Abstract
Deficiencies in respiratory-chain complexes lead to a variety of clinical phenotypes resulting from inadequate energy production by the mitochondrial oxidative phosphorylation system. Defective expression of mtDNA-encoded genes, caused by mutations in either the mitochondrial or nuclear genome, represents a rapidly growing group of human disorders. By whole-exome sequencing, we identified two unrelated individuals carrying compound heterozygous variants in TRMT5 (tRNA methyltransferase 5). TRMT5 encodes a mitochondrial protein with strong homology to members of the class I-like methyltransferase superfamily. Both affected individuals presented with lactic acidosis and evidence of multiple mitochondrial respiratory-chain-complex deficiencies in skeletal muscle, although the clinical presentation of the two affected subjects was remarkably different; one presented in childhood with failure to thrive and hypertrophic cardiomyopathy, and the other was an adult with a life-long history of exercise intolerance. Mutations in TRMT5 were associated with the hypomodification of a guanosine residue at position 37 (G37) of mitochondrial tRNA; this hypomodification was particularly prominent in skeletal muscle. Deficiency of the G37 modification was also detected in human cells subjected to TRMT5 RNAi. The pathogenicity of the detected variants was further confirmed in a heterologous yeast model and by the rescue of the molecular phenotype after re-expression of wild-type TRMT5 cDNA in cells derived from the affected individuals. Our study highlights the importance of post-transcriptional modification of mitochondrial tRNAs for faithful mitochondrial function.
Main Text
Mitochondria require unique and highly specialized mechanisms to maintain and express their genome (mtDNA). The mitochondrial genome encodes 13 essential subunits of the mitochondrial oxidative phosphorylation system (OXPHOS) and a set of tRNAs and rRNAs required for their translation. All protein components of the mitochondrial translation apparatus, including the mitochondrial ribosomal proteins, translation factors, aminoacyl tRNA synthetases, RNA modifying enzymes, and other auxiliary factors are encoded by nuclear genes and, after their synthesis in the cytoplasm, are delivered to mitochondria. Defective mtDNA expression, caused by mutations in either the mitochondrial or nuclear genomes, is associated with a diverse group of human disorders characterized by impaired mitochondrial respiration.1–3
The 22 mitochondrially encoded tRNAs (mt-tRNAs) act as crucial intermediaries between the mRNAs transcribed from mtDNA and the 13 subunits of OXPHOS that they encode. As with all known tRNAs, they are required to undergo numerous post-transcriptional nucleotide modifications prior to becoming active elements in protein translation in order to ensure efficiency and stringent accuracy. Mitochondrial tRNA processing and modifying enzymes represent an expanding group of mitochondrial disease-causing factors.4 Recent research describes mitochondrial dysfunction resulting from mutations in genes encoding the tRNA processing enzymes HSD10 (also known as MRPP2 [MIM: 300256])5 and ELAC2 (MIM: 605367),6 as well as tRNA modifiers, including PUS1 (MIM: 608109),7 TRIT1,8 TRMU (also known as MTU1 [MIM: 610230]),9 TRNT1 (MIM: 612907),10,11 MTO1 (MIM: 614667),12 and GTPBP3 (MIM: 608536).13 Furthermore, primary mtDNA mutations in mt-tRNA genes, which are a frequent cause of human respiratory-chain deficiencies, can also affect mt-tRNA modification.14–16
The tRNA anticodon loop position 37 (3′ of and adjacent to the anticodon) has risen to prominence with regard to maintaining translational fidelity and efficiency.17 Almost all tRNAs, regardless of organism, are modified at this site. Sophisticated purine modifications are found at position 37 (for example, N6-(dimethylallyl)adenosine, N6-2-methylthio-N6-threonylcarbamoyladenosine, and methylwyosine),17 demonstrating the critical role for this site. In Bacteria, N1-methylation of the guanosine at tRNA position 37 (m1G37) is performed by TrmD-type enzymes.18 In Archaea and Eukarya, m1G37 is introduced by evolutionarily and functionally unrelated Trm5-type proteins.19 In the yeast Saccharomyces cerevisiae, Trm5p is responsible for m1G37 methylation of cytoplasmic and mitochondrial tRNAs.20,21 The human ortholog,22 TRMT5 (tRNA methyltransferase 5), catalyzes the formation of m1G37 in vitro. The presence of m1G37 has been identified in mitochondrial tRNALeu(CUN)23 and tRNAPro24; however, the involvement of human TRMT5 has yet to be confirmed in vivo. In vitro methylation assays with human TRMT5 demonstrated low activity by using a mitochondrial substrate relative to a cytosolic substrate.22 Mitochondrial localization of TRMT5 within human cells has not been studied.
Subject 73901 (F1, II.3; Figure 1A) had a life-long history of exercise intolerance with prominent exertional dyspnea. A detailed clinical and physiological characterization of this subject has been reported in the literature.25 She presented at the age of 25 years with prolonged dyspnea associated with lactic acidosis following an episode of prolonged walking. Subsequent evaluation led to the diagnosis of a mitochondrial myopathy associated with a marked histochemical and biochemical deficiency of cytochrome c oxidase (COX), along with a defect in complex III activity.25 At the time of this evaluation she predominantly had exercise intolerance but also had slight gait unsteadiness and equivocal extensor plantar reflexes. She was able to perform her duties as a nurse. In her early 30s, she developed signs of exocrine pancreatic failure and malabsorption. Neurological evaluation at the age of 35 years revealed hyperreflexia and extensor plantar reflexes with some clinical spasticity, as well as mild distally reduced pallesthesia. When evaluated at the age of 40 years, she reported prominent dyspnea with trivial exercise and felt somewhat short of breath much of the time. She had developed weakness and difficulty getting up from low chairs or climbing steps unless there was a handrail, and she could not get up from the floor without help. Her pancreatic disorder worsened. There was an apparent renal tubular defect with 4+ glycosuria despite normal blood glucose levels. She developed glucose intolerance with an HbA1c concentration as high as 7.5% (normal < 5.7%) and began experiencing nausea and vomiting two to three times a month. She had no symptoms of sensory loss and her cognitive function was normal. An abdominal computed tomography scan at the age of 53 years showed cirrhosis, and she began developing ascites, hence she was evaluated for a possible liver transplant. She had increasing weakness, especially in her legs, and difficulty standing. However, neck weakness was absent and she had neither difficulty chewing or swallowing nor dysarthria. There was no known cardiac involvement apart from tachycardia during minor exercise.25 A more recent muscle biopsy revealed a severe histochemical defect of COX (Figure S1) and decreased activities of respiratory complexes I, III, and IV (Table 1), whereas fibroblast studies showed normal activities. Interestingly, serum FGF21 levels determined at the time of biopsy were markedly increased (2,851 pg/ml; mean ± SD [and normal range] for healthy subjects = 175 ± 155 pg/ml, range = 58–569 pg/ml, n = 10). She died during her sleep at the age of 55. There was no family history of mitochondrial disease. She has three brothers and two sisters, who are all healthy. Both parents are deceased (Figure 1A).
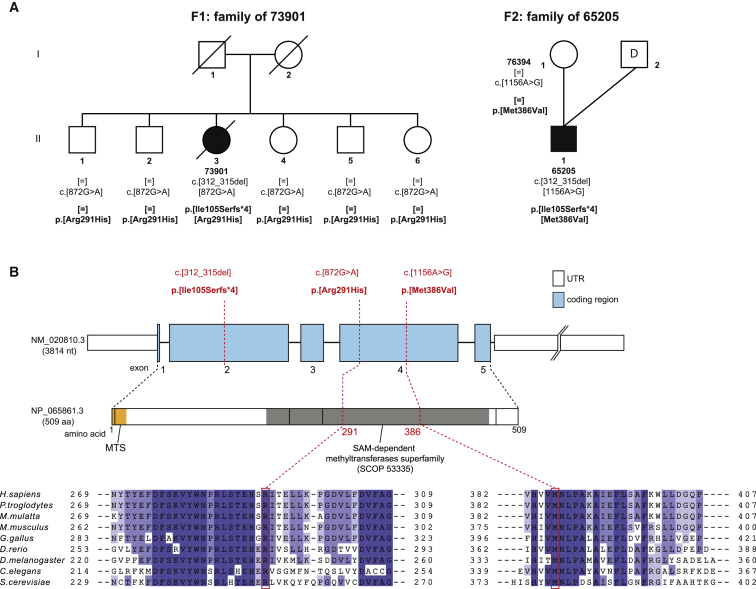
Figure 1.
TRMT5 Variants and Gene Structure
(A) Pedigrees of the two families identified with recessively inherited TRMT5 variants. “D” indicates anonymous sperm donor.
(B) Gene structure of TRMT5 with known protein domains of the gene product and location and conservation of amino-acid residues affected by mutations (in red). Intronic regions are not drawn to scale. Shadowing in the sequence alignment represents the homology of amino-acid residues.
Table 1.
Genetic and Clinical Findings in Individuals with TRMT5 Variants
| ID | Sex | TRMT5 Variantsa |
OXPHOS Activities in Skeletal Muscle |
Clinical Features |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Respiratory-Chain Complex | Enzyme Activity | Absolute Values | Reference Range (Mean ± SD [range]) | Histo-chemical COX Defect | Age at Onset | Clinical Course | Other Features | |||
| 73901b | female | c.[312_315del;872G>A], p.[Ile105Serfs∗4;Arg291His] | I | 59% | 2.9 | 4.9 ± 0.9 (3.1–6.7) | >95% COX-deficient fibers | childhood | died at 55 years of age | life-long exercise intolerance, dyspnea, lactic acidosis, pancreatic disorder, spasticity/peripheral neuropathy, muscular weakness, renal tubulopathy, cirrhosis |
| II | 138% | 2.9 | 11.2 ± 2.7 (5.8 −16.6) | |||||||
| II+III | 47% | 5.3 | 2.1 ± 0.6 (0.9–3.4) | |||||||
| IV | 36% | 1.4 | 3.9 ± 1.3 (1.3–6.6) | |||||||
| 65205c | male | c.[312_315del;1156A>G], p.[Ile105Serfs∗4;Met386Val] | I | 32% | 88 | 272 ± 115 (84–559) | none | birth | alive, 7 years old | premature delivery, failure to thrive, growth retardation, intestinal pseudo-obstruction, hypertrophic non-obstructive cardiomyopathy, muscular hypotonia, demyelinating neuropathy, global developmental delay, lactic acidosis |
| II | ND | ND | ND | |||||||
| II+III | 136% | 154 | 110 ± 70 (37–285) | |||||||
| IV | 27% | 315 | 1150 ± 400 (520–2080) | |||||||
Abbreviation is as follows: ND, no data.
cDNA (GenBank: NM_020810.3), protein (GenBank: NP_065861.3).
For subject 73901, enzyme activities are expressed as percentage of control enzyme activity. Absolute values of residual enzyme activities are expressed as U/min/g wet weight, and the reference range (mean ± SD [range]) is shown for 46 control subjects.
For subject 65205, percentages of residual enzyme activities are normalized to citrate synthase activity. Absolute values are expressed as mU/U citrate synthase activity, and the reference range is shown for 52 (complex I) and 27 (complex II+III and IV) control subjects.
Subject 65205 (F2, II.1; Figure 1A,), is the only child of a healthy mother of northern European descent and an anonymous sperm donor; the family history is empty. Pregnancy was complicated by placental insufficiency and incipient preeclampsia. At 35 weeks and 4 days of gestation, he was born small for date (Z score for weight was −2.2). In the first month of life, he showed irritability, tremor, high-pitched cries, muscular hypertonia, feeding difficulties, and inadequate weight gain. At the age of 3 months, a delayed psychomotor development was noted. At the age of 6 months, follow-up of tachycardia revealed a hypertrophic non-obstructive cardiomyopathy (HNOCM). Furthermore, he showed slight dystrophy and dysmorphic signs (asymmetric plagiocephalus, triangular face with small mouth, blue sclerae, unilateral maxillary fused primary incisor, and unilateral partial syndactyly of toes two and three). Chromosome anomalies including del22q11.2 and Pompe disease were excluded. Selective metabolic screening showed slightly elevated lactate in blood and urine. Brain MRI at 9 months of age showed slight brain atrophy, a larger left hemisphere, and delayed myelination. Brain magnetic resonance spectroscopy showed normal values without a lactate peak. Given that marked hyporeflexia and delayed nerve conduction were evident, Krabbe disease and metachromatic leukodystrophy were excluded. A persistent mild hypercalcemia was noted. At 17 months of age, a norovirus infection led to decompensation of the HNOCM and a nasogastric tube feeding was required. Moderate elevation of serum lactate (3.3–5.7 mmol/l, rising to 9.2 mmol/l on another occasion; normal range = 0.7–2.1 mmol/l) was noted; furthermore, alanine, threonine, and glycine levels were elevated. Elevation of cerebrospinal fluid lactate and alanine was also noted; serum FGF21 levels were not determined. Histological examination of a muscle biopsy showed slight myopathic changes and lipid droplets but no ragged-red fibers. Electron microscopy showed normal mitochondrial morphology and no further abnormalities. Respiratory-chain analysis in frozen muscle tissue showed decreased activity of respiratory complex IV and borderline-low complex I activity (Table 1), whereas fibroblast studies showed normal values. Consequently, mitochondrial disease was confirmed, and a treatment with coenzyme Q10, creatine, thiamine, and ascorbic acid was initiated. Considerable tympanites and gastro-intestinal dysmotility with frequent vomiting were evident. A gastrostomy tube was placed at 2 years of age. Currently, at 7 years of age, he remains a cheerful boy with a moderate to severe developmental delay. Presenting with muscular hypotonia and hypermotoric activity, he moves around in a wheelchair or by rolling on the ground and is unable to sit, stand, or walk unsupported. He communicates by facial expressions, gestures, and sounds. There is no visual or hearing impairment. Growth impairment is reflected by low body weight (Z score is −3.8), length (Z score is −3.1), and head circumference (Z score is −3.6). Bone age is delayed by 2.5–4 years. The HNOCM is stable, showing hypertrophy of the left ventricle (Z score for the interventricular septum-wall diameter is +6.7, Z score for the posterior left ventricular-wall diameter is +8.0) and fractional shortening of 33%. His current medication comprises coenzyme Q10, thiamine, riboflavin, omeprazole, and domperidone. He is showing minor developmental progress from receiving physiotherapy and occupational and speech therapy and being supported by adaptive devices. Informed consent for diagnostic and research studies was obtained for both subjects in accordance with the Declaration of Helsinki protocols and approved by local institutional review boards.
Having excluded mtDNA copy-number depletion by qPCR,26,27 mtDNA rearrangements by long-range PCR28 and mtDNA point mutations by direct Sanger sequencing29 in both subjects, we proceeded to undertake whole-exome sequencing (WES) to elucidate the molecular basis of the disease in individuals 73901 and 65205 by using previously described methodologies and bioinformatic filtering pipelines2,8,30,31 from which both affected individuals were found to harbor variants in TRMT5. Subject 73901 harbored compound heterozygous TRMT5 variants (GenBank: NM_020810.3): a c.312_315del (p.Ile105Serfs∗4) frameshift and a c.872G>A (p.Arg291His) missense variant. Both were confirmed by Sanger sequencing in the affected individual, and all five healthy siblings are heterozygous carriers of the c.872G>A (p. Arg291His) variant; parental samples were not available for carrier testing (Figure 1A). Subject 65205 harbored two heterozygous TRMT5 variants: the same c.312_315 del (p.Ile105Serfs∗4) frameshift variant identified in individual 73901 and a c.1156A>G (p.Met386Val) missense variant. The presence of both variants was confirmed by Sanger sequencing. Carrier testing confirmed heterozygosity of the p.Met386Val variant in the mother. No material from the father was available for testing. The p.Arg291 and p.Met386 residues show absolute evolutionary conservation between human and yeast (Figure 1B). None of the identified variants are present in our combined exome database, which contains > 5,700 samples. In the Exome Aggregation Consortium (ExAC) Browser, we found two of the three variants. The c.312_315del variant is reported with a minor-allele frequency (MAF) of 0.0009 (112/121,378 alleles), whereas the c.872G>A variant was detected five times (MAF = 0.00004). Both missense changes are predicted to be deleterious according to PolyPhen-2 (p.Arg291His, probably damaging, score = 1.000 and p.Met386Val, probably damaging, score = 0.992) and SIFT (p.Arg291His, affects protein function, score = 0.00 and p.Met386Val, affects protein function, score = 0.00).
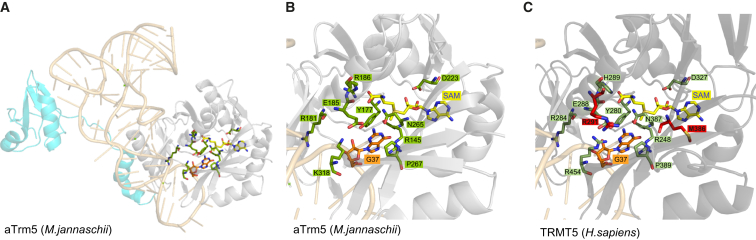
The p.Ile105Serfs∗4 frameshift mutation is upstream of the methyltransferase motif (Figure 1B), predicting a truncated protein that is expected to be non-functional. In order to evaluate the consequences of p.Arg291His and p.Met386Val variants on the enzyme function, we generated a homology model for TRMT5 by using SWISS-MODEL (Figure 2 and Figure S2A). For the modeling of the TRMT5 fragment corresponding to the SAM-dependent methyltransferase domain (residues 193–470, SCOP: 53335), we used the structure of the corresponding domain from the homologous Methanocaldococcus jannaschii aTrm5 (residues 92–334, PDB: 2NNZ) as a template. Recent research has shown that human TRMT5 is an active methyltransferase and that it is generally similar to aTrm5 in catalysis.33 Arg291 is in close vicinity to the strictly conserved and catalytically critical Glu288. Substitution of Glu288 in the human enzyme reduces the reaction rate by over 300-fold.33 It has been proposed that Glu288 participates in an acid-base proton-transfer reaction from the N1 of G37.32 The structural analysis performed here indicates that Arg291 forms two hydrogen bonds with Glu288 (Figure S2B). Substitution of Arg291 is expected to affect the interaction with Glu288, destabilizing the active-site structure. The Met386 residue is adjacent to Asn387. Asn387 forms hydrogen bonds with G37 and SAM, bringing them in close proximity to each other and allowing the catalysis of N1-methylation.32 Although Met386 is not directly involved in catalysis, its substitution to a small aliphatic amino acid is predicted to affect the structure of the catalytic pocket by altering the position of Asn387 and preventing proper hydrogen-bond formation. In conclusion, homology modeling implies that presence of the Met386Val and Arg291His variants would interfere with catalysis of TRMT5.
Figure 2.
3D Model of Human TRMT5
(A) Ribbon presentation of the Methanocaldococcus jannaschii aTrm5–tRNACys(GCA)–AdoMet complex structure. The fragment of structure typical for the AdoMet-dependent methyltransferases superfamily (SCOP: SSF53335) is colored gray (the remaining portion of aTrm5 is colored cyan). tRNACys is colored light orange, and G37 is presented as a stick model. SAM are depicted as stick models and carbon atoms are yellow. The residues that are key for catalysis32 are indicated in green.
(B) Detailed view of the structural model of the catalytic site of aTrm5. Color coding is the same as in panel (A).
(C) Detailed view of the 3D model of the predicted catalytic site of human TRMT5. TRMT5 residues corresponding to the catalytically important amino acids in aTrm5 are indicated in dark green. Seven out of nine catalytically important residues are absolutely conserved in TRMT5 and aTrm5, and the remaining two, His289 (Methanocaldococcus jannaschii Arg186) and Arg454 (Methanocaldococcus jannaschii Lys318), share very similar chemical properties between the two proteins. The residues mutated in individuals 73901 and 65205, Arg291 and Met389, respectively, are indicated in red.
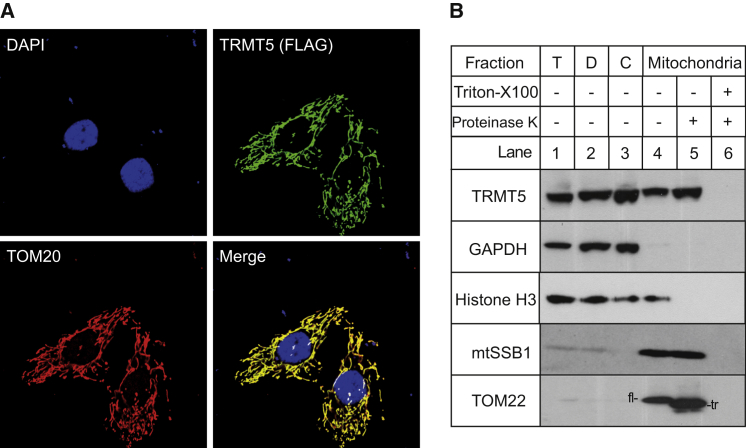
The subcellular localization of TRMT5 has not been studied thus far. Given the mitochondrial respiratory-chain deficiencies detected in individuals with potentially pathogenic TRMT5 variants, we set out to determine whether TRMT5 is a mitochondrial protein. Mitochondrial localization of TRMT5 was analyzed immunocytochemically with a FLAG-tagged version of the protein (TRMT5-FLAG, mRNA [GenBank: NM_020810.3] coding for a 509-aa protein). The corresponding cDNA was transiently expressed in HeLa cells as described previously.34 Signals from an anti-FLAG antibody and an antibody targeted against the mitochondrial protein TOM20 were shown to co-localize (Figure 3A). Cellular fractionation experiments, performed as described previously,35 with HeLa cells indicated that the endogenous TRMT5 is enriched within the mitochondrial fraction, along with a known mitochondrial matrix protein, mtSSB1. Both TRMT5 and mtSSB1 were resistant to proteinase K treatment of the mitochondrial fraction, whereas in similar conditions the outer-membrane protein TOM22 was truncated by proteinase K digestion (Figure 3B), confirming that TRMT5 is localized within mitochondria in human cells.
Figure 3.
Mitochondrial Localization of TRMT5
(A) Localization of TRMT5 by immunofluorescence in HeLa cells following the transient expression of a C-terminal FLAG-tagged TRMT5 construct (TRMT5-FLAG). Cell nuclei were stained with DAPI (top left). TRMT5-FLAG was detected by an anti-FLAG antibody (top right). We detected mitochondria by targeting a known mitochondrial protein, TOM20, with an anti-TOM20 antibody (bottom left). We digitally merged the preceding images, and signal overlap between TRMT5-FLAG and TOM20 appears yellow (bottom right).
(B) Localization of TRMT5 in sub-cellular fractions. HeLa cells were fractionated into debris (D, lane 2), cytosol (C, lane 3), and mitochondria (C, lanes 4–6). The mitochondrial fraction was treated with 25 μg/ml proteinase K in the absence (C, lane 5) or presence (C, lane 6) of 1% Triton X-100. The fractions were analyzed by western blotting with antibodies against TRMT5 (HPA000943, Sigma). The distribution of TRMT5 was compared with that of the following marker proteins: mtSSB1 (mitochondrial matrix), TOM22 (mitochondrial outer membrane), GAPDH (cytosol), and Histone H4 (nucleus). Abbreviations are as follows: T, total cell lysate; fl, full-length TOM22; tr, truncated TOM22.
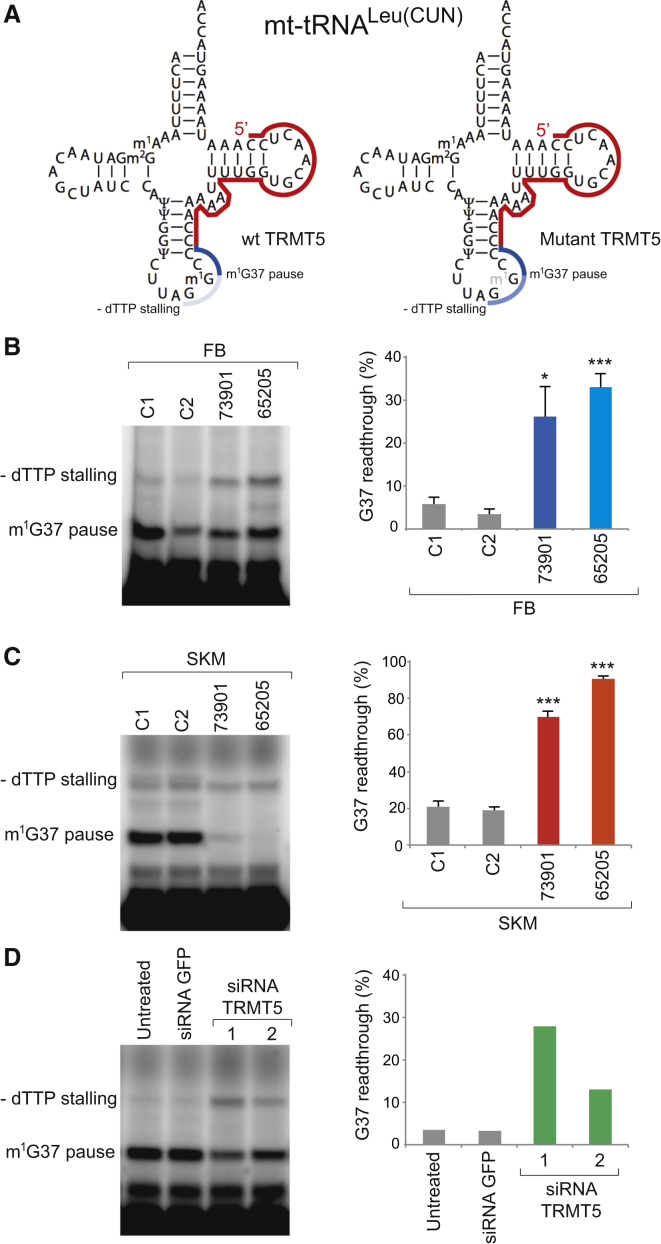
Next we intended to determine whether the identified TRMT5 variants result in diminished G37 modification of a mitochondrial tRNA. The detection of N1-methylguanosine by reverse-transcription primer extension (RT-PEx) makes use of the modification’s ability to interfere with standard Watson-Crick base pairing, thereby causing the reverse transcriptase to pause when incorporating cytosine and preventing the extension reaction from proceeding past the modification site (Figure S3). Reverse-transcription reactions were performed in the absence of a particular dinucleotide triphosphate (dNTP) selected to cause reaction stalling shortly downstream from the modification site, in our case deoxythymidine triphosphate (dTTP) for mitochondrial tRNALeu(CUN) (Figure 4A). The ratio between these two extension products is therefore proportional to the abundance of the modification (Figure S3). On the basis of previously published data reporting the presence of m1G37 in mitochondrial tRNALeu(CUN)23, a suitable primer was designed to anneal to this mt-tRNA (Figure 4A). This primer was subject to radioactive labeling and used in the RT-PEx assay using total RNA extracted from primary skin fibroblasts, as described previously.34 RT-PEx reactions performed in control cell lines demonstrate that only a small percentage (∼5%) of primers are able to readthrough G37, indicating high modification levels in the mt-tRNALeu(CUN) in control cells. This high activity of TRMT5 on its substrate is consistent with the previously described essential role of m1G37. However, mt-tRNALeu(CUN) was found to have considerably decreased levels of the m1G37 modification in primary fibroblasts from both individuals, as demonstrated by increased readthrough past G37 (Figure 4B). The loss of m1G37 does not appear to impact tRNA stability, given that similar steady-state levels of mt-tRNALeu(CUN) are found in control and affected fibroblast cells (Figure S4A). Remarkable tissue specificity for the effect of TRMT5 variants on G37 modification was identified when mt-tRNA from skeletal muscle was analyzed in the same assay (Figure 4C). RT-PEx reactions performed on total RNA preparations isolated from skeletal-muscle biopsy samples of affected individuals demonstrated severely diminished pausing at G37, in particular for the 65205 sample as compared to the control samples (Figure 4C).
Figure 4.
Hypomodification of G37 in Mitochondrial tRNA in Affected Individuals and upon Downregulation of TRMT5 Expression
(A) A radioactively labeled, complementary primer (red) is annealed to mt-tRNALeu(CUN) and subjected to a RT-PEx reaction. The presence of m1G37 results in RT-PEx pausing, producing a shorter product (dark blue). However, in the absence of m1G37, the extension is able to progress until stalling due to the lack of a dNTP (dTTP in the above case, light blue), producing a longer product.
(B) Separation and detection of products of RT-PEx reactions preformed on RNA extracted from skin fibroblasts (FB) of healthy controls (C1 and C2) and the affected individuals (65205 and 73901; left panel). The gel shows a representative result of at least three biological replicates (right panel). Quantification values represent percentage of RT-PEx reaction readthrough the G37 site as the average of the dTTP-induced stalling intensity to the total stalling (dTTP-induced stalling + m1G37-induced stalling). Error bars = 1 SEM; n = 5, ∗p < 0.05, ∗∗∗p < 0.001, unpaired two-tailed Student’s t test with C1.
(C) RT-PEx experiment performed as in (B) on RNA extracted from skeletal muscle (SKM) of healthy control individuals (C1 and C2) and affected individuals (65205 and 73901). Error bars = 1 SEM; n = 3, ∗∗∗p < 0.001, unpaired two-tailed Student’s t test with C1.
(D) Separation and detection of RT-PEx reactions on HeLa cells-derived RNA following a 6-day siRNA-mediated depletion of TRMT5. Two different siRNA were used (HSS126475 and HSS126476, Life Technologies). Untransfected cells and cells transfected with siRNA against GFP (Dsc-1001, Lonza) were used as controls. Quantification values displayed as per (B).
In order to further corroborate a direct role of TRMT5 in mt-tRNA modification, we downregulated its amount by using RNA interference in HeLa cells (Figure 4D)36 and confirmed the reduction of TRMT5 amounts by western blotting (Figure S4B). The siRNA-mediated depletion of TRMT5 resulted in G37 hypomodification in mt-tRNALeu(CUN) in the RT-PEx assay, as observed for the samples derived from the affected individuals (Figure 4D). Taken together, our results thus far establish TRMT5 as the methyltransferase responsible for m1G37 modification in human mitochondrial tRNAs and suggest the variants detected in subjects 65205 and 73901 as responsible for their respiratory-chain abnormalities.
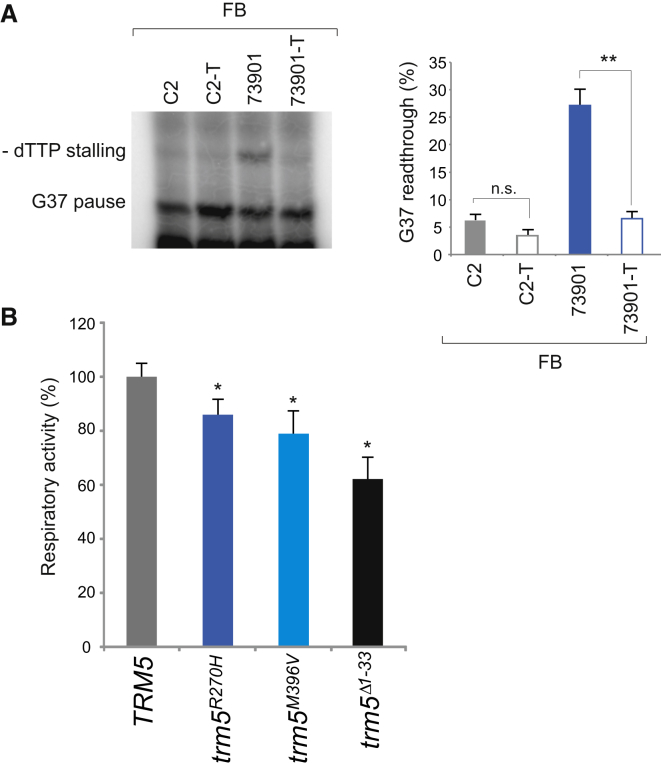
In order to confirm that defects in TRMT5 are the cause of the aberrant modification of G37 in mt-tRNALeu(CUN), we transduced the 65205, 73901, and control primary fibroblasts with a wild-type copy of TRMT5 cDNA by using a lentiviral vector (pLenti 6.3/V5 TOPO, Life Technologies) as previously described.37 The transduction had no noticeable effect on the G37 modification status of mt-tRNALeu(CUN) in the control-cell line (C2-T, Figure 5A). However, in transduced 73901 fibroblasts, the expression of wild-type TRMT5 was found to reverse the hypomodification effect observed in fibroblasts from the affected individuals, resulting in m1G37 amounts matching those of the control individual (73901-T, Figure 5A). We were unable to recover any viable cells from subject 65205 after the transduction procedure.
Figure 5.
Causal Role for p.Arg291His and p.Met386Val TRMT5 Variants in mt-tRNA Modification and Oxidative Metabolism Deficiency
(A) RT-PEx assay performed on RNA derived from individual 73901 and control (C2) skin fibroblasts (FB) with and without the expression of wild-type TRMT5. Quantification values are displayed as per Figure 4B. Error bars = 1 SEM; n = 4, ∗∗p < 0.01, unpaired two-tailed Student’s t test.
(B) Complementation assays in yeast. TRM5 was cloned under its natural promoter upon PCR amplification. PCR-based mutagenesis was performed to obtain the trm5R270H and trm5M368V mutant alleles. The trm5Δ1-33 allele was constructed as previously described.21 Disruption of genomic TRM5 gene was performed in the presence of TRM5 on the pFL38 plasmid, given that the deletion is lethal. The TRM5, trm5R270H, trm5M368V, and trm5Δ1-33 alleles cloned into the pFL39 vector were introduced into this strain, and then pFL38-TRM5 was lost through plasmid shuffling. Oxygen consumption rate was recorded on intact cells grown at 28°C in synthetic complete medium without tryptophan, supplemented with 0.5% glucose. Values were normalized to the rate of oxygen consumption of the TRM5 transformant and represented as the mean of at least three values. Error bars = 1 SD; ∗p < 0.05, paired Student’s t test.
To further substantiate the pathogenicity of the identified TRMT5 variants, we utilized a yeast model to analyze the Trm5p p.Arg270His and p.Met396Val substitutions corresponding to the human p.Arg291His and p.Met386Val variants, respectively (Figure 1B). In yeast, Trm5p has a dual, cytoplasmic and mitochondrial localization; consequently, the null mutant is lethal. The strain that lacks the Trm5p mitochondrial targeting sequence (amino acids 1–33) fails to localize Trm5p to the mitochondria, resulting in a respiratory deficiency.21 To obtain the “null mitochondrial” mutant, we constructed a gene encoding a protein lacking amino acids 1–33 (Δ1–33). This mutant displayed a 20% reduction of respiratory activity. In order to accentuate this effect, we expressed trm5Δ1-33 in a ParR strain that exhibits destabilized mt-tRNA-mitoribosome interaction, as a result of paromomycin-resistant mutation in its mitochondrial 15S rRNA gene, which renders the 15S RNA site A structurally similar to the human 12S RNA site A.12 In this genetic background, the trm5Δ1-33 mutant showed ∼40% reduction of respiratory activity (Figure 5B). The respiratory phenotype was rescued by expression of a wild-type copy of TRM5 (Figure 5B). However, expression of alleles carrying either the trm5R270H or the trm5M396V mutation did not lead to a full recovery of the decreased mitochondrial respiratory activity (Figure 5B). In conclusion, these data confirm a causal role for the p.Arg291His and p.Met386Val TRMT5 variants in the mt-tRNA modification and oxidative metabolism deficiency in the reported individuals.
Our experiments have shown that TRMT5 is responsible for the m1G37 modification in human mitochondrial tRNA molecules. Position 37 of a tRNA is almost exclusively a purine, and the G or A is modified to m1G or i6A in most cases. Both of these modifications have been shown to significantly enhance translational efficiency and accuracy in yeast, and the loss of i6A has been recently linked to pathogenicity in individuals with severe combined mitochondrial respiratory-chain defects.8 The methylation of G37 to form m1G acts to sterically block Watson-Crick base pairing and thereby both maintain an open loop conformation, by blocking base pairing with nucleotides elsewhere in the anticodon loop, and protect against frame shifting by preventing its interaction with the mRNA. The absence of m1G has been shown to lead to an increase in +1 frameshifting,38 altering the subsequent protein sequence and commonly resulting in a truncated product upon encountering a stop codon. The loss of m1G37 has also been linked to a reduced stringency of aminoacyl-tRNA selection at the ribosome,39 a reduced rate of polypeptide elongation,40 and an increase in the misacylation of the tRNA.41 Furthermore, studies of the TRMT5 orthologs in Saccharomyces cerevisiae21 and Trypanosoma brucei42 have revealed that the protein is responsible for introducing m1G37 in mt-tRNA and that it plays an essential role in mitochondrial protein synthesis. Our data support an analogous activity of TRMT5 in human mitochondria given that we identified a reduced modification of G37 in mt-tRNALeu in both cell lines with TRMT5 variants and in cells with RNAi-mediated downregulation of TRMT5 expression. Methylation of G37 has also been detected in mt-tRNAPro26. However, in our RT-PEx experiments, no changes in the mt-tRNAPro G37 modification were detected in fibroblasts from the analyzed subjects (Figure S5). This result indicates that either TRMT5 is not responsible for m1G37 in human mt-tRNAPro, or the p.Arg291His and p.Met386Val variants affect the enzymatic activity in a tRNA-specific manner and with a greater effect on mt-tRNALeu(CUN). Further studies would be required to address whether m1G37 is involved either in maintaining the proper ribosomal reading frame or in mitochondrial tRNA aminoacylation.
The two reported subjects both presented with lactic acidosis and evidence of multiple mitochondrial respiratory-chain-complex deficiencies in skeletal muscle, although the clinical presentation was remarkably different. The female individual (73901) presented mainly with a history of life-long exercise intolerance with no cardiac involvement, whereas the male individual (65205) presented with dysmorphic signs, failure to thrive, growth retardation, hypertrophic non-obstructive cardiomyopathy, peripheral neuropathy, and moderate to severe developmental delay in childhood. Interestingly, clinically relevant MTO1 or GTPBP3 variants responsible for a different modification in the mt-tRNA anticodon loop (position 34, “wobble base”) have also been associated with lactic acidosis, multiple respiratory-chain deficiencies, and hypertrophic cardiomyopathy.12,13 The presence of hypertrophic cardiomyopathy as a prominent and common phenotype in all three disorders indicates tissue-specific threshold effects; the heart is specifically vulnerable for a diminished mitochondrial translation rate. The increased sensitivity of cardiac tissue as a consequence of a perturbed mitochondrial translation has recently been recapitulated in an MTO1 mouse model.43,44 It is possible that the higher degree of mt-tRNA hypomodification detected in individual 65205 (Figure 4) and the consequent impairment of mitochondrial translation accounts for the more severe symptoms in this case (Table 1). The latter is also supported by a modestly lowered respiration rate in the 65205 fibroblast (Figure S6), but no detectable changes in oxygen consumption in the 73901 fibroblast. Such non-overlapping phenotypes between individuals harboring variants in the same gene involved in mt-tRNA maturation have been previously reported.45 The spectrum of clinical phenotypes associated with mt-tRNA modification deficiencies, including cases without cardiac disorder,9 is constantly growing.
In summary, through the identification of two individuals with mitochondrial disease-bearing compound heterozygous variants in TRMT5, our research further expands the list of mitochondrial-tRNA-modifying enzymes vital for proper mitochondrial function. Methylation of guanine 37 in mt-tRNALeu(CUN) is diminished in fibroblasts and skeletal muscle of affected individuals and recapitulated by siRNA-induced depletion of TRMT5 in HeLa cells, consistent with mitochondrial localization of the protein. The involvement of TRMT5 in m1G formation and mitochondrial respiratory function was confirmed through the rescue of tRNALeu(CUN) m1G37 modification by complementation with wild-type TRMT5 in human fibroblasts and by validation of the pathogenic variants in a yeast model. Given that 7% of all mammalian mt-tRNA residues undergo post-transcriptional modification, and that over 30 different modified mt-tRNA positions have been so far described,46 it is anticipated that future exome sequencing analyses of individuals with clinically diagnosed mitochondrial disease will reveal further pathogenic variants within mt-tRNA modifying factors.
Acknowledgments
This work was supported by grants from the Medical Research Council UK (MC_U105697135 to J.R., C.A.P., A.D., and M.M.), the German Bundesministerium für Bildung und Forschung through funding of the E-Rare project GENOMIT (01GM1207 to T.M. and H.P.), the German Network or Mitochondrial Disorders (01GM1113C and 01GM0866 to T.M., H.P., and M.S.), the Juniorverbund in der Systemmedizin “mitOmics” (FKZ 01ZX1405C to T.B.H.), the German Center for Heart Research (Z76010017300 and Z56010015300 to T.M.), the Wellcome Trust Strategic Award (096919/Z/11/Z to R.W.T. and P.F.C.), the Medical Research Council Centre for Neuromuscular Diseases (G0601943 to R.W.T. and P.F.C.), the UK National Health Service Highly Specialised “Rare Mitochondrial Disorders of Adults and Children” Service (C.L.A. and R.W.T.), the Lily Foundation (R.W.T.), a National Institute for Health Research doctoral fellowship (NIHR-HCS-D12-03-04 to C.L.A.), and Telethon Italy grant GGP11011 (C. Dallabona, C. Donnini, and I.F.). P.F.C. is a Wellcome Trust Senior Fellow in Clinical Science (101876/Z/13/Z) and a UK National Institute for Health Research Senior Investigator.
Published: July 16, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Data include six figures and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.06.011.
Contributor Information
Holger Prokisch, Email: prokisch@helmholtz-muenchen.de.
Michal Minczuk, Email: michal.minczuk@mrc-mbu.cam.ac.uk.
Web Resources
The URLs for data presented herein are as follows:
ExAC Browser, http://exac.broadinstitute.org
OMIM, http://www.omim.org
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
SIFT, http://sift.jcvi.org/
SWISS MODEL, http://swissmodel.expasy.org
Supplemental Data
References
- 1.Nicholls T.J., Rorbach J., Minczuk M. Mitochondria: mitochondrial RNA metabolism and human disease. Int. J. Biochem. Cell Biol. 2013;45:845–849. doi: 10.1016/j.biocel.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Taylor R.W., Pyle A., Griffin H., Blakely E.L., Duff J., He L., Smertenko T., Alston C.L., Neeve V.C., Best A. Use of whole-exome sequencing to determine the genetic basis of multiple mitochondrial respiratory chain complex deficiencies. JAMA. 2014;312:68–77. doi: 10.1001/jama.2014.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayr J.A., Haack T.B., Freisinger P., Karall D., Makowski C., Koch J., Feichtinger R.G., Zimmermann F.A., Rolinski B., Ahting U. Spectrum of combined respiratory chain defects. J. Inherit. Metab. Dis. 2015 doi: 10.1007/s10545-015-9831-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Powell C.A., Nicholls T.J., Minczuk M. Nuclear-encoded factors involved in post-transcriptional processing and modification of mitochondrial tRNAs in human disease. Front. Genet. 2015;6:79. doi: 10.3389/fgene.2015.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deutschmann A.J., Amberger A., Zavadil C., Steinbeisser H., Mayr J.A., Feichtinger R.G., Oerum S., Yue W.W., Zschocke J. Mutation or knock-down of 17β-hydroxysteroid dehydrogenase type 10 cause loss of MRPP1 and impaired processing of mitochondrial heavy strand transcripts. Hum. Mol. Genet. 2014;23:3618–3628. doi: 10.1093/hmg/ddu072. [DOI] [PubMed] [Google Scholar]
- 6.Haack T.B., Kopajtich R., Freisinger P., Wieland T., Rorbach J., Nicholls T.J., Baruffini E., Walther A., Danhauser K., Zimmermann F.A. ELAC2 mutations cause a mitochondrial RNA processing defect associated with hypertrophic cardiomyopathy. Am. J. Hum. Genet. 2013;93:211–223. doi: 10.1016/j.ajhg.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bykhovskaya Y., Casas K., Mengesha E., Inbal A., Fischel-Ghodsian N. Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA) Am. J. Hum. Genet. 2004;74:1303–1308. doi: 10.1086/421530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yarham J.W., Lamichhane T.N., Pyle A., Mattijssen S., Baruffini E., Bruni F., Donnini C., Vassilev A., He L., Blakely E.L. Defective i6A37 modification of mitochondrial and cytosolic tRNAs results from pathogenic mutations in TRIT1 and its substrate tRNA. PLoS Genet. 2014;10:e1004424. doi: 10.1371/journal.pgen.1004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeharia A., Shaag A., Pappo O., Mager-Heckel A.M., Saada A., Beinat M., Karicheva O., Mandel H., Ofek N., Segel R. Acute infantile liver failure due to mutations in the TRMU gene. Am. J. Hum. Genet. 2009;85:401–407. doi: 10.1016/j.ajhg.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakraborty P.K., Schmitz-Abe K., Kennedy E.K., Mamady H., Naas T., Durie D., Campagna D.R., Lau A., Sendamarai A.K., Wiseman D.H. Mutations in TRNT1 cause congenital sideroblastic anemia with immunodeficiency, fevers, and developmental delay (SIFD) Blood. 2014;124:2867–2871. doi: 10.1182/blood-2014-08-591370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasarman F., Thiffault I., Weraarpachai W., Salomon S., Maftei C., Gauthier J., Ellazam B., Webb N., Antonicka H., Janer A. The 3′ addition of CCA to mitochondrial tRNASer(AGY) is specifically impaired in patients with mutations in the tRNA nucleotidyl transferase TRNT1. Hum. Mol. Genet. 2015;24:2841–2871. doi: 10.1093/hmg/ddv044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghezzi D., Baruffini E., Haack T.B., Invernizzi F., Melchionda L., Dallabona C., Strom T.M., Parini R., Burlina A.B., Meitinger T. Mutations of the mitochondrial-tRNA modifier MTO1 cause hypertrophic cardiomyopathy and lactic acidosis. Am. J. Hum. Genet. 2012;90:1079–1087. doi: 10.1016/j.ajhg.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kopajtich R., Nicholls T.J., Rorbach J., Metodiev M.D., Freisinger P., Mandel H., Vanlander A., Ghezzi D., Carrozzo R., Taylor R.W. Mutations in GTPBP3 cause a mitochondrial translation defect associated with hypertrophic cardiomyopathy, lactic acidosis, and encephalopathy. Am. J. Hum. Genet. 2014;95:708–720. doi: 10.1016/j.ajhg.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yarham J.W., Elson J.L., Blakely E.L., McFarland R., Taylor R.W. Mitochondrial tRNA mutations and disease. Wiley Interdiscip Rev RNA. 2010;1:304–324. doi: 10.1002/wrna.27. [DOI] [PubMed] [Google Scholar]
- 15.Wei F.Y., Zhou B., Suzuki T., Miyata K., Ujihara Y., Horiguchi H., Takahashi N., Xie P., Michiue H., Fujimura A. Cdk5rap1-mediated 2-methylthio modification of mitochondrial tRNAs governs protein translation and contributes to myopathy in mice and humans. Cell Metab. 2015;21:428–442. doi: 10.1016/j.cmet.2015.01.019. [DOI] [PubMed] [Google Scholar]
- 16.Van Haute L., Pearce S.F., Powell C.A., D’Souza A.R., Nicholls T.J., Minczuk M. Mitochondrial transcript maturation and its disorders. J. Inherit. Metab. Dis. 2015;38:655–680. doi: 10.1007/s10545-015-9859-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helm M., Alfonzo J.D. Posttranscriptional RNA Modifications: playing metabolic games in a cell’s chemical Legoland. Chem. Biol. 2014;21:174–185. doi: 10.1016/j.chembiol.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Björk G.R., Wikström P.M., Byström A.S. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science. 1989;244:986–989. doi: 10.1126/science.2471265. [DOI] [PubMed] [Google Scholar]
- 19.Christian T., Hou Y.M. Distinct determinants of tRNA recognition by the TrmD and Trm5 methyl transferases. J. Mol. Biol. 2007;373:623–632. doi: 10.1016/j.jmb.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Björk G.R., Jacobsson K., Nilsson K., Johansson M.J., Byström A.S., Persson O.P. A primordial tRNA modification required for the evolution of life? EMBO J. 2001;20:231–239. doi: 10.1093/emboj/20.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee C., Kramer G., Graham D.E., Appling D.R. Yeast mitochondrial initiator tRNA is methylated at guanosine 37 by the Trm5-encoded tRNA (guanine-N1-)-methyltransferase. J. Biol. Chem. 2007;282:27744–27753. doi: 10.1074/jbc.M704572200. [DOI] [PubMed] [Google Scholar]
- 22.Brulé H., Elliott M., Redlak M., Zehner Z.E., Holmes W.M. Isolation and characterization of the human tRNA-(N1G37) methyltransferase (TRM5) and comparison to the Escherichia coli TrmD protein. Biochemistry. 2004;43:9243–9255. doi: 10.1021/bi049671q. [DOI] [PubMed] [Google Scholar]
- 23.Kirino Y., Yasukawa T., Marjavaara S.K., Jacobs H.T., Holt I.J., Watanabe K., Suzuki T. Acquisition of the wobble modification in mitochondrial tRNALeu(CUN) bearing the G12300A mutation suppresses the MELAS molecular defect. Hum. Mol. Genet. 2006;15:897–904. doi: 10.1093/hmg/ddl007. [DOI] [PubMed] [Google Scholar]
- 24.Brulé H., Holmes W.M., Keith G., Giegé R., Florentz C. Effect of a mutation in the anticodon of human mitochondrial tRNAPro on its post-transcriptional modification pattern. Nucleic Acids Res. 1998;26:537–543. doi: 10.1093/nar/26.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haller R.G., Lewis S.F., Estabrook R.W., DiMauro S., Servidei S., Foster D.W. Exercise intolerance, lactic acidosis, and abnormal cardiopulmonary regulation in exercise associated with adult skeletal muscle cytochrome c oxidase deficiency. J. Clin. Invest. 1989;84:155–161. doi: 10.1172/JCI114135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Acham-Roschitz B., Plecko B., Lindbichler F., Bittner R., Mache C.J., Sperl W., Mayr J.A. A novel mutation of the RRM2B gene in an infant with early fatal encephalomyopathy, central hypomyelination, and tubulopathy. Mol. Genet. Metab. 2009;98:300–304. doi: 10.1016/j.ymgme.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 27.Blakely E., He L., Gardner J.L., Hudson G., Walter J., Hughes I., Turnbull D.M., Taylor R.W. Novel mutations in the TK2 gene associated with fatal mitochondrial DNA depletion myopathy. Neuromuscul. Disord. 2008;18:557–560. doi: 10.1016/j.nmd.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 28.Blakely E.L., He L., Taylor R.W., Chinnery P.F., Lightowlers R.N., Schaefer A.M., Turnbull D.M. Mitochondrial DNA deletion in “identical” twin brothers. J. Med. Genet. 2004;41:e19. doi: 10.1136/jmg.2003.011296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blakely E.L., Yarham J.W., Alston C.L., Craig K., Poulton J., Brierley C., Park S.M., Dean A., Xuereb J.H., Anderson K.N. Pathogenic mitochondrial tRNA point mutations: nine novel mutations affirm their importance as a cause of mitochondrial disease. Hum. Mutat. 2013;34:1260–1268. doi: 10.1002/humu.22358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haack T.B., Haberberger B., Frisch E.M., Wieland T., Iuso A., Gorza M., Strecker V., Graf E., Mayr J.A., Herberg U. Molecular diagnosis in mitochondrial complex I deficiency using exome sequencing. J. Med. Genet. 2012;49:277–283. doi: 10.1136/jmedgenet-2012-100846. [DOI] [PubMed] [Google Scholar]
- 31.Synofzik M., Haack T.B., Kopajtich R., Gorza M., Rapaport D., Greiner M., Schönfeld C., Freiberg C., Schorr S., Holl R.W. Absence of BiP co-chaperone DNAJC3 causes diabetes mellitus and multisystemic neurodegeneration. Am. J. Hum. Genet. 2014;95:689–697. doi: 10.1016/j.ajhg.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christian T., Lahoud G., Liu C., Hoffmann K., Perona J.J., Hou Y.M. Mechanism of N-methylation by the tRNA m1G37 methyltransferase Trm5. RNA. 2010;16:2484–2492. doi: 10.1261/rna.2376210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christian T., Gamper H., Hou Y.M. Conservation of structure and mechanism by Trm5 enzymes. RNA. 2013;19:1192–1199. doi: 10.1261/rna.039503.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rorbach J., Boesch P., Gammage P.A., Nicholls T.J., Pearce S.F., Patel D., Hauser A., Perocchi F., Minczuk M. MRM2 and MRM3 are involved in biogenesis of the large subunit of the mitochondrial ribosome. Mol. Biol. Cell. 2014;25:2542–2555. doi: 10.1091/mbc.E14-01-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minczuk M., Kolasinska-Zwierz P., Murphy M.P., Papworth M.A. Construction and testing of engineered zinc-finger proteins for sequence-specific modification of mtDNA. Nat. Protoc. 2010;5:342–356. doi: 10.1038/nprot.2009.245. [DOI] [PubMed] [Google Scholar]
- 36.Nicholls T.J., Zsurka G., Peeva V., Schöler S., Szczesny R.J., Cysewski D., Reyes A., Kornblum C., Sciacco M., Moggio M. Linear mtDNA fragments and unusual mtDNA rearrangements associated with pathological deficiency of MGME1 exonuclease. Hum. Mol. Genet. 2014;23:6147–6162. doi: 10.1093/hmg/ddu336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kornblum C., Nicholls T.J., Haack T.B., Schöler S., Peeva V., Danhauser K., Hallmann K., Zsurka G., Rorbach J., Iuso A. Loss-of-function mutations in MGME1 impair mtDNA replication and cause multisystemic mitochondrial disease. Nat. Genet. 2013;45:214–219. doi: 10.1038/ng.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urbonavicius J., Qian Q., Durand J.M., Hagervall T.G., Björk G.R. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 2001;20:4863–4873. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J., Esberg B., Curran J.F., Björk G.R. Three modified nucleosides present in the anticodon stem and loop influence the in vivo aa-tRNA selection in a tRNA-dependent manner. J. Mol. Biol. 1997;271:209–221. doi: 10.1006/jmbi.1997.1176. [DOI] [PubMed] [Google Scholar]
- 40.Hagervall T.G., Ericson J.U., Esberg K.B., Li J.N., Björk G.R. Role of tRNA modification in translational fidelity. Biochim. Biophys. Acta. 1990;1050:263–266. doi: 10.1016/0167-4781(90)90178-5. [DOI] [PubMed] [Google Scholar]
- 41.Pütz J., Florentz C., Benseler F., Giegé R. A single methyl group prevents the mischarging of a tRNA. Nat. Struct. Biol. 1994;1:580–582. doi: 10.1038/nsb0994-580. [DOI] [PubMed] [Google Scholar]
- 42.Paris Z., Horáková E., Rubio M.A., Sample P., Fleming I.M., Armocida S., Lukes J., Alfonzo J.D. The T. brucei TRM5 methyltransferase plays an essential role in mitochondrial protein synthesis and function. RNA. 2013;19:649–658. doi: 10.1261/rna.036665.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Becker L., Kling E., Schiller E., Zeh R., Schrewe A., Hölter S.M., Mossbrugger I., Calzada-Wack J., Strecker V., Wittig I. MTO1-deficient mouse model mirrors the human phenotype showing complex I defect and cardiomyopathy. PLoS ONE. 2014;9:e114918. doi: 10.1371/journal.pone.0114918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tischner C., Hofer A., Wulff V., Stepek J., Dumitru I., Becker L., Haack T., Kremer L., Datta A.N., Sperl W. MTO1 mediates tissue specificity of OXPHOS defects via tRNA modification and translation optimization, which can be bypassed by dietary intervention. Hum. Mol. Genet. 2015;24:2247–2266. doi: 10.1093/hmg/ddu743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Euro L., Konovalova S., Asin-Cayuela J., Tulinius M., Griffin H., Horvath R., Taylor R.W., Chinnery P.F., Schara U., Thorburn D.R. Structural modeling of tissue-specific mitochondrial alanyl-tRNA synthetase (AARS2) defects predicts differential effects on aminoacylation. Front. Genet. 2015;6:21. doi: 10.3389/fgene.2015.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki T., Suzuki T. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2014;42:7346–7357. doi: 10.1093/nar/gku390. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.