Abstract
The gene(s) whose expression is regulated by allergy risk variants is unknown for many loci identified through genome-wide association studies. Addressing this knowledge gap might point to new therapeutic targets for allergic disease. The aim of this study was to identify the target gene(s) and the functional variant(s) underlying the association between rs7009110 on chromosome 8q21 and allergies. Eight genes are located within 1 Mb of rs7009110. Multivariate association analysis of publicly available exon expression levels from lymphoblastoid cell lines (LCLs) identified a significant association between rs7009110 and the expression of a single gene, PAG1 (p = 0.0017), 732 kb away. Analysis of histone modifications and DNase I hypersensitive sites in LCLs identified four putative regulatory elements (PREs) in the region. Chromosome conformation capture confirmed that two PREs interacted with the PAG1 promoter, one in allele-specific fashion. To determine whether these PREs were functional, LCLs were transfected with PAG1 promoter-driven luciferase reporter constructs. PRE3 acted as a transcriptional enhancer for PAG1 exclusively when it carried the rs2370615:C allergy predisposing allele, a variant in complete linkage disequilibrium with rs7009110. As such, rs2370615, which overlaps RelA transcription factor (TF) binding in LCLs and was found to disrupt Foxo3a binding to PRE3, represents the putative functional variant in this locus. Our studies suggest that the risk-associated allele of rs2370615 predisposes to allergic disease by increasing PAG1 expression, which might promote B cell activation and have a pro-inflammatory effect. Inhibition of PAG1 expression or function might have therapeutic potential for allergic diseases.
Main Text
To date, genome-wide association studies (GWASs) have identified 41 genetic associations with allergic diseases (Table S1), including asthma (MIM: 600807), hay fever or allergic rhinitis (MIM: 607154), and atopic dermatitis or eczema (MIM: 603165). The identification of allergy risk variants is expected to provide new insights into the molecular pathways involved in disease pathophysiology and, in this way, facilitate the development of new disease treatments. However, these expectations have been hard to meet and this represents a major bottleneck in the field. There are two main reasons for this. First, for many loci, there is no functional evidence linking the risk variant with changes in the expression or protein sequence of any nearby genes; the likely target gene(s) is therefore unknown. This is the case for 27 of the 41 allergy risk loci discovered to date; of note, for 10 of these 27 loci, experimental mouse models of allergic disease implicate nearby genes in disease pathophysiology. Second, often there is little or no information available to determine whether and how disruption of the expression or sequence of the target gene impacts cellular function and, ultimately, disease pathophysiology. Addressing these knowledge gaps is critical to help translate GWAS findings into clinically useful information.
In a recent GWAS, we compared 6,685 individuals with both asthma and hay fever against 14,091 control subjects free of asthma and hay fever and identified two new risk loci for allergic disease: 8q21 and 16p13.1 Variants in both were associated with the individual risks of asthma and hay fever, but the association was stronger with the combined asthma and hay fever phenotype. In the 16p13 locus, evidence from gene expression studies suggests that DEXI is the most likely target gene of this association;2,3 functional studies of this gene are now warranted to understand how variation in its expression might affect disease risk. Currently, the gene whose expression is regulated by allergy risk variants in the 8q21 locus is unknown. Therefore, the aim of this study was to use both population genetics and functional approaches to identify the target gene(s) and likely causal variant(s) underlying the 8q21 association with allergy risk. The study procedures used were approved by the Human Research Ethics Committee from the QIMR Berghofer Medical Research Institute.
Eight genes and one miRNA are located within 1 Mb of the sentinel SNP rs7009110, which has a 36% risk allele frequency in Europeans and was associated with a 1.14 per-allele odds of disease,1 the nearest gene being ZBTB10 (Figure 1A). rs7009110 was the variant with the strongest association with disease risk after imputation of unmeasured variants from the 1000 Genomes Project.1 Forty-three gene expression quantitative trait loci (eQTLs) have been reported in this 2 Mb region across six relevant tissue or cell types, but rs7009110 is not in linkage disequilibrium (LD) with any of these (Table S2). However, rs7009110 is in modest LD with two nearby risk variants associated with eczema (rs7000782, r2 = 0.51)4 and rheumatoid arthritis (MIM: 180300; rs998731, r2 = 0.20);5 in Europeans, the risk alleles for these three variants (rs7009110:T, rs7000782:A, and rs998731:T) are on the same haplotype. The associations at this locus with asthma, hay fever, eczema, and rheumatoid arthritis suggest that the underlying disrupted molecular mechanism affects a component of the immune system that is shared between allergic and auto-immune diseases. The most plausible candidate target genes are TPD52 (MIM: 604068), which is involved in B cell maturation;6 PAG1 (MIM: 605767), involved in T and B cell activation;7–10 and ZBTB10, a putative repressor of the Sp1 transcription factor (TF),11 which regulates multiple immune-related genes.12,13
Figure 1.
Allergy Risk SNPs Are Associated with PAG1 Expression and Overlap Putative Regulatory Elements
(A) Location of the core region of association (blue rectangle) and genes located within 1 Mb of the GWAS sentinel SNP, rs7009110.
(B) Results from multivariate association analysis (n = 373) between exon expression levels and variants in the core region of association. The color of each variant reflects linkage disequilibrium with rs7009110 (purple square).
(C) Location of four PREs, based on histone marks and DNase I hypersensitive sites in an LCL analyzed by the ENCODE project.
(D) Location of the 3C interacting fragments, the two fragments cloned in the luciferase assays, and the SNPs that are in LD (r2 > 0.6) with rs7009110 and overlap the cloned fragments.
We first considered the possibility that the lack of a published association between rs7009110 and the expression of nearby genes could represent a false-negative finding, arising because (1) a true association did not exceed the false discovery rate threshold in the original eQTL studies and so was not reported; and/or (2) most published eQTL studies surveyed used expression microarrays, which have incomplete coverage of gene expression patterns. To test this possibility, we analyzed RNA-seq data obtained by the Geuvadis Project14 for lymphoblastoid cell line (LCL) samples of 373 individuals of European descent from the 1000 Genomes Project.15 LCLs are derived from peripheral blood B cells and therefore represent a practical and effective in vitro model to study gene expression patterns relevant to immune-related conditions.
Genotype and RNA-seq data were downloaded from EBI ArrayExpress: E-GEUV-1 and E-GEUV-2. As in Lappalainen,14 we selected the exon (not the gene) as the quantification unit and restricted the analysis to exons expressed in >90% of all the individuals. Read counts normalized by library depth and with technical variation removed by PEER normalization14 were available for all exons of six of the eight genes located within a 1 Mb region around rs7009110. Read counts were quantile normalized and adjusted for ancestry informative covariates and genotype imputation status, as in Lappalainen.14 For each of these six genes, the association between rs7009110 allelic dosage and variation in the expression of all exons was tested simultaneously via a multivariate test of association that improves power over the alternative strategy of testing each exon individually.16 The remaining two genes, ZNF704 and STMN2 (MIM: 600621), were expressed in only 56% and 12% of samples, respectively. Given their low relative abundance and because there was no association between dichotomized read counts (expressed versus not expressed) and rs7009110 genotype (not shown), both genes were excluded from further analysis. Likewise, the expression of miR5708 was not associated with rs7009110 (not shown) and so this miRNA was not considered further.
Multivariate association was tested between rs7009110 and the expression levels of five exons in each of HEY1 (MIM: 602953), MRPS28 (MIM: 611990), and FABP5 (MIM: 605168); seven in ZBTB10; nine in PAG1; and ten in TPD52. In this analysis, rs7009110 was significantly associated with the expression of PAG1 (p = 0.0017) but with no other gene (Table S3). The weights attributed to individual PAG1 exons in the multivariate analysis were consistent with an effect of rs7009110 on the expression of exons 1, 2, and 3; this observation was confirmed with individual univariate analyses of these three exons (Table S4). The rs7009110:T allele that increases the risk of allergic disease was associated with an increased expression of PAG1 (Figure S1), explaining 2.6% of the observed variation.
To help fine-map the eQTL results for PAG1, we extended the multivariate association analysis to an additional 167 common variants (MAF ≥ 5%, call rate > 95%, Hardy-Weinberg equilibrium test p value > 10−6) located in the 69.4 kb core region of association with allergic disease, delimited by the left-most (rs7822328, chr8: 81,246,659) and right-most (rs4739746, chr8: 81,316,034) variants in LD (r2 ≥ 0.6) with rs7009110. This is the region most likely to include the underlying causal variant(s); most variants were in LD with rs7009110 (70% with r2 ≥ 0.6). Multiple SNPs in high LD (r2 ≥ 0.8) with rs7009110 were associated with the expression of PAG1 (Figure 1B). In general, the strength of the association increased with the physical proximity to rs7009110 but did not exceed that seen with this SNP. However, no SNPs in LD (r2 ≥ 0.6) with rs7009110 showed an association with the expression of the other five genes tested (p > 0.05, Figure S2).
Although the association between 8q21 allergy risk variants and PAG1 expression in LCLs was relatively modest, it pointed to the possibility that a regulatory element in this region might control the expression of PAG1. Thus, we next queried genome-wide maps of epigenetic profiles to search for putative regulatory elements (PREs) in the 8q21 core region of association. Analysis of histone modifications associated with regulatory activity (e.g., H3K4me1/2) and DNase I hypersensitive sites assayed by the ENCODE Project17 in an LCL (GM12878) identified four PREs in this region, named PRE1 to PRE4 (Figure 1C). Consistent with these results, Seumois et al.18 detected a significant gain in H3K4me2, which marks both active and poised enhancers, in PRE1 and PRE2 when comparing primary human CD4+ memory T cells against naive CD4+ T cells. Similarly, Hnisz et al.19 predicted PRE3 to be an enhancer in multiple human immune cell types. Of the 118 SNPs that are in LD (r2 ≥ 0.6) with rs7009110, 35 overlap one of the four PREs identified (Table S5); rs7009110 overlaps PRE3. We therefore hypothesized that (1) one (or more) of these four PREs in this region regulates the expression of PAG1 and (2) rs7009110 or a correlated variant disrupts the function of the PRE.
To test the first hypothesis, we used Chromosome Conformation Capture (3C) as described previously20 to quantify the frequency of long-range chromatin interactions that take place between the core region of association and the promoter of PAG1, 710 kb apart. In brief, 3C libraries were created by cross-linking the chromatin from LCLs of individuals homozygous for the rs7009110:T risk allele (n = 2) or the C protective allele (n = 2); DNA was then digested with EcoRI, which flanks 17 contiguous fragments that cover the core region of association, as well as the PAG1 promoter (Table S6); DNA was religated and decrosslinked; and quantitative PCR (qPCR) with primers for the bait (PAG1 promoter) and interactors (17 fragments) was then performed to detect the presence of ligation products, which represent gene loops. BAC clones covering the regions of interest were used to normalize for PCR efficiency.
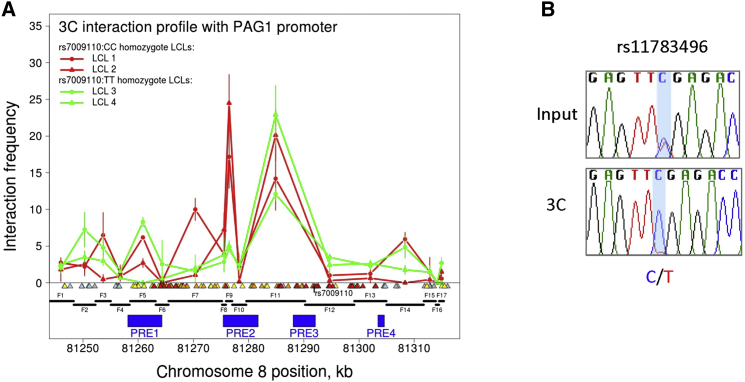
The frequency of chromatin interactions between the PAG1 promoter and 15 of the 17 fragments that covered the core region of association was low and consistent with being due to chance (Figure 2A). However, for two fragments—numbered F9 and F11—the interaction frequency was significantly above the background level. The size of the interaction products amplified was confirmed by gel electrophoresis and their sequence verified by Sanger sequencing; similar results were also obtained in two additional independent replicates of each sample (Figure S3). Fragment F9 is 1.1 kb long and overlaps the centromeric end of PRE2, including the peak region of H3K4me2 gain observed by Seumois et al.18 in memory CD4+ T cells. Interactions with fragment F9 were observed in LCLs homozygote for the rs7009110:C protective allele but not (or less frequently) for the T predisposing allele. Consistent with this, allele-specific 3C from two LCLs heterozygous for rs11783496, a SNP near fragment F9 and in LD with rs7009110 (r2 = 0.75), indicate that the protective C allele of rs11783496 is more strongly associated with looping of PRE2 to the PAG1 promoter (Figures 2B and S4). These results suggest that an allergy protective allele (rs11783496:C or another on the same haplotype) is associated with the establishment of long-range chromatin interactions between PRE2 and the PAG1 promoter. The interaction observed with fragment F11—which is 10.6 kb long and overlaps both the telomeric end of PRE2 and the centromeric end of PRE3—was detected in all four samples, irrespective of rs7009110 genotype. This suggests that allergy-associated variants do not influence the establishment of chromatin looping between the PAG1 promoter and PRE3.
Figure 2.
Chromatin Interactions at the 8q21 Risk Region with PAG1
(A) 3C interaction profiles indicating frequent interactions between PRE2 and PRE3 and the PAG1 promoter region. 3C libraries were generated with EcoRI, with the anchor point set at the PAG1 promoter. A physical map of the region interrogated by 3C is shown below, with the blue bars representing the position of the PREs and the black lines representing the EcoRI fragments interrogated. Triangles show position of variants in linkage disequilibrium (LD) with rs7009110, with color gradient representing the strength of association with PAG1 expression levels (cf. Figure 1B), from most associated in red to least associated in gray. Graph shows 3C profiles generated from four different LCLs assayed in duplicate. Error bars denote SD.
(B) Allele-specific chromatin looping between PRE2 and the PAG1 promoter. 3C followed by direct sequencing of the PCR product was performed in two heterozygous LCLs with rs11783496 as the surrogate marker. The rs11783496:C and rs7009110:C alleles are in phase (r2 = 0.75). Chromatograms represent one of two independent 3C libraries generated and sequenced.
Having established that two independent DNA fragments overlapping PRE2 and PRE3 physically interact with the promoter of PAG1, next we tested the hypothesis that the regulatory ability of these two PREs is modulated by allergy risk variants. To assess this, a PAG1 promoter-driven luciferase reporter construct was generated by inserting a 1,660 bp fragment containing the PAG1 promoter into pGL3-basic, as described previously.21 A fragment overlapping PRE2 (1,893 bp) or PRE3 (1,560 bp) and containing the major (i.e., allergy protective) allele for SNPs in LD (r2 > 0.6) with rs7009110 was inserted downstream of luciferase. The coordinates of these fragments were selected such that they coincided with the two interaction peaks observed in the 3C experiments (fragments F9 and F11) and, within these regions, with the highest density of histone modifications and DNase I hypersensitive sites from ENCODE (Figures 1D and S5). To assess the effect of individual SNPs on PRE activity, the minor (i.e., allergy predisposing) allele for SNPs in LD with rs7009110 was individually incorporated into PRE2 (rs11783496, rs11786685, rs11786704, or rs13275449) and PRE3 (rs4739737, rs10957979, or rs2370615) (Figure 1D) via standard DNA cloning. All constructs were sequenced to confirm variant incorporation (AGRF). Primers used to generate all constructs are listed in Table S7. LCLs were electroporated with the reporter plasmids and luciferase activity was measured 24 hr post-transfection. To correct for any differences in transfection efficiency or cell lysate preparation, Firefly luciferase activity was normalized to Renilla luciferase. The activity of each test construct was calculated relative to PAG1 promoter construct.
Our 3C studies provided evidence for allele-specific chromatin looping between PRE2 and PAG1. Despite this, PRE2 did not enhance or silence the PAG1 promoter in reporter assays (Figure 3A), making it unclear of the functional consequence of allele-specific chromatin looping. For PRE3, which interacted with the PAG1 promoter irrespective of genotype, we found that the construct containing the major (allergy protective) allele at the three SNPs (rs4739737, rs10957979, and rs2370615) in LD with rs7009110 also did not increase PAG1 promoter activity (Figure 3B). A similar lack of regulatory activity was observed for PRE3 when the fragments cloned contained the minor (allergy predisposing) allele for rs4739737 or rs10957979. However, the PRE3 construct containing the minor allele for rs2370615 increased PAG1 promoter activity by 1.8-fold when compared to the promoter-only construct (p = 0.0051) and by 2.3-fold when compared to the construct with the promoter plus the enhancer containing the major alleles (p = 0.0005). These results demonstrate that PRE3 acts as a transcriptional enhancer in the presence of the rs2370615:C allergy predisposing allele. Consistent with this effect, rs2370615:C was associated with increased expression of PAG1 in the eQTL analyses described above (p = 0.0018); this variant is in complete LD (r2 = 1) with rs7009110. Collectively, these results confirm that PAG1 is a target gene of 8q21 allergy risk variants and suggest that rs2370615 represents the underlying putative functional variant.
Figure 3.
PRE3 Containing the rs2370615 Risk Allele Acts as an Enhancer on the PAG1 Promoter
PRE2 (A) or PRE3 (B) was cloned upstream of a PAG1 promoter-driven luciferase reporter with and without the candidate causal SNPs. 1 μg of each plasmid was electroporated into 1 × 106 cells with Amaxa NucleofectorII with the SF buffer and the program EH-100. Luciferase activity was measured after 24 hr via the Dual-Glo luciferase assay system on a Beckman-Coulter DTX-800 plate reader. Graphs represent three independent experiments assayed in duplicate. Error bars denote 95% CI and p values were determined with a two-way ANOVA followed by Dunnett’s multiple comparisons test (∗p < 0.01, ∗∗p < 0.001).
Based on ENCODE ChIP-seq data for LCLs,17 two transcription factors (TFs) bind to PRE3 and overlap the putative functional variant rs2370615: the RelA (or p65 [MIM: 164014]) subunit of the nuclear factor κB (NF-κB) TF, which is critical for innate and adaptive immune responses,22 and the POU domain class 2 transcription factor 2 (POU2F2 or Oct-2 [MIM: 164176]), which regulates B-cell-specific genes.23 These TFs have been shown to co-occur in LCLs24 and to synergistically regulate enhancer activity in B cells.25 Furthermore, TNF-α (MIM: 191160)-induced recruitment of RelA to enhancers involved in long-range looping interactions is associated with transcriptional induction of target genes.26 These observations suggest that binding of RelA and Oct-2 to PRE3 might be required for long-range activation of PAG1 transcription. However, despite RelA binding to PRE3 being highest precisely over rs2370615 (Figure S5), this T/C polymorphism is not predicted to disrupt the binding motifs reported for either RelA or Oct-2.24,27 Based on these observations, it is possible that the rs2370615:C allele alters the binding affinity of another transcription factor, which is involved in RelA recruitment to PRE3 and so promotes long-range activation of PAG1 transcription.
Notably, the rs2370615:C allele is predicted to disrupt the binding motif (TTGTTTAC) for five Forkhead box (Fox) TFs,28 namely Foxo3a (MIM: 602681), an NF-κB antagonist that inhibits lymphocyte activation and proliferation.29 To test this prediction, we performed chromatin immunoprecipitation (ChIP) using an anti-Foxo3a rabbit mAb (2497S, Cell Signaling Technology) to determine Foxo3a binding to the PRE3 region that overlaps rs2370615 (Table S8), in LCLs with the rs2370615:TT, CT, or CC genotype. When compared to an IgG control antibody (SC-8334, Santa Cruz Biotechnology), we observed a significant enrichment in Foxo3a binding to PRE3 for the TT (3.4-fold, p = 0.001) and TC (1.3-fold, p = 0.006) but not CC (0.94-fold, p = 0.820) LCLs (Figure 4A). Differences in IgG-normalized Foxo3a binding to PRE3 were statistically significant when comparing TT to either the TC (p = 0.0018) or CC (p = 0.0003) LCLs. This pattern of Foxo3a binding was not observed in a control region 4 kb upstream of PRE3 that does not contain the canonical Fox binding motif (Table S8). Although there was a slight enrichment of Foxo3a binding across the three LCLs in this region (Figure 4B), suggesting that Foxo3a might bind weakly to it, there were no significant differences between genotypes after correction for multiple testing. These results confirm that Foxo3a binds with high affinity to the PRE3 region that overlaps rs2370615 and that the C allele of this polymorphism reduces Foxo3a binding to PRE3.
Figure 4.
Binding of the Foxo3a Transcription Factor to PRE3 Is Disrupted by the rs2370615:C Allele
Chromatin immunoprecipitation was performed as previously described.39 LCLs (1 × 107) with the indicated genotypes were crosslinked, sonicated, and immunoprecipitated with either anti-Foxo3a antibody or control IgG antibody. Immunoprecipitates were washed and DNA was extracted after reverse-crosslinking. The enrichment of Foxo3a binding to PRE3 (A) and control region 4 kb upstream of PRE3 (B) was examined by performing qPCR. The binding of Foxo3a is shown as fold enrichment over IgG (horizontal line). Graphs represent three independent experiments assayed in triplicate. Error bars denote 95% CI and p values were determined by unpaired t test.
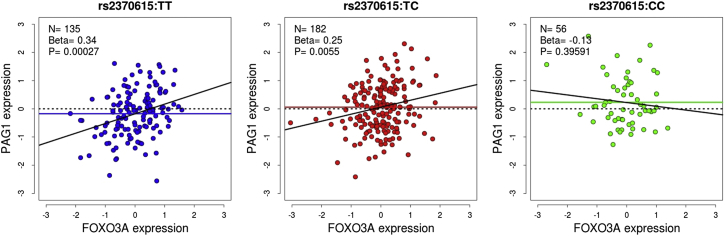
Lastly, given that Foxo3a was confirmed to bind to PRE3, which regulates PAG1 expression, we hypothesized that FOXO3A expression in LCLs might be correlated with PAG1 expression. To test this hypothesis, we used gene-expression results for LCLs of 373 individuals obtained by the Geuvadis Project,14 as described above, to measure the association between FOXO3A and PAG1 (exon 2) expression, stratified by rs2370615 genotype (Figure 5). There was a significant positive correlation between FOXO3A and PAG1 expression in TT (r = 0.31, 95% CI 0.15–0.45, p = 0.0003) and TC (r = 0.21, 95% CI 0.06–0.34, p = 0.0055) but not CC (r = −0.12, 95% CI −0.37–0.15, p = 0.3960) LCLs, consistent with the disruptive effect of the rs2370615:C allele on Foxo3a binding to PRE3. This interaction effect between FOXO3A levels and rs2370615 genotype on PAG1 expression was statistically significant (p = 0.0157).
Figure 5.
The Association between FOXO3A and PAG1 Expression in LCLs Is Modulated by rs2370615 Genotype
Gene expression levels were normalized via a rank-based transformation and adjusted for potential confounders (including ancestry and 16 principal components that capture the effects of unobserved technical confounders on gene expression) and the association tested by linear regression. Horizontal solid lines show the mean expression of PAG1 for a given genotype class (cf. Figure S1).
Collectively, our results are consistent with a model that includes the following. (1) PRE3 is a poised regulatory element for PAG1 in all individuals, irrespective of genotype, and promotes PAG1 transcription. (2) PRE2 is a more dynamic regulatory element, with as yet unconfirmed induction/suppression capacity for PAG1. To become regulatory active, PRE2 might require, for example, the presence of signals from PRE3 (e.g., PRE2 might suppress PRE3), appropriate cellular activation (e.g., allergen or viral stimulation), or transcription factors that have low/no expression in the LCL tested. (3) Individuals with the rs2370615:C allergy risk allele at PRE3 have overall higher PAG1 expression, perhaps because TFs such as RelA are able to bind to PRE3 and drive high levels of PAG1 transcription (Figure S6A). (4) Individuals with the rs2370615:T allergy protective allele at PRE3 have overall lower PAG1 expression, perhaps because Foxo3a binds to PRE3 and drives lower levels of transcription (when compared to for example RelA) and/or because PRE2, which physically interacts with the PAG1 promoter in these individuals, might suppress PRE3 activity (Figure S6B). Further studies that characterize in greater detail the molecular mechanisms underlying the regulation of PAG1 expression by PRE2 and PRE3 are warranted.
Importantly, our results suggest that increased PAG1 transcription is associated with increased risk of allergic disease. To our knowledge, there are no published studies investigating the contribution of PAG1 to the pathophysiology of allergic diseases. PAG1 encodes the phosphoprotein associated with glycosphingolipid microdomains (or Csk-binding protein, Cbp), a transmembrane adaptor protein localized to lipid rafts that has highest expression in the immune system, notably in T and B cells.30–32 One of the main cellular functions of PAG1 is the regulation of Csk activity,30 with direct effects on immunoreceptor signaling.33 The role of PAG1 in the development of immune responses has been studied extensively using in vitro and in vivo experimental systems, with conflicting results (reviewed in Hrdinka and Horejsi33). In brief, in vitro studies suggest that PAG1 overexpression inhibits T cell, B cell, and mast cell activation7–10,34 and so, by extension, would be expected to have an anti-inflammatory effect. In contrast, in vivo studies have shown that T and B cell development and function are normal in Pag1-deficient mice.35,36
Our finding that allergy predisposing alleles increase the transcription of PAG1 in human B cell lines is not consistent with results from these experimental studies; instead, it suggests that PAG1 overexpression might promote B cell activation and so have a pro-inflammatory effect. Interestingly, this possibility was proposed by Hrdinka and Horejsi33 to explain the observation that PAG1 is phosphorylated upon antigen stimulation in B cells but not T cells. This notion is also supported by the recent observation that PAG1 inhibition in mast cells can suppress degranulation and the production of pro-inflammatory cytokines if cells are activated via an allergen-dependent mechanism.37 Given the potential role of the NF-κB pathway in PAG1 transcription and the observation that this pathway is activated by allergen and/or pathogen engagement of Toll-like receptors,38 characterization of the immune response to allergen or viral challenge in Pag1-deficient mice might help resolve the conflicting observations reported in experimental studies.
A limitation of our work is that we cannot exclude the possibility that other genes in the region are also targets of the same variants. We focused our experimental work on PAG1 because this was the only gene for which we found a significant association between allergy risk SNPs and variation in gene expression levels. However, it is possible that despite no current available eQTL evidence, the expression of genes such as ZBTB10 or TPD52, which are expressed in relevant immune cells, might also be regulated by the PREs we identified, perhaps in different cell types.
In conclusion, we showed that an allergy risk allele on chromosome 8q21 increases PAG1 transcription by activating a poised enhancer located 732 kb away from the gene promoter. The significance of these results is two-fold. First, they highlight the fact that the target gene(s) underlying an observed genetic association can be a considerable distance away from the GWAS sentinel SNP. Second, our results suggest that inhibition of PAG1 expression or function might have therapeutic potential to treat allergic diseases.
Acknowledgments
We would like to thank Prof. Peter Visscher and Prof. Grant Montgomery for providing the lymphoblastoid cell lines used in this study.
Published: July 23, 2015
Footnotes
Supplemental Data include six figures and eight tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.06.010.
Contributor Information
Juliet D. French, Email: juliet.french@qimrberghofer.edu.au.
Manuel A.R. Ferreira, Email: manuel.ferreira@qimrberghofer.edu.au.
Web Resources
The URLs for data presented herein are as follows:
ArrayExpress, http://www.ebi.ac.uk/arrayexpress/
OMIM, http://www.omim.org/
Supplemental Data
References
- 1.Ferreira M.A., Matheson M.C., Tang C.S., Granell R., Ang W., Hui J., Kiefer A.K., Duffy D.L., Baltic S., Danoy P., Australian Asthma Genetics Consortium Collaborators Genome-wide association analysis identifies 11 risk variants associated with the asthma with hay fever phenotype. J. Allergy Clin. Immunol. 2014;133:1564–1571. doi: 10.1016/j.jaci.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeller T., Wild P., Szymczak S., Rotival M., Schillert A., Castagne R., Maouche S., Germain M., Lackner K., Rossmann H. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PLoS ONE. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davison L.J., Wallace C., Cooper J.D., Cope N.F., Wilson N.K., Smyth D.J., Howson J.M., Saleh N., Al-Jeffery A., Angus K.L., Cardiogenics Consortium Long-range DNA looping and gene expression analyses identify DEXI as an autoimmune disease candidate gene. Hum. Mol. Genet. 2012;21:322–333. doi: 10.1093/hmg/ddr468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paternoster L., Standl M., Chen C.M., Ramasamy A., Bonnelykke K., Duijts L., Ferreira M.A., Alves A.C., Thyssen J.P., Albrecht E. Meta-analysis of genome-wide association studies identifies three new risk loci for atopic dermatitis. Nat. Genet. 2011;44:187–192. doi: 10.1038/ng.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okada Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Yoshida S., RACI consortium. GARNET consortium Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tiacci E., Orvietani P.L., Bigerna B., Pucciarini A., Corthals G.L., Pettirossi V., Martelli M.P., Liso A., Benedetti R., Pacini R. Tumor protein D52 (TPD52): a novel B-cell/plasma-cell molecule with unique expression pattern and Ca(2+)-dependent association with annexin VI. Blood. 2005;105:2812–2820. doi: 10.1182/blood-2004-07-2630. [DOI] [PubMed] [Google Scholar]
- 7.Smida M., Cammann C., Gurbiel S., Kerstin N., Lingel H., Lindquist S., Simeoni L., Brunner-Weinzierl M.C., Suchanek M., Schraven B., Lindquist J.A. PAG/Cbp suppression reveals a contribution of CTLA-4 to setting the activation threshold in T cells. Cell Commun. Signal. 2013;11:28. doi: 10.1186/1478-811X-11-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itoh K., Sakakibara M., Yamasaki S., Takeuchi A., Arase H., Miyazaki M., Nakajima N., Okada M., Saito T. Cutting edge: negative regulation of immune synapse formation by anchoring lipid raft to cytoskeleton through Cbp-EBP50-ERM assembly. J. Immunol. 2002;168:541–544. doi: 10.4049/jimmunol.168.2.541. [DOI] [PubMed] [Google Scholar]
- 9.Davidson D., Bakinowski M., Thomas M.L., Horejsi V., Veillette A. Phosphorylation-dependent regulation of T-cell activation by PAG/Cbp, a lipid raft-associated transmembrane adaptor. Mol. Cell. Biol. 2003;23:2017–2028. doi: 10.1128/MCB.23.6.2017-2028.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalland M.E., Solheim S.A., Skånland S.S., Taskén K., Berge T. Modulation of proximal signaling in normal and transformed B cells by transmembrane adapter Cbp/PAG. Exp. Cell Res. 2012;318:1611–1619. doi: 10.1016/j.yexcr.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 11.Tillotson L.G. RIN ZF, a novel zinc finger gene, encodes proteins that bind to the CACC element of the gastrin promoter. J. Biol. Chem. 1999;274:8123–8128. doi: 10.1074/jbc.274.12.8123. [DOI] [PubMed] [Google Scholar]
- 12.Tone M., Powell M.J., Tone Y., Thompson S.A., Waldmann H. IL-10 gene expression is controlled by the transcription factors Sp1 and Sp3. J. Immunol. 2000;165:286–291. doi: 10.4049/jimmunol.165.1.286. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi K., Hayashi N., Shimokawa T., Umehara N., Kaminogawa S., Ra C. Cooperative regulation of Fc receptor gamma-chain gene expression by multiple transcription factors, including Sp1, GABP, and Elf-1. J. Biol. Chem. 2008;283:15134–15141. doi: 10.1074/jbc.M800498200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lappalainen T., Sammeth M., Friedländer M.R., ’t Hoen P.A., Monlong J., Rivas M.A., Gonzàlez-Porta M., Kurbatova N., Griebel T., Ferreira P.G., Geuvadis Consortium Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferreira M.A., Purcell S.M. A multivariate test of association. Bioinformatics. 2009;25:132–133. doi: 10.1093/bioinformatics/btn563. [DOI] [PubMed] [Google Scholar]
- 17.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seumois G., Chavez L., Gerasimova A., Lienhard M., Omran N., Kalinke L., Vedanayagam M., Ganesan A.P., Chawla A., Djukanović R. Epigenomic analysis of primary human T cells reveals enhancers associated with TH2 memory cell differentiation and asthma susceptibility. Nat. Immunol. 2014;15:777–788. doi: 10.1038/ni.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., Young R.A. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghoussaini M., Edwards S.L., Michailidou K., Nord S., Cowper-Sal Lari R., Desai K., Kar S., Hillman K.M., Kaufmann S., Glubb D.M., Australian Ovarian Cancer Management Group. Australian Ovarian Cancer Management Group Evidence that breast cancer risk at the 2q35 locus is mediated through IGFBP5 regulation. Nat. Commun. 2014;4:4999. doi: 10.1038/ncomms5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.French J.D., Ghoussaini M., Edwards S.L., Meyer K.B., Michailidou K., Ahmed S., Khan S., Maranian M.J., O’Reilly M., Hillman K.M., GENICA Network. kConFab Investigators Functional variants at the 11q13 risk locus for breast cancer regulate cyclin D1 expression through long-range enhancers. Am. J. Hum. Genet. 2013;92:489–503. doi: 10.1016/j.ajhg.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonizzi G., Karin M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 23.Matthias P. Lymphoid-specific transcription mediated by the conserved octamer site: who is doing what? Semin. Immunol. 1998;10:155–163. doi: 10.1006/smim.1998.0117. [DOI] [PubMed] [Google Scholar]
- 24.Zhao B., Barrera L.A., Ersing I., Willox B., Schmidt S.C., Greenfeld H., Zhou H., Mollo S.B., Shi T.T., Takasaki K. The NF-κB genomic landscape in lymphoblastoid B cells. Cell Rep. 2014;8:1595–1606. doi: 10.1016/j.celrep.2014.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sepulveda M.A., Emelyanov A.V., Birshtein B.K. NF-kappa B and Oct-2 synergize to activate the human 3′ Igh hs4 enhancer in B cells. J. Immunol. 2004;172:1054–1064. doi: 10.4049/jimmunol.172.2.1054. [DOI] [PubMed] [Google Scholar]
- 26.Jin F., Li Y., Dixon J.R., Selvaraj S., Ye Z., Lee A.Y., Yen C.A., Schmitt A.D., Espinoza C.A., Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staudt L.M., Clerc R.G., Singh H., LeBowitz J.H., Sharp P.A., Baltimore D. Cloning of a lymphoid-specific cDNA encoding a protein binding the regulatory octamer DNA motif. Science. 1988;241:577–580. doi: 10.1126/science.3399892. [DOI] [PubMed] [Google Scholar]
- 28.Ward L.D., Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin L., Hron J.D., Peng S.L. Regulation of NF-kappaB, Th activation, and autoinflammation by the forkhead transcription factor Foxo3a. Immunity. 2004;21:203–213. doi: 10.1016/j.immuni.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 30.Brdicka T., Pavlistová D., Leo A., Bruyns E., Korínek V., Angelisová P., Scherer J., Shevchenko A., Hilgert I., Cerný J. Phosphoprotein associated with glycosphingolipid-enriched microdomains (PAG), a novel ubiquitously expressed transmembrane adaptor protein, binds the protein tyrosine kinase csk and is involved in regulation of T cell activation. J. Exp. Med. 2000;191:1591–1604. doi: 10.1084/jem.191.9.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inomata M., Shimada Y., Hayashi M., Shimizu J., Ohno-Iwashita Y. Impairment in a negative regulatory system for TCR signaling in CD4+ T cells from old mice. FEBS Lett. 2007;581:3039–3043. doi: 10.1016/j.febslet.2007.05.063. [DOI] [PubMed] [Google Scholar]
- 32.Tedoldi S., Paterson J.C., Hansmann M.L., Natkunam Y., Rüdiger T., Angelisova P., Du M.Q., Roberton H., Roncador G., Sanchez L. Transmembrane adaptor molecules: a new category of lymphoid-cell markers. Blood. 2006;107:213–221. doi: 10.1182/blood-2005-06-2273. [DOI] [PubMed] [Google Scholar]
- 33.Hrdinka M., Horejsi V. PAG--a multipurpose transmembrane adaptor protein. Oncogene. 2014;33:4881–4892. doi: 10.1038/onc.2013.485. [DOI] [PubMed] [Google Scholar]
- 34.Ohtake H., Ichikawa N., Okada M., Yamashita T. Cutting Edge: Transmembrane phosphoprotein Csk-binding protein/phosphoprotein associated with glycosphingolipid-enriched microdomains as a negative feedback regulator of mast cell signaling through the FcepsilonRI. J. Immunol. 2002;168:2087–2090. doi: 10.4049/jimmunol.168.5.2087. [DOI] [PubMed] [Google Scholar]
- 35.Xu S., Huo J., Tan J.E., Lam K.P. Cbp deficiency alters Csk localization in lipid rafts but does not affect T-cell development. Mol. Cell. Biol. 2005;25:8486–8495. doi: 10.1128/MCB.25.19.8486-8495.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dobenecker M.W., Schmedt C., Okada M., Tarakhovsky A. The ubiquitously expressed Csk adaptor protein Cbp is dispensable for embryogenesis and T-cell development and function. Mol. Cell. Biol. 2005;25:10533–10542. doi: 10.1128/MCB.25.23.10533-10542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Draberova L., Bugajev V., Potuckova L., Halova I., Bambouskova M., Polakovicova I., Xavier R.J., Seed B., Draber P. Transmembrane adaptor protein PAG/CBP is involved in both positive and negative regulation of mast cell signaling. Mol. Cell. Biol. 2014;34:4285–4300. doi: 10.1128/MCB.00983-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Medzhitov R. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 39.Lee J.S., Kim Y., Kim I.S., Kim B., Choi H.J., Lee J.M., Shin H.J., Kim J.H., Kim J.Y., Seo S.B. Negative regulation of hypoxic responses via induced Reptin methylation. Mol. Cell. 2010;39:71–85. doi: 10.1016/j.molcel.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.