Abstract
Pubertal timing is influenced by complex interactions among genetic, nutritional, environmental, and socioeconomic factors. The role of MKRN3, an imprinted gene located in the Prader-Willi syndrome critical region (chromosome 15q11-q13), in pubertal initiation was first described in 2013 after the identification of deleterious MKRN3 mutations in five families with central precocious puberty (CPP) using whole-exome sequencing analysis. Since then, additional loss-of-function mutations of MKRN3 have been associated with the inherited premature sexual development phenotype in girls and boys from different ethnic groups. In all of these families, segregation analysis clearly demonstrated autosomal dominant inheritance with complete penetrance, but with exclusive paternal transmission, consistent with the monoallelic expression of MKRN3 (a maternally imprinted gene). Interestingly, the hypothalamic Mkrn3 mRNA expression pattern in mice correlated with a putative inhibitory input on puberty initiation. Indeed, the initiation of puberty depends on a decrease in factors that inhibit the release of GnRH combined with an increase in stimulatory factors. These recent human and animal findings suggest that MKRN3 has an inhibitory role in the reproductive axis to represent a new pathway in pubertal regulation.
Introduction
Puberty, a complex biologic process involving sexual maturation and accelerated linear growth, is initiated when the pulsatile secretion of gonadotropin-releasing hormone (GnRH) increases after a quiescent period during childhood. The regulation of puberty initiation remains one of the great mysteries of human biology and it is thought that a conjunction of factors play a role to initiate puberty.
Environmental and metabolic factors are important regulators of pubertal development, but these influences are superimposed upon substantial genetic control. Similar timing of puberty shared by mothers and her children, and within racial groups, suggests a genetic component to the onset of puberty (Herman-Giddens, et al. 1997; Zacharias and Wurtman 1969). Substantial efforts have been made to identify genes that are responsible for the initiation of puberty (Ojeda and Lomniczi 2014a). The identification of these genes is critical for advancing the understanding of the neuroendocrine regulation of puberty initiation.
In an attempt to identify genes operating within the neuroendocrine brain that ultimately regulate the reproductive axis, researchers have been studying patients with pubertal disorders. The hypothalamic-pituitary-gonadal axis regulates puberty initiation and reproduction. GnRH is produced and secreted by the hypothalamus in a pulsatile fashion during the embryonic and neonatal phases of life. GnRH secretion is actively inhibited during infancy and puberty begins with the reactivation of its secretion. GnRH deficiency results in hypogonadotropic hypogonadism, in which patients fail to develop puberty and are usually infertile. Conversely, early reactivation of GnRH secretion results in central precocious puberty (CPP). Whilst several genes have been detected in association with GnRH deficiency and have contributed to the current knowledge of GnRH regulation (Bianco and Kaiser 2009; Semple and Topaloglu 2010), genes linked to CPP have until recently only been identified subsequent to their association with hypogonadotropic hypogonadism, such as KISS1 and KISS1R. However, only rare genetic defects in KISS1 and its receptor have been identified in patients with CPP (Silveira, et al. 2010; Teles, et al. 2008).
The advances in sequencing methods permitted the detection of new genes implicated in the neuroendocrine regulation of puberty. Using exome sequencing analysis and studying familial cases of CPP, genetic defects in a gene with no previous link to the hypothalamic-pituitary-gonadal axis, MKRN3, were identified as the cause of premature sexual development in one-third of the families (Abreu, et al. 2013). MKRN3 is located on the long arm of chromosome 15 in the Prader-Willi syndrome critical region and is maternally imprinted (discussed below). Subsequently, we showed that mutations in MKRN3 are also the cause of CPP in apparently sporadic cases (Macedo, et al. 2014). These findings were significant contributions to the field and will advance the understanding of the pubertal process. We will review the genetic defects identified in patients with CPP and their clinical implications, and discuss here the possible role of MKRN3 within the reproductive axis.
Loss-of-function mutations of MKRN3 cause familial CPP
The role of MKRN3 in pubertal initiation was first described in 2013 after a comprehensive genetic study of several families with CPP (Abreu et al. 2013). In this study, the authors investigated 40 members of 15 families with CPP from different ethnic groups (12 Brazilian, 2 American and 1 Belgian families), and applying whole exome sequencing four deleterious MKRN3 mutations - three frameshift and a missense mutation (Figure 1) - were detected in five of these families (33%). Both sexes were equally affected by MKRN3 mutations (8 girls and 7 boys) (Table 1).
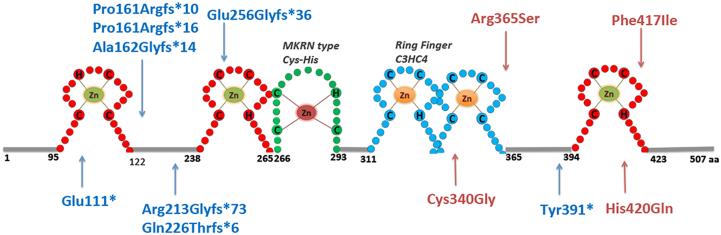
Figure 1. MKRN3 protein structure and mutations identified in patients with central precocious puberty.
(Zn) zinc; (H) histidine; (C) cysteine. The three C3H zinc finger motifs are shown in red, the C3HC4 RING finger motif is in blue, and the MKRN-specific Cys–His domain is shown in green. The numbers correspond to the amino acid positions in the protein. Blue mutation labels and arrows indicate the location of frameshift mutations; pink mutation labels and arrows indicate the location of the missense mutations.
Table 1.
MKRN3 mutations described in children with CPP from different origins.
| Family number |
Origin | Patient number |
Initial clinical manifestation (Age in Years) |
Diagnosis | MKRN3 mutation | References | |||
|---|---|---|---|---|---|---|---|---|---|
| Age (Years) |
Breast/ Genital development Tanner stage |
Bone age (Years) |
DNA | Protein | |||||
| 1 | USA | I | Thelarche (5.7) | 6.5 | 2 | 7.7 | 637delC | Arg213Glyfs*73 | Abreu AP et al, 2013 |
| II | Gonadarche (8.0) | 8.7 | 3 | 11 | 637delC | Arg213Glyfs*73 | |||
| III | Thelarche (6.5) | 6.7 | 2 | 7.8 | 637delC | Arg213Glyfs*73 | |||
| 2 | Brazil | I | Thelarche (6.2) | 7.0 | 3 | 7.0 | 1171_1172insA | Tyr391* | Abreu AP et al, 2013 |
| II | Thelarche (5.7) | 6.0 | 3 | 6.0 | 1171_1172insA | Tyr391* | |||
| 3 | Brazil | I | Gonadarche and pubarche (5.9) |
8.1 | 3 | 10 | 475_476insC | Ala162Glyfs*14 | Abreu AP et al, 2013 |
| II | Gonadarche and pubarche unknown |
9.7 | 3 | 9.7 | 475_476insC | Ala162Glyfs*14 | |||
| 4 | USA | I | Gonadarche and pubarche (8.5) |
8.8 | 3 | 11 | 475_476insC | Ala162Glyfs*14 | Abreu AP et al, 2013 |
| II | Thelarche (5.0) | 6.5 | 2 | 8.3 | 475_476insC | Ala162Glyfs*14 | |||
| 5 | Brazil | I | Thelarche and pubarche (6.4) |
7.8 | 3 | 11 | 482delC | Pro161Argfs*10 | Macedo DB et al, 2014 |
| 6 | Brazil | I | Thelarche (5.4) | 6.1 | 3 | 8.8 | 482_483insC | Pro161Argfs*16 | Macedo DB et al, 2014 |
| 7 | Brazil | I | Thelarche (6.0) | 7.9 | 4 | 11 | 482_483insC | Pro161Argfs*16 | Macedo DB et al, 2014 |
| 8 | Brazil | I | Thelarche (3.0) and pubarche (6.0) |
6.7 | 3 | 8.8 | 482delC | Pro161Argfs*10 | Macedo DB et al, 2014 |
| 9 | Brazil | I | Thelarche (4.0) | 6.8 | 4 | 12 | 675_676insA | Gln226Thrfs*6 | Macedo DB et al, 2014 |
| 10 | Brazil | I | Thelarche and pubarche (6.0) |
6.6 | 3 | 7.8 | 766_767delA | Glu256Glyfs*36 | Macedo DB et al, 2014 |
| 11 | Brazil | I | Thelarche (6.0) and pubarche (6.3) |
6.9 | 3 | 9.3 | 482_483insC | Pro161Argfs*16 | Macedo DB et al, 2014 |
| 12 | Belgium | I | Gonadarche and pubarche unknown |
9.7 | 3 | 12 | 1095G>T | Arg365Ser | Abreu AP et al, 2013 |
| II | Thelarche (6.2) | 6.4 | 2 | 9.4* | 1095G>T | Arg365Ser | |||
| III | Thelarche and pubarche (5.4) |
5.7 | 2 | 8.5 | 1095G>T | Arg365Ser | |||
| 13 | Brazil | I | Thelarche (6.0) | 7.1 | 3 | 8.8 | 1249T>A | Phe417Ile | Macedo DB et al, 2014 |
| 14 | Greece | I | Thelarche (6.0) | 7.1 | 3 | 10 | 1018T>G | Cys340Gly | Settas N et al, 2014 |
| II | Gonadarche unknown | 9.2 | 2 | 10 | 1018T>G | Cys340Gly | |||
| 15 | Germany | I | Gonadarche and pubarche (9.0) |
10 | 3 | - | 331G>T | Glu111* | Schreiner F et al, 2014 |
| II | Thelarche (6.1) | 7.5 | 3 | - | 331G>T | Glu111* | |||
| 16 | Germany | I | Thelarche (7.5) | 7.5 | - | - | 475_476insC | Ala162Glyfs*14 | Schreiner F et al, 2014 |
| II | Thelarche (6.8) | 7.1 | 2 | - | 475_476insC | Ala162Glyfs*14 | |||
| 17 | Israel | I | Thelarche and pubarche (5.5) |
5.8 | 3 | 6.8 | 1260T>G | His420Gln | De Vries L et al, 2014 |
| II | Gonadarche and pubarche unknown |
9.8 | 3 | 11.5 | 1260T>G | His420Gln | |||
| III | Thelarche and pubarche (5.5) |
6.3 | 3 | 8.8 | 1260T>G | His420Gln | |||
| IV | Thelarche (4.5) | 4.5 | 2 | 6.0 | 1260T>G | His420Gln | |||
E2: estradiol; T: testosterone
Gonadarche or testicular enlargement. Breast/Genital development Tanner stage and bone age assessed at time of diagnosis.
Bone age at 7.4yr.
More recently, Macedo et al. (Macedo, et al. 2014) studied 215 unrelated children (207 girls and 8 boys) with CPP from three different University Medical Centers and identified five novel heterozygous mutations in 8 unrelated Brazilian girls, including 4 frameshift variants and one missense variant (Figure 1, Table 1). No family history of premature sexual development was reported in the majority of these studied patients. However, segregation analysis was performed for five of these 8 girls, and in all cases demonstrated that the mutant allele was paternally inherited in all families with MKRN3 mutations.
In order to investigate if the CPP phenotype could arise from loss of MKRN3 expression by the paternal allele due to a de novo deletion, maternal uniparental disomy, or an imprinting defect, Macedo et al. (Macedo et al. 2014) investigated 52 patients with familial and sporadic CPP, without known MKRN3 sequence defects, by methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA) of chromosome 15q11. No copy number changes or methylation abnormalities were detected in the 15q11 locus, suggesting that epigenetic defects involving this locus are likely rare mechanisms in this disorder (Macedo et al. 2014).
Recently, other investigators have also reported MKRN3 defects associated with familial CPP. Settas et al. (Settas, et al. 2014) described a novel heterozygous missense mutation (p.Cys340Gly) in MKRN3 in two Greek siblings, a girl with CPP and a boy with early puberty. Soon thereafter, Schreiner et al. (Schreiner, et al. 2014) identified two heterozygous MKRN3 mutations (p.Glu111* and p.Ala162Glyfs*14) in affected members of 2 German families. Lastly, de Vries et al (de Vries, et al. 2014) identified a novel missense mutation (p.His420Gln) in four siblings, including a boy, from an Ashkenazi-Sephardic Jewish family (Figure 1, Table 1). These studies further expanded the MKRN3 mutational spectrum.
Clinical Features of CPP associated with MKRN3 mutations
All patients with CPP carrying loss-of-function mutations in MKRN3 exhibited typical clinical and hormonal features of premature activation of the reproductive axis, including early pubertal signs, such as breast, testes, and pubic hair development, accelerated linear growth, advanced bone age, and elevated basal and/or GnRH-stimulated LH levels (Table 1). Except for two related patients (one girl and her brother) who presented with esotropia (Abreu et al. 2013), which is a minor criterion for Prader-Willi syndrome (PWS), no other signs of PWS were detected in any patient with CPP caused by MKRN3 defects. Another female patient had mild nonspecific syndromic features, including a high-arched palate, dental abnormalities, clinodactyly and hyperlordosis (Macedo et al. 2014). Regarding the therapeutic response of CPP patients with MKRN3 defects, six of eight patients have demonstrated adequate responses to conventional treatment with depot GnRH agonists to date (Macedo et al. 2014).
In patients with CPP due to MKRN3 defects, the median age of puberty onset was 6.0 years in girls (ranging from 3.0 to 7.5 years) and 8.25 years in boys (ranging from 5.9 to 9.0 years) (Abreu et al. 2013; de Vries et al. 2014; Schreiner et al. 2014; Settas et al. 2014), suggesting that the MKRN3 mutations may affect girls more severely than boys (Table 1). Given the median age of pubertal onset of affected patients with MKRN3 mutations, it is speculated that the prepubertal inhibitory tonus on GnRH secretion took place normally, but was lost prematurely in patients with MKRN3 mutations. This clinical observation suggests that MKRN3 may not be crucial for GnRH suppression after the mini-puberty of early infancy, but that the downregulation of MKRN3 plays a relevant role for the re-emergence of GnRH pulses in the pubertal onset (Macedo et al. 2014).
MKRN3 mutations and polymorphisms
Currently, 12 distinct loss-of-function mutations of MKRN3 have been described in 30 patients (22 girls and 8 boys) with CPP from 17 families from different ethnicities (Figure 1, Table 1). Remarkably, eight of these mutations encode either premature stop codons or frameshift mutations (Table 1). The four missense mutations (p.Cys340Gly, p.Arg365Ser, p.Phe417Ile and p.His420Gln) were located within a zinc finger motif or a RING finger motif (Figure 1), regions predicted to be involved in RNA binding and ubiquitin ligase activity of the protein respectively, and essential for protein function. All missense mutations are predicted to be pathogenic by in silico analysis. Additionally, ab initio modeling of the mutations p.Arg365Ser and p.Cys340Gly predicts that these mutations lead to significant structural perturbations in the 3D structure of the RING finger motif (Settas et al. 2014). The substitution of histidine 420 with glutamine in the MKRN3 protein is predicted reduce the affinity between the Zn ion binding site and the relevant Zn, disrupting the binding pocket leading to unfolding of the finger (de Vries et al. 2014). Interestingly, most of the MKRN3 mutations (64%) were located in the amino-terminal region of the protein, which is encoded by a poly C rich sequence, suggesting that this area may be a potential hotspot.
The important role of MKRN3 in human puberty initiation was reinforced recently by large genome-wide and custom-genotyping arrays in up to 182,416 women of European descent (57 studies) (Perry, et al. 2014). There was evidence of 123 signals at 106 genomic loci associated with age at menarche. Three of these loci were located in imprinted regions, including the MKRN3 locus, demonstrating parent-of-origin-specific associations concordant with known parental expression patterns (Perry et al. 2014). This study suggests that not only are rare variants in MKRN3 associated with CPP, but that more common variants/polymorphisms may be associated with changes in the timing of puberty (as reflected by age of menarche) within the normal range and within the general population. This was a remarkable extension of the findings of rare variants in a disease such as CPP to more common variants in a polygenic trait such as age of menarche. The lack of parent-of-origin-specific analysis in previous GWAS studies may explain why MKRN3 has not been previously associated with age of menarche (Elks, et al. 2010; He, et al. 2009).
MKRN3 Gene and Protein Structure and Expression
Makorin ring finger 3 (MKRN3) was first cloned in 1999 by Jong et al. (Jong, et al. 1999b) during a study of the Prader-Willi/Angelman syndrome (PW/AS) critical region. They identified a cDNA in the PW/AS region encoding a zinc finger protein, initially named ZNF127 (zinc finger protein 127) and later renamed MKRN3. The functional and physiological relevance of MKRN3 is not known and despite its location in the PWS critical region, its role in this syndrome is also unclear. An antisense transcript was concomitantly identified and named ZNF127AS. The antisense gene is not translated and is expressed weakly during fetal development and at very low levels in adult brain regions (Jong et al. 1999b). Similar to other antisense genes, ZNF127AS may regulate the expression of the “sense” gene (MKRN3).
MKRN3 is located on human chromosome 15q11–13 (chromosome 7C in mouse) in a region that contains a cluster of imprinted genes associated with two different neurobehavioral disorders (Lalande 1996; Nicholls, et al. 1998). Mutations or loss of expression of the maternally expressed gene UBE3A lead to Angelman syndrome (AS), while PWS is thought to be a contiguous gene syndrome arising from the loss of expression of multiple paternally expressed genes, including MKRN3. The report of two patients with PWS with a deletion of the 15q11–13 locus that did not include MKRN3 suggested that this gene is not required for the development of the syndrome, but we still cannot rule out a role for MKRN3 in some of the clinical features of PWS (Kanber, et al. 2009).
MKRN3 differential allele expression occurs through silencing of the MKRN3 maternal allele, which is associated with 5’ CpG island methylation. (Hershko, et al. 1999; Jong, et al. 1999a). Methylation of MKRN3 and the other imprinted genes located within the PW/AS region of chromosome 15 is coordinately regulated by a bipartite imprinting control center (IC), composed of a sequence around the SNRPN promoter, the PWS-IC, and an 880 bp sequence located 35 kb upstream, the AS-IC (Rabinovitz, et al. 2012). The molecular mechanisms of IC function are not well understood, but it has been postulated, based on studies in humans and mouse models, that the PWS-IC is a positive regulatory element and activates paternal-specific gene expression (Bielinska, et al. 2000; Bressler, et al. 2001), whereas the AS-IC functions in the maternal imprint by allele-specific repression of the PWS-IC to prevent the paternal imprinting program (Ohta, et al. 1999; Rodriguez-Jato, et al. 2005; Shemer, et al. 2000) (Jong et al. 1999a).
MKRN3 is an intronless gene, and likely arose by germline retrotransposition from the Makorin ring finger protein 1 gene (MKRN1), a highly transcribed, intron-containing gene, 180-90 million years ago (Rapkins, et al. 2006). MKRN1 is the ancestral founder of the Makorin gene family (Gray, et al. 2000). Several other retrocopies of MKRN1 have been identified in mammalian genomes, most of them probably corresponding to pseudogenes. Nine MKRN family loci distributed throughout the human genome have been identified so far, with only three functional genes - MKRN1, MKRN2 and MKRN3 - identified in vertebrates (Bohne, et al. 2010; Gray et al. 2000).
The MAKORIN family
The MKRN gene family encodes putative ribonucleoproteins with a distinctive array of zinc-finger motifs including several C3H zinc fingers, a makorin-specific Cys-His arrangement, and a RING zinc finger (Jong et al., 1999a; b). In particular, MKRN3 has a centrally located RING finger motif (C3HC4), two amino-terminal C3H zinc finger motifs followed by the unique pattern of conserved Cys-His residues called a Makorin zinc finger motif, and a carboxy-terminal C3H zinc finger motif (Jong et al. 1999b) (Figure 1). MKRN3 is highly conserved among species, and the mouse and human MKRN3 amino acid sequences share 69% identity and 82% similarity (Jong et al. 1999b). Mice and humans usually do not have conserved untranslated regions (UTRs), yet the MKRN3 3’-UTR has 90% identity between these two species, implying a functional significance to this region of MKRN3. MKRN3 is ubiquitously expressed in adult human tissue, with highest expression levels in the testis. In the fetus, it is highly expressed in the central nervous system and is expressed in post-meiotic sperm germ cells, particularly in round spermatids (Jong et al. 1999b).
The characteristic arrangement of cysteine (Cys or C) and histidine (His or H) residues in the zinc finger proteins in MKRN3 can allow some predictions of its function. C3H zinc fingers have been identified in RNA-binding proteins (Barabino, et al. 1997; Murray, et al. 1997). RING zinc fingers have been shown to mediate protein:protein interactions (Schwabe and Klug 1994). More recent evidence suggests that the RING zinc finger is a signature domain for E3 ligases, a category of enzymes mediating the transfer of ubiquitin from an E2 ubiquitin-conjugating enzyme to target protein substrates. (Deshaies and Joazeiro 2009). Ubiquitination can have diverse effects on the protein substrate, varying from proteasome-dependent proteolysis to modulation of protein function and/or localization (Behrends and Harper 2011; Deshaies and Joazeiro 2009). The multiple types and number of zinc finger motifs in makorin proteins suggest possible multiple cellular functions for this protein.
Makorin proteins share a highly homologous amino acid sequence, particularly in the zinc finger domains (Figure 2), suggesting that they may share similar functions or regulatory mechanisms. Although there are not many studies on MKRN3 function to date, MKRN1 function has been better explored. A possible important cellular function for MKRN1 is supported by the high identity (92%) between the human and murine orthologs and the ubiquitous protein expression in human and mouse tissues (Gray et al. 2000). MKRN1 acts as an E3 ubiquitin ligase, inducing degradation of human telomerase reverse transcriptase (hTERT), viral capsid proteins, p53 and p21 cell cycle regulators, and peroxisome proliferator activated receptor gamma (PPARγ) (Kim, et al. 2014; Kim, et al. 2005; Lee, et al. 2009).
Figure 2. MKRN1 and MKRN3 protein sequence alignment.
The two protein sequences share high homology and similarity, especially in the RING finger domains. Bold letters represent conserved amino acids, and squares similar amino acids. Sections highlighted in red represent the three zinc fingers C3H motifs, in yellow the MKRN-specific Cys-His motif, and in green the RING finger domain. MKRN1 NCBI Reference Sequence NP_038474, UniProtKB/Swiss-Prot: Q9UHC7.3. MKRN3 NP_005655.1 and UniProtKB/Swiss-Prot Q13064.1.
An alternative mechanism of action for MKRN1 was described when it was identified as a repressor of c-Jun transcriptional activity using a yeast assay. Through regulation of RNA polymerase II-dependent transcription, MKRN1 can have either negative or positive effects on gene expression (Omwancha, et al. 2006). Interestingly, it was shown that the disruption of ubiquitin ligase activity of MKRN1 did not affect its inhibitory transcriptional activity, suggesting that this function is independent of protein ubiquitination (Omwancha et al. 2006).
MKRN2 is also ubiquitously expressed (Gray et al. 2000). Like other members of this family, MKRN2 is highly conserved throughout evolution. It has been shown that mkrn2 negatively regulates neurogenesis via PI3K/Akt signaling in Xenopus embryos; however, the detailed molecular mechanisms of this effect and the potential functions of mammalian MKRN2 remain to be studied (Yang, et al. 2008).
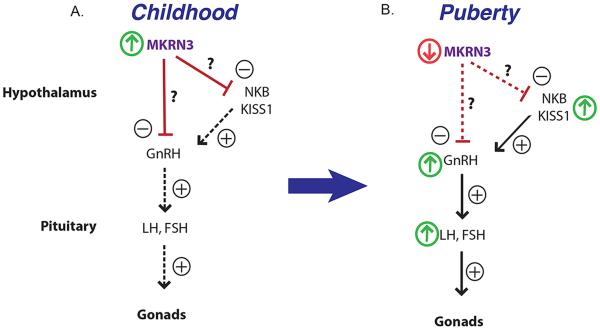
MKRN3 presumptively possesses ubiquitin-protein isopeptide ligase (E3) activity, intimated by the presence of the highly conserved C3HC4 RING finger domain. The tandem repeat of C3H zinc fingers may provide high specificity RNA binding (Hudson, et al. 2004), and the unique Cys-His makorin motif has been suggested to be a DNA binding domain. Taken together, the studies of MKRN1 and MKRN2 imply that MKRN3 can likewise act as an E3 ligase, based on the high homology of the RING finger domain (Figure 2). A previous mouse study showed that Mkrn3 is highly expressed in the hypothalamic arcuate nucleus during the infantile and early juvenile periods, with a reduction in expression at postnatal days 12 to 15, prior to puberty initiation (Abreu et al. 2013). The arcuate nucleus is the site of expression of critical regulators of GnRH secretion, such as kisspeptin, neurokinin B and dynorphin (Navarro, et al. 2011). Puberty is initiated with a loss of inhibitory inputs and a gain in excitatory inputs (Ojeda and Lomniczi 2014b). The Mkrn3 expression pattern in the hypothalamic arcuate nucleus suggests that this peptide plays a role in the inhibition of GnRH secretion during the prepubertal quiescent period (Figure 3). The decrease in Mkrn3 expression is hypothesized to be associated with an increase in GnRH stimulatory factors and/or GnRH expression, and it can be postulated that MKRN3 may be acting at the hypothalamic level as an E3 ligase to inhibit stimulatory inputs, so that loss-of-function mutations of MKRN3 result in early activation of the hypothalamic-pituitary-gonadal axis, expressed phenotypically as central precocious puberty. It is also possible that MKRN3 can act as a transcriptional regulator, as has been demonstrated for MKRN1. Data from the Human Protein Atlas shows that MKRN3 is located primarily in the plasma membrane and cytoplasm, but is also located in the nucleus (www.proteinatlas.org). Based on its location in the plasma membrane, we can speculate that MKRN3 may also be involved in endocytosis and downregulation of receptors, as has been demonstrated for some other E3 ligases (Hershko and Ciechanover 1998). The genetic findings from patients with CPP are in agreement with the hypothesis that MKRN3 may act as a ‘brake’ or inhibitor of GnRH secretion during childhood (Hughes 2013). Further studies are needed to elucidate the precise mechanism(s) of action of MKRN3.
Figure 3. Schematic representation of MKRN3 mechanism of action.
Human and mouse studies suggest that MKRN3 acts as an inhibitor of GnRH secretion during childhood (diagram on A), and that a decrease in MKRN3 expression is associated with an increase in GnRH stimulatory factors and/or GnRH resulting in puberty initiation (diagram on B). NKB, neurokinin B; − inhibition, + stimulation. ↑ increase ↓ decrease.
Conclusion
Age of puberty initiation is associated with causes of substantial morbidity and mortality. Early age of menarche is associated with breast cancer and cardiovascular disease (Kvale 1992; Lakshman, et al. 2009). Therefore it is important to understand the mechanisms controlling puberty initiation. In an attempt to identify genes that will broaden the knowledge on GnRH regulation to bring new genetic screening, and diagnostic and treatment tools, researchers have been trying to identify genes associated with CPP for several years. By studying individuals with hypogonadotropic hypogonadism, a genetic disorder caused by absence or deficiency of GnRH secretion, we learned about important stimulators of GnRH secretion (Bianco and Kaiser 2009; Semple and Topaloglu 2010). Because of the complexity of the mechanisms involved in puberty initiation and the initial failure to identify genes associated with CPP, it has been hypothesized that CPP was not caused by mutations in a single gene, but rather was a consequence of complex interactions among environment, metabolic factors and polygenic defects. However, with the development of new sequencing methodologies, the combination of whole exome sequencing analysis with detailed phenotypic characterization and the careful selection of the correct cohort of patients led to the identification of MKRN3 as an important regulator of pubertal development. MKRN3 is the first gene with a probable inhibitory effect on GnRH secretion, with mutations identified with in humans. To date, MKRN3 is the most common genetic defect associated with CPP. The identification of loss-of-function mutations in this gene can contribute to the diagnosis of CPP, especially in boys in whom the signs of puberty initiation are not easily detectable, thereby helping to make the diagnosis earlier and facilitating treatment decisions. In addition, the presence of MKRN3 mutations can contribute to early diagnosis of CPP in familial cases and guide genetic counseling. It is not clear if there is a gender difference in the effect of MKRN3 mutations, but it is evident that mutations in this gene accelerate puberty initiation in both sexes. Although the precise mechanism of regulation of GnRH secretion by MKRN3 is not yet understood, its importance in the hypothalamic-pituitary-axis is indisputable. The studies of Mkrn3 expression in the hypothalamic arcuate nucleus of mice support the findings that loss-of-function mutations in humans lead to early puberty initiation and strengthen the hypothesis that MKRN3 acts as an inhibitor of GnRH secretion during childhood (Figure 3). Ultimately, the recent GWAS findings linking MKRN3 with age of menarche have consolidated the involvement of this gene in pubertal timing.
The identification of a putative inhibitory factor in the hypothalamic-pituitary-gonadal axis has opened an exciting new arena in the neuroendocrine field. Further studies will elucidate the precise mechanism of action of this important regulator of GnRH secretion.
Acknowledgments
Fundação de Amparo do Estado de Sao Paulo FAPESP (Brazil). Grant 2013-06391-1, to DBM; and 2013-03236-5, to ACL.
Eunice Kennedy Shriver NICHD/NIH through cooperative agreement U54HD28138 as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research and R01HD019938, to UK; and 1F05HD072773-03, to APA.
Footnotes
Conflicts of interest
We declare that we have no conflicts of interest.
Contributor Information
Ana Paula Abreu, Division of Endocrinology, Diabetes and Hypertension, Brigham and Women‘s Hospital and Harvard Medical School, Boston, Massachusetts, USA.
Delanie B. Macedo, Unidade de Endocrinologia do Desenvolvimento, Laboratório de Hormônios e Genética Molecular, LIM 42, Hospital das Clínicas, Disciplina de Endocrinologia, Faculdade de Medicina da Universidade de São Paulo, São Paulo, Brasil
Vinicius N. Brito, Unidade de Endocrinologia do Desenvolvimento, Laboratório de Hormônios e Genética Molecular, LIM 42, Hospital das Clínicas, Disciplina de Endocrinologia, Faculdade de Medicina da Universidade de São Paulo, São Paulo, Brasil
Ursula B. Kaiser, Division of Endocrinology, Diabetes and Hypertension, Brigham and Women‘s Hospital and Harvard Medical School, Boston, Massachusetts, USA
Ana Claudia Latronico, Unidade de Endocrinologia do Desenvolvimento, Laboratório de Hormônios e Genética Molecular, LIM 42, Hospital das Clínicas, Disciplina de Endocrinologia, Faculdade de Medicina da Universidade de São Paulo, São Paulo, Brasil.
REFERENCES
- Abreu AP, Dauber A, Macedo DB, Noel SD, Brito VN, Gill JC, Cukier P, Thompson IR, Navarro VM, Gagliardi PC, et al. Central precocious puberty caused by mutations in the imprinted gene MKRN3. N Engl J Med. 2013;368:2467–2475. doi: 10.1056/NEJMoa1302160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabino SM, Hubner W, Jenny A, Minvielle-Sebastia L, Keller W. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev. 1997;11:1703–1716. doi: 10.1101/gad.11.13.1703. [DOI] [PubMed] [Google Scholar]
- Behrends C, Harper JW. Constructing and decoding unconventional ubiquitin chains. Nat Struct Mol Biol. 2011;18:520–528. doi: 10.1038/nsmb.2066. [DOI] [PubMed] [Google Scholar]
- Bianco SD, Kaiser UB. The genetic and molecular basis of idiopathic hypogonadotropic hypogonadism. Nat Rev Endocrinol. 2009;5:569–576. doi: 10.1038/nrendo.2009.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielinska B, Blaydes SM, Buiting K, Yang T, Krajewska-Walasek M, Horsthemke B, Brannan CI. De novo deletions of SNRPN exon 1 in early human and mouse embryos result in a paternal to maternal imprint switch. Nat Genet. 2000;25:74–78. doi: 10.1038/75629. [DOI] [PubMed] [Google Scholar]
- Bohne A, Darras A, D'Cotta H, Baroiller JF, Galiana-Arnoux D, Volff JN. The vertebrate makorin ubiquitin ligase gene family has been shaped by large-scale duplication and retroposition from an ancestral gonad-specific, maternal-effect gene. BMC Genomics. 2010;11:721. doi: 10.1186/1471-2164-11-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressler J, Tsai TF, Wu MY, Tsai SF, Ramirez MA, Armstrong D, Beaudet AL. The SNRPN promoter is not required for genomic imprinting of the Prader-Willi/Angelman domain in mice. Nat Genet. 2001;28:232–240. doi: 10.1038/90067. [DOI] [PubMed] [Google Scholar]
- de Vries L, Gat-Yablonski G, Dror N, Singer A, Phillip M. A novel MKRN3 missense mutation causing familial precocious puberty. Human reproduction. 2014;29:2838–2843. doi: 10.1093/humrep/deu256. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Elks CE, Perry JR, Sulem P, Chasman DI, Franceschini N, He C, Lunetta KL, Visser JA, Byrne EM, Cousminer DL, et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat Genet. 2010;42:1077–1085. doi: 10.1038/ng.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray TA, Hernandez L, Carey AH, Schaldach MA, Smithwick MJ, Rus K, Marshall Graves JA, Stewart CL, Nicholls RD. The ancient source of a distinct gene family encoding proteins featuring RING and C(3)H zinc-finger motifs with abundant expression in developing brain and nervous system. Genomics. 2000;66:76–86. doi: 10.1006/geno.2000.6199. [DOI] [PubMed] [Google Scholar]
- He C, Kraft P, Chen C, Buring JE, Pare G, Hankinson SE, Chanock SJ, Ridker PM, Hunter DJ, Chasman DI. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat Genet. 2009;41:724–728. doi: 10.1038/ng.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman-Giddens ME, Slora EJ, Wasserman RC, Bourdony CJ, Bhapkar MV, Koch GG, Hasemeier CM. Secondary sexual characteristics and menses in young girls seen in office practice: a study from the Pediatric Research in Office Settings network. Pediatrics. 1997;99:505–512. doi: 10.1542/peds.99.4.505. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hershko A, Razin A, Shemer R. Imprinted methylation and its effect on expression of the mouse Zfp127 gene. Gene. 1999;234:323–327. doi: 10.1016/s0378-1119(99)00192-4. [DOI] [PubMed] [Google Scholar]
- Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol. 2004;11:257–264. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- Hughes IA. Releasing the brake on puberty. N Engl J Med. 2013;368:2513–2515. doi: 10.1056/NEJMe1306743. [DOI] [PubMed] [Google Scholar]
- Jong MT, Carey AH, Caldwell KA, Lau MH, Handel MA, Driscoll DJ, Stewart CL, Rinchik EM, Nicholls RD. Imprinting of a RING zinc-finger encoding gene in the mouse chromosome region homologous to the Prader-Willi syndrome genetic region. Hum Mol Genet. 1999a;8:795–803. doi: 10.1093/hmg/8.5.795. [DOI] [PubMed] [Google Scholar]
- Jong MT, Gray TA, Ji Y, Glenn CC, Saitoh S, Driscoll DJ, Nicholls RD. A novel imprinted gene, encoding a RING zinc-finger protein, and overlapping antisense transcript in the Prader-Willi syndrome critical region. Hum Mol Genet. 1999b;8:783–793. doi: 10.1093/hmg/8.5.783. [DOI] [PubMed] [Google Scholar]
- Kanber D, Giltay J, Wieczorek D, Zogel C, Hochstenbach R, Caliebe A, Kuechler A, Horsthemke B, Buiting K. A paternal deletion of MKRN3, MAGEL2 and NDN does not result in Prader-Willi syndrome. Eur J Hum Genet. 2009;17:582–590. doi: 10.1038/ejhg.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Park KW, Lee EW, Jang WS, Seo J, Shin S, Hwang KA, Song J. Suppression of PPARgamma through MKRN1-mediated ubiquitination and degradation prevents adipocyte differentiation. Cell Death Differ. 2014;21:594–603. doi: 10.1038/cdd.2013.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Park SM, Kang MR, Oh SY, Lee TH, Muller MT, Chung IK. Ubiquitin ligase MKRN1 modulates telomere length homeostasis through a proteolysis of hTERT. Genes Dev. 2005;19:776–781. doi: 10.1101/gad.1289405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvale G. Reproductive factors in breast cancer epidemiology. Acta oncologica. 1992;31:187–194. doi: 10.3109/02841869209088901. [DOI] [PubMed] [Google Scholar]
- Lakshman R, Forouhi NG, Sharp SJ, Luben R, Bingham SA, Khaw KT, Wareham NJ, Ong KK. Early age at menarche associated with cardiovascular disease and mortality. The Journal of clinical endocrinology and metabolism. 2009;94:4953–4960. doi: 10.1210/jc.2009-1789. [DOI] [PubMed] [Google Scholar]
- Lalande M. Parental imprinting and human disease. Annu Rev Genet. 1996;30:173–195. doi: 10.1146/annurev.genet.30.1.173. [DOI] [PubMed] [Google Scholar]
- Lee EW, Lee MS, Camus S, Ghim J, Yang MR, Oh W, Ha NC, Lane DP, Song J. Differential regulation of p53 and p21 by MKRN1 E3 ligase controls cell cycle arrest and apoptosis. EMBO J. 2009;28:2100–2113. doi: 10.1038/emboj.2009.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macedo DB, Abreu AP, Reis AC, Montenegro LR, Dauber A, Beneduzzi D, Cukier P, Silveira LF, Teles MG, Carroll RS, et al. Central precocious puberty that appears to be sporadic caused by paternally inherited mutations in the imprinted gene makorin ring finger 3. J Clin Endocrinol Metab. 2014;99:E1097–1103. doi: 10.1210/jc.2013-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MV, Turnage MA, Williamson KJ, Steinhauer WR, Searles LL. The Drosophila suppressor of sable protein binds to RNA and associates with a subset of polytene chromosome bands. Mol Cell Biol. 1997;17:2291–2300. doi: 10.1128/mcb.17.4.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro VM, Gottsch ML, Wu M, Garcia-Galiano D, Hobbs SJ, Bosch MA, Pinilla L, Clifton DK, Dearth A, Ronnekleiv OK, et al. Regulation of NKB pathways and their roles in the control of Kiss1 neurons in the arcuate nucleus of the male mouse. Endocrinology. 2011;152:4265–4275. doi: 10.1210/en.2011-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls RD, Saitoh S, Horsthemke B. Imprinting in Prader-Willi and Angelman syndromes. Trends Genet. 1998;14:194–200. doi: 10.1016/s0168-9525(98)01432-2. [DOI] [PubMed] [Google Scholar]
- Ohta T, Buiting K, Kokkonen H, McCandless S, Heeger S, Leisti H, Driscoll DJ, Cassidy SB, Horsthemke B, Nicholls RD. Molecular mechanism of angelman syndrome in two large families involves an imprinting mutation. Am J Hum Genet. 1999;64:385–396. doi: 10.1086/302232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojeda SR, Lomniczi A. Puberty in 2013: Unravelling the mystery of puberty. Nat Rev Endocrinol. 2014a;10:67–69. doi: 10.1038/nrendo.2013.233. [DOI] [PubMed] [Google Scholar]
- Ojeda SR, Lomniczi A. Puberty in 2013: Unravelling the mystery of puberty. Nature reviews. Endocrinology. 2014b;10:67–69. doi: 10.1038/nrendo.2013.233. [DOI] [PubMed] [Google Scholar]
- Omwancha J, Zhou XF, Chen SY, Baslan T, Fisher CJ, Zheng Z, Cai C, Shemshedini L. Makorin RING finger protein 1 (MKRN1) has negative and positive effects on RNA polymerase II-dependent transcription. Endocrine. 2006;29:363–373. doi: 10.1385/ENDO:29:2:363. [DOI] [PubMed] [Google Scholar]
- Perry JR, Day F, Elks CE, Sulem P, Thompson DJ, Ferreira T, He C, Chasman DI, Esko T, Thorleifsson G, et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014;514:92–97. doi: 10.1038/nature13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovitz S, Kaufman Y, Ludwig G, Razin A, Shemer R. Mechanisms of activation of the paternally expressed genes by the Prader-Willi imprinting center in the Prader-Willi/Angelman syndromes domains. Proc Natl Acad Sci U S A. 2012;109:7403–7408. doi: 10.1073/pnas.1116661109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapkins RW, Hore T, Smithwick M, Ager E, Pask AJ, Renfree MB, Kohn M, Hameister H, Nicholls RD, Deakin JE, et al. Recent assembly of an imprinted domain from non-imprinted components. PLoS genetics. 2006;2:e182. doi: 10.1371/journal.pgen.0020182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Jato S, Nicholls RD, Driscoll DJ, Yang TP. Characterization of cis- and trans-acting elements in the imprinted human SNURF-SNRPN locus. Nucleic Acids Res. 2005;33:4740–4753. doi: 10.1093/nar/gki786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner F, Gohlke B, Hamm M, Korsch E, Woelfle J. MKRN3 Mutations in Familial Central Precocious Puberty. Horm Res Paediatr. 2014;82:122–126. doi: 10.1159/000362815. [DOI] [PubMed] [Google Scholar]
- Schwabe JW, Klug A. Zinc mining for protein domains. Nat Struct Biol. 1994;1:345–349. doi: 10.1038/nsb0694-345. [DOI] [PubMed] [Google Scholar]
- Semple RK, Topaloglu AK. The recent genetics of hypogonadotrophic hypogonadism - novel insights and new questions. Clin Endocrinol (Oxf) 2010;72:427–435. doi: 10.1111/j.1365-2265.2009.03687.x. [DOI] [PubMed] [Google Scholar]
- Settas N, Dacou-Voutetakis C, Karantza M, Kanaka-Gantenbein C, Chrousos GP, Voutetakis A. Central Precocious Puberty in a Girl and Early Puberty in her Brother Caused by a Novel Mutation in the MKRN3 Gene. J Clin Endocrinol Metab. 2014:jc20134084. doi: 10.1210/jc.2013-4084. [DOI] [PubMed] [Google Scholar]
- Shemer R, Hershko AY, Perk J, Mostoslavsky R, Tsuberi B, Cedar H, Buiting K, Razin A. The imprinting box of the Prader-Willi/Angelman syndrome domain. Nat Genet. 2000;26:440–443. doi: 10.1038/82571. [DOI] [PubMed] [Google Scholar]
- Silveira LG, Noel SD, Silveira-Neto AP, Abreu AP, Brito VN, Santos MG, Bianco SD, Kuohung W, Xu S, Gryngarten M, et al. Mutations of the KISS1 gene in disorders of puberty. J Clin Endocrinol Metab. 2010;95:2276–2280. doi: 10.1210/jc.2009-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teles MG, Bianco SD, Brito VN, Trarbach EB, Kuohung W, Xu S, Seminara SB, Mendonca BB, Kaiser UB, Latronico AC. A GPR54-activating mutation in a patient with central precocious puberty. N Engl J Med. 2008;358:709–715. doi: 10.1056/NEJMoa073443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PH, Cheung WK, Peng Y, He ML, Wu GQ, Xie D, Jiang BH, Huang QH, Chen Z, Lin MC, et al. Makorin-2 is a neurogenesis inhibitor downstream of phosphatidylinositol 3-kinase/Akt (PI3K/Akt) signal. J Biol Chem. 2008;283:8486–8495. doi: 10.1074/jbc.M704768200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias L, Wurtman RJ. Age at menarche. Genetic and environmentalinfluences. N Engl J Med. 1969;280:868–875. doi: 10.1056/NEJM196904172801606. [DOI] [PubMed] [Google Scholar]