Abstract
Objective
The triglyceride (TG) response to a high-fat meal (postprandial lipemia, PPL) affects cardiovascular disease risk and is influenced by genes and environment. Genes involved in lipid metabolism have dominated genetic studies of PPL TG response. We sought to elucidate common genetic variants through a genome-wide association (GWA) study in the Genetics of Lipid Lowering Drugs and Diet Network (GOLDN).
Methods
The GOLDN GWAS discovery sample consisted of 872 participants within families of European ancestry. Genotypes for 2,543,887 variants were measured or imputed from HapMap. Replication of our top results was performed in the Heredity and Phenotype Intervention (HAPI) Heart Study (n=843). PPL TG response phenotypes were constructed from plasma TG measured at baseline (fasting, 0 hour), 3.5 and 6 hours after a high-fat meal, using a random coefficient regression model. Association analyses were adjusted for covariates and principal components, as necessary, in a linear mixed model using the kinship matrix; additional models further adjusted for fasting TG were also performed. Meta-analysis of the discovery and replication studies (n=1,715) was performed on the top SNPs from GOLDN.
Results
GOLDN revealed 111 suggestive (p<1E-05) associations, with two SNPs meeting GWA significance level (p<5E-08). Of the two significant SNPs, rs964184 demonstrated evidence of replication (p=1.20E-03) in the HAPI Heart Study and in a joint analysis, was GWA significant (p=1.26E-09). Rs964184 has been associated with fasting lipids (TG and HDL) and is near ZPR1 (formerly ZNF259), close to the APOA1/C3/A4/A5 cluster. This association was attenuated upon additional adjustment for fasting TG.
Conclusion
This is the first report of a genome-wide significant association with replication for a novel phenotype, namely PPL TG response. Future investigation into response phenotypes is warranted using pathway analyses, or newer genetic technologies such as metabolomics.
Keywords: Triglyceride, postprandial lipemia, gwas, genetics
1. INTRODUCTION
Fasting and postprandial plasma triglyceride (TG) levels are known risk factors for cardiovascular disease (CVD) [1–4]. Common genetic variants associated with fasting TG, assessed through single nucleotide polymorphisms (SNPs), have been studied extensively [5–10]. The results of a meta-analysis of lipid genome wide association (GWA) studies by Teslovich et al [5] implicated 24 loci associated with fasting TG, with the most significant single nucleotide polymorphism (SNP), rs964184. While fasting TG remains the gold standard of TG measurement by physicians and is most studied in relation to CVD risk [11], humans spend a majority of their waking hours in the postprandial state [1, 3, 4, 11].
Postprandial lipemia (PPL) encompasses the changes in plasma TG and lipid profile as well as the duration of these changes due to the ingestion of a high-fat meal [11]. An elevated or elongated PPL leads to the production of atherogenic TG-rich lipoproteins and activation of thrombotic processes [11]. Atherogenic TG-rich lipoproteins that remain in the circulation for extended periods of time are independently related to progression of coronary heart disease (CHD) and thus are a risk factor for CVD [3, 11–15].
Meal challenges that induce PPL produce large and highly variable changes in circulating TG’s and TG rich lipoproteins. Within a population the PPL TG response is more variable than fasting TG levels and is believed to be modulated by both genes and environment [15]. To date, there has been one genomewide association study (GWAS) of PPL in an extended pedigree which found a rare mutation [16], while the majority of genetic PPL studies have investigated candidate genes with known involvement in lipid metabolism (i.e. APOA1/C3/A4/A5 cluster, ABCA1, CETP, GCKR, IL6, LPL, PLIN1, TCF7L2, etc.) [17, 18]. Some of these studies demonstrated genetic variation associated with PPL, however they suffer from small sample sizes, differing meal challenges, and limited replication [17].
We sought to elucidate the genetic determinants of PPL TG response to a standardized high-fat meal by performing a GWAS among participants from the Genetics of Lipid Lowering Drugs and Diet Network (GOLDN). We used the Hereditary and Phenotype Intervention (HAPI) Heart study for replication of our top findings, and also performed a joint meta-analysis (discovery and replication results) for the top discovery findings from GOLDN.
2. MATERIALS AND METHODS
2.1 Study Design
The GOLDN study was designed to characterize the genetic basis of TG response to two environmental contexts: one to raise triglycerides (consumption of a high-fat meal, PPL); and one to lower TG (a 3-week treatment with 160 g/day of fenofibrate). The specific methodology of GOLDN is reported elsewhere [19]. The study population consisted of 189 families, recruited from 3-generational pedigrees at two genetically homogeneous European ancestry field centers of the NHLBI Family Heart Study (FamHS: Minneapolis, MN, and Salt Lake City, UT) [20]. Inclusion criteria were: ≥ 18 years of age, fasting triglycerides (TGs) < 1500 mg/dL, willingness to participate in the study and attend the scheduled clinic exams, being part of a family with at least 2 members in a sibship, AST and ALT tests within normal range, and creatinine ≤ 2.0. Only subjects not using lipid-lowering agents (pharmaceuticals or nutraceuticals) for at least 4 weeks after the screening visit were eligible. The Institutional Review Boards at the University of Alabama at Birmingham, the University of Minnesota, the University of Utah, and Tufts University approved the study protocol. Informed consent was obtained on all participants.
This analysis focused on plasma TG concentration in response to the standardized high fat challenge (PPL; n=872), which followed the protocol of Patsch et al [21]. The caloric intake of the intervention meal was determined by body surface area, containing 700 kilocalories per m2 of body surface area (2.93 MJ/m2 body surface area). The meal composition was 83% of calories from fat, 14% from carbohydrates, and 3% from protein. The meal was formulated to have a cholesterol content of 240 mg and a polyunsaturated:saturated fat ratio of 0.06. Based on these guidelines, the average individual ingested 175 mL of heavy whipping cream (39.5% fat) combined with 7.5 mL powdered, instant, non-fat, dry milk, and blended with ice. To increase palatability of the drink, 15 mL of chocolate or strawberry-flavored syrup was also added. Participants had 15 minutes to ingest this meal and were required to fast a minimum of 12 hours prior to the meal. Immediately before ingestion, we drew blood samples on all participants (0 hr, fasting), and then again at 3.5 and 6 hours after the high-fat meal. Participants rested and remained otherwise fasting during the whole PPL challenge.
2.2 Clinical and Biochemical Measurements
Participants were asked to abstain from using alcohol for at least 24 hours and fast for at least 12 hours before the PPL challenge. We obtained anthropometric measurements, assessed current medications, blood pressure, medical history, dietary history, and personal history. Blood was collected to obtain DNA for use in genotyping. Additional blood samples were collected before and during the PPL challenge for use in obtaining biochemical measurements. All samples (serum and EDTA-anticoagulant tubes) were centrifuged within 20 minutes of collection at 2000 × g for 15 min at 4°C and stored frozen at −70°C until time of use. Analysis was completed on all stored samples at the end of the study and all samples for an individual were processed in the same batch to reduce measurement error. TG measurements were calculated using the glycerol-blanked enzymatic method on the Roche COBAS FARA centrifugal analyzer (Roche Diagnostics Corporation, Basel, Switzerland).
2.3 Genotyping data and quality control
All GOLDN participants were phenotyped for PPL under the GOLDN protocol. However, a small subset of GOLDN participants (n=116) were genotyped by the parent study, FamHS (GOLDN-FamHS); and the remaining 756 participants were genotyped by the GOLDN study for a total discovery sample size of 872.
2.3.1 GOLDN genotyping
For participants genotyped under GOLDN, genotyping procedures are fully described in Aslibekyan et al [22]. In brief, a total of 906,600 SNPs were genotyped on the Affymetrix Genome-Wide Human SNP Array 6.0 using the Birdseed calling algorithm [23]. After quality control, consisting of participant overall SNP call rate, monomorphic SNP exclusion, SNP call rate <96% exclusion, Mendelian error checks, minor allele frequency ≤0.01 exclusion, and Hardy-Weinberg equilibrium p-value <1E-06 exclusion, a total of 584,060 genotyped autosomal SNPs remained for use in the imputation. We employed MACH (version1.0.16, http://www.sph.umich.edu/csg/abecasis/mach/) [24, 25] to impute genotype data for untyped SNPs using the CEU Human Genome release 22, build 36 as the reference. Excepting 31 genotyped SNPs passing quality control with unknown strand orientation, all SNPs (typed and imputed) were combined and strand oriented to the mlinfo file from MACH. Our final hybrid dataset contained 2,543,887 SNPs of which 584,029 were genotyped.
2.3.2 GOLDN-FamHS genotyping
GOLDN-FamHS participants were genotyped on one of three Illumina platforms: (i) Illumina HumMap 550K chip with 547,353 SNP markers, (ii) Illumina Human 610-Quadv1 chip with 576,888 SNP markers, or (iii) Illumina Human 1M-Duov3 chip with 1,111,639 SNP markers [26]. Quality control was performed before imputation by checking pedigree relationships (using GRR software), and excluding SNPs with low call rates (< 98%). A framework map for imputation was defined by choosing SNPs on all platforms, and restricting to those with minor allele frequency between 1% and 50%, and excluding SNPs with significant (p< 1.0E-06) departure from Hardy-Weinberg equilibrium. This panel was used to impute ~ 2.5 M SNPs based on phased haplotypes from HapMap (release 22, build 36, CEU, http://hapmap.ncbi.nlm.nih.gov/downloads/phasing/2007-08_rel22/phased/) using MACH (version 1.0.16, http://www.sph.umich.edu/csg/abecasis/mach/). Genetic analyses for these subjects was similar to the subjects genotyped by GOLDN, however additional adjustment was performed to control for differences due to the use of different genotyping platforms (550K, 610, and 1M).
2.4 Replication Dataset
2.4.1 The HAPI Heart Study
The sample consisted of 843 healthy Amish participants in the Heredity and Phenotype Intervention (HAPI) Heart Study [27]. The HAPI Heart Study began in 2003 to identify genes that interact with environmental exposures to influence risk for CVD. The study protocol was approved by the Institutional Review Board at the University of Maryland and all subjects gave written informed consent. The replication study described here was performed in 843 Old Order Amish participants (of a total of 868 HAPI Heart participants) who underwent a high fat feeding intervention and were successfully genotyped using the Affymetrix GeneChip® Human Mapping 500K Array Set.
All subjects could be connected to a single 14-generation pedigree [28, 29]. HAPI participants underwent a medical history interview including assessment of CVD risk factors, prescription and nonprescription medication usage and questions about prior history of CVD. Physical examinations were conducted at the Amish Research Clinic in Strasburg, PA. Those taking lipid-lowering medications at enrollment (1.3%) discontinued usage 7 days prior to the examination.
The high fat challenge, administered to the HAPI Heart subjects, was nearly identical to that used in GOLDN. A whipping cream milk shake was standardized to consist of 782 calories per m2 of body surface with 77.6% of calories from fat, 19.2% from carbohydrate, and 3.1% from protein, and was administered after an overnight fast. Blood was drawn just prior to (time = 0 hours) and 1, 2, 3, 4, and 6 hours after ingestion of the fat load to assess the TG excursion. The subject rested and remained otherwise fasting during the 6 hours post-fat challenge. Total cholesterol, TG and HDL-C levels were measured at each time point.
2.4.2 HAPI Heart genotyping
HAPI Heart Study participants were genotyped using the Affymetrix GeneChip® Human Mapping 500K Array Set which consisted of 500,568 SNP genotyping calls on each participant using the BRLMM genotype-calling algorithm. The mean genotype call rate was 98.3%. After quality control, which consisted of Mendelian error checks, Hardy-Weinberg equilibrium (p>1E-06 included), and minor allele frequency (>1% included), a total of 369,241 SNPs remained for use in the imputation to the HapMap population (release 22, build36, CEU, http://hapmap.ncbinlm.nih.gov/downloads/phasing/2007-08_rel22/phased/) using MACH (version 1.0.16, http://www.sph.umich.edu/csg/abecasis/mach/).
2.5 Statistical Analyses
2.5.1 Phenotype definitions
TG was log transformed to obtain a normal distribution. We assessed four PPL response phenotypes: uptake, clearance, area under the curve (AUC), and area under the increase (AUI). Uptake was defined as the slope of the line of TG response from 0 to 3.5 hours after meal ingestion, as by 3.5 hours most individuals have absorbed the majority of fat from the meal into their plasma [11, 17]. Clearance was defined as the slope of the line from 3.5 to 6 hours after meal ingestion, which we considered to be the time in which metabolic processes are working to eliminate the excess fat from the plasma [11]. The AUC was calculated using the trapezoid rule, and the AUI was calculated by subtracting the baseline area from AUC. All PPL response phenotypes were estimated from growth curve models of TG as a function of time. This method leverages variation within the individual as well as the population.
We ran two adjustment models for each phenotype: a minimal model and a minimal plus baseline TG model. In both models we stratified by sex and forced field center into the adjustment. Within each sex strata, we used a stepwise regression approach, offering age, age2,age3, and 10 principal components (PCs, to adjust for potential cryptic population stratification, estimated using EIGENSTRAT [30]) to both models as potential covariates (entry p-value=.10, retention p-value=.05) . The second model (minimal plus baseline TG) added baseline TG to the list of potential covariates. The adjusted phenotypic residuals were standardized to a mean of 0 and standard deviation of 1 and were used as phenotypes for the genetic association analyses.
HAPI Heart and GOLDN phenotypes were harmonized using the same SAS code to identically derive the four PPL TG response phenotypes (AUC, AUI, uptake slope, and clearance slope) using the above stated methods, with one exception: HAPI Heart did not include PCs because all individuals are from one large pedigree.
2.5.2 GOLDN GWAS Discovery
The association of hybrid data SNPs with the standardized phenotypic residuals was tested using a linear mixed model in R to account for familial dependencies described by a pedigree-based kinship matrix as a random effect, assuming additivy to model genotypes. Due to genotyping heterogeneity from different manufacturers, we performed the GWAS independently for GOLDN and GOLDN-FamHS followed by an uncorrelated beta meta-analysis of the two GWAS results. The results from the meta-analysis represent the discovery results. We used a Bonferroni correction to adjust for multiple testing, yielding a significant genome-wide p-value < 5E-08 for the discovery meta-analysis. A genome-wide p-value <1E-05 was considered ‘suggestive’ [31] and followed for replication in HAPI Heart. Quantile-quantile (QQ) plots were generated to assess deviations from the expected distribution and Manhattan plots were created. Assuming an additive genetic model, we had >80% power (α=0.05) to observe an effect size of at least R2=0.03, where R2 represents the marginal proportion of variance in the phenotype explained.
2.5.3 HAPI Heart Replication
TG PPL phenotypes were created in HAPI Heart as described above. The association of GOLDN discovery meta-analysis ‘top SNPs’ (suggestive and GWA significant) with the standardized phenotypic residuals in HAPI Heart was tested using a linear mixed model accounting for familial dependencies described by a pedigree-based kinship matrix as a random effect, and an additive genetic model assumption.
2.5.4 Joint Meta-Analysis of Discovery SNPs in GOLDN and HAPI
Due to our unique study design and the small number of available replication studies, we also performed a joint meta-analysis of GOLDN top hits. This was an inverse variance-weighted meta-analysis with fixed effects to estimate summary effects (METAL software, http://www.sph.umich.edu/csg/abecasis/metal/index.html) for the association of allelic dosage at each replication SNP and phenotype combination for the combined studies (GOLDN and HAPI Heart; n=1,715) [32]. Heterogeneity among studies was assessed using a χ2 test, and was limited for PPL TG phenotypes among our results for these SNPs. For the joint meta-analysis, we defined replication as evidence of an effect in the same direction for all studies and a meta-p-value less than the discovery p-value.
3. RESULTS
3.1 Descriptive Analyses
Men and women had similar body mass indices (BMI) in the discovery cohort; however there was a significant difference in BMI between men and women in the replication cohort (HAPI Heart), Table 1. Similarly, men and women had similar ages in GOLDN but significant age difference by gender in HAPI Heart. GOLDN men and women were slightly older than HAPI Heart participants. The HAPI Heart participants had baseline TG levels that were significantly lower than GOLDN and the values did not differ between men and women in the HAPI Heart Study. Additionally, the TG values of HAPI Heart participants were lower than the general population used in NHANES [33]. In contrast, GOLDN men and women demonstrated significant gender differences in baseline TG levels. For both discovery and replication cohorts, the PPL TG response phenotypes had similar mean values for AUI, uptake and clearance slopes between the minimal and baseline TG adjusted models. As expected, the AUC phenotype was lower in the baseline TG adjusted model than in the minimal adjusted model.
Table 1.
Mean (standard deviation) of discovery sample (GOLDN) and replication data (HAPI Heart).
| Discovery | Replication | |||||||
|---|---|---|---|---|---|---|---|---|
| GOLDN Study | HAPI Heart Study | |||||||
| Everyone (N=872) |
Men (N=429) | Women (N=443) |
Everyone (N=843) |
Men (N=453) |
Women (N=390) |
|||
| Age, yrs | 50.0 (16.2) | 50.0 (16.2) | 49.9 (16.2) | 44 (14) | 42 (14) | 45 (14) | ¶ | |
| Body Mass Index, kg/m2 | 28.5 (5.6) | 28.5 (4.6) | 28.5 (6.4) | 27 (4) | 26 (3) | 28 (5) | * | |
| Triglycerides, mg/dL | 139.4 (96.7) | 150.0 (107.5) | 129.0 (83.7) | ¶ | 69 (42) | 69 (40) | 69 (44) | |
| Phenotypes--Minimal model | ||||||||
| TG Uptake Slope | 0.18 (0.03) | 0.18 (0.03) | 0.18 (0.03) | * | 0.30 (0.00) | 0.30 (0.00) | 0.30 (0.00) | |
| TG Clearance Slope | −0.06 (0.05) | −0.06 (0.05) | −0.07 (0.05) | ¶ | −0.06 (0.08) | −0.05 (0.08) | −0.07 (0.07) | |
| TG Area Under the Curve | 31.1 (3.4) | 31.7 (3.3) | 30.5 (3.3) | * | 28.6 (2.9) | 28.8 (2.6) | 28.4 (3.2) | |
| TG Area Under the Increase | 2.5 (0.6) | 2.5 (0.6) | 2.4 (0.5) | * | 4.3 (0.29) | 4.3 (0.3) | 4.2 (0.3) | |
| Phenotypes—Adjusted for baseline | ||||||||
| TG model | ||||||||
| Uptake Slope | 0.19 (0.03) | 0.19 (0.03) | 0.18 (0.03) | 0.30 (0.00) | 0.30 (0.00) | 0.30 (0.00) | ||
| Clearance Slope | −0.07 (0.05) | −0.06 (0.05) | −0.07 (0.05) | ¶ | −0.06 (0.02) | −0.06 (0.02) | −0.05 (0.02) | |
| AUC | 28.0 (8.9) | 28.7 (8.8) | 27.3 (8.8) | 28.6 (2.6) | 28.4 (2.4) | 28.9 (2.9) | ||
| AUI | 2.5 (0.6) | 2.6 (0.6) | 2.5 (0.6) | 4.3 (0.05) | 4.3 (0.05) | 4.3 (0.06) | ||
p-value for difference between men and women (within each study) significant with p<0.0001
p-value for difference between men and women (within each study) significant with p<0.005
The lipid profiles during PPL for GOLDN and HAPI Heart are presented in Supplementary Table S1. As illustrated in this table, during the PPL challenge the lipid profiles varied over time. In particular, in both cohorts the TG concentration was highest at the 3.5 hour time-point, and by 6 hours after the meal TG levels remained higher than fasting. Thus, in both studies, participants were still removing excess TG from their blood 6 hours after its ingestion, which is not unexpected as a return to baseline values often requires from 8 to 10 hours [34].
3.2 Genome-Wide Association (GWA) Results
Discovery GWA Results
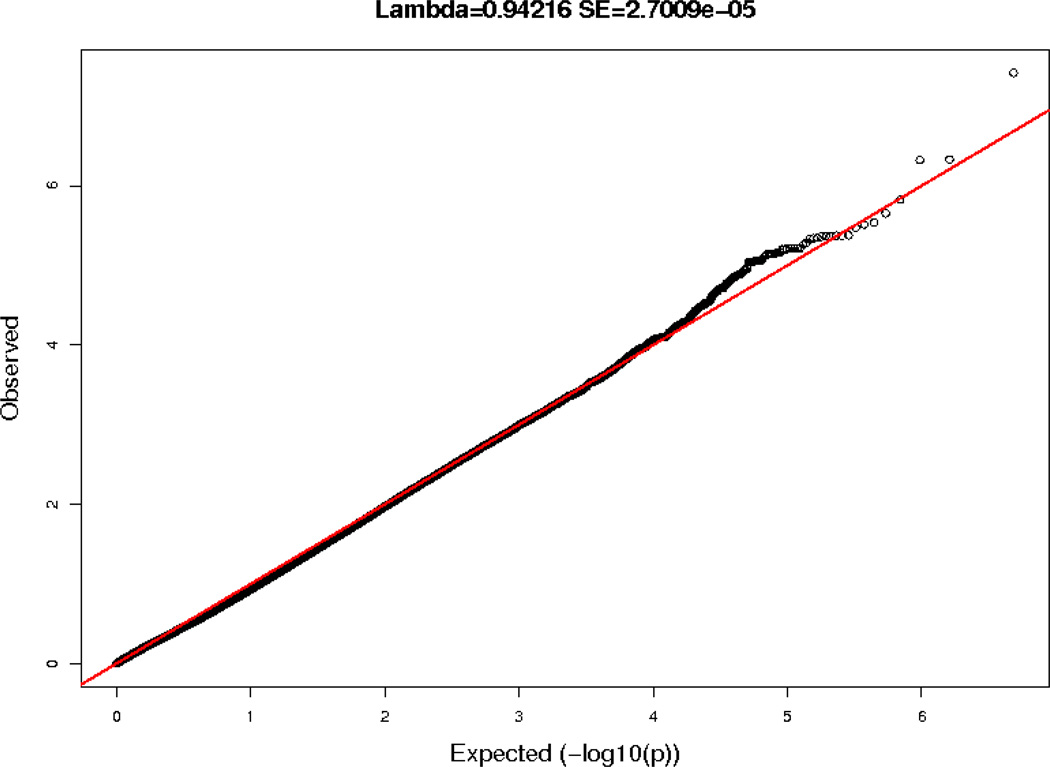
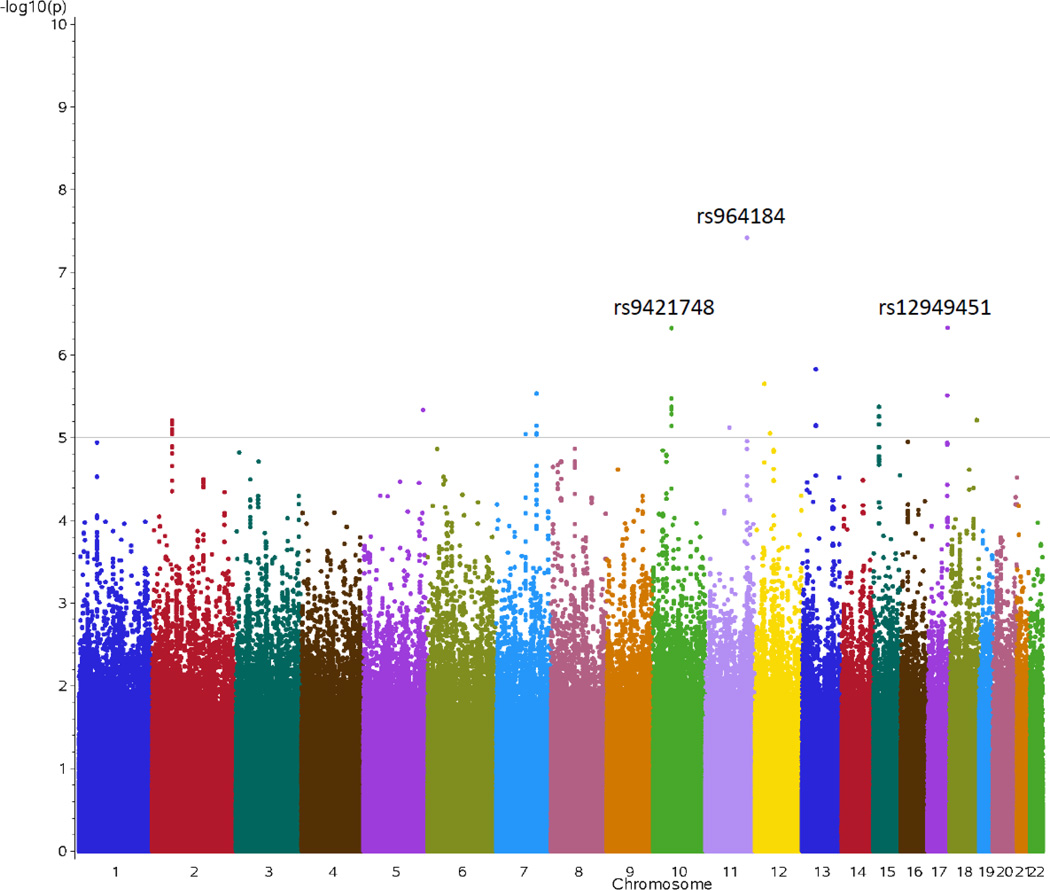
The QQ plot for the PPL TG AUC phenotype for the GOLDN discovery sample is shown in Figure 1. Supplementary Figure S1A displays the QQ plots for all the minimally adjusted PPL TG phenotypes (AUC, AUI, uptake, clearance) while the minimal plus baseline TG adjusted PPL TG phenotypes (AUC, AUI, uptake, clearance) are in Supplementary Figure S1B. The discovery results did not significantly deviate from the expected results, with genomic control lambda values between 0.88439 and 0.94216. Figure 2 displays the Manhattan plot for the discovery GWA results for the TG AUC phenotype. Results from the minimally adjusted PPL TG phenotypes are summarized in Supplementary Figure S2A. Results from the minimal plus baseline TG adjusted PPL TG phenotypes are provided in Supplementary Figure S2B.
Figure 1.
Quantile-quantile plot TG AUC GWAS results in the GOLDN study (1% allele frequency filter).
Figure 2.
Distribution of p-values from the genome-wide analysis of genotyped plus imputed SNPs associated with AUC PPL TG phenotype in GOLDN study (Manhattan plot). Two loci reached genome-wide significance. Loci above the line reflecting a p-value of 1 × 10−5 are suggestive for significance. Charted SNPs passed a 1% allele frequency filter.
Overall, we observed 111 ‘top discovery SNPs’ across all phenotypes. Two SNPs, rs10243693 (AUC adjusted for baseline TG) and rs964184 (AUC) met the GWA significant p-value threshold (p<5E-08) and 109 SNP-phenotype associations (85 unique SNPs) were considered ‘suggestive’ (p<1E-05) across all phenotypes. Of these 85 ‘top SNPs’ most were intronic or intergenic. Fifty-seven of these suggestive SNPs were only identified in our minimal model (Table 2 and Supplementary Table S2) while 20 were unique to the minimal plus baseline TG adjusted model (Table 3 and Supplementary Table S3). Eight SNPs demonstrated suggestive associations with both models, and twelve SNPs had suggestive associations with more than one phenotype. Furthermore, the 85 ‘top SNPs’ actually represent 43 independent SNPs when linkage disequilibrium (LD) is taken into account. The SNP annotation and the quality control of each SNP for each study are documented in Supplementary Table S4. The proportion of variance explained by each of these top SNPs (R2) is between 0.01 and 0.04.
Table 2.
Top SNPs Associated with Triglyceride Response to a high-fat meal in the GOLDN and the HAPI Heart Studies.
| Discovery | Replication | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phenotype | GOLDN (n=872) | HAPI Heart (n=843) | Joint Meta-Analysis (GOLDN and HAPI, n=1,715) | |||||||||||
| AUC | Chr | Gene | Role | EA | EAF* | Beta (SE) | p-value | Beta (SE) | p- value |
EA | EAF | Beta (SE) | p-value | Direction |
| rs964184 | 11:116648917 |
ZPR1 (FORMERLY ZNF259) |
downstream- 500 Bases |
C | 0.8738 | −0.4148 (0.0754) | 3.82E-08 | −0.2154 (0.0663) | 0.0012 | C | 0.833 | −0.3023 (0.0498) | 1.259E-09 | -- |
| rs12949451 | 17:73032319 | A | 0.9125 | −0.4390 (0.0871) | 4.66E-07 | −0.1725 (0.1423) | 0.2258 | A | 0.9152 | −0.439 (0.0871) | 4.677E-07 | -- | ||
| rs9421748 | 10:48499271 | A | 0.5292 | −0.2526 (0.0501) | 4.74E-07 | 0.0551 (0.0519) | 0.2887 | A | 0.5279 | −0.2526 (0.0501) | 4.786E-07 | -+ | ||
| AUI | ||||||||||||||
| rs10866682 | 5:174036233 | C | 0.1039 | 0.4356 (0.0870) | 5.59E-07 | −0.0905 (0.0983) | 0.3572 | C | 0.1051 | 0.4356 (0.087) | 5.623E-07 | +- | ||
| rs7712993 | 5:174037081 | A | 0.1039 | 0.4359 (0.0871) | 5.65E-07 | −0.0904 (0.0990) | 0.3615 | A | 0.1051 | 0.4359 (0.0871) | 5.623E-07 | +- | ||
| rs13096657 | 3:121300728 | ARGFX | intron | C | 0.8569 | 0.3634 (0.0739) | 8.75E-07 | 0.0757 (0.1405) | 0.5902 | C | 0.8601 | 0.3634 (0.0739) | 8.71E-07 | ++ |
| Uptake Slope | ||||||||||||||
| rs13096657 | 3:121300728 | ARGFX | intron | C | 0.8569 | 0.3713 (0.0732) | 3.93E-07 | −0.0687 (0.1386) | 0.6204 | C | 0.8603 | 0.3713 (0.0732) | 3.89E-07 | +- |
| rs3749872 | 6:107388504 | BEND3 | utr-3-prime | C | 0.0450 | 0.5892 (0.1167) | 4.48E-07 | −0.3145 (0.1436) | 0.0288 | C | 0.0451 | 0.5892 (0.1167) | 4.467E-07 | +- |
| rs10866682 | 5:174036233 | C | 0.1039 | 0.4323 (0.0861) | 5.10E-07 | −0.1286 (0.0964) | 0.1824 | C | 0.1052 | 0.4323 (0.0861) | 5.129E-07 | +- | ||
| rs7712993 | 5:174037081 | A | 0.1039 | 0.4326 (0.0862) | 5.15E-07 | −0.1295 (0.0971) | 0.1827 | A | 0.1051 | 0.4326 (0.0862) | 5.129E-07 | +- | ||
| rs13065635 | 3:121302410 | ARGFX | intron | G | 0.8776 | 0.3971 (0.0808) | 8.95E-07 | −0.0786 (0.1606) | 0.6247 | G | 0.879 | 0.3971 (0.0808) | 8.913E-07 | +- |
| Clearance Slope | ||||||||||||||
| none | ||||||||||||||
EAF, effect allele frequency
Table 3.
Top SNPs Associated with Triglyceride Response to a high-fat meal adjusted for baseline TG Level in the GOLDN and HAPI Heart Studies.
| Phenotype | Discovery GOLDN (n=872) |
Replication HAPI Heart (n=843) |
Joint Meta-Analysis (GOLDN and HAPI, n=1,715) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AUC | Chr | Gene | Role | EA | EAF | Beta (SE) | p-value | Beta (SE) | p- value |
EA | EAF | Beta (SE) | p-value | Direction |
| rs10243693 | 7:68904887 | A | 0.3642 | 0.2788 (0.0506) | 3.50E-08 | −0.0966 (0.0637) | 0.1298 | A | 0.3392 | 0.1338 (0.0396) | 7.24E-04 | +− | ||
| rs6959964 | 7:68905738 | C | 0.3724 | 0.2574 (0.0502) | 2.86E-07 | −0.1194 (0.0645) | 0.0642 | C | 0.369 | 0.2574 (0.0502) | 2.88E-07 | +− | ||
| AUI | ||||||||||||||
| rs10866682 | 5:174036233 | C | 0.1039 | 0.4331 (0.0869) | 6.31E-07 | −0.0820 (0.0982) | 0.4044 | C | 0.1051 | 0.4331 (0.0869) | 6.31E-07 | +− | ||
| rs7712993 | 5:174037081 | A | 0.1039 | 0.4334 (0.0870) | 6.38E-07 | −0.0818 (0.0990) | 0.4091 | A | 0.1051 | 0.4334 (0.087) | 6.31E-07 | +− | ||
| Uptake Slope | ||||||||||||||
| rs10866682 | 5:174036233 | C | 0.1039 | 0.4271 (0.0861) | 6.99E-07 | −0.1538 (0.0967) | 0.1122 | C | 0.1052 | 0.4271 (0.0861) | 6.92E-07 | +− | ||
| rs7712993 | 5:174037081 | A | 0.1039 | 0.4274 (0.0862) | 7.05E-07 | −0.1550 (0.0975) | 0.1121 | A | 0.1051 | 0.4274 (0.0862) | 7.08E-07 | +− | ||
| Clearance Slope | ||||||||||||||
| none | ||||||||||||||
3.3 HAPI Heart Replication
The replication results for these ‘top discovery SNPs’ in the independent HAPI Heart Study are provided in Table 2 and Supplemental Table S2 for the minimal model, and Table 3 and Supplemental Table S3 for the minimal plus baseline TG adjusted model. For the majority of these signals, there was little evidence of replication across the PPL TG phenotypes. However, rs964184 near ZPR1 (formerly ZNF259) gene and mapping quite close to the APOA1/C3/A4/A5 cluster displayed modest evidence of replication (p=1.20E-03; Bonferroni corrected p-value=0.05/43=1.20E-03) for the same phenotype as in our discovery (Table 2), and also across additional phenotypes in the HAPI Heart Study (Table 4).
Table 4.
Association results between rs964184 and all PPL triglyceride response phenotypes.
| Discovery | Replication | Joint Meta-Analysis | ||||
|---|---|---|---|---|---|---|
| GOLDN (n=872) | HAPI Heart (n=843) | GOLDN Discovery and HAPI Heart Meta- Analysis |
||||
| PPL Measurement | beta (SE) | p-value | beta (SE) | p-value | beta (SE) | p-value |
| Area under the curve | −0.4148 (0.0754) | 3.82E-08 | −0.2154 (0.0663) | 1.20E-03 | −0.302 (0.0498) | 1.26E-09 |
| Area under the increase | 0.0199 (0.0851) | 8.15E-01 | −0.2056 (0.0674) | 2.36E-03 | −0.1057 (0.0508) | 3.72E-02 |
| Uptake slope | 0.0767 (0.0762) | 3.14E-01 | −0.1124 (0.0672) | 9.48E-02 | −0.0297 (0.0504) | 5.50E-01 |
| Clearance slope | −0.0951 (0.0767) | 2.15E-01 | −0.2239 (0.0672) | 9.04E-04 | −0.168 (0.0506) | 8.91E-04 |
| Adjusted for baseline TG | ||||||
| Area under the curve | −0.1253 (0.0763) | 1.01E-01 | −0.1681 (0.0662) | 1.13E-02 | −0.1497 (0.05) | 2.75E-03 |
| Area under the increase | −0.0288 (0.0771) | 7.09E-01 | −0.2455 (0.0672) | 2.77E-04 | −0.1519 (0.0507) | 2.75E-03 |
| Uptake slope | −0.0166 (0.0763) | 8.27E-01 | −0.2199 (0.0683) | 1.33E-03 | −0.1295 (0.0509) | 1.10E-02 |
| Clearance slope | −0.0758 (0.0768) | 3.24E-01 | −0.2487 (0.0671) | 2.25E-04 | −0.1739 (0.0506) | 5.75E-04 |
| Baseline TG | −0.4063 (0.0727) | 2.99E-08 | ||||
3.4 GOLDN Discovery and HAPI Heart Meta-Analyzed ‘Top Discovery SNPs’
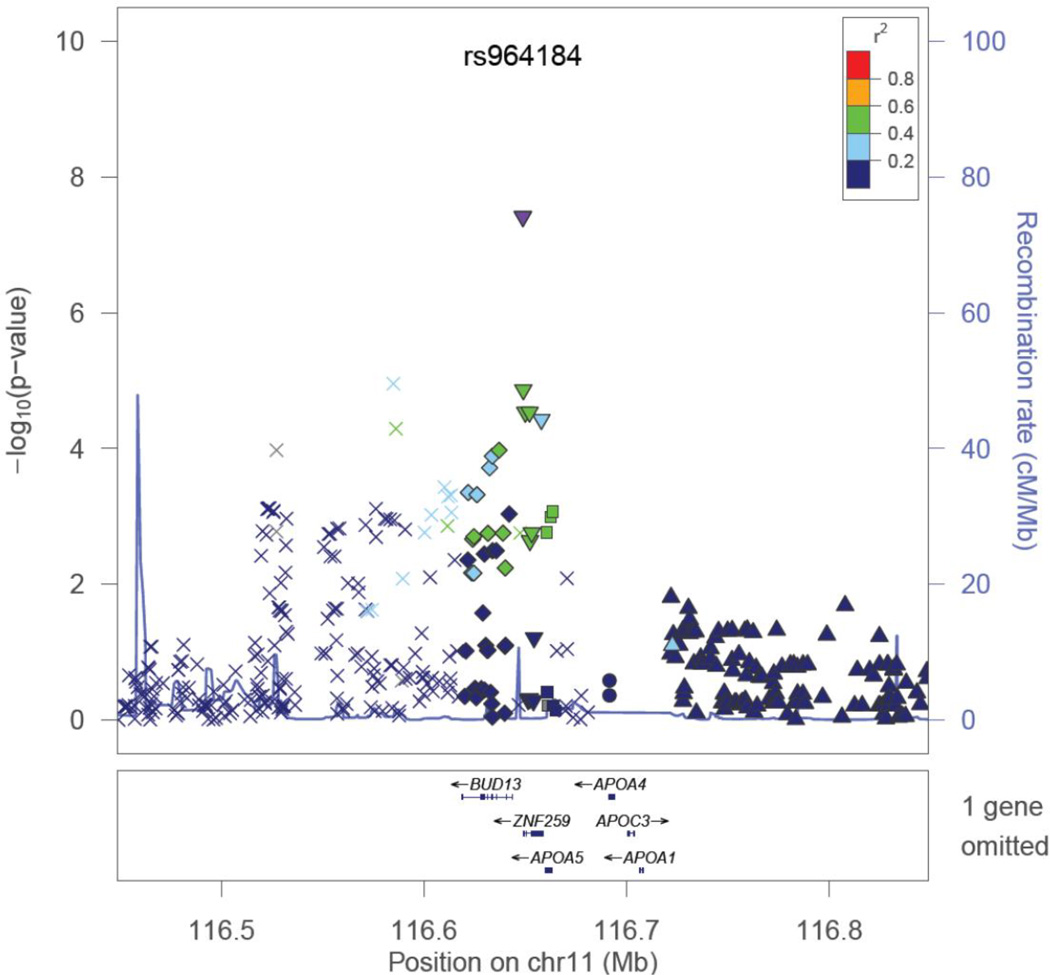
For the ‘top discovery SNPs’, we performed a joint meta-analysis for results from GOLDN discovery and HAPI Heart (Tables 2 and Supplemental Table S2 for the minimal model; Tables 3 and Supplemental Table S3 for the minimal plus baseline TG adjusted model). With this approach, we observed little replication of these ‘top SNPs’. Again one SNP, rs964184, satisfied this replication definition and in the meta-analysis was genome-wide significant (p<5E-08), Table 2. Figure 3 demonstrates the association results of SNPs around rs964184, and their LD with rs964184, created with LocusZoom [35]. This variant (rs964184) is not in high LD with other SNPs in our data, but lies in close proximity to the APOA1/C3/A4/A5 cluster, which harbors numerous variants with associations to baseline TG [36], and postprandial lipid metabolism parameters [37] as well as numerous gene-environment interactions involving TG as phenotype and fat intake as a modifying environmental factor [38]. The association of this SNP with all fasting and PPL phenotypes is provided in Table 4. Because rs964184 was not among the ‘top discovery SNPs’ for the AUC adjusted for baseline TG phenotype, we tested an association of this SNP with baseline TG in GOLDN to assess if the SNP is more strongly associated with PPL or baseline TG. As displayed in Table 4, the signal for this SNP is mostly responsible for an association with baseline TG in GOLDN. In HAPI Heart, the signal for this SNP is not attenuated when adjustment for baseline TG is included. Thus, we sought to determine if the R19X APOC3 mutation in HAPI Heart is driving their association signal, although LD is not strong between these two variants. When HAPI Heart was further adjusted for R19X, the signal was only slightly attenuated (β=−0.1780, p-value=0.0078 with R19X adjustment; β=−0.2154, p-value=0.0012 without R19X adjustment), thus R19X is not driving their association, as the association persisted after R19X adjustment.
Figure 3.
Regional plot of ZPR1 (formerly ZNF259) on chromosome 11 for the replicated SNP, rs964184 [35]. Symbol key: downward triangle, ZNF259; upward triangle, SIK3; diamond, BUD13; square, APOA5; circle, APOA4; X, SNP not assigned to any gene.
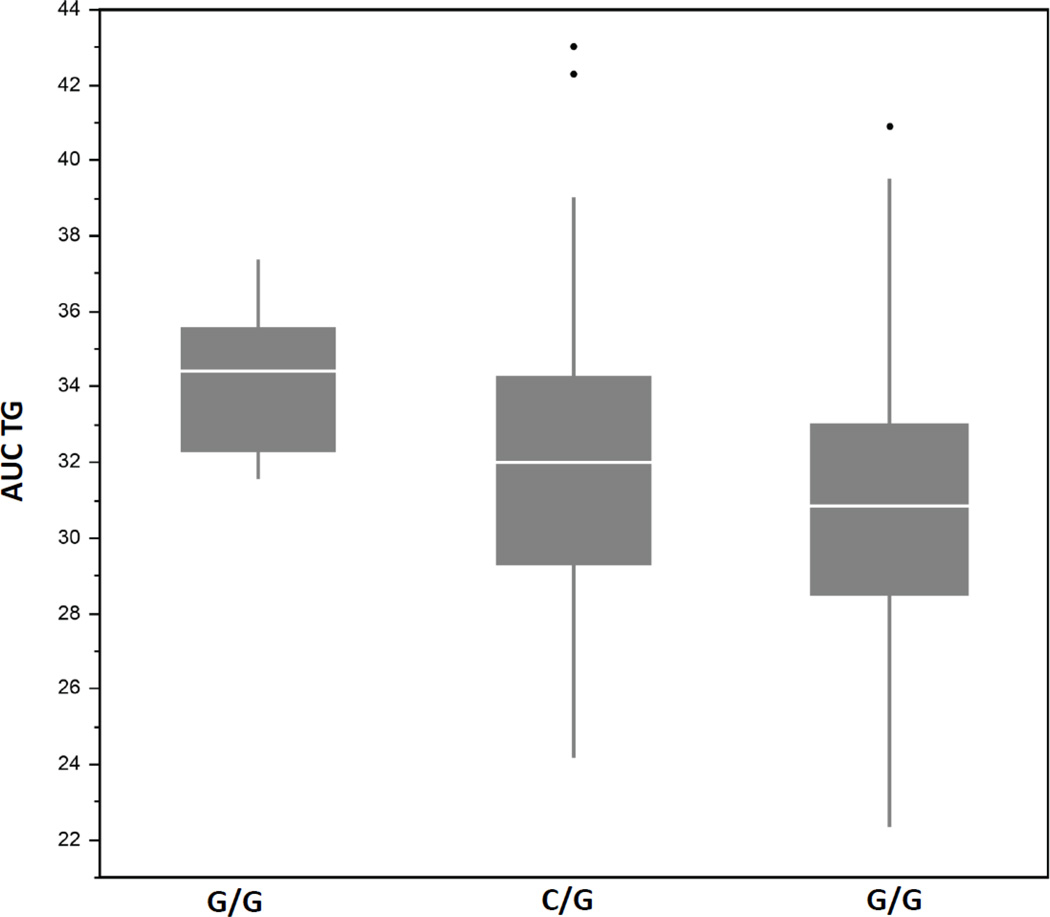
3.5 Effect of rs964184 genotype on plasma TG levels
We assessed the genotypic effect on plasma TG levels at 0, 3.5, and 6 hours PPL, as well as mean AUC TG for our top SNP, rs964184, as depicted in Table 5 and Figure 4. Presence of one minor allele (G) increases all plasma TG values across all time points compared to the reference allele (C). Further, participants homozygous for the minor allele display even larger increases in plasma TG values. This is also demonstrated using AUC TG (ANOVA p-value=1.98052E-05), and holds when stratified by men and women.
Table 5.
Genotype Specific Effects for rs964184 on fasting TG (mean and standard deviation) in GOLDN
| Everyone | Sample Size | Baseline TG Mean (SD) | 3.5 Hr TG Mean (SD) | 6 Hr TG Mean (SD) | AUC TG Mean (SD) |
|---|---|---|---|---|---|
| C/C | 569 | 130.25 (81.96) | 246.73 (144.49) | 228.49 (176.79) | 30.83 (3.31) |
| C/G | 177 | 169.54 (132.46) | 287.49 (178.26) | 275.3 (211.98) | 31.95 (3.41) |
| G/G | 8 | 224.75 (110.86) | 320.63 (90.38) | 351.75 (150.25) | 34.22 (1.93) |
| Men | |||||
| C/C | 287 | 139.46 (84.13) | 272.5 (152.24) | 258.26 (185.79) | 31.46 (3.23) |
| C/G | 81 | 188.05 (164.51) | 318.19 (208.52) | 305.73 (245.08) | 32.49 (3.53) |
| G/G | 5 | 256 (127.63) | 350.8 (94.87) | 401.2 (169.34) | 34.75 (2.16) |
| Women | |||||
| C/C | 282 | 120.87 (78.74) | 220.5 (131.31) | 198.18 (161.92) | 30.19 (3.27) |
| C/G | 96 | 153.92 (95.8) | 261.59 (144.14) | 248.8 (175.32) | 31.47 (3.25) |
| G/G | 3 | 172.67 (62.66) | 270.33 (67.25) | 269.33 (73.24) | 33.33 (1.38) |
Figure 4.
Rs964184 genotype specific effects on mean AUC TG, ANOVA p-value=1.98052E-05.
4. DISCUSSION
We analyzed the GOLDN data for PPL TG phenotypes to identify new genes influencing the PPL TG response and by extension, lipid metabolism. PPL TG response phenotypes are unique because humans spend the majority of their day in the PPL state, thereby these results may be more informative than fasting TG results. In the GOLDN sample for PPL TG, two genome-wide significant SNPs were documented; however, only one demonstrated evidence of replication in the HAPI Heart Study. In the joint meta-analysis (discovery and replication samples) this SNP (rs964184) had a more significant association than it did in the GOLDN analysis alone. The replication cohort did not demonstrate significant replication for the additional significant SNP (rs10243693) from the discovery. The joint meta-analysis did produce a genome-wide significant result but upon further analyses, it appears that the association of rs964184, is mainly due to its association with baseline TG rather than PPL TG response.
From the GOLDN discovery SNPs for the PPL TG response, only one SNP, rs964184, reached genomewide significance in the combined joint meta-analysis. This SNP is located near the ZPR1 (formerly ZNF259) gene, close to the APOA5/APOA4/APOC3/APOA1 gene cluster and is associated with fasting TG [5], metabolic syndrome [26, 39], TG fenofibrate response [40], coronary artery disease [41], and a dietary gene-environment interaction [42]. Kraja et al [26] plotted the LD blocks of the APOA1-gene cluster, which included our lead SNP. This SNP was part of a LD block including an APOA5 variant, with which it is in high LD. Thus, it is still unknown which variant in this region is the functional or biologically relevant variant, but the data support the involvement of variants in this LD region in TG metabolism. Since baseline TG is the best predictor of PPL TG, we further adjusted our phenotype for baseline TG. Unfortunately this attenuated the association with this SNP and it no longer significant, leading to the conclusion that rs964184 is more strongly associated with fasting TG than the PPL TG response. Van de Woestijne et al [43] documented an association between fasting TG and rs964184 among patients with known vascular disease which was modified by the presence of metabolic syndrome. While we were unable to assess this in our population because we did not measure clinical vascular disease, it corroborates the importance of studying lipids in relation to CVD.
Variants associating with PPL TG phenotypes that map within or near genes showing consistent differential expression across a panel of high-fat feeding or caloric manipulation experiments in rodent livers would be of interest as would genes that are members of the PPARA or PPARG regulatory networks. Variants within such diet-sensitive genes and/or variants responsible for the regulation of their expression might lead to a differential TG response under conditions of high-fat feeding. Data from over 9 different experiments in which gene expression differences in rat or mouse liver were examined after high-fat feeding, caloric restriction or other similar manipulations, or identified as responsive to the PPARA/PPARG lipid homeostasis networks [44–53] indicated that orthologs of C9orf72, CYFIP1, EXT2, NIPA2, and APOA1 cluster genes ZNF259, APOA5, APOA4 and APOA1 each showed changes in expression in five or more experiments. Alternatively, these genes and their variants might also be constituents of gene-environment interactions. In this regard, APOA4 SNP rs5110 has been shown to associate with postprandial lipemia in a European population in a BMI-dependent manner [54]. Other similar gene-diet interactions have been catalogued [38]; although none of the lead SNPs or SNPs in LD with the lead SNPs identified here have been reported for other gene-diet interactions.
Certain suggestive SNPs identified by this study or their proxies in strong LD (r2 > 0.8) map to regions identified by ENCODE and other datasets and are predicted to affect gene expression. Such allele-specific differences in gene regulation can arise from binding of transcription factors, microRNAs or altering epigenetic marks in liver cells. Three cases of such predicted function are highlighted here as plausible examples of variants that could affect the PPL TG response. One, SNP rs964184 has been thoroughly discussed above and its proximity to other variants within the APOA1 gene cluster and their associations with fasting TG levels have been well documented. Additionally, we note that this variant also directs an expression QTL (eQTL) for TAGLN [55], a gene 360 kilo base pairs distal to this region and harboring variants associated with fasting TG (-log10 p-value 9.7) [56]. Two, we observed that three LD groups drawn from the top SNPs associated with PPL phenotypes in GOLDN (Table S4) contain SNPs (rs10202351, rs6764992 and rs7895237) with alleles predicted to alter binding of serum response factor SRF. Interestingly, mice carrying a liver Srf knockout have altered TG levels [57] and so the altered ability of this transcription factor to regulate certain genes may have effects on PPL. Three, chromosome 2 SNP rs7588818, with suggestive association to AUC after baseline TG adjustment (Table S4), shows a predicted allele-specific binding of transcription factor TFAP2B. It has been demonstrated in murine adipocytes that postprandial activation of protein kinase Cµ (PRKD1) regulates the expression of adipocytokines CCL2 (MCP-1), IL6 and adiponectin (ADIPOQ) via TFAP2B [58]. Although we observed numerous instances from ENCODE epigenetic data where a SNP allele would destroy an observed liver methylated CpG or maps within hepatic cell histone acetylation or methylation peaks, we can only speculate on the relevance of such to the PPL condition. Nonetheless, these and other data we mined from various sources give credence to the possibility that these suggestive SNPs do exert influence on the PPL TG response but did not reach genome wide significance for reasons outlined above. Thus, these would be worthwhile candidate variants under a focused approach.
These experiments in mice and rodents on our top human PPL TG associations provide complementary evidence that the genetics of PPL TG is complicated and not fully understood. It also suggests that non-fasting TG is important to study and measure as well as fasting TG. Measurement of PPL lipids in the future may be more informative and predictive of CVD risk [4], and different genetic mechanisms may affect PPL TG versus fasting TG. Thus while more investigation is needed into mechanisms, pathophysiology, and genetics of PPL TG, it is becoming clear that non-fasting lipids are very important in the management of CVD and a better understanding of their role is needed.
One of the main strengths of GOLDN is its interventional nature and its unique design. The HAPI Heart Study provided an opportunity for replication, however, there was limited evidence for replication of our main results despite attempts to harmonize the PPL intervention and TG outcomes. In an attempt to overcome the small sample sizes, we performed a joint analysis with our discovery and replication cohorts [32], which did enrich some of our initial signals. Furthermore the GOLDN study examined Caucasians of European ancestry and thus the results may not be applicable to other ethnicities or a more heterogeneous sample. Although study participants were instructed to fast overnight, it is possible that the composition of the diet in the 12–48 hours preceding the onset of that fast did affect the TG response to the high-fat challenge [59, 60]. Nonetheless, the unique design of GOLDN is a strength as it allows for the investigation of unique questions about lipid metabolism and genes influencing lipid metabolism and the postprandial state.
To summarize, PPL TG response is a known CVD risk factor and in contrast to fasting TG, PPL TG is the usual state of triglycerides in humans. The response is highly variable among individuals and is influenced by both genes and environment. Using the GOLDN study we performed a GWAS on PPL TG response phenotypes in hopes of identifying potential new genes influencing lipid metabolism following a high-fat meal. While the results supported previous work on the genetics of fasting TG, there were no genome-wide significant results for the PPL TG response phenotypes. Future work may be enhanced by using a pathway analysis or newer genetic technologies more specific to our study, such as interrogating the metabolome.
Supplementary Material
Acknowledgments
We thank the GOLDN Study participants, collaborators & staff. The HAPI Heart Study acknowledges the Amish liaisons, field workers, and the cooperation and support of the Amish community.
Funding
The GOLDN Study was funded by grant HL091357 (Arnett PI) from the National Heart, Lung and Blood Institute. The HAPI Heart Study was supported by research grants P30DK072488, U01HL072515, and U01HL084756 from the National Institutes of Health.
Abbreviations
- GOLDN
Genetics of Lipid Lowering Drugs and Diet Network
- TG
triglyceride
- PPL
postprandial lipemia
- GWA
genome-wide association
- HAPI
Heredity and Phenotype Intervention
- SNP
single nucleotide polymorphism
- HDL
high density lipoprotein
- CVD
cardiovascular disease
- CHD
coronary heart disease
- FamHS
Family Heart Study
- AUC
area under the curve
- AUI
area under the increase
- PC
principal component
quartile-quartile
- LD
linkage disequilibrium
- EA
effect allele
- EAF
effect allele frequency
- Chr
chromosome
- SE
standard error
- SD
standard deviation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Design and Conduct Study: M.K.W., M.A.P., I.B.B.
Data Collection: T.I.P., M.A.P., J.M.O., A.R.S., D.K.A., I.B.B.
Data Analysis: M.K.W. T.I.P., C.Q.L., J.R.O’C., Q.G., K.A.R., H.K.T.
Data Interpretation: M.K.W., L.D.P., M.F.F., A.C.F-W., S.A., J.M.O., A.R.S., D.K.A., I.B.B.
Manuscript Writing: M.K.W., L.D.P, all co-authors read, commented, and approved manuscript.
Conflict of interest: All of the authors declare that they have no relevant conflicts of interest. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. The USDA is an equal opportunity provider and employer.
REFERENCES
- 1.Jackson KG, Poppitt SD, Minihane AM. Postprandial lipemia and cardiovascular disease risk: Interrelationships between dietary, physiological and genetic determinants. Atherosclerosis. 2012;220(1):22–33. doi: 10.1016/j.atherosclerosis.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 2.Nordestgaard BG, Varbo A. Triglycerides and cardiovascular disease. Lancet. 2014;384(9943):626–635. doi: 10.1016/S0140-6736(14)61177-6. [DOI] [PubMed] [Google Scholar]
- 3.Alcala-Diaz JF, Delgado-Lista J, Perez-Martinez P, Garcia-Rios A, Marin C, Quintana-Navarro GM, Gomez-Luna P, Camargo A, Almaden Y, Caballero J, et al. Hypertriglyceridemia influences the degree of postprandial lipemic response in patients with metabolic syndrome and coronary artery disease: from the CORDIOPREV study. PloS one. 2014;9(5):e96297. doi: 10.1371/journal.pone.0096297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hegele RA, Ginsberg HN, Chapman MJ, Nordestgaard BG, Kuivenhoven JA, Averna M, Boren J, Bruckert E, Catapano AL, Descamps OS, et al. The polygenic nature of hypertriglyceridaemia: implications for definition, diagnosis, and management. The lancet Diabetes & endocrinology. 2014;2(8):655–666. doi: 10.1016/S2213-8587(13)70191-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asselbergs FW, Guo Y, van Iperen EP, Sivapalaratnam S, Tragante V, Lanktree MB, Lange LA, Almoguera B, Appelman YE, Barnard J, et al. Large-scale gene-centric meta-analysis across 32 studies identifies multiple lipid loci. American journal of human genetics. 2012;91(5):823–838. doi: 10.1016/j.ajhg.2012.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nature genetics. 2009;41(1):56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waterworth DM, Ricketts SL, Song K, Chen L, Zhao JH, Ripatti S, Aulchenko YS, Zhang W, Yuan X, Lim N, et al. Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arteriosclerosis, thrombosis, and vascular biology. 2010;30(11):2264–2276. doi: 10.1161/ATVBAHA.109.201020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johansen CT, Hegele RA. Allelic and phenotypic spectrum of plasma triglycerides. Biochimica et biophysica acta. 2012;1821(5):833–842. doi: 10.1016/j.bbalip.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 10.Johansen CT, Kathiresan S, Hegele RA. Genetic determinants of plasma triglycerides. Journal of lipid research. 2011;52(2):189–206. doi: 10.1194/jlr.R009720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pirillo A, Norata GD, Catapano AL. Postprandial lipemia as a cardiometabolic risk factor. Current medical research and opinion. 2014;30(8):1489–1503. doi: 10.1185/03007995.2014.909394. [DOI] [PubMed] [Google Scholar]
- 12.Nordestgaard BG, Benn M, Schnohr P, Tybjaerg-Hansen A. Nonfasting triglycerides and risk of myocardial infarction, ischemic heart disease, and death in men and women. Jama. 2007;298(3):299–308. doi: 10.1001/jama.298.3.299. [DOI] [PubMed] [Google Scholar]
- 13.Bansal S, Buring JE, Rifai N, Mora S, Sacks FM, Ridker PM. Fasting compared with nonfasting triglycerides and risk of cardiovascular events in women. Jama. 2007;298(3):309–316. doi: 10.1001/jama.298.3.309. [DOI] [PubMed] [Google Scholar]
- 14.Freiberg JJ, Tybjaerg-Hansen A, Jensen JS, Nordestgaard BG. Nonfasting triglycerides and risk of ischemic stroke in the general population. Jama. 2008;300(18):2142–2152. doi: 10.1001/jama.2008.621. [DOI] [PubMed] [Google Scholar]
- 15.Rosenson RS, Davidson MH, Hirsh BJ, Kathiresan S, Gaudet D. Genetics and causality of triglyceride-rich lipoproteins in atherosclerotic cardiovascular disease. Journal of the American College of Cardiology. 2014;64(23):2525–2540. doi: 10.1016/j.jacc.2014.09.042. [DOI] [PubMed] [Google Scholar]
- 16.Pollin TI, Damcott CM, Shen H, Ott SH, Shelton J, Horenstein RB, Post W, McLenithan JC, Bielak LF, Peyser PA, et al. A null mutation in human APOC3 confers a favorable plasma lipid profile and apparent cardioprotection. Science. 2008;322(5908):1702–1705. doi: 10.1126/science.1161524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boren J, Matikainen N, Adiels M, Taskinen MR. Postprandial hypertriglyceridemia as a coronary risk factor. Clinica chimica acta; international journal of clinical chemistry. 2014;431:131–142. doi: 10.1016/j.cca.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 18.Matikainen N, Burza MA, Romeo S, Hakkarainen A, Adiels M, Folkersen L, Eriksson P, Lundbom N, Ehrenborg E, Orho-Melander M, et al. Genetic variation in SULF2 is associated with postprandial clearance of triglyceride-rich remnant particles and triglyceride levels in healthy subjects. PloS one. 2013;8(11):e79473. doi: 10.1371/journal.pone.0079473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai CQ, Arnett DK, Corella D, Straka RJ, Tsai MY, Peacock JM, Adiconis X, Parnell LD, Hixson JE, Province MA, et al. Fenofibrate effect on triglyceride and postprandial response of apolipoprotein A5 variants: the GOLDN study. Arteriosclerosis, thrombosis, and vascular biology. 2007;27(6):1417–1425. doi: 10.1161/ATVBAHA.107.140103. [DOI] [PubMed] [Google Scholar]
- 20.Higgins M, Province M, Heiss G, Eckfeldt J, Ellison RC, Folsom AR, Rao DC, Sprafka JM, Williams R. NHLBI Family Heart Study: objectives and design. American journal of epidemiology. 1996;143(12):1219–1228. doi: 10.1093/oxfordjournals.aje.a008709. [DOI] [PubMed] [Google Scholar]
- 21.Patsch JR, Miesenbock G, Hopferwieser T, Muhlberger V, Knapp E, Dunn JK, Gotto AM, Jr, Patsch W. Relation of triglyceride metabolism and coronary artery disease. Studies in the postprandial state. Arteriosclerosis and thrombosis : a journal of vascular biology / American Heart Association. 1992;12(11):1336–1345. doi: 10.1161/01.atv.12.11.1336. [DOI] [PubMed] [Google Scholar]
- 22.Aslibekyan S, Kabagambe EK, Irvin MR, Straka RJ, Borecki IB, Tiwari HK, Tsai MY, Hopkins PN, Shen J, Lai CQ, et al. A genome-wide association study of inflammatory biomarker changes in response to fenofibrate treatment in the Genetics of Lipid Lowering Drug and Diet Network. Pharmacogenetics and genomics. 2012;22(3):191–197. doi: 10.1097/FPC.0b013e32834fdd41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, Cawley S, Hubbell E, Veitch J, Collins PJ, Darvishi K, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nature genetics. 2008;40(10):1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annual review of genomics and human genetics. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genetic epidemiology. 2010;34(8):816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraja AT, Vaidya D, Pankow JS, Goodarzi MO, Assimes TL, Kullo IJ, Sovio U, Mathias RA, Sun YV, Franceschini N, et al. A bivariate genome-wide approach to metabolic syndrome: STAMPEED consortium. Diabetes. 2011;60(4):1329–1339. doi: 10.2337/db10-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell BD, McArdle PF, Shen H, Rampersaud E, Pollin TI, Bielak LF, Jaquish C, Douglas JA, Roy-Gagnon MH, Sack P, et al. The genetic response to short-term interventions affecting cardiovascular function: rationale and design of the Heredity and Phenotype Intervention (HAPI) Heart Study. American heart journal. 2008;155(5):823–828. doi: 10.1016/j.ahj.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agarwala R, Biesecker LG, Hopkins KA, Francomano CA, Schaffer AA. Software for constructing and verifying pedigrees within large genealogies and an application to the Old Order Amish of Lancaster County. Genome research. 1998;8(3):211–221. doi: 10.1101/gr.8.3.211. [DOI] [PubMed] [Google Scholar]
- 29.Agarwala R, Schaffer AA, Tomlin JF. Towards a complete North American Anabaptist Genealogy II: analysis of inbreeding. Human biology. 2001;73(4):533–545. doi: 10.1353/hub.2001.0045. [DOI] [PubMed] [Google Scholar]
- 30.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature genetics. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 31.Asking for more. Nature genetics. 2012;44(7):733. doi: 10.1038/ng.2345. [DOI] [PubMed] [Google Scholar]
- 32.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nature genetics. 2006;38(2):209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 33.Miller M, Stone NJ, Ballantyne C, Bittner V, Criqui MH, Ginsberg HN, Goldberg AC, Howard WJ, Jacobson MS, Kris-Etherton PM, et al. Triglycerides and cardiovascular disease: a scientific statement from the American Heart Association. Circulation. 2011;123(20):2292–2333. doi: 10.1161/CIR.0b013e3182160726. [DOI] [PubMed] [Google Scholar]
- 34.Schrezenmeir J, Keppler I, Fenselau S, Weber P, Biesalski HK, Probst R, Laue C, Zuchhold HD, Prellwitz W, Beyer J. The phenomenon of a high triglyceride response to an oral lipid load in healthy subjects and its link to the metabolic syndrome. Annals of the New York Academy of Sciences. 1993;683:302–314. doi: 10.1111/j.1749-6632.1993.tb35721.x. [DOI] [PubMed] [Google Scholar]
- 35.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai CQ, Parnell LD, Ordovas JM. The APOA1/C3/A4/A5 gene cluster, lipid metabolism and cardiovascular disease risk. Current opinion in lipidology. 2005;16(2):153–166. doi: 10.1097/01.mol.0000162320.54795.68. [DOI] [PubMed] [Google Scholar]
- 37.Delgado-Lista J, Perez-Jimenez F, Ruano J, Perez-Martinez P, Fuentes F, Criado-Garcia J, Parnell LD, Garcia-Rios A, Ordovas JM, Lopez-Miranda J. Effects of variations in the APOA1/C3/A4/A5 gene cluster on different parameters of postprandial lipid metabolism in healthy young men. Journal of lipid research. 2010;51(1):63–73. doi: 10.1194/jlr.M800527-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parnell LD, Blokker BA, Dashti HS, Nesbeth PD, Cooper BE, Ma Y, Lee YC, Hou R, Lai CQ, Richardson K, et al. CardioGxE, a catalog of gene-environment interactions for cardiometabolic traits. BioData mining. 2014;7:21. doi: 10.1186/1756-0381-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kristiansson K, Perola M, Tikkanen E, Kettunen J, Surakka I, Havulinna AS, Stancakova A, Barnes C, Widen E, Kajantie E, et al. Genome-wide screen for metabolic syndrome susceptibility Loci reveals strong lipid gene contribution but no evidence for common genetic basis for clustering of metabolic syndrome traits. Circulation Cardiovascular genetics. 2012;5(2):242–249. doi: 10.1161/CIRCGENETICS.111.961482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aslibekyan S, Goodarzi MO, Frazier-Wood AC, Yan X, Irvin MR, Kim E, Tiwari HK, Guo X, Straka RJ, Taylor KD, et al. Variants identified in a GWAS meta-analysis for blood lipids are associated with the lipid response to fenofibrate. PloS one. 2012;7(10):e48663. doi: 10.1371/journal.pone.0048663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nature genetics. 2011;43(4):333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X, Qi Q, Bray GA, Hu FB, Sacks FM, Qi L. APOA5 genotype modulates 2-y changes in lipid profile in response to weight-loss diet intervention: the Pounds Lost Trial. The American journal of clinical nutrition. 2012;96(4):917–922. doi: 10.3945/ajcn.112.040907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van de Woestijne AP, van der Graaf Y, de Bakker PI, Asselbergs FW, Spiering W, Visseren FL, Group SS. Rs964184 (APOA5-A4-C3-A1) is related to elevated plasma triglyceride levels, but not to an increased risk for vascular events in patients with clinically manifest vascular disease. PloS one. 2014;9(6):e101082. doi: 10.1371/journal.pone.0101082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie Z, Li H, Wang K, Lin J, Wang Q, Zhao G, Jia W, Zhang Q. Analysis of transcriptome and metabolome profiles alterations in fatty liver induced by high-fat diet in rat. Metabolism: clinical and experimental. 2010;59(4):554–560. doi: 10.1016/j.metabol.2009.08.022. [DOI] [PubMed] [Google Scholar]
- 45.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444(7117):337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kennedy AR, Pissios P, Otu H, Roberson R, Xue B, Asakura K, Furukawa N, Marino FE, Liu FF, Kahn BB, et al. A high-fat, ketogenic diet induces a unique metabolic state in mice. American journal of physiology Endocrinology and metabolism. 2007;292(6):E1724–E1739. doi: 10.1152/ajpendo.00717.2006. [DOI] [PubMed] [Google Scholar]
- 47.Bouchard L, Rabasa-Lhoret R, Faraj M, Lavoie ME, Mill J, Perusse L, Vohl MC. Differential epigenomic and transcriptomic responses in subcutaneous adipose tissue between low and high responders to caloric restriction. The American journal of clinical nutrition. 2010;91(2):309–320. doi: 10.3945/ajcn.2009.28085. [DOI] [PubMed] [Google Scholar]
- 48.Ghosh S, Dent R, Harper ME, Stuart J, McPherson R. Blood gene expression reveal pathway differences between diet-sensitive and resistant obese subjects prior to caloric restriction. Obesity. 2011;19(2):457–463. doi: 10.1038/oby.2010.209. [DOI] [PubMed] [Google Scholar]
- 49.Bouwman FG, de Roos B, Rubio-Aliaga I, Crosley LK, Duthie SJ, Mayer C, Horgan G, Polley AC, Heim C, Coort SL, et al. 2D-electrophoresis and multiplex immunoassay proteomic analysis of different body fluids and cellular components reveal known and novel markers for extended fasting. BMC medical genomics. 2011;4:24. doi: 10.1186/1755-8794-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rakhshandehroo M, Knoch B, Muller M, Kersten S. Peroxisome proliferator-activated receptor alpha target genes. PPAR research. 2010;2010 doi: 10.1155/2010/612089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perera RJ, Marcusson EG, Koo S, Kang X, Kim Y, White N, Dean NM. Identification of novel PPARgamma target genes in primary human adipocytes. Gene. 2006;369:90–99. doi: 10.1016/j.gene.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 52.Taleb S, Lacasa D, Bastard JP, Poitou C, Cancello R, Pelloux V, Viguerie N, Benis A, Zucker JD, Bouillot JL, et al. Cathepsin S, a novel biomarker of adiposity: relevance to atherogenesis. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2005;19(11):1540–1542. doi: 10.1096/fj.05-3673fje. [DOI] [PubMed] [Google Scholar]
- 53.Zhong H, Yang X, Kaplan LM, Molony C, Schadt EE. Integrating pathway analysis and genetics of gene expression for genome-wide association studies. American journal of human genetics. 2010;86(4):581–591. doi: 10.1016/j.ajhg.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fisher RM, Burke H, Nicaud V, Ehnholm C, Humphries SE. Effect of variation in the apo A-IV gene on body mass index and fasting and postprandial lipids in the European Atherosclerosis Research Study II. EARS Group. Journal of lipid research. 1999;40(2):287–294. [PubMed] [Google Scholar]
- 55.Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, Maouche S, Germain M, Lackner K, Rossmann H, et al. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PloS one. 2010;5(5):e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Middelberg RP, Ferreira MA, Henders AK, Heath AC, Madden PA, Montgomery GW, Martin NG, Whitfield JB. Genetic variants in LPL, OASL and TOMM40/APOE-C1-C2-C4 genes are associated with multiple cardiovascular-related traits. BMC medical genetics. 2011;12:123. doi: 10.1186/1471-2350-12-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun K, Battle MA, Misra RP, Duncan SA. Hepatocyte expression of serum response factor is essential for liver function, hepatocyte proliferation and survival, and postnatal body growth in mice. Hepatology. 2009;49(5):1645–1654. doi: 10.1002/hep.22834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kondo M, Ugi S, Morino K, Fuke T, Obata T, Yoshizaki T, Nishio Y, Maeda S, Kashiwagi A, Maegawa H. Postprandial activation of protein kinase Cmicro regulates the expression of adipocytokines via the transcription factor AP002D2beta. International journal of molecular medicine. 2011;28(1):95–100. doi: 10.3892/ijmm.2011.651. [DOI] [PubMed] [Google Scholar]
- 59.Pellis L, van Erk MJ, van Ommen B, Bakker GC, Hendriks HF, Cnubben NH, Kleemann R, van Someren EP, Bobeldijk I, Rubingh CM, et al. Plasma metabolomics and proteomics profiling after a postprandial challenge reveal subtle diet effects on human metabolic status. Metabolomics : Official journal of the Metabolomic Society. 2012;8(2):347–359. doi: 10.1007/s11306-011-0320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perez-Caballero AI, Alcala-Diaz JF, Perez-Martinez P, Garcia-Rios A, Delgado-Casado N, Marin C, Yubero-Serrano E, Camargo A, Caballero J, Malagon MM, et al. Lipid metabolism after an oral fat test meal is affected by age-associated features of metabolic syndrome, but not by age. Atherosclerosis. 2013;226(1):258–262. doi: 10.1016/j.atherosclerosis.2012.10.052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.