Abstract
The circadian clock governs biological timekeeping on a systemic level, helping to regulate and maintain physiological processes, including endocrine and metabolic pathways with a periodicity of 24-hours. Disruption within the circadian clock machinery has been linked to numerous pathological conditions, including cancer, suggesting that clock-dependent regulation of the cell cycle is an essential control mechanism. This review will highlight recent advances on the ‘gating’ controls of the circadian clock at various checkpoints of the cell cycle and also how the cell cycle can influence biological rhythms. The reciprocal influence that the circadian clock and cell cycle exert on each other suggests that these intertwined biological circuits are essential and multiple regulatory/control steps have been instated to ensure proper timekeeping.
The circadian clock is a remarkable timekeeping system that regulates numerous biological processes with a high degree of specificity, to ensure proper 24-hour rhythms. Light is the major zeitgeber, or time giver, that provides synchrony to the organism. The central circadian clock resides in the brain, within the suprachiasmatic nucleus (SCN), which is able to transmit signals to the peripheral clocks, such as liver, muscle, skin, that maintain their rhythms in a tissue-specific manner1, 2. The clock system is remarkably plastic and adapt to external cues, such as temperature and food intake, which are able to reset the clock system and maintain homeostasis of the organism.
The molecular circadian clock is driven by the transcription factors, circadian locomotor output cycles kaput (CLOCK) and aryl hydrocarbon receptor nuclear translocator-like (ARNTL or BMAL1), which are responsible for directing the positive arm of transcription, and are subsequently antagonized by the circadian transcriptional repressors period (PER) and cryptochrome (CRY) family members3. In addition to transcriptional control, the circadian clock is regulated at the level of translational and post-translational modifications4, 5 that are able to fine tune the circadian system. Similar to the circadian cycle, the cell cycle is a tightly regulated system with checkpoints that exist at key transitions in the cell cycle to regulate fidelity of DNA replication and mitosis. How do these cycles converge and regulate the other? Are there cues in place, such as metabolic signals, that can direct the regulatory checkpoints? Given, the excellent reviews on cell cycle and the circadian clock6-8, this review will focus on new and unexplored areas of circadian and cell cycle coordinated control and how these pathways can be influenced by external cues.
Gating controls of the circadian clock and the cell cycle
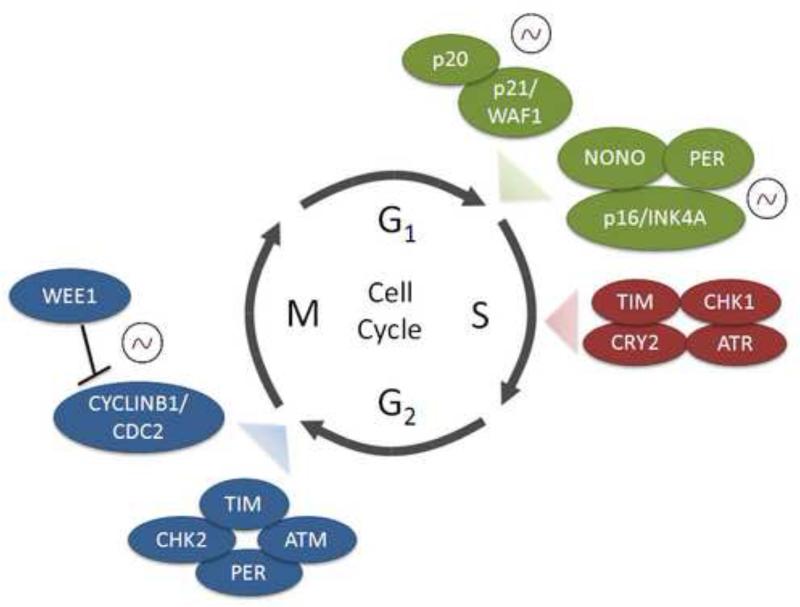
It has been previously shown in plants9, cyanobacteria10, 11, zebrafish12-14, and mouse15 that the circadian clock specifically regulates or ‘gates’ key phases of the cell cycle. Gating of the cell cycle by the circadian clock has been reported at the entry to S-phase12, 16, though the defined mechanism(s) of this phenomenon are unclear. Circadian clock-dependent regulation of S-phase is important for vulnerability to UV-induced DNA damage in proliferating skin cells17. Also, it has been reported that time-of-day dependent susceptibility to UV radiation that has been linked to skin cancer in mice and is dependent on DNA damage repair through the xeroderma pigmentosum group A (XPA) protein18. Also, recent data has revealed that p20 and p21, important regulators of the G1/S transition, are clock-controlled genes that are temporally expressed in a tissue-specific manner19, 20 (Figure 1).
Figure 1. Gating of the cell cycle by the circadian clock.
The circadian clock machinery regulates the cell cycle at G1/S, S-phase and G2/M checkpoints of the cell cycle. Expression profiles of certain genes have been reported to oscillate in a circadian manner, and are indicated (~). Clock and cell cycle complexes that regulate the G1/S checkpoint are indicated in green, S-phase proteins shown in red and G2/M checkpoint regulatory complexes are presented in blue. Other factors such as Cyclin D1 that controls the G1/S checkpoint and c-Myc that regulates proliferation via the G0/G1 transition are also important clock targets not shown.
Similarly, the circadian clock has been reported to specifically gate the G2/M checkpoint in proliferating hair follicles21 and a proliferative liver regeneration mouse model, whereby impaired liver regeneration was observed in arrhythmic Cry1/Cry2-deficient mice15. Furthermore, WEE1 kinase is responsible for regulating the CYCLIN B1/CDC2 (cell division control protein 2 also known as cyclin-dependent kinase 1, CDK1) complex and subsequent entry into mitosis15. Wee1 mRNA and protein expression was found to be highly circadian, along with its kinase activity that regulates mitotic entry15. The Wee1 promoter contains three E-box sequences15, the sites to which CLOCK and BMAL1 bind. Of particular interest, the Wee1 promoter is subject to chromatin remodeling and hyperacetylation at the histone H3 tail (H3K14) in response to light13. Not only are cell cycle genes such as Wee1 (and others such as c-Myc, Cyclin D1, Cyclin B1 and Cdc2) subject to circadian oscillation in expression, but the extent to which chromatin modifications play a role in light-dependent remodeling at cell cycle gene promoters is unclear. The CLOCK complex is known to have intrinsic histone acetyltransferase (HAT) activity, which contributes to BMAL1 and H3 tail acetylation22, 23. Similarly, the histone methyltransferase mixed-lineage leukemia (MLL1) is responsible for H3K4 tri-methylation, an active mark for gene expression which permits circadian oscillation24. What is the role that MLL1 plays in chromatin remodeling and subsequent recruitment of the CLOCK/BMAL1 transcriptional complex to clock-dependent cell cycle gene promoters? It would be expected that dynamic and circadian changes in both H3 methylation and acetylation establish a permissive chromatin state for transcription of cell cycle genes.
The control that the circadian clock exerts on the cell cycle seems to be extensive. Yet, little is known about cell cycle control over the circadian clock. Nagoshi et al. reported that oscillations in gene expression persisted during the cell cycle and that daughter cells maintained the circadian rhythm of the maternal cell after mitosis was complete25. Also, it was shown that the circadian period length of proliferating NIH3T3 fibroblasts is longer than the period of non-proliferating fibroblasts, suggesting the cell cycle can influence the period of the circadian clock25. The authors suggest that cell division could alter PER/CRY repressor protein concentrations and subsequently alter circadian rhythms, though the precise mechanisms are not understood. Recent data shows that an RNA binding protein called p54nrb or NONO partnered with PER proteins to activate the p16-Ink4A cell cycle checkpoint gene in a circadian manner26. Furthermore, it was noted that WT fibroblasts demonstrated circadian variation in the proportion of cells in S-phase. A greater than 2-fold variation in the number of cells that had entered S-phase was observed between circadian time (CT) 7 and CT 1926. Remarkably, this diurnal effect was abolished in NONO-null fibroblasts and Per1/Per2 mutant cells, which divided equally during the day and night, indicating a strong relationship between these proteins and circadian control of cell cycle gating. What remains to be determined is the extent to which this coordination occurs; are there cell cycle proteins that could work in concert with the circadian clock machinery, and vice-versa?
Common circadian and cell cycle proteins
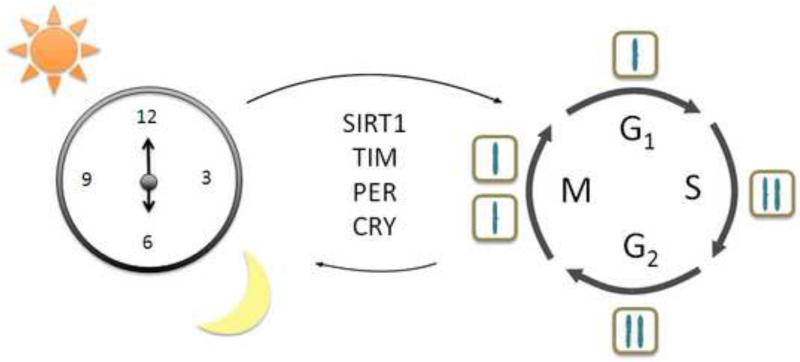
The idea that proteins can be conserved among different pathways is not new and most likely is taking place between the circadian clock molecular machinery and that of the cell cycle. The NONO/PER coordination is one such example. What about other factors that have been reported to participate in both biological circuits? The mammalian Timeless (TIM) protein shares homology with the reported drosophila clock protein TIM27, but is most closely related to cell cycle regulators and involved in DNA damage response. Mammalian TIM was found to interact with the circadian protein CRY2, checkpoint kinase 1 (CHK1) and ataxia telangiectasia and rad3 related (ATR), resulting in regulation of S phase and DNA damage response28. Furthermore, mammalian TIM was found to be important for ataxia telangiectasia mutated (ATM)-dependent activation of checkpoint kinase 2 (CHK2) and subsequent regulation of the G2/M checkpoint in response to DNA damage29, and CHK2 and ATM were also reported to interact with PER130. As the role of TIM in the mammalian circadian clock machinery appears pleiotropic31, 32, is it possible that this protein has evolved from the circadian circuit to regulate the cell cycle? Potentially, this could explain why TIM retains interaction with the clock machinery via PER and CRY and coordinates DNA damage response (Figure 2).
Figure 2. Common proteins that function in both circadian and cell cycles.
The circadian clock is regulated by the light/dark cycle that helps maintain periodicity of the system to approximately 24 hours. The cell cycle, comprised of G1, S, G2 and M phases, whereby the cell makes the decision to replicate its DNA and subsequently divide, is also under strict timing cues. Many proteins seem to have overlapping functions in both biological circuits. Shown are the mammalian Timeless protein (TIM), the mammalian Sirtuin (SIRT1), and Period (PER) and Cryptochrome (CRY) regulatory proteins that are involved in modulation of multiple points of both circadian and cell cycles.
The histone deacetylase (HDAC) SIRT1 is an NAD+-dependent mammalian sirtuin that deacetylates PER2, BMAL1 and the H3 tail and is important for maintaining circadian gene expression33, 34. Remarkably, SIRT1 was reported to be phosphorylated by CYCLINB1/CDK1 (CDC2) which modulates its HDAC activity and subsequent progression through the cell cycle, yet the targets of SIRT1 during mitosis remain to be determined35. Also, SIRT1 deacetylates p5336, 37 which is involved in DNA damage response and apoptosis, and forkhead box O3 (FOXO3)38 which is involved in G1/S and G2/M checkpoints, DNA damage response and apoptosis. The function of SIRT1 as an HDAC is extensive, targeting both circadian and cell cycle proteins. Yet, a crosstalk may exist in that SIRT1 can target the cell cycle machinery, which can feedback and phosphorylate SIRT1. There is also an additional layer of complexity involving NAD+, which will be discussed in the next section. Overall, there seems to be common regulators between the circadian and cell cycles and bidirectional regulation is in place involving both biological circuits, potentially to attain proper cellular homeostasis depending on stress response or metabolic state.
Influence of metabolism on the clock/cell cycle
It has been suggested that the circadian gating mechanism of the cell cycle may not be restricted to specific checkpoints, but could be an overarching mechanism with multiple points of control39. This brings into question the metabolic state of the cell and how this can influence the decision of a cell to replicate its DNA and divide. This concept has been highlighted in yeast where an ultradian rhythm has been reported for the cellular metabolic state which cycles between a reductive and non-respiratory state to an oxidative/respiratory phase40. Microarray analysis revealed that expression of genes with similar functions was compartmentalized into specific phases of the yeast metabolic cycle. Genes involved in DNA replication and cell cycle are coordinately expressed and the cell cycle occurred specifically during the reductive phase of the yeast metabolic cycle40. More recently, it was also proposed that circadian gating of the cell cycle may follow complex pathways in mammalian cells. Cry1/2-deficient fibroblasts display differences in their rates of proliferation as compared to WT cells, but the authors propose that this is due to global cellular changes, as many alterations in gene expression were observed in BMAL1-dependent targets41. The authors also highlight differences in proliferation rates in Cry1/2-deficient cultured fibroblasts41 versus previously reported Cry1/2-deficient mice15 and therefore suggest that systemic cues likely play a larger and unappreciated role in global circadian control of the cell cycle. Similarly, Sancar and colleagues reported that DNA damage response maybe very different in cells grown in culture, unlike an intact organism that relies on various inputs to regulate homeostasis42.
Based on these ideas that systemic cues likely play an important role in understanding circadian control of the cell cycle, it is imperative to extend our understanding of cellular metabolites that are under circadian control. It has been shown that nicotinamide adenine dinucleotide (NAD+) oscillates over the circadian cycle and its levels are directly controlled by the clock machinery43, 44. As previously discussed, SIRT1 is NAD+-dependent and plays an important role in regulating the clock machinery and is itself regulated by cell cycle factors. Given that the metabolic state of the cell can influence the cell cycle and that many metabolites are under control of the circadian clock, this suggests that the level of gating or control by the circadian clock could be quite extensive. The question is to what extent do these metabolites influence cell cycle commitment and progression? For example, a key metabolite, ATP, oscillates in a circadian manner45. Kinases require ATP and the G2/M checkpoint that is controlled by WEE1 could be tightly regulated by circadian oscillations in ATP levels. While experimental evidence is lacking, it could be speculated that cyclic levels of ATP may influence the many cyclin-dependent kinases that rely on ATP to phosphorylate and regulate their targets throughout the cell cycle. The extent to which the circadian clock regulates the cellular metabolome is becoming evident46-48, therefore it will be intriguing to understand how metabolic state fully influences the cell cycle.
The circadian clock and cell cycle are two very different biological circuits with many layers of coordinated regulation. Timing for both the circadian and cell cycles is critical and this maybe the reason for such elaborate levels of control. The circadian clock controls multiple aspects of the cell cycle, conferring gating mechanisms at key checkpoints of the cell cycle to ensure fidelity in DNA replication and cell division. How this is accomplished is not fully understood. Yet, it is clear that the circadian clock controls oscillatory expression of several cell cycle genes. The role of epigenetic control by the circadian clock is relatively unexplored, but most likely plays a critical role in dynamic changes in cell cycle gene expression. Also, common factors, such as the HDAC SIRT1, are involved in both circadian clock and cell cycle regulation. An additional layer of complexity exists with metabolites that could influence both circadian and cell cycles and the extent to which metabolic state is critical in gating mechanisms remains to be determined. Overall, the specific mechanisms in place that regulate both the clock and cell cycles, including bi-directional modulation between these two pathways, might be essential to protect the organism from rampant cycling that could lead to diseases such as cancer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol. 2010;72:551–77. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919–22. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- 3.Masri S, Sassone-Corsi P. The circadian clock: a framework linking metabolism, epigenetics and neuronal function. Nat Rev Neurosci. 2013;14:69–75. doi: 10.1038/nrn3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–48. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 5.Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–76. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 6.Hunt T, Sassone-Corsi P. Riding tandem: circadian clocks and the cell cycle. Cell. 2007;129:461–4. doi: 10.1016/j.cell.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 7.Johnson CH. Circadian clocks and cell division: what's the pacemaker? Cell Cycle. 2010;9:3864–73. doi: 10.4161/cc.9.19.13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khapre RV, Samsa WE, Kondratov RV. Circadian regulation of cell cycle: Molecular connections between aging and the circadian clock. Ann Med. 2010;42:404–15. doi: 10.3109/07853890.2010.499134. [DOI] [PubMed] [Google Scholar]
- 9.Salter MG, Franklin KA, Whitelam GC. Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature. 2003;426:680–3. doi: 10.1038/nature02174. [DOI] [PubMed] [Google Scholar]
- 10.Mori T, Binder B, Johnson CH. Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 hours. Proc Natl Acad Sci U S A. 1996;93:10183–8. doi: 10.1073/pnas.93.19.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong G, et al. Elevated ATPase activity of KaiC applies a circadian checkpoint on cell division in Synechococcus elongatus. Cell. 2010;140:529–39. doi: 10.1016/j.cell.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dekens MP, et al. Light regulates the cell cycle in zebrafish. Curr Biol. 2003;13:2051–7. doi: 10.1016/j.cub.2003.10.022. [DOI] [PubMed] [Google Scholar]
- 13.Hirayama J, Cardone L, Doi M, Sassone-Corsi P. Common pathways in circadian and cell cycle clocks: light-dependent activation of Fos/AP-1 in zebrafish controls CRY-1a and WEE-1. Proc Natl Acad Sci U S A. 2005;102:10194–9. doi: 10.1073/pnas.0502610102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamai TK, Young LC, Cox CA, Whitmore D. Light acts on the zebrafish circadian clock to suppress rhythmic mitosis and cell proliferation. J Biol Rhythms. 2012;27:226–36. doi: 10.1177/0748730412440861. [DOI] [PubMed] [Google Scholar]
- 15.Matsuo T, et al. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–9. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 16.Bjarnason GA, Jordan R. Rhythms in human gastrointestinal mucosa and skin. Chronobiol Int. 2002;19:129–40. doi: 10.1081/cbi-120002595. [DOI] [PubMed] [Google Scholar]
- 17.Geyfman M, et al. Brain and muscle Arnt-like protein-1 (BMAL1) controls circadian cell proliferation and susceptibility to UVB-induced DNA damage in the epidermis. Proc Natl Acad Sci U S A. 2012;109:11758–63. doi: 10.1073/pnas.1209592109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaddameedhi S, Selby CP, Kaufmann WK, Smart RC, Sancar A. Control of skin cancer by the circadian rhythm. Proc Natl Acad Sci U S A. 2011;108:18790–5. doi: 10.1073/pnas.1115249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grechez-Cassiau A, Rayet B, Guillaumond F, Teboul M, Delaunay F. The circadian clock component BMAL1 is a critical regulator of p21WAF1/CIP1 expression and hepatocyte proliferation. J Biol Chem. 2008;283:4535–42. doi: 10.1074/jbc.M705576200. [DOI] [PubMed] [Google Scholar]
- 20**.Laranjeiro R, et al. Cyclin-dependent kinase inhibitor p20 controls circadian cell-cycle timing. Proc Natl Acad Sci U S A. 2013;110:6835–40. doi: 10.1073/pnas.1217912110. [This paper illustrates the circadian expression of p20 and its critical control over the G1/S transition of the cell cycle.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plikus MV, et al. Local circadian clock gates cell cycle progression of transient amplifying cells during regenerative hair cycling. Proc Natl Acad Sci U S A. 2013;110:E2106–15. doi: 10.1073/pnas.1215935110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 23.Hirayama J, et al. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450:1086–90. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- 24.Katada S, Sassone-Corsi P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat Struct Mol Biol. 2010;17:1414–21. doi: 10.1038/nsmb.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagoshi E, et al. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 26**.Kowalska E, et al. NONO couples the circadian clock to the cell cycle. Proc Natl Acad Sci U S A. 2012;110:1592–9. doi: 10.1073/pnas.1213317110. [This study illustrates the novel role of NONO, in connection with PER proteins, in the circadian activation of p16-Ink4A cell cycle checkpoint gene.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sehgal A, Price JL, Man B, Young MW. Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science. 1994;263:1603–6. doi: 10.1126/science.8128246. [DOI] [PubMed] [Google Scholar]
- 28.Unsal-Kacmaz K, Mullen TE, Kaufmann WK, Sancar A. Coupling of human circadian and cell cycles by the timeless protein. Mol Cell Biol. 2005;25:3109–16. doi: 10.1128/MCB.25.8.3109-3116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X, Wood PA, Hrushesky WJ. Mammalian TIMELESS is required for ATM-dependent CHK2 activation and G2/M checkpoint control. J Biol Chem. 2010;285:3030–4. doi: 10.1074/jbc.M109.050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gery S, et al. The circadian gene per1 plays an important rolef in cell growth and DNA damage control in human cancer cells. Mol Cell. 2006;22:375–82. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 31.Barnes JW, et al. Requirement of mammalian Timeless for circadian rhythmicity. Science. 2003;302:439–42. doi: 10.1126/science.1086593. [DOI] [PubMed] [Google Scholar]
- 32.Gotter AL, et al. A time-less function for mouse timeless. Nat Neurosci. 2000;3:755–6. doi: 10.1038/77653. [DOI] [PubMed] [Google Scholar]
- 33.Asher G, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–28. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 34.Nakahata Y, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–40. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sasaki T, et al. Phosphorylation regulates SIRT1 function. PLoS One. 2008;3:e4020. doi: 10.1371/journal.pone.0004020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaziri H, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–59. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 37.Luo J, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–48. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 38.Brunet A, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–5. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 39.Yang Q, Pando BF, Dong G, Golden SS, van Oudenaarden A. Circadian gating of the cell cycle revealed in single cyanobacterial cells. Science. 2010;327:1522–6. doi: 10.1126/science.1181759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–8. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 41*.Destici E, Oklejewicz M, Saito S, van der Horst GT. Mammalian cryptochromes impinge on cell cycle progression in a circadian clock-independent manner. Cell Cycle. 2011;10:3788–97. doi: 10.4161/cc.10.21.17974. [This paper highlights that circadian clock control of the cell cycle could be dictated by systemic cues that we currently do not fully understand.] [DOI] [PubMed] [Google Scholar]
- 42*.Gaddameedhi S, Reardon JT, Ye R, Ozturk N, Sancar A. Effect of circadian clock mutations on DNA damage response in mammalian cells. Cell Cycle. 2012;11:3481–91. doi: 10.4161/cc.21771. [Systemic cues provided by the organism may be very important in directing DNA damage response.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–7. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramsey KM, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–4. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Womac AD, Burkeen JF, Neuendorff N, Earnest DJ, Zoran MJ. Circadian rhythms of extracellular ATP accumulation in suprachiasmatic nucleus cells and cultured astrocytes. Eur J Neurosci. 2009;30:869–76. doi: 10.1111/j.1460-9568.2009.06874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. The human circadian metabolome. Proc Natl Acad Sci U S A. 2012;109:2625–9. doi: 10.1073/pnas.1114410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eckel-Mahan KL, et al. Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci U S A. 2012;109:5541–6. doi: 10.1073/pnas.1118726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minami Y, et al. Measurement of internal body time by blood metabolomics. Proc Natl Acad Sci U S A. 2009;106:9890–5. doi: 10.1073/pnas.0900617106. [DOI] [PMC free article] [PubMed] [Google Scholar]