Abstract
Export of mRNA from the cell nucleus to the cytoplasm is essential for protein synthesis, a process vital to all living eukaryotic cells. mRNA export is highly conserved and ubiquitous. Mutations affecting mRNA and mRNA processing or export factors, which cause aberrant retention of mRNAs in the nucleus, are thus emerging as contributors to an important class of human genetic disorders. Here, we report that variants in THOC2, which encodes a subunit of the highly conserved TREX mRNA-export complex, cause syndromic intellectual disability (ID). Affected individuals presented with variable degrees of ID and commonly observed features included speech delay, elevated BMI, short stature, seizure disorders, gait disturbance, and tremors. X chromosome exome sequencing revealed four missense variants in THOC2 in four families, including family MRX12, first ascertained in 1971. We show that two variants lead to decreased stability of THOC2 and its TREX-complex partners in cells derived from the affected individuals. Protein structural modeling showed that the altered amino acids are located in the RNA-binding domains of two complex THOC2 structures, potentially representing two different intermediate RNA-binding states of THOC2 during RNA transport. Our results show that disturbance of the canonical molecular pathway of mRNA export is compatible with life but results in altered neuronal development with other comorbidities.
Main Text
Human neuronal development is an extremely complex process. It requires proper function and dosage of thousands of genes, as demonstrated by the complexity of currently known genetic architecture of neurodevelopmental disorders (NDDs) such as intellectual disability (ID), epilepsy, or autism.1 The recent revolution in systematic DNA sequencing and its application to large cohorts has dramatically accelerated identification of a plethora of potentially causative NDD variants. In-depth functional investigations on these variants are essential for timely and meaningful translation of this knowledge for short (diagnosis) and long-term (prognosis and treatment) clinical benefit.
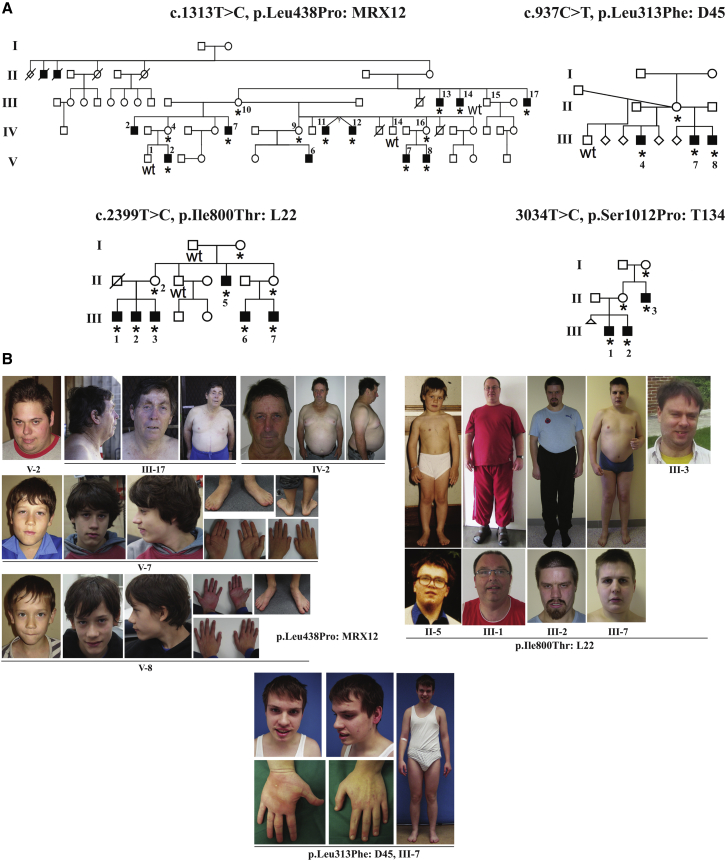
Variants in excess of 100 human X chromosome genes have been identified as causing X-linked NDDs, also known as X-linked ID, or XLID. Many more such disease-causing variants remain to be discovered, especially for non-syndromic forms.2 Here, we implicate THOC2 variants in XLID. As part of systematic X chromosome exome re-sequencing of ID-affected individuals from 405 families,3 we identified THOC2 variants in four multigenerational families. These variants are not present in >60,000 individuals from the 1000 Genomes Project and Exome Aggregation Consortium (ExAC) databases, resources which are unlikely to contain variants from individuals with NDDs. All four variants lead to missense substitutions, a class of mutations for which it is notoriously difficult to assign functional effect. The variants in THOC2 co-segregate with the phenotype in extended pedigrees (Figures 1A and 1B and Figure S1), affect highly conserved amino acids (Figure S2), and are predicted to be pathogenic by different bioinformatic tools (Table S1). The variant identified in the large Australian family, MRX12,4 (c.1313T>C [p.Leu438Pro], [GenBank: NM_001081550.1]) initially mapped outside the published linkage interval. However, upon reanalysis and inclusion of additional family members, we were able to redefine the linkage interval between markers DXS8067 and DXS8009 (chrX: 119,360,400–126,174,300; UCSC Genome Browser hg19); this region overlaps THOC2 (data not shown). Other THOC2 variants were identified in the European families D45 (c.937C>T [p.Leu313Phe], [GenBank: NM_001081550.1]), L22, (previously published as MRX355; c.2399T>C [p.Ile800Thr], [GenBank: NM_001081550.1]), and T134 (c.3034T>C [p.Ser1012Pro], [GenBank: NM_001081550.1]). The calculated LOD scores for MRX12, D45, L22, and T134 were 3.6, 1.2, 2.1, and 1.2 (combined LOD = 8.1), respectively. All procedures followed were in accordance with the ethical standards of the Women’s and Children’s Health Network Human Research Ethics Committee, and proper informed consent was obtained.
Figure 1.
THOC2 Missense Variants Identified By X Chromosome Exome Sequencing
(A) Pedigrees of the families affected by THOC2 missense variants. Sanger sequencing confirmed co-segregation of the variants with the phenotype. The pedigree of family MRX12 has been updated since it was first published.4 Roman numerals on the left of each pedigree indicate individual generation. Abbreviations are as follows: ∗, has a mutation; WT, does not have a mutation.
(B) Photographs of MRX12, D45, and L22 (previously MRX35) affected family members who harbor THOC2 variants.
All affected males in this study had ID, but this varied in severity from borderline to severe. Common additional clinical features of the affected individuals (n = 20) include speech delay, short stature, elevated BMI, and a truncal obesity pattern in older males in two of the four families (Table 1 and Table S2). A variety of neurological symptoms, including tremors, gait disturbance, and seizure disorder, were observed. Neuroradiological findings detected in the limited number of individuals who had had CNS imaging included mild ventriculomegaly, gliosis, inferior cerebellar vermis dysplasia, and cervical cord compression. Less common clinical features included microcephaly and microorchidism and/or microphallus. We did not recognize a distinctive facial gestalt (Figure 1B) or overlapping pattern of congenital anomalies. One of the boys was diagnosed with growth-hormone deficiency and is being successfully treated with growth-hormone replacement. One female from family L22 (II-2 in Figure 1A) had borderline-mild ID; otherwise, carrier females were not noted to have any NDDs. Taken together, our genetic and clinical data implicate these THOC2 variants in the causation of NDDs. This conclusion is also supported by the recent publication of an individual with cognitive impairment and who carries a de novo chromosome translocation that includes PTK2-THOC2 fusion genes.6 A de novo THOC2 missense variant (c.1550A>G [p.Tyr517Cys], [GenBank: NM_001081550.1]) was recently identified in a female with moderate-severe ID, speech problems, epileptic encephalopathy, cortical visual impairment, and gait disturbances7 (Epi4k Consortium and the Epilepsy Phenome/Genome Project investigators, personal communication).
Table 1.
Clinical Features of Individuals with THOC2 Mutations
|
Family T134 (p.Ser1012Pro) |
Family MRX12 (p.Leu438Pro) |
Family L22 (p.Ile800Thr) |
Family D45 (p.Leu313Phe) |
Summary: Affected/Total (Percentage) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| II-3 | III-1 | III-2 | III-13 | III-17 | IV-2 | IV-11 | IV-12 | V-2 | V-7 | V-8 | II-5 | III-1 | III-2 | III-3 | III-6 | III-7 | III-4 | III-7 | III-8 | ||
| Gender | male | male | male | male | male | male | male | male | male | male | male | male | male | male | male | male | male | male | male | male | NA |
| Age (year) | 9 | 5 | 21 | 77 | 61 | 63 | 51 | 51 | 30 | 14 | 9 | 44 | 43 | 42 | 34 | 30 | 28 | 24 | 30 | 38 | NA |
| Perinatal Features | |||||||||||||||||||||
| Prematurity | − | − | − | + | + | − | − | − | + | − | − | − | − | − | − | − | − | − | − | + | 4/20 (20%) |
| Low birth weight | − | − | + | − | − | − | + | − | + | − | + | − | − | − | − | − | − | − | − | − | 4/20 (20%) |
| Neurologic Features | |||||||||||||||||||||
| Intellectual disability | mod | bord | mod | mod | mod-sev | mod | mild | mild | mild | mod | mild | mild-mod | mod-sev | mod | mod | mod | mod | sev | mild-mod | mild | NA |
| Speech delay | + | − | + | + | − | + | + | + | + | + | + | + | + | + | + | − | − | + | − | − | 14/20 (70%) |
| Hypotonia | + | + | + | + | + | − | − | − | + | + | + | − | + | − | − | − | − | + | − | − | 10/20 (50%) |
| Hyperkinesia | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | + | + | + | − | − | 4/20 (20%) |
| Tremor | − | − | − | − | + | + | + | + | + | − | − | − | − | − | − | − | − | + | + | + | 8/20 (40%) |
| Epilepsy | + | − | + | − | − | − | − | − | − | − | − | − | + | − | − | − | − | + (GTC) | + | − | 5/20 (25%) |
| Gait disturbances | + | − | − | − | + | + | − | − | − | − | − | − | + | − | − | − | − | + | −b | −b | 5/20 (25%) |
| Behavior problems | + (ASD) | − | − | − | − | + | − | − | − | − | − | + | + | − | + | + | + | + (SM) | + (SM) | − | 9/20 (45%) |
| Anxiety problems | − | − | − | − | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 2/20 (10%) |
| Depression | − | − | − | − | − | + | − | + | − | − | − | − | − | − | − | − | − | − | − | − | 2/20 (10%) |
| Brain MRI and/or CT | ND | ND | ND | ND | CCC, glio | ND | ND | ND | N | ND | N | ND | VM | ND | ND | ND | ND | VM, CH | CVD | ND | 4/6 (67%) |
| Growth Parameters | |||||||||||||||||||||
| Microcephaly | − | − | + | − | + | − | + | − | − | + | + | − | − | − | − | − | − | − | − | − | 5/20 (25%) |
| Short stature (≤P3) | − | − | − | + | + | + | + | + | + | + | +a | + | + | + | + | − | + | − | − | − | 13/20 (65%) |
| Overweight (BMI ≥ 25) | ND | ND | ND | − | + | + | − | + | + | − | − | + | + | + | + | + | − | − | + | − | 10/17 (59%) |
| Dysmorphisms | |||||||||||||||||||||
| Broad high forehead | − | − | − | − | + | − | − | − | + | − | − | − | − | − | − | + | + | − | − | − | 4/20 (20%) |
| High palate | − | − | − | + | + | + | + | − | − | + | + | − | − | − | − | − | − | ND | ND | ND | 6/17 (35%) |
| Large ears (>2 SDs) | − | − | − | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 3/20 (15%) |
| Small penis and/or microorchidism | − | − | − | ND | ND | ND | ND | ND | ND | − | − | − | + | ND | + | + | + | ND | − | ND | 4/11 (36%) |
| Truncal obesity | − | − | − | + | + | + | + | + | + | − | − | + | + | + | + | + | + | − | + | − | 13/20 (65%) |
| Other | |||||||||||||||||||||
| Physical features | − | − | − | − | − | − | − | F | − | − | SP, F | − | strab, nys | − | − | HT | − | STM, ULS | − | ULS | NA |
| Medical conditions | − | − | − | IP, MI | − | − | − | LV | − | − | GH | − | − | − | − | − | − | − | − | CHD | NA |
Abbreviations are as follows: +, present; −, not present; ASD, autism spectrum disorder; bord, borderline; CCC, mild cervical cord compression; CH, cranial hyperostosis; CHD, congenital hip dysplasia; CT, computer tomography; CVD, inferior cerebellar vermis dysplasia; F, pes planus and hind foot valgus deformity; GH, growth hormone deficiency; glio, gliosis; GTC, generalized tonic clonic; HT, hypertelorism; IP, idiopathic pulmonary fibrosis; LV, increased left ventricular wall thickness; MI, myocardial infarction; mod, moderate; N, normal; NA, not applicable; ND, no data; sev, severe; strab, strabismus; nys, nystagmus; SM, self-mutilation; SP, single palmar crease; STM, stereotypic movements; ULS, evolving upper-limb spasticity; VM, ventriculomegaly.
On growth hormone treatment; height < P3 before treatment.
Gait disturbances were noted in childhood, but gait is normal in adulthood.
Human THOC2 encodes a 1,593-aa, 183-kDa nuclear protein with 98% amino acid identity to mouse THOC2. THOC2 forms part of the THO sub-complex (THOC1–3 and THOC5–7), which is an essential component of the large TREX complex (THO, UAP56, Aly, CIP29, PDIP3, ZC11A, and Chtop).8,9 THO proteins are related only by name and do not share significant sequence similarity. THOC2 is in high abundance in the developing and mature human10–12 and adult mouse brains.6 We observed THOC2 in primary mouse hippocampal and cortical neurons and human cerebral cortex and hippocampus (Figure S3). THOC2 function is critical for many living organisms as indicated by studies in yeast, roundworms, fruit flies, and vertebrates.6,13–17 THOC2 depletion has been shown to (1) interfere with mRNA export, chromosome alignment, mitotic progression, and genomic stability in humans13,15 and (2) stimulate neurite outgrowth in primary rat hippocampal neurons.6 Depletion of other TREX subunits also interferes with mRNA export, resulting in nuclear retention of mRNAs.15,16,18 Mouse Thoc2, along with Thoc5, is required for epithelial stem cell self-renewal and differentiation.19 Thoc2 depletion in Drosophila Schneider 2 (S2) cells results in significant mRNA nuclear retention, inhibition of protein synthesis and cell proliferation, and chromosome misalignment.16,17 thoc2-knockout Caenorhabditis elegans are slow growing, sterile, have functional defects in specific sensory neurons, and die prematurely from defective progression of meiosis.6,14 Thoc2 is an essential gene for zebrafish embryonic development because Thoc2 inactivation causes multiple anatomical abnormalities.20
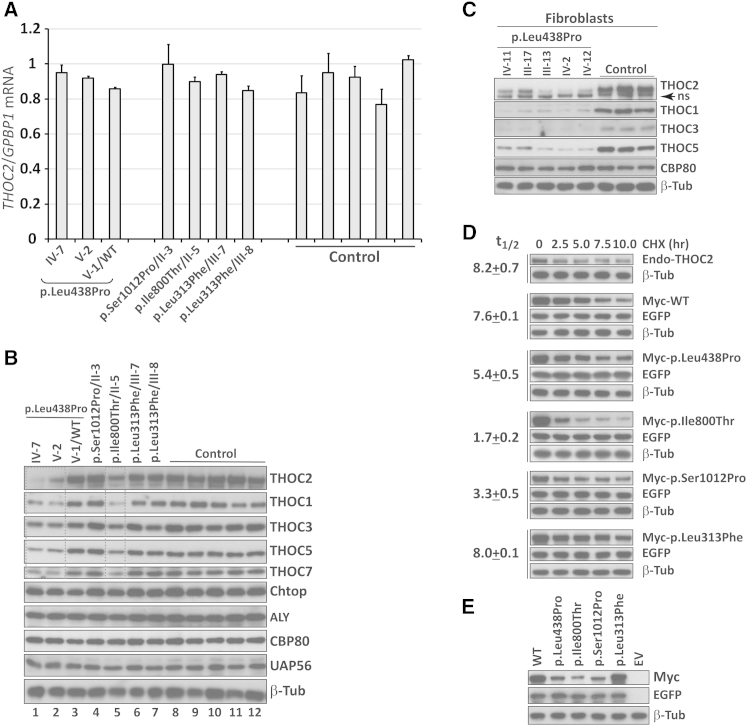
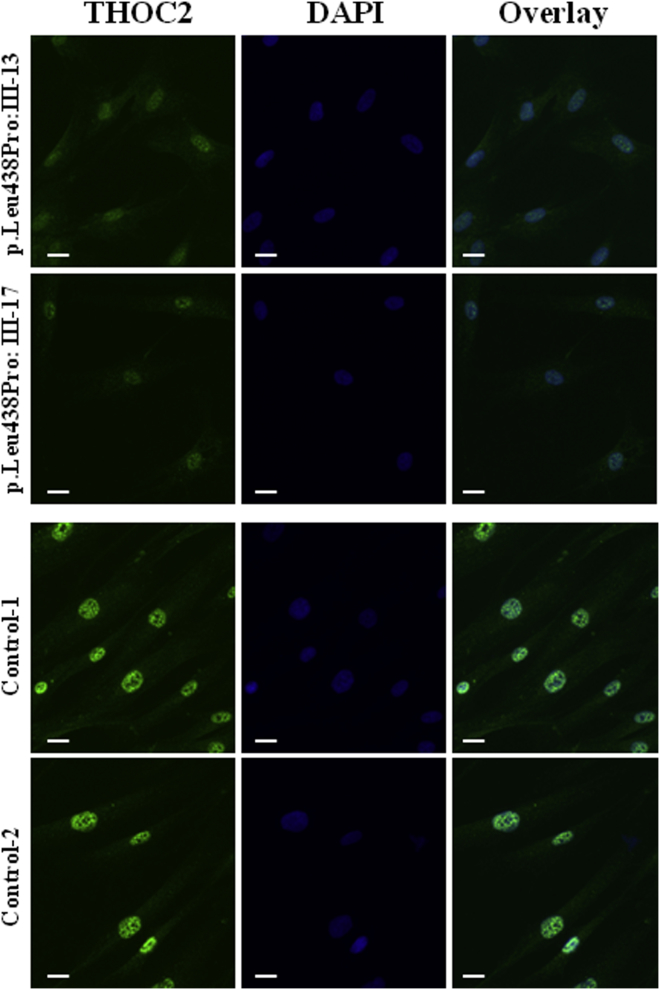
To investigate the functional effect of THOC2 variants, we used lymphoblastoid cell lines (LCLs; immortalized B lymphocytes) from at least one affected individual from each family. LCLs have been successfully used for inferring the biological relevance of pathway mutations causing neurological disease.21,22 We sought to determine the effect of THOC2 variants on protein localization or stability. Using immunofluorescence staining assays, we saw no mislocalization of ectopic Myc-tagged altered THOC2 in HeLa cells and primary mouse cortical neurons (E18; data not shown). We also observed no impact on THOC2 mRNA expression in mutant LCLs by qRT-PCR (Figure 2A). However, when compared to that of controls, the amount of altered THOC2 was significantly reduced in two LCLs (with the c.1313T>C [p.Leu438Pro] and c.2399T>C [p.Ile800Thr] variants) but were unchanged in LCLs harboring the c.3034T>C (p.Ser1012Pro) and c.937C>T (p.Leu313Phe) variants (Figure 2B). Given that THOC2 depletion destabilizes the THO sub-complex proteins THOC1, THOC3, THOC5, and THOC7 of the TREX complex in HeLa cells,8 we investigated the amounts of these complex partners in affected cells. We detected reduced amounts of these THO subunits in LCL lysates from the c.1313T>C (p.Leu438Pro) and c.2399T>C (p.Ile800Thr) mutants, but not in those of the c.3034T>C (p.Ser1012Pro) and c.937C>T (p.Leu313Phe) mutants. Protein amounts of the other TREX subunits, Aly, UAP56, and Chtop, remained unchanged (Figure 2B). THO sub-complex instability was confirmed with five different primary skin fibroblasts from the affected individuals of the MRX12 family with the c.1313T>C (p.Leu438Pro) mutation (Figure 2C). Immunofluorescence showed very low amounts of diffused THOC2 nuclear staining in c.1313T>C (p.Leu438Pro) fibroblasts in comparison to bright speckled staining in controls (Figure 3 and Figure S4). Reduced THOC2 amounts in LCLs and skin fibroblasts from the c.1313T>C (p.Leu438Pro)-affected individuals (MRX12) and in LCLs from the c.2399T>C (p.Ile800Thr)-affected individuals (L22) strongly suggest compromised THOC2 function.
Figure 2.
Mutations Affect THOC2 Stability, Whereas mRNA Expression Is Unaltered
(A) THOC2 mRNA expression remained unaltered in LCLs derived from affected individuals in comparison to expression in LCLs from control individuals. Amounts of THOC2 mRNA were assayed by qRT-PCR and normalized to the housekeeping gene GPBP1’s mRNA. Primers are listed in Table S3. Error bars show SDs, which were calculated from three independent experiments.
(B) THOC2 (and THOC1, THOC3, THOC5, and THOC7) amounts were lower in p.Leu438Pro and p.Ile800Thr altered cells (boxed), but not in p.Ser1012Pro or p.Leu313Phe LCLs, than in control LCLs. Protein lysates from control and mutant LCLs were analyzed by western blotting. β-tubulin probing was used as a loading control. LCL proteins were extracted and western blotting was performed as reported previously.23 Western blots were probed with the following antibodies: rabbit anti-THOC2 (303-629A), rabbit anti-NCBP1/CBP80, rabbit anti-THOC1, rabbit anti-Aly, rabbit anti-SRAG/Chtop (Bethyl laboratories), rabbit anti-THOC7, rabbit anti-THOC3 (Sigma-Aldrich), rabbit anti-CIP29 (Thermo Scientific), and rabbit β-tubulin (Abcam).
(C) Amounts of p.Leu438Pro THOC2 were reduced in fibroblasts from five different affected individuals from the MRX12 family. Protein lysates from control and mutant fibroblasts were analyzed by western blotting with the antibodies shown. Probing for β-tubulin was used as a loading control.
(D) Mutations affect THOC2 turnover. For generating Myc-tagged human THOC2 expression plasmids, THOC2 coding sequence was PCR amplified from brain cDNA with hTHOC2-XbaI-XhoI-F and hTHOC2-NotI-R primers (Table S3) and cloned at XhoI-NotI sites in the pCMV-Myc mammalian expression vector. Each mutation was introduced by the overlap PCR method using the primers listed in Table S3. All constructs were validated by Sanger sequencing. Untransfected HEK293T cells or HEK293T cells transfected with wild-type or altered Myc-tagged THOC2 and EGFP expression plasmid were treated with the translation inhibitor cycloheximide (CHX) and harvested at different time points after CHX addition. Cell lysates were analyzed by western blotting with an antibody to the Myc tag, EGFP (transfection control), or β-tubulin (loading control). Whereas amounts of the endogenous, wild-type, or p.Leu313Phe THOC2 proteins declined slowly over a period of 8.0 hr, p.Leu438Pro (5.4 hr), p.Ile800Thr (1.7 hr), or p.Ser1012Pro (3.3 hr) amounts declined more rapidly. The amounts of THOC2 and loading-control β-tubulin were measured by quantification of the band intensities with ImageJ software. THOC2 amounts were relativized to a β-tubulin loading control for determining the half-life of proteins. Each experiment was repeated three times, and the results of one representative experiment are shown. Data shown are the mean ± SD. Note that western blot signals for p.Leu438Pro, p.Ile800Thr, and p.Ser1012Pro at 0 hr look different than those in Figure 2E. This is because blots were exposed for durations different to those shown in Figure 2E.
(E) THOC2 variants affect stability of ectopic recombinant Myc-tagged THOC2 proteins. Total lysates of HEK293T cells transfected with wild-type THOC2 (WT), altered Myc-tagged THOC2, or empty vector (EV) and EGFP expression plasmid were analyzed by western blotting with an antibody to the Myc tag, EGFP (transfection control), or β tubulin (loading control).
Figure 3.
THOC2 Immunofluorescence Staining Is Significantly Reduced in THOC2 p.Leu438Pro Fibroblasts
Fibroblasts seeded on coverslips precoated with BD BioCoat were fixed with 4% paraformaldehyde for 10 min at room temperature and permeabilized with 0.2% Triton X-100 for 10 min at room temperature. Cells were then incubated with rabbit anti-THOC2 antibody (303-630A; Bethyl Laboratories) at 500 ng/ml in 10% horse serum in 1 × PBS overnight at 4°C and then in a 1:2,500 dilution of Alexa Fluor 488 donkey anti-rabbit IgG (H+L) antibody in 10% horse serum in 1 × PBS for 1 hr at room temperature and mounted in ProLong Gold Antifade Mountant with DAPI. Cells were imaged with an Axiocam camera (AxioCam MRm). Scale bars represent 20 μm. See also Figure S4.
Because THOC2 is known to be ubiquitylated,24,25 the reduced amounts of p.Leu438Pro and p.Ile800Thr proteins are most likely the result of enhanced proteasome-mediated degradation. To test this, we subsequently compared the turnover rates of endogenous and ectopic Myc-tagged wild-type or altered recombinant THOC2 in the presence of the translational inhibitor cycloheximide. Whereas the endogenous wild-type and tagged p.Leu313Phe THOC2 proteins have half-lives of about 8.0 hr, both the p.Leu438Pro and p.Ile800Thr altered proteins showed significantly shorter half-lives of 5.4 hr and 1.7 hr, respectively (Figure 2D). This is consistent with our results obtained on endogenous altered THOC2 in the cells derived from the affected individuals (see Figures 2B and 2C). These results were also consistent with the reduced amounts of ectopic THOC2 p.Leu438Pro and p.Ile800Thr in untreated HEK293T cells (Figure 2E). Interestingly however, the amount of altered p.Ser1012Pro also declined rapidly (3.3 hr) in the presence of cycloheximide (Figure 2D), which might be due to the addition of an epitope tag to the altered THOC2. No change in the amount of EGFP from the co-transfected plasmid over the course of cycloheximide sampling indicated that the differences in amount of THOC2 at various time points were not due to differences in the transfection efficiency of THOC2 expression plasmids (Figures 2D and 2E).
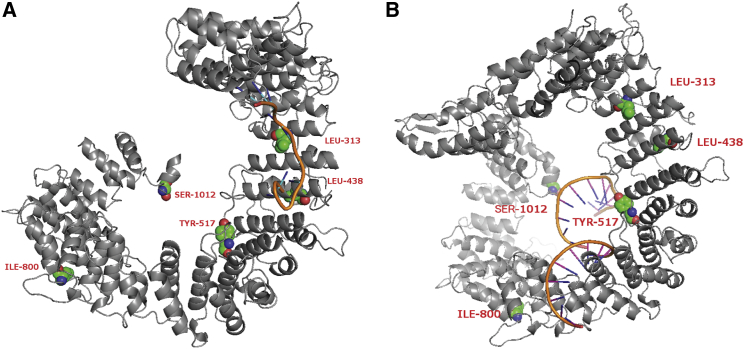
We also investigated the predicted impact of altered amino acid residues on THOC2 by using a structural model of THOC2 (SPARKS-X program26,27). We built THOC2 complex models by aligning the predicted model to known protein-RNA complex structures (Figure S5). Predicted THOC2 structures showed that the p.Leu313Phe and p.Leu438Pro variants identified in this study and the previously reported p.Tyr517Cys variant are located in the RNA-binding regions. The predicted complex structures suggest two potential intermediate RNA-binding states of THOC2 in transporting RNAs (Figure 4).
Figure 4.
Models of the Predicted Complex Structure of the THOC2-RNA Complex
(A) Complex structure was built on the basis of the template 1m8y. Variants p.Leu313Phe and p.Leu438Pro are located in mRNA-binding sites.
(B) Complex structure was built on the basis of the template 3a6p. Variant Tyr517Cys is in an mRNA-binding site.
The workflow used for predicting the THOC2-RNA interaction is shown (Figure S4). First, the sequence of THOC2 was analyzed by SPARKS-X, which predicts several potential protein structures. Then, the best structure model was compared to a library of 1,164 RNA-binding proteins, and the two best-aligned complex-structure models were generated on the basis of chains 3a6pA and 1m8yA by SPalign28 and had SP-scores of 0.68 and 0.65, respectively. A score > 0.5 indicates homologous structures, which were selected for building the complex models. From the complex models, a protein residue and a RNA base were considered in contact if the shortest distance between any pair of heavy atoms from them was within 4.5Å. For the first complex structure, variants p.Leu313Phe and p.Leu438Pro were located in the mRNA-binding domain, but p.Leu313Phe interacted with RNA. The other two variants, p.Ile800Thr and p.Ser1012Pro, were located on another domain, which was annotated with DNA-binding function by Pfam.29
The process of mRNA export is tightly regulated and highly conserved from yeast to mammals.30,31 Mutations that cause mRNA-export problems are rare because of their substantial impact on cells, organs, and individuals.32,33 For example, mutations in the human mRNA-export mediator GLE1 (MIM: 603371) result in a severe fetal motor neuron disease34 (lethal congenital contracture syndrome 1 [MIM: 253310]) and amyotrophic lateral sclerosis.35 Impaired RNA transport out of the nucleus can be caused by a splice-site mutation in COL1A1 (MIM: 120150) in some affected individuals with osteogenesis imperfect, type I36 (MIM: 166200). Impaired RNA transport out of the nucleus can also be caused by toxic CUG expansion in the 3′ UTR of the dystrophia myotonica-protein kinase mRNA in individuals affected by myotonic dystrophy 1 (MIM: 160900).33,37 In addition, a missense mutation (c.136G>A [p.Gly46Arg]) has been described as causing THOC6 (MIM: 615403) loss of function and protein mislocalization to the cytoplasm and was associated with a syndromic form of autosomal-recessive ID.38 More recently, mutations in DCPS, encoding an mRNA decapping enzyme (MIM: 610534), and EDC3, encoding enhancer of mRNA decapping 3 (MIM: 609842), have been implicated in ID.39,40
All human cells require mRNA export for efficient protein synthesis, yet tissue-specific defects due to mutations in general mRNA-export factors have been observed.33 In mice, Thoc5 depletion interferes with the maintenance of hematopoiesis,41,42 and Thoc1 deficiency interferes with testis development.43 Embryonic lethality of Thoc1- and Thoc5-knockout mice also suggests a requirement for the THO complex during early development.30 It appears that different tissues might require different amounts of specific export factors for their function.30,33 Given that THOC2 is highly expressed in the developing and mature human brain,10–12 it is not surprising that dysregulation of THOC2 function leads to neurodevelopmental clinical outcomes.
THOC2, THOC1, and THOC7 depletion results in severe mRNA-export blockage.8 Complete absence of THOC2 is most likely lethal. The correspondence between the abundance of THOC2 and the extent of mRNA-export blockage suggests that THOC2 is an essential mRNA-export factor. 8 In addition, evidence from lower-animal studies predicts that complete Thoc2 deficiency would cause severe developmental defects.6,14,16,17,20 This is consistent with the observation that THOC2 is intolerant to variation (residual variation intolerance score = −0.53, 18.95th percentile)44 and is a highly constrained human gene.45 We therefore assume that the clinical manifestations in our affected individuals are most likely the consequence of a partial rather than complete loss of THOC2 function. This is consistent with the reduced amount of THOC2 (and consequently reduced stability of THOC1, THOC3, THOC5, and THOC7) rather than complete absence of THOC2. Whether TREX export activity is altered by reduced THOC2 amounts alone or by the simultaneous reduction of other THO subunits is not clear at present. Notably, however, THOC2 p.Leu438Pro and p.Ile800Thr instability was associated with short stature (in 13/14 individuals) in the two families. This might be due to reduced cell proliferation, as also observed in Thoc2-depleted Drosophila S2 cells16 resulting from impaired mRNA export. The mechanism of perturbed p.Leu313Phe and p.Ser1012Pro THOC2 function is not clear at present, although these variants might have an effect on THOC2 interaction with RNA or other proteins, which subsequently results in altered mRNA export.
In conclusion, our data indicate that partial loss-of-function THOC2 variants cause NDD with a broad phenotypic spectrum. We predict that mRNA export is altered in cells harboring THOC2 mutations, which most likely impair protein synthesis, and thus underpins clinical presentations in the affected individuals. Taken together, our results identify THOC2 and the nuclear mRNA-export complex as crucial factors for proper neuronal development.
Acknowledgments
This work was supported by the National Health and Medical Research Council (grants 628952 and 1041920 to J.G.), the Channel 7 Children’s Research Foundation (R.K., M.F., and J.G.), the European Commission 7th Framework Programme project Genetic and Epigenetic Networks in Cognitive Dysfunction (grant 241995 to H.H. and V.M.K.), the MS McLeod Research Fellowship from the Women’s and Children’s Hospital Foundation (to M.A.C.), and the Ter Meulen Fonds stipendium (to B.W.M.v.B). We thank Amanda Springer, Sharon Grosvenor, and Julie Rogerson for their help with the MRX12 family. The authors are also grateful to the families who participated in the study.
Published: July 9, 2015
Footnotes
Supplemental Data include five figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.05.021.
Web Resources
The URLs for data presented herein are as follows:
ExAC Browser, http://exac.broadinstitute.org
MutationTaster, http://www.mutationtaster.org/
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
PROVEAN, http://provean.jcvi.org
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
References
- 1.Hu W.F., Chahrour M.H., Walsh C.A. The diverse genetic landscape of neurodevelopmental disorders. Annu. Rev. Genomics Hum. Genet. 2014;15:195–213. doi: 10.1146/annurev-genom-090413-025600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gécz J., Shoubridge C., Corbett M. The genetic landscape of intellectual disability arising from chromosome X. Trends Genet. 2009;25:308–316. doi: 10.1016/j.tig.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Hu H., Haas S.A., Chelly J., Van Esch H., Raynaud M., de Brouwer A.P., Weinert S., Froyen G., Frints S.G., Laumonnier F. X-exome sequencing of 405 unresolved families identifies seven novel intellectual disability genes. Mol. Psychiatry. 2015 doi: 10.1038/mp.2014.193. Published online February 3, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kerr B., Gedeon A., Mulley J., Turner G. Localization of non-specific X-linked mental retardation genes. Am. J. Med. Genet. 1992;43:392–401. doi: 10.1002/ajmg.1320430160. [DOI] [PubMed] [Google Scholar]
- 5.Gu X.X., Decorte R., Marynen P., Fryns J.P., Cassiman J.J., Raeymaekers P. Localisation of a new gene for non-specific mental retardation to Xq22-q26 (MRX35) J. Med. Genet. 1996;33:52–55. doi: 10.1136/jmg.33.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Gregorio E., Bianchi F.T., Schiavi A., Chiotto A.M., Rolando M., Verdun di Cantogno L., Grosso E., Cavalieri S., Calcia A., Lacerenza D. A de novo X;8 translocation creates a PTK2-THOC2 gene fusion with THOC2 expression knockdown in a patient with psychomotor retardation and congenital cerebellar hypoplasia. J. Med. Genet. 2013;50:543–551. doi: 10.1136/jmedgenet-2013-101542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allen A.S., Berkovic S.F., Cossette P., Delanty N., Dlugos D., Eichler E.E., Epstein M.P., Glauser T., Goldstein D.B., Han Y., Epi4K Consortium. Epilepsy Phenome/Genome Project De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chi B., Wang Q., Wu G., Tan M., Wang L., Shi M., Chang X., Cheng H. Aly and THO are required for assembly of the human TREX complex and association of TREX components with the spliced mRNA. Nucleic Acids Res. 2013;41:1294–1306. doi: 10.1093/nar/gks1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang C.T., Hautbergue G.M., Walsh M.J., Viphakone N., van Dijk T.B., Philipsen S., Wilson S.A. Chtop is a component of the dynamic TREX mRNA export complex. EMBO J. 2013;32:473–486. doi: 10.1038/emboj.2012.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X., Somel M., Tang L., Yan Z., Jiang X., Guo S., Yuan Y., He L., Oleksiak A., Zhang Y. Extension of cortical synaptic development distinguishes humans from chimpanzees and macaques. Genome Res. 2012;22:611–622. doi: 10.1101/gr.127324.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson M.B., Kawasawa Y.I., Mason C.E., Krsnik Z., Coppola G., Bogdanović D., Geschwind D.H., Mane S.M., State M.W., Sestan N. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 13.Paulsen R.D., Soni D.V., Wollman R., Hahn A.T., Yee M.C., Guan A., Hesley J.A., Miller S.C., Cromwell E.F., Solow-Cordero D.E. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol. Cell. 2009;35:228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castellano-Pozo M., García-Muse T., Aguilera A. R-loops cause replication impairment and genome instability during meiosis. EMBO Rep. 2012;13:923–929. doi: 10.1038/embor.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamazaki T., Fujiwara N., Yukinaga H., Ebisuya M., Shiki T., Kurihara T., Kioka N., Kambe T., Nagao M., Nishida E., Masuda S. The closely related RNA helicases, UAP56 and URH49, preferentially form distinct mRNA export machineries and coordinately regulate mitotic progression. Mol. Biol. Cell. 2010;21:2953–2965. doi: 10.1091/mbc.E09-10-0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rehwinkel J., Herold A., Gari K., Köcher T., Rode M., Ciccarelli F.L., Wilm M., Izaurralde E. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat. Struct. Mol. Biol. 2004;11:558–566. doi: 10.1038/nsmb759. [DOI] [PubMed] [Google Scholar]
- 17.Somma M.P., Ceprani F., Bucciarelli E., Naim V., De Arcangelis V., Piergentili R., Palena A., Ciapponi L., Giansanti M.G., Pellacani C. Identification of Drosophila mitotic genes by combining co-expression analysis and RNA interference. PLoS Genet. 2008;4:e1000126. doi: 10.1371/journal.pgen.1000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lei H., Dias A.P., Reed R. Export and stability of naturally intronless mRNAs require specific coding region sequences and the TREX mRNA export complex. Proc. Natl. Acad. Sci. USA. 2011;108:17985–17990. doi: 10.1073/pnas.1113076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L., Miao Y.L., Zheng X., Lackford B., Zhou B., Han L., Yao C., Ward J.M., Burkholder A., Lipchina I. The THO complex regulates pluripotency gene mRNA export and controls embryonic stem cell self-renewal and somatic cell reprogramming. Cell Stem Cell. 2013;13:676–690. doi: 10.1016/j.stem.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amsterdam A., Nissen R.M., Sun Z., Swindell E.C., Farrington S., Hopkins N. Identification of 315 genes essential for early zebrafish development. Proc. Natl. Acad. Sci. USA. 2004;101:12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen L.S., Jolly L., Shoubridge C., Chan W.K., Huang L., Laumonnier F., Raynaud M., Hackett A., Field M., Rodriguez J. Transcriptome profiling of UPF3B/NMD-deficient lymphoblastoid cells from patients with various forms of intellectual disability. Mol. Psychiatry. 2012;17:1103–1115. doi: 10.1038/mp.2011.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishimura Y., Martin C.L., Vazquez-Lopez A., Spence S.J., Alvarez-Retuerto A.I., Sigman M., Steindler C., Pellegrini S., Schanen N.C., Warren S.T., Geschwind D.H. Genome-wide expression profiling of lymphoblastoid cell lines distinguishes different forms of autism and reveals shared pathways. Hum. Mol. Genet. 2007;16:1682–1698. doi: 10.1093/hmg/ddm116. [DOI] [PubMed] [Google Scholar]
- 23.Johansson P., Jeffery J., Al-Ejeh F., Schulz R.B., Callen D.F., Kumar R., Khanna K.K. SCF-FBXO31 E3 ligase targets DNA replication factor Cdt1 for proteolysis in the G2 phase of cell cycle to prevent re-replication. J. Biol. Chem. 2014;289:18514–18525. doi: 10.1074/jbc.M114.559930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopitz-Otsoa F., Rodriguez-Suarez E., Aillet F., Casado-Vela J., Lang V., Matthiesen R., Elortza F., Rodriguez M.S. Integrative analysis of the ubiquitin proteome isolated using Tandem Ubiquitin Binding Entities (TUBEs) J. Proteomics. 2012;75:2998–3014. doi: 10.1016/j.jprot.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 25.Wagner S.A., Beli P., Weinert B.T., Nielsen M.L., Cox J., Mann M., Choudhary C. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.013284. M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Y., Faraggi E., Zhao H., Zhou Y. Improving protein fold recognition and template-based modeling by employing probabilistic-based matching between predicted one-dimensional structural properties of query and corresponding native properties of templates. Bioinformatics. 2011;27:2076–2082. doi: 10.1093/bioinformatics/btr350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao H., Yang Y., Zhou Y. Highly accurate and high-resolution function prediction of RNA binding proteins by fold recognition and binding affinity prediction. RNA Biol. 2011;8:988–996. doi: 10.4161/rna.8.6.17813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao H., Yang Y., Zhou Y. Structure-based prediction of RNA-binding domains and RNA-binding sites and application to structural genomics targets. Nucleic Acids Res. 2011;39:3017–3025. doi: 10.1093/nar/gkq1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katahira J. mRNA export and the TREX complex. Biochim. Biophys. Acta. 2012;1819:507–513. doi: 10.1016/j.bbagrm.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 31.Reed R., Cheng H. TREX, SR proteins and export of mRNA. Curr. Opin. Cell Biol. 2005;17:269–273. doi: 10.1016/j.ceb.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 32.Carmody S.R., Wente S.R. mRNA nuclear export at a glance. J. Cell Sci. 2009;122:1933–1937. doi: 10.1242/jcs.041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hurt J.A., Silver P.A. mRNA nuclear export and human disease. Dis. Model. Mech. 2008;1:103–108. doi: 10.1242/dmm.000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nousiainen H.O., Kestilä M., Pakkasjärvi N., Honkala H., Kuure S., Tallila J., Vuopala K., Ignatius J., Herva R., Peltonen L. Mutations in mRNA export mediator GLE1 result in a fetal motoneuron disease. Nat. Genet. 2008;40:155–157. doi: 10.1038/ng.2007.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaneb H.M., Folkmann A.W., Belzil V.V., Jao L.E., Leblond C.S., Girard S.L., Daoud H., Noreau A., Rochefort D., Hince P. Deleterious mutations in the essential mRNA metabolism factor, hGle1, in amyotrophic lateral sclerosis. Hum. Mol. Genet. 2015;24:1363–1373. doi: 10.1093/hmg/ddu545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson C., Primorac D., McKinstry M., McNeil J., Rowe D., Lawrence J.B. Tracking COL1A1 RNA in osteogenesis imperfecta. splice-defective transcripts initiate transport from the gene but are retained within the SC35 domain. J. Cell Biol. 2000;150:417–432. doi: 10.1083/jcb.150.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brook J.D., McCurrach M.E., Harley H.G., Buckler A.J., Church D., Aburatani H., Hunter K., Stanton V.P., Thirion J.P., Hudson T. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;69:385. doi: 10.1016/0092-8674(92)90418-c. [DOI] [PubMed] [Google Scholar]
- 38.Beaulieu C.L., Huang L., Innes A.M., Akimenko M.A., Puffenberger E.G., Schwartz C., Jerry P., Ober C., Hegele R.A., McLeod D.R., FORGE Canada Consortium Intellectual disability associated with a homozygous missense mutation in THOC6. Orphanet J. Rare Dis. 2013;8:62. doi: 10.1186/1750-1172-8-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahmed I., Buchert R., Zhou M., Jiao X., Mittal K., Sheikh T.I., Scheller U., Vasli N., Rafiq M.A., Brohi M.Q. Mutations in DCPS and EDC3 in autosomal recessive intellectual disability indicate a crucial role for mRNA decapping in neurodevelopment. Hum. Mol. Genet. 2015;24:3172–3180. doi: 10.1093/hmg/ddv069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ng C.K., Shboul M., Taverniti V., Bonnard C., Lee H., Eskin A., Nelson S.F., Al-Raqad M., Altawalbeh S., Séraphin B., Reversade B. Loss of the scavenger mRNA decapping enzyme DCPS causes syndromic intellectual disability with neuromuscular defects. Hum. Mol. Genet. 2015;24:3163–3171. doi: 10.1093/hmg/ddv067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guria A., Tran D.D., Ramachandran S., Koch A., El Bounkari O., Dutta P., Hauser H., Tamura T. Identification of mRNAs that are spliced but not exported to the cytoplasm in the absence of THOC5 in mouse embryo fibroblasts. RNA. 2011;17:1048–1056. doi: 10.1261/rna.2607011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mancini A., Niemann-Seyde S.C., Pankow R., El Bounkari O., Klebba-Färber S., Koch A., Jaworska E., Spooncer E., Gruber A.D., Whetton A.D., Tamura T. THOC5/FMIP, an mRNA export TREX complex protein, is essential for hematopoietic primitive cell survival in vivo. BMC Biol. 2010;8:1. doi: 10.1186/1741-7007-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X., Chinnam M., Wang J., Wang Y., Zhang X., Marcon E., Moens P., Goodrich D.W. Thoc1 deficiency compromises gene expression necessary for normal testis development in the mouse. Mol. Cell. Biol. 2009;29:2794–2803. doi: 10.1128/MCB.01633-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnström K., Mallick S., Kirby A. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.