA in plant immunity. Proc Natl Acad Sci U S A. 2006;103:18002–18007. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li F, Pignatta D, Bendix C, Brunkard JO, Cohn MM, Tung J, Sun H, Kumar P, Baker B. MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci U S A. 2012;109:1790–1795. doi: 10.1073/pnas.1118282109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shivaprasad PV, Chen HM, Patel K, Bond DM, Santos BA, Baulcombe DC. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell. 2012;24:859–874. doi: 10.1105/tpc.111.095380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29•.Zhai J, Jeong DH, De Paoli E, Park S, Rosen BD, Li Y, Gonzalez AJ, Yan Z, Kitto SL, Grusak MA, et al. MicroRNAs as master regulators of the plant NB LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011;25:2540–2553. doi: 10.1101/gad.177527.111. [This study revealed the production of phased 21 siRNAs for the regulation of genes belonging to the class of NB-LRRs in Medicago and other plant species. The production of phased siRNAs is initiated by a class of conserved miRNAs, and has been confirmed in following publications to play a pivotal role in plant immunity.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu QH, Fan L, Liu Y, Xu H, Llewellyn D, Wilson I. miR482 regulation of NBS LRR defense genes during fungal pathogen infection in cotton. PLoS One. 2013;8:e84390. doi: 10.1371/journal.pone.0084390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Navarro L, Jay F, Nomura K, He SY, Voinnet O. Suppression of the microRNA pathway by bacterial effector proteins. Science. 2008;321:964–967. doi: 10.1126/science.1159505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- 33.Vaucheret H. Plant ARGONAUTES. Trends Plant Sci. 2008;13:350–358. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 34.Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X, Niu D, Carbonell A, Wang A, Lee A, Tun V, Wang Z, Carrington JC, Chang CE, Jin H. ARGONAUTE PIWI domain and microRNA duplex structure regulate small RNA sorting in Arabidopsis. Nat Commun. 2014;5:5468. doi: 10.1038/ncomms6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA. 2010;16:43–56. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang SM, Choi JW, Hong SH, Lee HJ. Up-Regulation of microRNA* Strands by Their Target Transcripts. Int J Mol Sci. 2013;14:13231–13240. doi: 10.3390/ijms140713231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ellendorff U, Fradin EF, de Jonge R, Thomma BP. RNA silencing is required for Arabidopsis defence against Verticillium wilt disease. J Exp Bot. 2009;60:591–602. doi: 10.1093/jxb/ern306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boccara M, Sarazin A, Thiebeauld O, Jay F, Voinnet O, Navarro L, Colot V. The Arabidopsis miR472-RDR6 silencing pathway modulates PAMP- and effector-triggered immunity through the post-transcriptional control of disease resistance genes. PLoS Pathog. 2014;10:e1003883. doi: 10.1371/journal.ppat.1003883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boccara M, Sarazin A, Thiebeauld O, Jay F, Voinnet O, Navarro L, Colot V. Correction: The Arabidopsis miR472-RDR6 Silencing Pathway Modulates PAMP- and Effector-Triggered Immunity through the Post-transcriptional Control of Disease Resistance Genes. PLoS Pathog. 2015;11:e1004814. doi: 10.1371/journal.ppat.1004814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41••.Dowen RH, Pelizzola M, Schmitz RJ, Lister R, Dowen JM, Nery JR, Dixon JE, Ecker JR. Widespread dynamic DNA methylation in response to biotic stress. Proc Natl Acad Sci U S A. 2012;109:E2183–2191. doi: 10.1073/pnas.1209329109. [This work demonstrated that dynamic changes in DNA methylation level occur at distinct genomic regions, such as transposons, upon pathogen challenge or salicylic acid (SA) treatment. Several coding genes co-localizing with transposons at the genomic level showed co-expressional regulation upon SA treatment, indicating that plant immunity genes are regulated by DNA methylation/de methylation through the RNA-mediated RNA methylation pathway.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu A, Lepere G, Jay F, Wang J, Bapaume L, Wang Y, Abraham AL, Penterman J, Fischer RL, Voinnet O, et al. Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc Natl Acad Sci U S A. 2013;110:2389–2394. doi: 10.1073/pnas.1211757110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Le TN, Schumann U, Smith NA, Tiwari S, Au PC, Zhu QH, Taylor JM, Kazan K, Llewellyn DJ, Zhang R, et al. DNA demethylases target promoter transposable elements to positively regulate stress responsive genes in Arabidopsis. Genome Biol. 2014;15:458. doi: 10.1186/s13059-014-0458-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lopez A, Ramirez V, Garcia-Andrade J, Flors V, Vera P. The RNA silencing enzyme RNA polymerase v is required for plant immunity. PLoS Genet. 2011;7:e1002434. doi: 10.1371/journal.pgen.1002434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luna E, Bruce TJ, Roberts MR, Flors V, Ton J. Next-generation systemic acquired resistance. Plant Physiol. 2012;158:844–853. doi: 10.1104/pp.111.187468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Filiatrault MJ, Stodghill PV, Bronstein PA, Moll S, Lindeberg M, Grills G, Schweitzer P, Wang W, Schroth GP, Luo S, et al. Transcriptome analysis of Pseudomonas syringae identifies new genes, noncoding RNAs, and antisense activity. J Bacteriol. 2010;192:2359–2372. doi: 10.1128/JB.01445-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang RP, Tang DJ, Chen XL, He YQ, Feng JX, Jiang BL, Lu GT, Lin M, Tang JL. Identification of four novel small non-coding RNAs from Xanthomonas campestris pathovar campestris. BMC Genomics. 2010;11:316. doi: 10.1186/1471-2164-11-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang H, Zhao YT, Zhang JQ, Wang XJ, Fang RX, Jia YT. Identification and functional characterization of small non-coding RNAs in Xanthomonas oryzae pathovar oryzae. BMC Genomics. 2011;12:87. doi: 10.1186/1471-2164-12-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmidtke C, Findeiss S, Sharma CM, Kuhfuss J, Hoffmann S, Vogel J, Stadler PF, Bonas U. Genome-wide transcriptome analysis of the plant pathogen Xanthomonas identifies sRNAs with putative virulence functions. Nucleic Acids Res. 2012;40:2020–2031. doi: 10.1093/nar/gkr904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilms I, Overloper A, Nowrousian M, Sharma CM, Narberhaus F. Deep sequencing uncovers numerous small RNAs on all four replicons of the plant pathogen Agrobacterium tumefaciens. RNA Biol. 2012;9:446–457. doi: 10.4161/rna.17212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51••.Schmidtke C, Abendroth U, Brock J, Serrania J, Becker A, Bonas U. Small RNA sX13: a multifaceted regulator of virulence in the plant pathogen Xanthomonas. PLoS Pathog. 2013;9:e1003626. doi: 10.1371/journal.ppat.1003626. [This article describes for the first time a conserved non-coding RNA, sX13, as a master regulator of virulence in a phytopathogenic bactierium, Xanthomonas campestris. sX13 regulates and interferes with Hrp genes, the key components of the type-3-secretion-system. The authors also provide structural insights of non-coding RNA/target mRNA interaction criteria.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chao NX, Wei K, Chen Q, Meng QL, Tang DJ, He YQ, Lu GT, Jiang BL, Liang XX, Feng JX, et al. The rsmA-like gene rsmA(Xcc) of Xanthomonas campestris pv. campestris is involved in the control of various cellular processes, including pathogenesis. Mol Plant Microbe Interact. 2008;21:411–423. doi: 10.1094/MPMI-21-4-0411. [DOI] [PubMed] [Google Scholar]
- 53.Wilms I, Moller P, Stock AM, Gurski R, Lai EM, Narberhaus F. Hfq influences multiple transport systems and virulence in the plant pathogen Agrobacterium tumefaciens. J Bacteriol. 2012;194:5209–5217. doi: 10.1128/JB.00510-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nunes CC, Gowda M, Sailsbery J, Xue M, Chen F, Brown DE, Oh Y, Mitchell TK, Dean RA. Diverse and tissue-enriched small RNAs in the plant pathogenic fungus, Magnaporthe oryzae. BMC Genomics. 2011;12:288. doi: 10.1186/1471-2164-12-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raman V, Simon SA, Romag A, Demirci F, Mathioni SM, Zhai J, Meyers BC, Donofrio NM. Physiological stressors and invasive plant infections alter the small RNA transcriptome of the rice blast fungus, Magnaporthe oryzae. BMC Genomics. 2013;14:326. doi: 10.1186/1471-2164-14-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fahlgren N, Bollmann SR, Kasschau KD, Cuperus JT, Press CM, Sullivan CM, Chapman EJ, Hoyer JS, Gilbert KB, Grunwald NJ, et al. Phytophthora have distinct endogenous small RNA populations that include short interfering and microRNAs. PLoS One. 2013;8:e77181. doi: 10.1371/journal.pone.0077181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vetukuri RR, Asman AK, Tellgren-Roth C, Jahan SN, Reimegard J, Fogelqvist J, Savenkov E, Soderbom F, Avrova AO, Whisson SC, et al. Evidence for small RNAs homologous to effector-encoding genes and transposable elements in the oomycete Phytophthora infestans. PLoS One. 2012;7:e51399. doi: 10.1371/journal.pone.0051399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whisson S, Vetukuri R, Avrova A, Dixelius C. Can silencing of transposons contribute to variation in effector gene expression in Phytophthora infestans? Mob Genet Elements. 2012;2:110–114. doi: 10.4161/mge.20265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59•.Qutob D, Chapman BP, Gijzen M. Transgenerational gene silencing causes gain of virulence in a plant pathogen. Nat Commun. 2013;4:1349. doi: 10.1038/ncomms2354. [This report illustrates how an effector gene, Avr3a, in an oomycete plant pathogen, Phytophthora sojae, is controlled by 25 nt cis-acting sRNAs. Remarkably, silencing of Avr3a transcripts was inheritable and conferred virulence on plants carrying the Avr3a-recognizing R gene Rps3a, indicating that silencing of effector genes might be of advantage for pathogens in the context of corresponding R-genes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koch A, Kumar N, Weber L, Keller H, Imani J, Kogel KH. Host-induced gene silencing of cytochrome P450 lanosterol C14alpha-demethylase-encoding genes confers strong resistance to Fusarium species. Proc Natl Acad Sci U S A. 2013;110:19324–19329. doi: 10.1073/pnas.1306373110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nowara D, Gay A, Lacomme C, Shaw J, Ridout C, Douchkov D, Hensel G, Kumlehn J, Schweizer P. HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell. 2010;22:3130–3141. doi: 10.1105/tpc.110.077040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62•.LaMonte G, Philip N, Reardon J, Lacsina JR, Majoros W, Chapman L, Thornburg CD, Telen MJ, Ohler U, Nicchitta CV, et al. Translocation of sickle cell erythrocyte microRNAs into Plasmodium falciparum inhibits parasite translation and contributes to malaria resistance. Cell Host Microbe. 2012;12:187–199. doi: 10.1016/j.chom.2012.06.007. [In this report the authors provide data on miRNA species that mediate malaria resistance in erythrocyte sickle cells. Two human miRNAs were shown to be translocated into parasite Plasmodium cells. Although Plasmodium has no RNAi machinary, these miRNAs managed to silence target genes by blocking translation of a Plasmodium mRNA that leads to resistance, indicating that host produced sRNAs can induce cross-kingdom gene silencing in parasites as a host defense strategy. Most importantly, this study provided an excellent example that miRNA function is not always dependent on RNAi machinery.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63•.Liu H, Wang X, Wang HD, Wu J, Ren J, Meng L, Wu Q, Dong H, Wu J, Kao TY, et al. Escherichia coli noncoding RNAs can affect gene expression and physiology of Caenorhabditis elegans. Nat Commun. 2012;3:1073. doi: 10.1038/ncomms2071. [This study reports on bacteria-produced non-coding RNAs, which have physiological effects on Caenorhabditis elegans, when fed with Escherichia coli. This study provides yet another example of cross-kingdom RNA silencing, however, it remains elusive, what the evolutionary/ecological context of non-coding RNAs evolution in E. coli that manipulate gene expression in C. elegans might be.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64•.Mayoral JG, Hussain M, Joubert DA, Iturbe-Ormaetxe I, O'Neill SL, Asgari S. Wolbachia small noncoding RNAs and their role in cross-kingdom communications. Proc Natl Acad Sci U S A. 2014;111:18721–18726. doi: 10.1073/pnas.1420131112. [This work showed that non-coding RNAs of the bacterial species Wolbachia can enter the host cell of mosquito and Drosophila spp. In addition, a Wolbachia non-coding RNA showed positive regulation of a mosquito host gene, indicating cross-kingdom RNA communication at play.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheng G, Luo R, Hu C, Cao J, Jin Y. Deep sequencing-based identification of pathogen-specific microRNAs in the plasma of rabbits infected with Schistosoma japonicum. Parasitology. 2013;140:1751–1761. doi: 10.1017/S0031182013000917. [DOI] [PubMed] [Google Scholar]
- 66.Garcia-Silva MR, das Neves RF, Cabrera-Cabrera F, Sanguinetti J, Medeiros LC, Robello C, Naya H, Fernandez-Calero T, Souto-Padron T, de Souza W, et al. Extracellular vesicles shed by Trypanosoma cruzi are linked to small RNA pathways, life cycle regulation, and susceptibility to infection of mammalian cells. Parasitol Res. 2014;113:285–304. doi: 10.1007/s00436-013-3655-1. [DOI] [PubMed] [Google Scholar]
- 67.Quintana JF, Makepeace BL, Babayan SA, Ivens A, Pfarr KM, Blaxter M, Debrah A, Wanji S, Ngangyung HF, Bah GS, et al. Extracellular Onchocerca-derived small RNAs in host nodules and blood. Parasit Vectors. 2015;8:58. doi: 10.1186/s13071-015-0656-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Katoch R, Sethi A, Thakur N, Murdock LL. RNAi for insect control: current perspective and future challenges. Appl Biochem Biotechnol. 2013;171:847–873. doi: 10.1007/s12010-013-0399-4. [DOI] [PubMed] [Google Scholar]
- 69.Lilley CJ, Davies LJ, Urwin PE. RNA interference in plant parasitic nematodes: a summary of the current status. Parasitology. 2012;139:630–640. doi: 10.1017/S0031182011002071. [DOI] [PubMed] [Google Scholar]
- 70.Zhang H, Li HC, Miao XX. Feasibility, limitation and possible solutions of RNAi-based technology for insect pest control. Insect Sci. 2013;20:15–30. doi: 10.1111/j.1744-7917.2012.01513.x. [DOI] [PubMed] [Google Scholar]
- 71.Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, Ilagan O, Johnson S, Plaetinck G, Munyikwa T, Pleau M, et al. Control of coleopteran insect pests through RNA interference. Nat Biotechnol. 2007;25:1322–1326. doi: 10.1038/nbt1359. [DOI] [PubMed] [Google Scholar]
- 72.Alakonya A, Kumar R, Koenig D, Kimura S, Townsley B, Runo S, Garces HM, Kang J, Yanez A, David-Schwartz R, et al. Interspecific RNA interference of SHOOT MERISTEMLESS-like disrupts Cuscuta pentagona plant parasitism. Plant Cell. 2012;24:3153–3166. doi: 10.1105/tpc.112.099994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73•.Kim G, LeBlanc ML, Wafula EK, dePamphilis CW, Westwood JH. Plant science. Genomic-scale exchange of mRNA between a parasitic plant and its hosts. Science. 2014;345:808–811. doi: 10.1126/science.1253122. [This study provides data on cross-species RNA transport during parasitic plant-plant interaction. Bi-directional translocation of host as well as parasite mRNAs occurs and mRNA spread can be detected seveal cm away from the parasitic junction. However, the biological role of RNA exchange between parasite and host plant is not clear.] [DOI] [PubMed] [Google Scholar]
- 74.LeBlanc M, Kim G, Patel B, Stromberg V, Westwood J. Quantification of tomato and Arabidopsis mobile RNAs trafficking into the parasitic plant Cuscuta pentagona. New Phytol. 2013;200:1225–1233. doi: 10.1111/nph.12439. [DOI] [PubMed] [Google Scholar]
- 75.Westwood JH, Roney JK, Khatibi PA, Stromberg VK. RNA translocation between parasitic plants and their hosts. Pest Manag Sci. 2009;65:533–539. doi: 10.1002/ps.1727. [DOI] [PubMed] [Google Scholar]
- 76.Aidoo M, Terlouw DJ, Kolczak MS, McElroy PD, ter Kuile FO, Kariuki S, Nahlen BL, Lal AA, Udhayakumar V. Protective effects of the sickle cell gene against malaria morbidity and mortality. Lancet. 2002;359:1311–1312. doi: 10.1016/S0140-6736(02)08273-9. [DOI] [PubMed] [Google Scholar]
- 77.Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, Wang LJ, Huang YP, Chen XY. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol. 2007;25:1307–1313. doi: 10.1038/nbt1352. [DOI] [PubMed] [Google Scholar]
- 78.Zhang J, Khan SA, Hasse C, Ruf S, Heckel DG, Bock R. Pest control. Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids. Science. 2015;347:991–994. doi: 10.1126/science.1261680. [DOI] [PubMed] [Google Scholar]
- 79.Ding Y, Robinson DG, Jiang L. Unconventional protein secretion (UPS) pathways in plants. Curr Opin Cell Biol. 2014;29:107–115. doi: 10.1016/j.ceb.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 80.Drakakaki G, Dandekar A. Protein secretion: how many secretory routes does a plant cell have? Plant Sci. 2013;203-204:74–78. doi: 10.1016/j.plantsci.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 81.Baisa GA, Mayers JR, Bednarek SY. Budding and braking news about clathrin-mediated endocytosis. Curr Opin Plant Biol. 2013;16:718–725. doi: 10.1016/j.pbi.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 82.Fan L, Li R, Pan J, Ding Z, Lin J. Endocytosis and its regulation in plants. Trends Plant Sci. 2015 doi: 10.1016/j.tplants.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 83.Huckelhoven R. Transport and secretion in plant-microbe interactions. Curr Opin Plant Biol. 2007;10:573–579. doi: 10.1016/j.pbi.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 84.Frei dit Frey N, Robatzek S. Trafficking vesicles: pro or contra pathogens? Curr Opin Plant Biol. 2009;12:437–443. doi: 10.1016/j.pbi.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 85.Kale SD, Tyler BM. Entry of oomycete and fungal effectors into plant and animal host cells. Cell Microbiol. 2011;13:1839–1848. doi: 10.1111/j.1462-5822.2011.01659.x. [DOI] [PubMed] [Google Scholar]
- 86.Mittelbrunn M, Sanchez-Madrid F. Intercellular communication: diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol. 2012;13:328–335. doi: 10.1038/nrm3335. [DOI] [PMC free article] [PubMed] [Google Scholar]