Abstract
Circadian rhythms occur in most of the living organisms, and with a 24 hour periodicity govern a number of physiological and metabolic functions. During the past few years, an important research effort has uncovered new trails that intersect between circadian rhythms and metabolic pathways. At a molecular level, the clock machinery is responsible for the establishment of a circadian epigenome, and this can be modulated by metabolic cues. Indeed, metabolic control by the circadian clock is manifest in the development of metabolic diseases when circadian rhythms are impaired. Thus, pharmacological modulation of circadian rhythms promises new avenues for the treatment of metabolic and sleep disorders.
Introduction
The 24-hour day-night cycles caused by the rotation of the Earth on its axis have accompanied and influenced the evolution of all life forms. Consequently, almost all organisms that we know so far, display some sort of adaptation to these daily light-dark cycles, which is manifest in a wide variety of biological cycles with 24 hour period [1]. These are known as circadian rhythms.
Chronobiologists have studied circadian rhythms for decades and have uncovered networks of interconnected biological processes that coordinate the cyclic nature of metabolism, physiology and behavior. Circadian rhythms are manifested as sleep-wake cycles, circadian hormone secretion, daily changes in body temperature and blood pressure, motor activity, feeding-fasting cycles or different levels of alertness around the clock. In mammals, the core of this network resides in the suprachiasmatic nucleus (SCN) of the hypothalamus in the brain. This tiny region is composed of around 20,000 “pacemaker” neurons whose activity oscillates in synchrony [2]. SCN neurons receive light information from retinal cells via the retinohypothalamic tract [3]. Therefore, light entrainment can adjust the phase of the SCN oscillator, ensuring a daily synchronization of the body’s circadian clock with geophysical time [4]. This central pacemaker dictates the phase of other oscillators that reside in the rest of the brain and in most of the peripheral tissues, via endocrine or neuronal connections, with light being the main zeitgeber (time giver) [4,5]. Strikingly, feeding-fasting cycles can profoundly affect peripheral oscillators in tissues such as liver, pancreas, kidney or heart. Thus, when the natural feeding-fasting cycles are shifted, i.e. by time restricted access to food, the phase of the clocks in these tissues is reset and uncoupled from the central pacemaker, and becomes entrained by food instead of light [6,7]. These evidences strongly suggest a key role for metabolites in the clock function.
A highly dynamic circadian gene transcription program
The molecular clock operates in most cells and is based on interlocked transcriptional-translational feedback loops, as revealed by genetics and molecular studies in Drosophila and mammals [8–11]. The mammalian core clock proteins CLOCK (Circadian Locomotor Output Cycles Kaput) and BMAL1 (Brain and Muscle ARNT-Like 1), are two basic helix-loop-helix (b-HLH)-PAS transcription activators that dictate the expression of many clock controlled genes (CCGs). These heterodimerize via their PAS domains and bind to E-boxes in the promoters of CCGs. Among the genes regulated by CLOCK:BMAL1 are Period1-3 (Per) and Cryptochrome1-2 (Cry) are transcribed. These encode the core clock proteins PERs and CRYs, which heterodimerize and associates with other partners in the nucleus and repress CLOCK-BMAL1-driven activation, thus a negative autoregulatory feedback loop is generated [10].
Some of the CCGs whose circadian expression is driven by CLOCK:BMAL1 are transcription factors, such as DBP (D-site Binding Protein), TEF (Thyrotroph Embryonic Factor), HLF (Hepatic Leukemia Factor) and E4BP4 (E4 Promoter–Binding Protein 4), RORα (Retinoic Acid-Related Orphan Receptor α), and REV-ERBα/β (Reverse Erithroblastosis Virus α and β). DBP, TEF, HLH and E4BP4 bind to D-boxes in the genome [12,13], whereas REV-ERBα/β and RORα bind to the Rev-Erb/ROR-binding element (RRE) [14,15], and subsequently drive cyclic expression of downstream genes.
Other interconnected transcriptional feedback loops provide additional plsticity to the circadian clock, and a very complex system of posttranslational modifications of clock proteins intertwine[16] to generate daily oscillations of a portion of the transcriptome, ranging from 3% to 30% depending on the tissue or cell line [17–20].
Drawing the circadian epigenome
The process of transcriptional regulation of clock gene expression requires the rhythmic assembly and recruitment to chromatin of multiprotein complexes in a circadian time dependent fashion. These events are accompanied by rhythmic changes in the epigenetic landscape that is in very much part drawn by chromatin modifying enzymes (Figure 1) [21,22]. The CLOCK:BMAL1 complex is recruited to chromatin in a circadian manner at the level of E-boxes. The protein CLOCK itself has histone acetyl-transferase (HAT) activity, with a preference for histone H3 K9 and K14 [23], possibly contributing to the opening of the chromatin fiber and promoting rhythmic transcription of CCGs [24]. Other HATs are rhythmically recruited by the CLOCK:BMAL1 complex; amongst them the CREB binding protein (CBP) and p300. While p300 appears to function as a coactivator that cooperates with CLOCK:BMAL1 by influencing the acetylation state of histones in a circadian fashion [25,26••], the role of CBP is not fully understood. CBP can transactivate circadian gene transcription in a similar manner as p300, but it also interacts with PER2, thus collaborating with the clock repressing loop [26••]. It would be interesting to determine whether these acetyltransferases can target non histone proteins of the clock machinery and influence their function. In this line, CLOCK was found to acetylate its own transcriptional partner BMAL1 at the single K537 residue, an essential event for circadian rhythmicity[27].
Figure 1. Chromatin remodeling in the circadian clock.
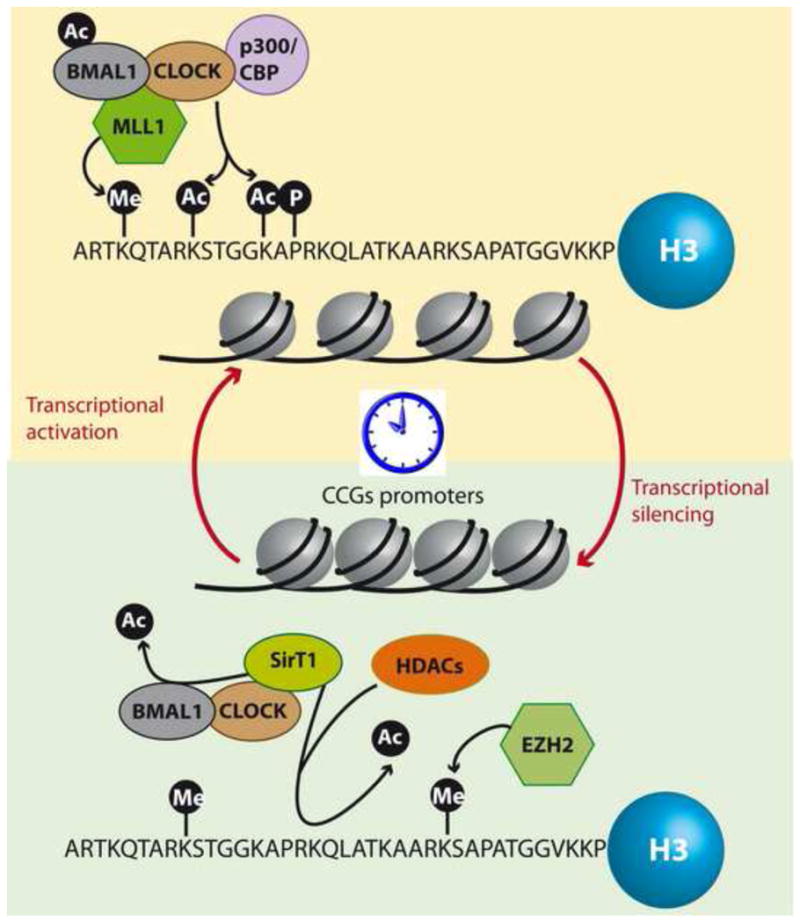
The epigenetic mechanisms that underlie clock controlled gene transcription are on the way to be uncovered. Chromatin modifying enzymes act in synchrony for the fine tuning necessary to achieve clock-controlled gene expression. Transcriptional activators coordinate rhythmic hyperacetylation and H3K4 trimethylation at circadian gene promoters that promote transcription. Conversely, repressors remove acetylation marks and promote a closed state of the chromatin fiber at the clock controlled gene promoters that inhibit transcription. Thus, activator and repressor enzymes act in a very precise synchrony that coordinates the circadian transcription of about 10% – 15% of all transcripts.
The methyltransferase mixed lineage leukemia 1 (MLL1) rhythmically interacts with CLOCK and specifically trimethylates histone H3K4 on circadian promoters, a mark that is associated with transcriptional activation [28••]. Conversely, EZH2 has been described to methylate H3K27 on circadian promoters, an event that has been associated to CRY dependent inhibition of transcription [29]. The histone demethylases Jarid1A and JMJD5 also participate in maintaining the circadian epigenome [30•, 31]. The elongation marks, H3K36me3 and H3K79me2, also exhibit circadian modulation although they present a much lower amplitude than for example H3K4me3 [26••]. Thus, other methyltransferases may have a role in the circadian system of gene expression.
Recent findings demonstrate a pivotal role for the histone deacetylase HDAC3 in maintaining the epigenetic landscape of circadian genes in the mouse liver [32,33••]. HDAC3 and its coactivator NCoR are rhythmically recruited to chromatin via the nuclear receptor Rev-Erbα, which is expressed in a circadian fashion [33••]. This event sustains circadian histone acetylation at genes regulating hepatic lipid metabolism, and its disruption significantly impairs normal liver metabolism [33••, 34]. These findings underscore the relevance of the clock directed epigenetic landscape in maintaining tissue homeostasis via coordinated regulation of a highly tuned program of gene expression.
Metabolic states influence the circadian epigenome
Chromatin plasticity relies on a variety of remodeling enzymes that use cellular metabolites as a source to elicit phosphorylation, acetylation, methylation etc. reactions, leading to specific histone post-translational modifications (PTMs, Figure 2) [35]. The dependence of many enzymes on the coenzyme NAD+ for their activities places this metabolite as a central connector between cellular metabolic state and the enzymatic reactions. There are two major groups of chromatin regulatory enzymes that depend on NAD+ for their function; the histone deacetylases (HDACs) class III, also known as sirtuins, and the poly-ADP ribose polymerases (PARPs). Of those, the sirtuin SIRT1 and PARP1 have been shown to modulate the circadian machinery [36–38], and their dependence on NAD+ places them as molecular links that communicate the metabolic state of the cell to the clock machinery (Figure 3).
Figure 2. Clock-regulated metabolite availability can be “sensed” by chromatin remodeling enzymes and effect circadian gene expression.
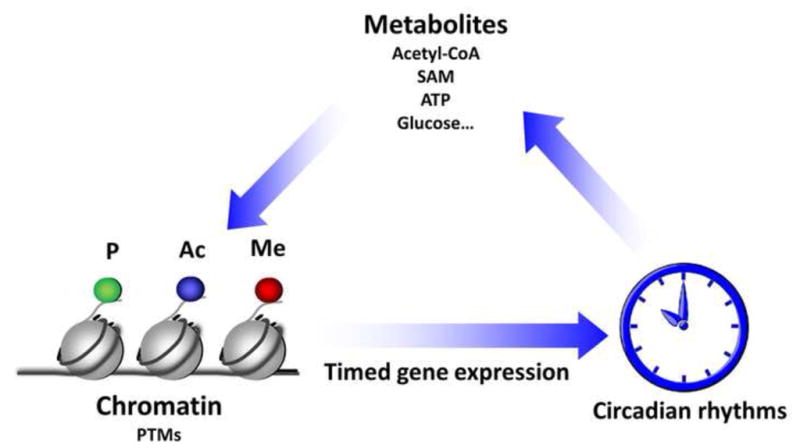
Clock-controlled program of gene expression dictates the circadian oscillation of a portion of the transcriptome. A number of these genes encode enzymes and proteins that exert control on metabolic pathways and metabolite availability. Some of these metabolites, such as NAD+, ATP, Acetyl-CoA or maybe glucose, could be used as cofactors by chromatin remodeling enzymes that modify histone tails leading to phosphorylation (P), acetylation (Ac) or methylation (Me). These epigenetic modifications are associated with changes in gene transcription and other chromatin functions. Thus, metabolite availability could affect the activity of chromatin modifiers and may constitute a regulatory mechanism for gene expression [35].
Figure 3. Clock-controlled NAD+ levels regulate enzymatic activity of SIRT1 and PARP1.
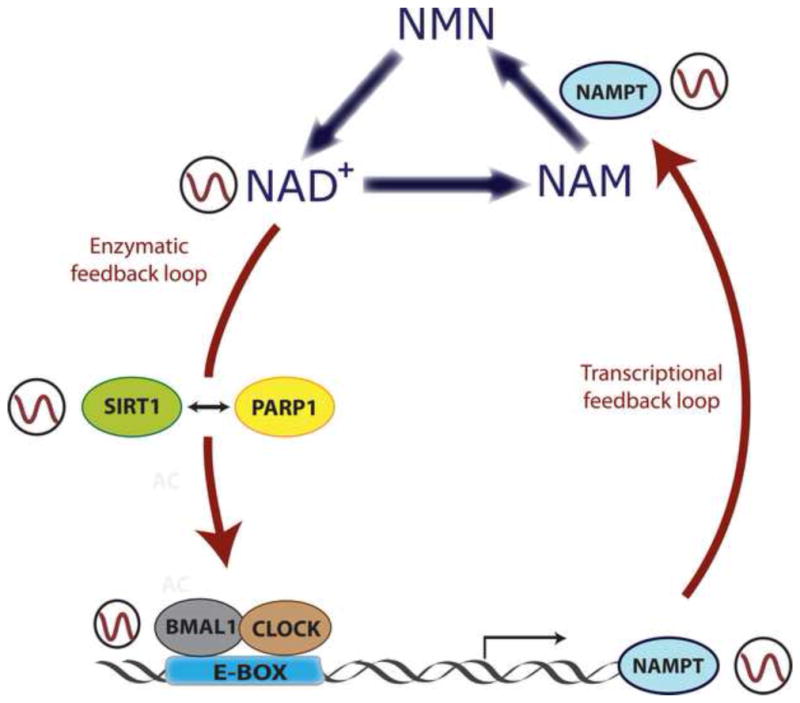
Intracellular levels of NAD+ oscillate in a circadian manner, and this oscillation is driven by the molecular clock. CLOCK:BMAL1 complexes rhythmically bind to E-boxes at the Nampt gene promoter, directing its circadian transcription. The resulting NAMPT protein is a key rate-limiting enzyme in the NAD+ salvage pathway, which dictates the circadian biosynthesis of NAD+. This metabolite feeds back into the clock by serving as a cofactor for SIRT1 and PARP1 enzymes, whose enzymatic activity is circadian. SIRT1 and PARP1 directly target CLOCK:BMAL1 complexes to change their transactivational activity. A molecular interplay between SIRT1 and PARP1 exists, and depends on NAD+ levels. By this mechanism, NAD+ intersects in between a transcriptional and an enzymatic feedback loop to regulate energy homeostasis.
The activity of PARP1 in the liver of the mice oscillates following the feeding-fasting cycles [37]. PARP1 interacts with CLOCK and BMAL1, and catalyzes the circadian ADP-rybosilation of CLOCK, reducing its binding to DNA and possibly altering circadian gene transcription. Moreover, Parp1 knockout mice present impaired ability to adapt to changes in the timing of food intake and longer circadian period, suggesting that PARP1 may feedback into the clock [37]. Interestingly, PARP1 has a role in regulating chromatin dynamics through modulation of the activity of the histone demethylase KDM5B [39]. An appealing avenue to consider is that PARP1 may affect circadian gene expression and metabolism through changes in chromatin modifications and transcription (Figure 3).
The deacetylase activity of SIRT1 has been shown to oscillate during the circadian cycle [38]. SIRT1 binds to the CLOCK:BMAL1 complex and is recruited to circadian genes, where it can directly deacetylate histones, thus counteracting the HAT activity of CLOCK [38]. Liver-specific genetic ablation of Sirt1 in the mouse or pharmacological treatment with SIRT1 inhibitors, such as nicotinamide or splitomicin, generates decreased amplitude in circadian gene expression [36,38]. In this scenario, it is remarkable the finding that the levels of NAD+ itself oscillate in a circadian fashion [40,41], an event that explains the circadian enzymatic activity of SIRT1. Moreover, NAD+ synthesis is directly regulated by the circadian clock machinery, via transcriptional control of the Nampt gene by CLOCK:BMAL1. The product of this gene is the enzyme nicotinamide phosphoribosyl-transferase (NAMPT) that catalyzes a key rate limiting step in the NAD+ salvage pathway. Indeed, pharmacologically or genetically inhibition of the NAMPT enzyme depletes the levels of NAD+ in the cells and impairs SIRT1 activity, that is translated into higher levels of acetylation of its targets H3K9/K14 at circadian gene promoters, as well as hyperacetylation of BMAL1, and subsequently disruption of circadian gene expression [38]. Interestingly, mice in which the endogenous levels of NAD+ are disrupted by a genetic mutation display disturbances in circadian behavior and metabolism[42].
A functional interplay between the nuclear proteins PARP1 and SIRT1 has been recently established [43•–45], where NAD+ availability links their activity ratios. Remarkably, PARP1 depletion enhances SIRT1 activity, but not SIRT2 or SIRT3 which are located in the cytoplasm and the mitochondria respectively [43•]. This finding supports the idea of an independent regulation of the metabolites availability in different subcellular locations. Further investigations are necessary to determine to which extent the clock controls the availability of cell metabolites in different subcellular compartments [35].
Oscillations in the redox state can sustain circadian rhythms
At the cellular level metabolic state can be manifest as redox state, described by the homeostasis of reactive free radicals, such as NAD+ and FAD, from reduction-oxidation reactions in metabolism [46]. The redox state is reflected in the balance of several metabolites, for example lactate and pyruvate, or beta-hydroxybutyrate and acetoacetate, whose interconversion depends on these ratios. An abnormal redox state can induce a variety of deleterious situations, such as hypoxia, shock, and sepsis. It is known that the circadian components CLOCK and NPAS2 can directly sense the redox state of the cell, thus modulating their DNA-binding activities to E-boxes [47]. Strikingly, recent advances underscore the importance of the redox state in maintaining circadian oscillatory rhythms in a transcription independent manner. For example, studies carried out in mammalian red blood cells, which lack nucleus and thus do not perform transcription, show that they do present circadian rhythms in their redox state [48••]. These are accompanied by a circadian redox rhythm in the proteins peroxiredoxins, a family of antioxidant proteins that control peroxide levels in the cell [48••]. Remarkably these proteins are highly conserved, and their circadian redox rhythm exists from prokaryotes to mammals [49]. However, redox rhythms on peroxiredoxins are also under the control of the molecular clock in mammalian nucleated cells, indicating that clock-controlled transcriptional loops intersect with the metabolic processes. To which extent the clock determines the redox state in mammalian cells is still not fully understood.
Circadian oscillation in the redox state is self-sustained in the mouse SCN, and this is required for the molecular clock function [50••]. Moreover, the redox oscillation affects the excitability of the SCN neurons via a non-transcriptional mechanism that involves the modulation of potassium channels [50••]. However, these oscillatory events depend on a functional molecular clock, since they are impaired on Bmal1−/− mice. Importantly, recent studies in Drosophila support the idea that changes in the electrical activity of the pacemaker neurons in the brain can indeed affect the circadian program of gene expression [51]. Taken together, these findings provide new insights into how energy metabolism can exert control over circadian physiology. Further experiments are necessary to how circadian transcriptional networks intersect with the cellular metabolic states.
Transcriptional networks and circadian metabolism. A key to understand diseases?
Accumulating evidence illustrates the circadian transcriptome for a number of cell lines and tissues, building up a powerful set of data that demonstrates the rhythmic transcription of a variety of metabolic genes in liver, adipose tissue, muscle, heart, pituitary gland or brain. A striking observation that arises from these studies is that many key rate-limiting enzymes of metabolic pathways are under circadian control [18,52]. This observation places the clock machinery in a strategic position within the regulation of metabolic processes. Moreover, disruption of the clock leads to a wide variety of metabolic disorders, including obesity and diabetes, both in mouse and human [53•–56]. Conversely, the deleterious effects over metabolic homeostasis of high fat diet-induced obesity in mice can be substantially improved by keeping a restricted time of feeding schedule, in which food is available only during a particular time of the day [57•,58]. Consequently, it is important to decipher the circuits between circadian and metabolic processes to further expand our understanding into the development of many metabolic diseases [59].
Recent studies in our laboratory reveal the interconnected network of the circadian transcriptome with the metabolome in the mouse liver [60••]. The diurnal oscillation of metabolic genes directly impinges on metabolite production, giving rise to a diurnal metabolome that is essential for liver function and general homeostasis. One example, relates to the circadian rhythmicity of the urea cycle, which eliminates ammonia to urea. The importance of this circadian regulation is apparent since failure to faithfully coordinate excretion of amino acids into urea can lead to severe pathophysiological consequences [61]. Importantly, this study presents the integration of the liver metabolome and transcriptome with other existing databases in a computational platform named “CircadiOmics” (http://circadiomics.igb.uci.edu/). This provides a system-wide picture and allows researchers to analyze circadian metabolism with a broader perspective [62].
A human circadian metabolome has also been described in blood plasma and saliva [63,64 •], revealing that a number of metabolites are under circadian control. Interestingly, these studies open the door for using the metabolic molecular timetables as a diagnostic tool [64•]. By using the levels of particular metabolites as molecular markers, the body time can be determined. Thus this study provides a powerful tool for detection of circadian rhythm disorders and for the development of personalized chronotherapy.
Finding new therapeutic targets that can modulate circadian physiology is highly relevant for the treatment of metabolic disorders. This has been proved evident by recent advances in the drug discovery field. Targeting REV-ERB with a synthetic agonist can alter the time of day in mice [65••]. Moreover, this compound can improve lipid homeostasis in diet-induced obese mice [65••]. Indeed, stabilization of CRY protein by interaction with a novel molecule named KL001 can modulate glucose homeostasis [66••]. Taken together, these results indicate that therapies based on molecules that target clock components can be very useful in the treatment of sleep disorders as well as metabolic diseases.
Conclusions
The epigenetic mechanisms that underlie clock-controlled gene expression are on the way of being elucidated. The availability of high throughput techniques has allowed researchers to uncover the circadian epigenome in a wide variety of systems and conditions, improving our understanding in the molecular clock function. Indeed, these many outcomes have highlighted the importance of metabolic homeostasis in the maintenance of the circadian epigenome. We are in turn beginning to understand the molecular links between circadian physiology and metabolic pathways, and it can be anticipated that more exciting discoveries will follow soon. The discovery of NAD+ as a master metabolite that connects cellular metabolism to the circadian epigenome underscores the relevance of deciphering how metabolite levels can be sensed by the clock machinery and sets the basis for the search of other metabolites that can present similar functions. In this line, further investigations are necessary to elucidate to which extent NAD+ levels direct circadian gene expression and vice-versa. In turn, as we begin to understand the molecular links between circadian physiology and metabolic pathways, the relevance of this crosstalk for maintaining metabolic homeostasis and preventing metabolic disorders is apparent. Systems biology approaches are becoming essential for our understanding of these complex networks and will provide with powerful tools for novel pharmacological strategies towards the treatment of metabolic pathologies.
Highlights.
A clock driven dynamic epigenome contributes to rhythmicity of the transcriptome
The molecular clock can sense metabolic states and modulate its function accordingly
Metabolic and non-transcriptional mechanisms can sustain circadian rhythms in mammals
Circadian rhythms affect metabolic homeostasis and its disturbance causes disease
Acknowledgments
The authors thank members of the Sassone-Corsi laboratory for discussions and comments. Work in the lab of PS-C is supported by grants from the National Institute of Health (NIH), the Institut National de la Santé et de la Recherche Médicale (INSERM), the Institut Merieux and Sirtris Pharmaceuticals, Inc. LA-A is a recipient of an EMBO long-term Postdoctoral Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111(7):919–922. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- 2.Ralph MR, Foster RG, Davis FC, Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science. 1990;247(4945):975–978. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- 3.Freedman MS, Lucas RJ, Soni B, von Schantz M, Munoz M, David-Gray Z, Foster R. Regulation of mammalian circadian behavior by non-rod, non-cone, ocular photoreceptors. Science. 1999;284(5413):502–504. doi: 10.1126/science.284.5413.502. [DOI] [PubMed] [Google Scholar]
- 4.Cermakian N, Sassone-Corsi P. Environmental stimulus perception and control of circadian clocks. Curr Opin Neurobiol. 2002;12(4):359–365. doi: 10.1016/s0959-4388(02)00347-1. [DOI] [PubMed] [Google Scholar]
- 5.Quintero JE, Kuhlman SJ, McMahon DG. The biological clock nucleus: A multiphasic oscillator network regulated by light. J Neurosci. 2003;23(22):8070–8076. doi: 10.1523/JNEUROSCI.23-22-08070.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellet MM, Sassone-Corsi P. Mammalian circadian clock and metabolism - the epigenetic link. J Cell Sci. 2010;123(Pt 22):3837–3848. doi: 10.1242/jcs.051649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14(23):2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cermakian N, Sassone-Corsi P. Multilevel regulation of the circadian clock. Nat Rev Mol Cell Biol. 2000;1(1):59–67. doi: 10.1038/35036078. [DOI] [PubMed] [Google Scholar]
- 9.King DP, Takahashi JS. Molecular genetics of circadian rhythms in mammals. Annu Rev Neurosci. 2000;23:713–742. doi: 10.1146/annurev.neuro.23.1.713. [DOI] [PubMed] [Google Scholar]
- 10.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418(6901):935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 11.Young MW, Kay SA. Time zones: A comparative genetics of circadian clocks. Nat Rev Genet. 2001;2(9):702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 12.Wuarin J, Schibler U. Expression of the liver-enriched transcriptional activator protein DBP follows a stringent circadian rhythm. Cell. 1990;63(6):1257–1266. doi: 10.1016/0092-8674(90)90421-a. [DOI] [PubMed] [Google Scholar]
- 13.Gachon F, Olela FF, Schaad O, Descombes P, Schibler U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006;4(1):25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 14.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor Rev-Erb alpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110(2):251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 15.Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals rora as a component of the mammalian circadian clock. Neuron. 2004;43(4):527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 16.Mehra A, Baker CL, Loros JJ, Dunlap JC. Post-translational modifications in circadian rhythms. Trends Biochem Sci. 2009;34(10):483–490. doi: 10.1016/j.tibs.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, Dunlap JC. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol. 2002;12(7):551–557. doi: 10.1016/s0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- 18.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109(3):307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 19.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417(6884):78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 20.Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, Iino M, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418(6897):534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 21.Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. Decoding the epigenetic language of neuronal plasticity. Neuron. 2008;60(6):961–974. doi: 10.1016/j.neuron.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masri S, Sassone-Corsi P. Plasticity and specificity of the circadian epigenome. Nat Neurosci. 2010;13(11):1324–1329. doi: 10.1038/nn.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator clock is a histone acetyltransferase. Cell. 2006;125(3):497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 24.Nakahata Y, Grimaldi B, Sahar S, Hirayama J, Sassone-Corsi P. Signaling to the circadian clock: Plasticity by chromatin remodeling. Curr Opin Cell Biol. 2007;19(2):230–237. doi: 10.1016/j.ceb.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 25.Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421(6919):177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 26••.Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012 doi: 10.1126/science.1226339. Using a combination of ChIP-seq and transcriptome analysis techniques, the authors describe the dynamics of the recruitment to chromatin of many of the clock-related transcriptional factors during the circadian cycle in mouse liver. Several histone postranslational modifications are also analyzed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y, Sassone-Corsi P. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450(7172):1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- 28••.Katada S, Sassone-Corsi P. The histone methyltransferase mll1 permits the oscillation of circadian gene expression. Nat Struct Mol Biol. 2010;17(12):1414–1421. doi: 10.1038/nsmb.1961. This study implicates for the first time a H3K4 methyltransferase in the transcriptional mechanism of circadian genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Etchegaray JP, Yang X, DeBruyne JP, Peters AH, Weaver DR, Jenuwein T, Reppert SM. The polycomb group protein EZH2 is required for mammalian circadian clock function. J Biol Chem. 2006;281(30):21209–21215. doi: 10.1074/jbc.M603722200. [DOI] [PubMed] [Google Scholar]
- 30•.DiTacchio L, Le HD, Vollmers C, Hatori M, Witcher M, Secombe J, Panda S. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science. 2011;333(6051):1881–1885. doi: 10.1126/science.1206022. This study describes for the first time the role of a H3K4 demethylase in clock-controlled gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones MA, Covington MF, DiTacchio L, Vollmers C, Panda S, Harmer SL. Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc Natl Acad Sci U S A. 2010;107(50):21623–21628. doi: 10.1073/pnas.1014204108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alenghat T, Meyers K, Mullican SE, Leitner K, Adeniji-Adele A, Avila J, Bucan M, Ahima RS, Kaestner KH, Lazar MA. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456(7224):997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33••.Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331(6022):1315–1319. doi: 10.1126/science.1198125. This article describes the genomic recruitment of HDAC3 by Rev-Erbα, which directs a circadian rhythm of histone acetylation and gene expression in mouse liver. The authors demonstrate that this event is required for normal hepatic lipid homeostasis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, Liddle C, et al. Regulation of circadian behaviour and metabolism by Rev-Erb-alpha and RevErb-beta. Nature. 2012;485(7396):123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katada S, Imhof A, Sassone-Corsi P. Connecting threads: Epigenetics and metabolism. Cell. 2012;148(1–2):24–28. doi: 10.1016/j.cell.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 36.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134(2):317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 37.Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, Hottiger MO, Schibler U. Poly(adp-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell. 2010;142(6):943–953. doi: 10.1016/j.cell.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 38.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates clock-mediated chromatin remodeling and circadian control. Cell. 2008;134(2):329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krishnakumar R, Kraus WL. PARP-1 regulates chromatin structure and transcription through a KDM5b-dependent pathway. Mol Cell. 2010;39(5):736–749. doi: 10.1016/j.molcel.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by clock-SIRT1. Science. 2009;324(5927):654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, Takahashi JS, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324(5927):651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahar S, Nin V, Barbosa MT, Chini EN, Sassone-Corsi P. Altered behavioral and metabolic circadian rhythms in mice with disrupted nad+ oscillation. Aging (Albany NY) 2011;3(8):794–802. doi: 10.18632/aging.100368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43•.Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, Schoonjans K, et al. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 2011;13(4):461–468. doi: 10.1016/j.cmet.2011.03.004. The authors demonstrate a connection between PARP-1 and SIRT1 activities via NAD+ availability. They show that in the absence of PARP-1, the enzyme SIRT1 becomes more active, leading to significant effects over metabolism and physiology of mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pang J, Gong H, Xi C, Fan W, Dai Y, Zhang TM. Poly(adp-ribose) polymerase 1 is involved in glucose toxicity through SIRT1 modulation in Hepg2 hepatocytes. J Cell Biochem. 2011;112(1):299–306. doi: 10.1002/jcb.22919. [DOI] [PubMed] [Google Scholar]
- 45.Kolthur-Seetharam U, Dantzer F, McBurney MW, de Murcia G, Sassone-Corsi P. Control of AIF-mediated cell death by the functional interplay of SIRT1 and PARP-1 in response to DNA damage. Cell Cycle. 2006;5(8):873–877. doi: 10.4161/cc.5.8.2690. [DOI] [PubMed] [Google Scholar]
- 46.Droge W. Free radicals in the physiological control of cell function. Physiol Rev. 2002;82(1):47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 47.Rutter J, Reick M, Wu LC, McKnight SL. Regulation of CLOCK and NPAS2 DNA binding by the redox state of NAD+ cofactors. Science. 2001;293(5529):510–514. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- 48••.O’Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469(7331):498–503. doi: 10.1038/nature09702. Using anucleated human red blood cells, the authors demonstrate the existence of circadian rhythms in the redox state of the proteins peroxiredoxins. These findings indicate that non-transcriptional mechanisms can sustain circadian rhythms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, Maywood ES, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485(7399):459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50••.Wang TA, Yu YV, Govindaiah G, Ye X, Artinian L, Coleman TP, Sweedler JV, Cox CL, Gillette MU. Circadian rhythm of redox state regulates excitability in suprachiasmatic nucleus neurons. Science. 2012;337(6096):839–842. doi: 10.1126/science.1222826. This study reveals that circadian cycles in the cellular redox state drive cycles of excitability in mammalian SCN in a non-translational mechanisms that involves oscillations in potassium currents. The cycles in the redox state of SCN neurons depends on the molecular clock. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mizrak D, Ruben M, Myers GN, Rhrissorrakrai K, Gunsalus KC, Blau J. Electrical activity can impose time of day on the circadian transcriptome of pacemaker neurons. Curr Biol. 2012 doi: 10.1016/j.cub.2012.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sahar S, Sassone-Corsi P. Regulation of metabolism: The circadian clock dictates the time. Trends Endocrinol Metab. 2012;23(1):1–8. doi: 10.1016/j.tem.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53•.Buxton OM, Cain SW, O’Connor SP, Porter JH, Duffy JF, Wang W, Czeisler CA, Shea SA. Adverse metabolic consequences in humans of prolonged sleep restriction combined with circadian disruption. Sci Transl Med. 2012;4(129):129ra143. doi: 10.1126/scitranslmed.3003200. In this clinical study performed with healthy adults, the participants are subjected to an experimental paradigm that disrupts their circadian rhythms. The authors show that the participants become primed for developing metabolic diseases such as obesity and diabetes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, Lopez JP, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466(7306):627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scott EM, Carter AM, Grant PJ. Association between polymorphisms in the clock gene, obesity and the metabolic syndrome in man. Int J Obes (Lond) 2008;32(4):658–662. doi: 10.1038/sj.ijo.0803778. [DOI] [PubMed] [Google Scholar]
- 56.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, Eckel RH, et al. Obesity and metabolic syndrome in circadian clock mutant mice. Science. 2005;308(5724):1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57•.Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JA, Ellisman MH, et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15(6):848–860. doi: 10.1016/j.cmet.2012.04.019. In this study, the authors show that high fat diet fed mice become healthier and protected against metabolic disorders such as obesity and hyperinsulinemia when they are fed in a time restricted manner. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sherman H, Genzer Y, Cohen R, Chapnik N, Madar Z, Froy O. Timed high-fat diet resets circadian metabolism and prevents obesity. Faseb J. 2012;26(8):3493–3502. doi: 10.1096/fj.12-208868. [DOI] [PubMed] [Google Scholar]
- 59.Eckel-Mahan K, Sassone-Corsi P. Metabolism control by the circadian clock and vice versa. Nat Struct Mol Biol. 2009;16(5):462–467. doi: 10.1038/nsmb.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60••.Eckel-Mahan KL, Patel VR, Mohney RP, Vignola KS, Baldi P, Sassone-Corsi P. Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci U S A. 2012;109(14):5541–5546. doi: 10.1073/pnas.1118726109. With a systems biology approach, the authors describe in this study the intersections between the circadian transcriptome and metabolic pathways in the mouse liver. A comprehensive circadian metabolome is also presented. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Braissant O. Current concepts in the pathogenesis of urea cycle disorders. Mol Genet Metab. 2010;100(Suppl 1):S3–S12. doi: 10.1016/j.ymgme.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 62.Patel VR, Eckel-Mahan K, Sassone-Corsi P, Baldi P. Circadiomics: Integrating circadian genomics, transcriptomics, proteomics and metabolomics. Nat Methods. 2012;9(8):772–773. doi: 10.1038/nmeth.2111. [DOI] [PubMed] [Google Scholar]
- 63.Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. The human circadian metabolome. Proc Natl Acad Sci U S A. 2012;109(7):2625–2629. doi: 10.1073/pnas.1114410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64•.Kasukawa T, Sugimoto M, Hida A, Minami Y, Mori M, Honma S, Honma K, Mishima K, Soga T, Ueda HR. Human blood metabolite timetable indicates internal body time. Proc Natl Acad Sci U S A. 2012;109(37):15036–15041. doi: 10.1073/pnas.1207768109. Using human blood samples, the authors elaborate a reference timetable of circadian oscillating metabolites. This resource is intended to determine body time in patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65••.Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, Lundasen T, Shin Y, Liu J, Cameron MD, Noel R, Yoo SH, et al. Regulation of circadian behaviour and metabolism by synthetic Rev-Erb agonists. Nature. 2012;485(7396):62–68. doi: 10.1038/nature11030. In this study the authors identify for the first time a synthetic Rev-Erb agonists with in vivo activity. Several experiments in mice reveal that these molecules have a potential benefit for the treatment of sleep and metabolic disorders. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66••.Hirota T, Lee JW, St John PC, Sawa M, Iwaisako K, Noguchi T, Pongsawakul PY, Sonntag T, Welsh DK, Brenner DA, Doyle FJ, 3rd , et al. Identification of small molecule activators of cryptochrome. Science. 2012;337(6098):1094. doi: 10.1126/science.1223710. Using a screening from a large library of small molecules, the authors identify a compound that can modulate circadian rhythms in human cells by stabilizing CRY protein. This molecule is shown to inhibit glucagon-induced gluconeogenesis in primary hepatocites. [DOI] [PMC free article] [PubMed] [Google Scholar]


