Summary
The evolutionary history of tumor cell populations can be reconstructed from patterns of genetic alterations. In contrast to stable genetic events, epigenetic states are reversible and sensitive to the microenvironment, prompting the question whether epigenetic information can similarly be used to discover tumor phylogeny. We examined the spatial and temporal dynamics of DNA methylation in a cohort of low-grade gliomas and their patient-matched recurrences. Genes transcriptionally upregulated through promoter hypomethylation during malignant progression to high-grade glioblastoma were enriched in cell cycle function, evolving in parallel with genetic alterations that deregulate the G1/S cell cycle checkpoint. Moreover, phyloepigenetic relationships robustly recapitulated phylogenetic patterns inferred from somatic mutations. These findings highlight widespread co-dependency of genetic and epigenetic events throughout brain tumor evolution.
INTRODUCTION
Cancers develop through a process of clonal evolution in which ongoing genetic and epigenetic diversification allows for repeated cycles of subclonal selection and expansion (Greaves and Maley, 2012; Nowell, 1976). As a result, human tumors can display substantial intratumoral heterogeneity, including discordant genetic alterations between initial tumors and their associated local recurrences or distant metastases (Gerlinger et al., 2012; Okosun et al., 2014; Wu et al., 2012; Yachida et al., 2010). While genomic profiling of spatially or temporally separated tumor samples can be used to reconstruct the evolutionary history and underlying clonal architectures of individual tumors (Gerlinger et al., 2014), this view is incomplete without a parallel analysis of the heterogeneity and evolution of the epigenome, an approach only rarely attempted (Brocks et al., 2014; Oakes et al., 2014).
In low-grade glioma, the course of tumor evolution is particularly clinically significant. World Health Organization (WHO) grade II gliomas (low-grade gliomas) are diffuse, infiltrative tumors that frequently recur and may unpredictably undergo malignant progression to a higher grade with a worse prognosis (Sanai et al., 2011). Recurrences that progress to highly malignant WHO grade IV glioblastoma (GBM) acquire genetic alterations in the RB and AKT-mTOR pathways (Cancer Genome Atlas Research Network, 2008; Johnson et al., 2014; Louis, 2006). In fact, adjuvant treatment with alkylating chemotherapeutics such as temozolomide (TMZ) can induce hypermutation that emerges in recurrent tumors (Hunter et al., 2006), and we recently linked treatment-associated driver mutations in these two pathways to malignant progression of grade II glioma to GBM (Johnson et al., 2014). It remains unknown, however, how epigenetic alterations contribute to the different courses of evolution of low-grade gliomas and how or if they relate to concurrent mutational evolution.
The critical role that epigenetic alterations play in the development and therapeutic response of gliomas is increasingly being appreciated (Fouse and Costello, 2009). Epigenetic mechanisms can alter gene expression, and have been shown to affect tumor suppressors and oncogenes in gliomas (Baeza et al., 2003; Costello et al., 1996; Kim et al., 2006; Nagarajan et al., 2014; Nakamura et al., 2001; Wiencke et al., 2007). Somatic mutations in IDH1 or IDH2 may be the first genetic driver in the development of many low-grade gliomas (Johnson et al., 2014; Lai et al., 2011; Watanabe et al., 2009). Genetic mutations in IDH genes induce a pattern of early epigenetic alterations known as the glioma CpG island methylator phenotype (G-CIMP) characterized by extensive remodeling of the DNA methylome (Hill et al., 2014; Noushmehr et al., 2010; Toyota et al., 1999; Turcan et al., 2012). The inactivation of other genes mutated in low-grade gliomas, such as ATRX (Jiao et al., 2012) and SMARCA4 (Johnson et al., 2014), is known to induce specific DNA methylation changes as well (Banine et al., 2005; Gibbons et al., 2000). Of clinical importance is DNA hypermethylation of the MGMT promoter, which is associated with loss of SP1 binding, closed chromatin and transcriptional silencing in GBM cells (Costello et al., 1994a; Costello et al., 1994b) and increased survival in GBM patients treated with TMZ (Hegi et al., 2005). Whether the DNA methylation status at this locus predicts the same survival benefit in low-grade glioma patients is unclear (Everhard et al., 2006; Kesari et al., 2009; Taal et al., 2011; van Thuijl et al., 2015; Wick et al., 2013). Although there has been extensive characterization of tumor methylomes using a single sampling per tumor, little is known about intratumoral heterogeneity at the epigenetic level or of temporal evolution of the low-grade glioma methylome and its relationship to the genome. An integrated model of the genomic and epigenomic evolutionary trajectory of initially low-grade gliomas may suggest strategies for delaying or treating recurrent disease, identify biomarkers for predicting the clinical course of a low-grade glioma, and also shed light on dynamic relationships between the genome and epigenome in other cancer types.
RESULTS
We profiled the DNA methylomes of 19 clinically-annotated initial grade II gliomas and their patient-matched recurrences (Table S1) using the Illumina HumanMethylation450 bead array (Illumina 450K) (Figure S1A). We also performed transcriptome sequencing on the initial and recurrent tumors of 13 patients (Table S2). All gliomas profiled here are IDH1-mutant (Johnson et al., 2014; van Thuijl et al., 2015) and are therefore expected to possess the characteristic methylation patterns associated with G-CIMP (Lu et al., 2012; Noushmehr et al., 2010; Turcan et al., 2012). From these methylation array data we confirmed that G-CIMP was present in all initial tumors and always maintained at recurrence (Figure S1B), highlighting that these epigenetic changes arise very early and are potentially tumor-initiating.
Patterns of DNA methylation are patient-specific and evolve in a manner specific to the grade of recurrence
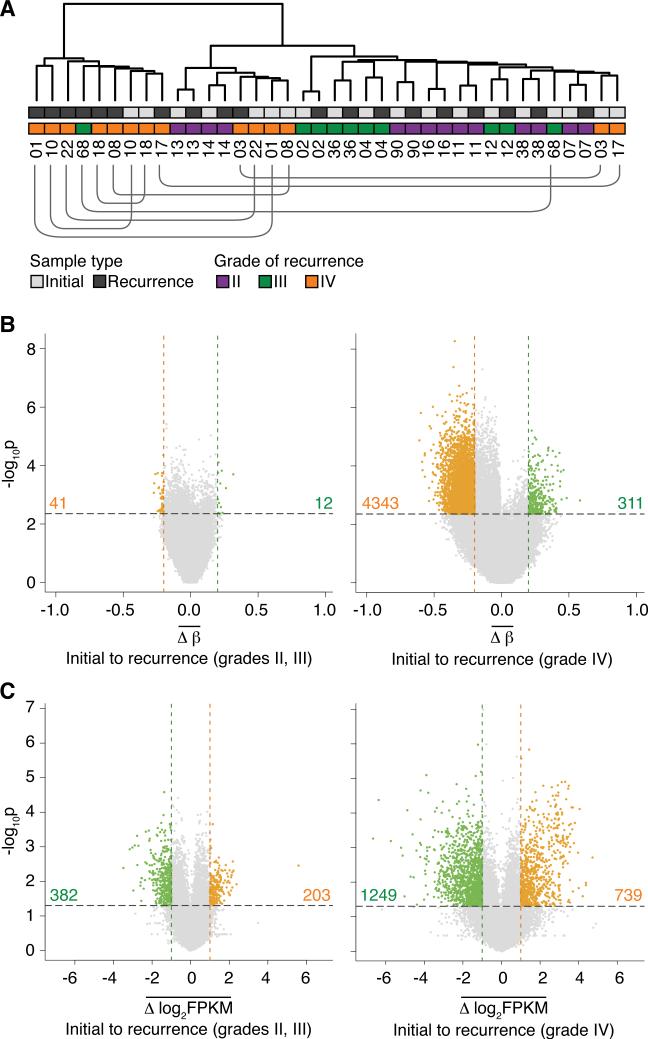
To determine the extent to which these tumors had altered methylomes beyond the ubiquitous G-CIMP methylation patterns, we identified the most variable CpG sites across all initial and recurrent gliomas and performed unsupervised hierarchical clustering. Initial and recurrent tumors from the same individual clustered together (Figure S1C). This result reflects patient-specific methylation patterns, consistent with a previous report on glioma (Laffaire et al., 2011), and may be indicative of normal inter-individual epigenetic variation, patient-specific aberrant methylation from early stages of gliomagenesis, or both. Within the clustering, six of the seven patients who recurred with GBM formed a distinct subgroup, suggesting there may be a shared methylation pattern associated with malignant progression to GBM relative to a lower grade of recurrence. To further evaluate this pattern, we performed unsupervised clustering with progressively more lenient selections of variable CpG sites to discover additional global DNA methylation patterns. At intermediate cutoffs, a gradual switch in clustering patterns was evident (Figure S1D). At the most lenient cutoff, the methylation patterns separated GBM recurrences, as well as two initial tumors that recurred as GBM, from the grade II and III gliomas (Figure 1A). This further supports a GBM recurrence-specific methylation pattern and suggests extensive evolution of the methylome during malignant progression to GBM (Figure S1E). This unique pattern of epigenome evolution was prominent across GBM recurrences that arose in the absence of adjuvant therapy as well as in GBMs that arose in a treatment-associated manner, adding to our prior genetic findings that spontaneous and treatment-associated progression to GBM have convergent genetic alterations (Johnson et al., 2014). Interestingly, clustering of the transcriptome segregated some of the grade III recurrences with GBM samples (Figures S1F and S1G), indicating transcriptional changes are complementary to, but not exclusively overlapping with, changes in the DNA methylome during malignant progression. Thus, integrating the methylome and transcriptome may provide important insight into the functional epigenetic events that underlie malignant progression to GBM.
Figure 1. Evolutionary dynamics of the methylome and transcriptome in initial and recurrent glioma pairs.
(A) Unsupervised hierarchical clustering of the top 50% most variable CpG sites. Annotations of sample type, grade of recurrence, and patient identification numbers are provided. The lines beneath the patient identification numbers connect initial and recurrent tumors from the same patient that are not adjacent to each other.
(B) The average methylation change from initial low-grade tumor to recurrence at each CpG site measured in patients that do not (left) or do (right) undergo malignant progression to GBM (grade IV). Colored dots represent CpG sites that show significant hypomethylation (orange dots, total count provided) or hypermethylation (green dots, total count provided) at recurrence (p valueadjust < 0.05 and |Δβ| > 0.2).
(C) Average gene-level expression changes from initial to recurrence in patients that do not (left) or do (right) undergo malignant progression to GBM. Significantly differentially expressed genes are highlighted in green (down-regulated at recurrence, total count provided) and orange (up-regulated at recurrence, total count provided) (p value < 0.05 and |Δlog2FPKM| > 1).
See also Figure S1 and Tables S1 and S2.
Identification of CpGs that lose methylation specifically during malignant progression to GBM
We next examined changes in the methylome and transcriptome to determine whether there is a signature of methylation or expression changes associated with recurrence. We calculated the change in methylation (β value, methylated fraction at a CpG site) from initial to recurrent tumor at each CpG site in each patient, and then identified CpG sites with consistent methylation changes upon recurrence across all patients. This powerful intra-patient approach controls for differences in DNA methylation that are age-related or reflect germline genetic effects, which confound inter-patient comparisons. DNA methylation differences between normal brain and glioma may be aberrant events in the tumor, or may reflect differences between the normal brain tissue sample and the methylation patterns of the tumor's cell of origin (Sproul et al., 2012; Witte et al., 2014). In contrast, the differences we report between initial and recurrent tumors are more likely to be aberrant changes attributable to tumor progression rather than cell of origin. We also applied an equivalent model to the transcriptome sequencing data and identified genes that commonly increase or decrease in expression from initial to recurrent glioma (Figure S1H). The separation by grade in the methylation clustering suggested that a specific pattern of DNA methylation changes may underlie malignant progression to GBM. To discover this pattern in detail, we stratified patients by grade of recurrence. There were few common methylation changes evident in patients that recurred at grade II or III, whereas a strong pattern of hypomethylation was associated with malignant progression to GBM (Figures 1B, 1C and S1I). Patients that recurred at grade II or III were combined into a single group for further analysis.
To determine which methylation changes were specific to recurrence as GBM, we compared the change in methylation from initial to recurrence in patients who recurred as GBM versus those that recurred at grades II or III. We identified 1,953 CpG sites that were specifically hypomethylated upon recurrence as GBM (Figure 2A and Table S3). Given the G-CIMP-associated hypermethylation in these tumors, we first set out to determine if the hypomethylation in GBM recurrences affected G-CIMP genes. Noushmehr et al. identified 50 genes that were hypermethylated and downregulated in a G-CIMP specific manner. Only two of those genes (ACSS3 and RAB36) showed GBM-specific hypomethylation, but in neither case did the genes show concurrent decreased expression. Further examination of these sites of decreasing methylation revealed a surprising enrichment for CpG sites that undergo age-related increased methylation in a comparison of normal fetal and adult brain (odds ratio 4.64, p value < 10-4, permutation test; see Supplemental Experimental Procedures). This is contrary to the typical pattern in cancer in which CpG sites that are hypermethylated during aging are also hypermethylated in cancer (Issa et al., 1994; Toyota et al., 1999). To further investigate whether the methylation changes alter gene regulation, we integrated active regulatory regions defined from histone H3K4me3, H3K4me1 and H3K27ac ChIP-seq in adult normal brain and primary GBM tissue (Figure S2A and Table S2) and found that sites of GBM-specific DNA hypomethylation were enriched for candidate active enhancers (odds ratio 1.68, p value < 10-4, permutation test). These hypomethylated loci thus may have gene regulatory effects. To enrich for functional methylation changes and exclude passenger events, we next integrated our transcriptome sequencing data with the DNA methylation analysis.
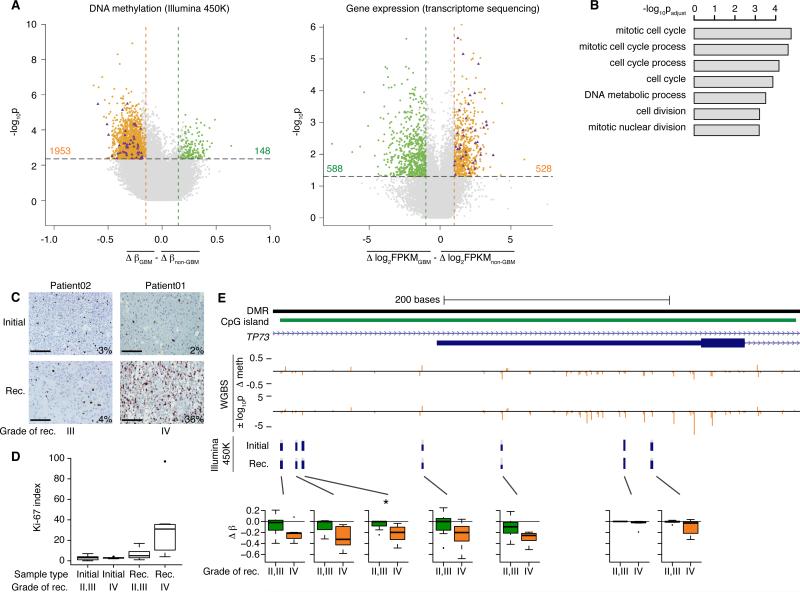
Figure 2. Cell cycle genes are hypomethylated and over-expressed specifically upon recurrence as GBM, coordinately with an increase in actively cycling cells.
(A) Left panel shows a scatter plot of differences between GBM and non-GBM recurrent tumors in methylation changes from initial grade II to recurrent gliomas. Right panel shows an equivalent representation of differences in expression changes between GBM and non-GBM recurrent tumors. Colored points indicate significant differences. Purple triangles highlight genes that become hypomethylated at promoter CpGs (left) and over-expressed (right) during malignant progression to GBM.
(B) Barplot of the top results of a gene ontology analysis of genes that are both significantly hypomethylated and over-expressed specifically upon recurrence as GBM.
(C) Representative staining for Ki-67 in a patient that recurred at grade III (left) and a patient that recurred at grade IV (right). Bars represent 100 μm.
(D) Boxplot representing the Ki-67 labeling index of tumors in the cohort (n=16 patients), subdivided by grade of recurrence (p value = 0.026, two-sided Wilcoxon rank sum test between GBM recurrences and recurrences at grades II or III). The box encompasses data points between the first and third quartiles, with a horizontal line indicating the median value. Whiskers extend to 1.5 × interquartile range, and any data points beyond that range are shown as individual dots.
(E) Whole-genome shotgun bisulfite sequencing data (WGBS) of Patient01 across an intragenic CpG island in the TP73 locus. From top to bottom, tracks represent: a differentially methylated region (DMR) reported in primary GBM (Nagarajan et al., 2014); CpG island; TP73 full-length and truncated transcripts; change in methylation level from initial to recurrent tumor by WGBS; statistical significance of the WGBS methylation changes, where positive values indicate hypermethylation at recurrence and negative values indicate hypomethylation; methylation levels from Illumina 450K array in Patient01 at the seven CpG sites assayed on the array. Box plots present the methylation change in all patients in the cohort across the same seven CpG sites. Boxplots are drawn as in panel D.
See also Figure S2 and Tables S2, S3 and S4.
Cell cycle genes are specifically hypomethylated upon malignant progression
We applied an analysis similar to that of the methylation data and identified 528 genes with GBM-specific over-expression (Figure 2A). Of these, 39 genes showed GBM-specific hypomethylation of at least one CpG site within their promoter regions (Figures S2B, S2C and Table S3). Among genes with GBM-specific promoter hypermethylation, only NTSR2 showed consistent transcriptional downregulation. We additionally identified four genes with consistent downregulation and gene body hypomethylation (Table S3). Strikingly, the set of 39 hypomethylated and over-expressed genes was significantly enriched for cell cycle genes (Figure 2B and Table S4). Ki-67 is a marker of actively proliferating cells, and staining in initial and recurrent tumors confirmed that a statistically significantly higher fraction of cells (p value = 0.026, two-sided Wilcoxon rank sum test) were actively proliferating among the GBM recurrences (Figures 2C and 2D). Increased proliferation is a hallmark of GBM. These results thus highlight an epigenetic mechanism that may contribute to increased proliferation, concurrent with genetic alterations in key members of the RB pathway (Johnson et al., 2014) that abrogate the G1/S cell cycle checkpoint.
Among the epigenetically modified cell cycle genes, we noted that the hypomethylation in TP73 was at an internal gene body promoter. Indeed, TP73 possesses a gene body CpG island (CGI) that we identified recently as recurrently hypomethylated in primary GBM (Nagarajan et al., 2014). The gene body CGI spans the transcription start site of a truncated, oncogenic form of TP73 (ΔNp73), which is correspondingly expressed in primary GBM. Similarly, increased expression (Figure S2D) and hypomethylation of ΔNp73 was observed only in GBM recurrences. Due to the limited resolution of the Illumina 450K array, we identified only one significantly hypomethylated CpG site in this gene body CGI specifically upon recurrence as GBM, although several nearby CpG sites showed a similar trend. To gain greater resolution across the full CGI and other regions genome-wide, we examined our whole-genome shotgun bisulfite sequencing (WGBS) data on the initial and recurrent tumors of Patient01 (Figure 2E and Table S2). These data show that the pattern of hypomethylation indeed extends across the local genomic region. Among other hypomethylated and over-expressed genes, significant probes from array-based data were similarly indicative of a local effect including multiple CpG sites (Figures S2E and S2F), consistent with previous literature showing that the methylation levels of CpG sites within 1kb are highly correlated (Eckhardt et al., 2006).
The functional effect of DNA hypomethylation of cell cycle genes specifically upon recurrence as GBM parallels the known GBM-specific genetic events that inactivate the G1/S cell cycle checkpoint (Cancer Genome Atlas Research Network, 2008; Johnson et al., 2014; Louis, 2006). These convergent genetic and epigenetic signals, in addition to the well-characterized functional relationships between genetic and epigenetic aberrations (Banine et al., 2005; Gibbons et al., 2000; Kerkel et al., 2008; Lu et al., 2012; Turcan et al., 2012), prompted us to explore evolutionary relationships among different tumor cell populations within a tumor, as has been previously done with genetic data, and then compare the relationships inferred from DNA methylation to those inferred from somatic mutation in the same samples.
Reconstruction of tumor evolution from intratumoral and longitudinal DNA methylation patterns
We first examined the evolutionary relationships of tumor samples that were previously genetically characterized (Johnson et al., 2014). We performed methylation profiling of seven spatially distinct pieces of tumor tissue from Patient17, three from the initial tumor and four from the recurrent tumor, and built a phyloepigenetic tree (Figure 3A, left; Table S3). The phyloepigenetic tree presented an intriguing model with early divergence between the initial and recurrent tumors, and more subtle divergences among the samples within each time point (initial A vs. initial B/C; recurrence A/C vs. recurrence B/D). We then used exome sequencing data of these same spatially distinct tumor samples to independently construct a phylogenetic tree (Figure 3A, right and Table S5) (Johnson et al., 2014). The genetically defined relationships among tumor cell clones were consistent with those determined from DNA methylation data. We quantified this similarity as the correlation between the distance matrices that were used to build the phyloepigenetic and phylogenetic trees (Spearman's rho = 0.90).
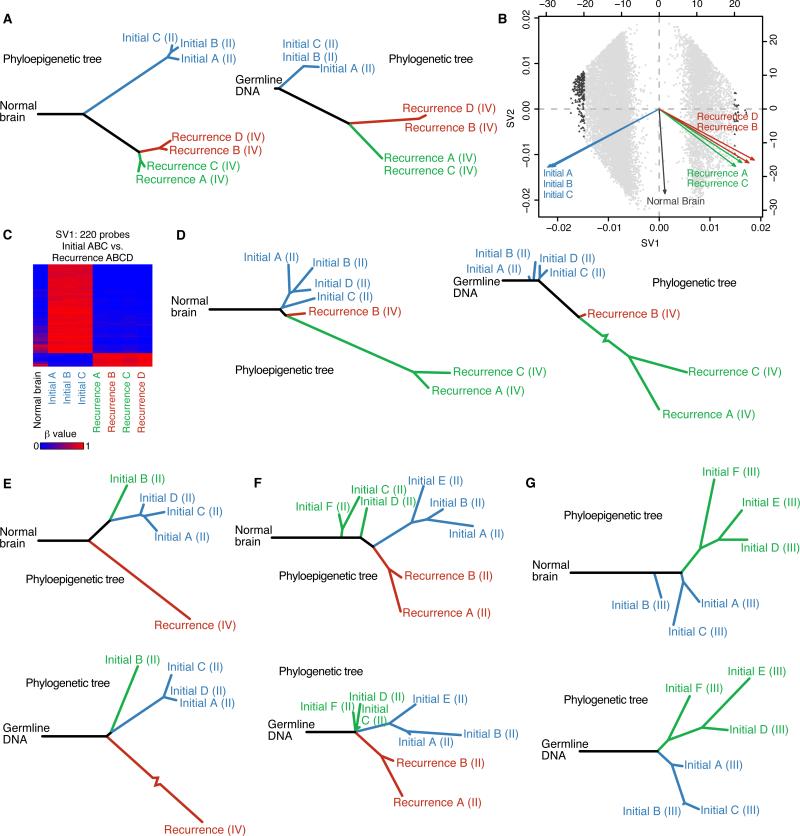
Figure 3. The spatial and temporal patterns of tumor evolution observed from DNA methylation dynamics and somatic mutations yield similar evolutionary histories.
(A) A phyloepigenetic tree constructed from seven samples from Patient17 (left) and a phylogenetic tree derived from somatic mutations from exome sequencing of the same DNA samples (right) (Spearman's rho = 0.90). Tumor grade is provided in parentheses after each sample name.
(B) Singular value decomposition biplot shows the probes involved in separating tumor samples. Each probe used to build the phyloepigenetic tree in (A) is plotted (grey dots). The most highly weighted probes are highlighted (triangles).
(C) A heatmap of the beta values at the 220 probes most highly weighted by SV1.
(D) A phyloepigenetic tree (left) and a phylogenetic tree (right) were constructed to infer the evolutionary relationships within and between the initial and recurrent tumors of Patient01 (Spearman's rho = 0.83). Tumor grade is provided in parentheses after each sample name.
(E-G) Phyloepigenetic (top) and phylogenetic trees (bottom) for Patient 18 (E, Spearman's rho = 0.90), Patient90 (F, Spearman's rho = 0.56) and Patient49 (G, Spearman's rho = 0.64). Tumor grade is provided in parentheses after each sample name. See also Figure S3 and Tables S3, S4, S5 and S6.
To identify the CpG sites underlying each branch point in the phyloepigenetic tree, we applied singular value decomposition to the methylation data from each patient to weigh the influence of individual CpG sites on separating particular subsets of samples (Figure S3A). For Patient17, the first singular vector (SV1), which accounts for the most methylation variability, mimicked the first major branch point of the phyloepigenetic tree (Figure 3B). We then selected the most influential CpGs for each singular vector and inferred that these underlie a particular branch point. The most highly weighted CpG sites within SV1 from Patient17 clearly showed differential methylation between the initial and recurrent tumor samples (Figure 3C). We examined the potential implications of these methylation changes by focusing on those affecting active promoters and enhancers in normal brain and primary GBM tissue and performed a gene ontology enrichment analysis. For Patient17, the CpG sites that underlie the first major branch point were enriched for a variety of developmental, biosynthetic and metabolic processes, indicating that methylation changes during tumor progression may influence cellular metabolic states, in parallel with the genetic events disrupting cell cycle that separate these two main branches on the phylogenetic tree (Table S4).
We then looked specifically at the evolutionary relationships of tumor samples from patients that underwent chemotherapy-associated malignant progression (Johnson et al., 2014; van Thuijl et al., 2015). We performed methylation profiling of four spatially distinct pieces of the initial tumor and three pieces of recurrent tumor from Patient01 and inferred a phyloepigenetic tree (Figure 3D, left; Figure S3B and Table S3). While the four pieces of the initial tumor clustered together, the recurrent tumor consisted of two distinct populations. Recurrence B was relatively closely related to the initial tumor, while a long branch separated it from recurrences A and C, indicating significant evolutionary distance. A phylogenetic tree from these same tumor pieces (Figure 3D, right; Tables S2 and S5) similarly demonstrates the large evolutionary distance between recurrence B and recurrences A and C (Spearman's rho = 0.83). In the phylogenetic tree, this longest branch corresponds to the development of a hypermutated population in the recurrent tumor. Intriguingly, this same branch is the longest in the phyloepigenetic tree, indicating that the hypermutated cells also have the greatest methylation change. Similarly, in Patient18, the phyloepigenetic tree identified three epigenetically similar pieces of the initial tumor, a piece of the initial tumor that branched off at an earlier evolutionary time point, and a recurrence that diverged even earlier – relationships that are accurately recapitulated in the phylogenetic tree (Spearman's rho = 0.90) (Figures 3E, S3C, Tables S3 and S5). Thus, even in extreme evolutionary events such as chemotherapy-associated hypermutation, both DNA methylation changes and mutational landscapes encode similar tumor evolutionary relationships. In these two cases with TMZ-associated hypermutation (Figures 3D and 3E), the longest branch length in both the phyloepigenetic and phylogenetic trees is the hypermutated recurrence. These results suggest a potentially quantitative relationship between the number of mutations and epimutations in each tumor cell clone.
To determine if the strong correlations between phylogenetic and phyloepigenetic trees depend on the large-scale hypomethylation during malignant progression to GBM, we next compared the evolutionary relationships only in lower grade initial and recurrent tumors. Six pieces of tissue from the initial tumor and two pieces of tissue from the grade II recurrence from Patient90 were subjected to DNA methylation profiling. Construction of a phyloepigenetic tree revealed three distinct clusters of samples, with the initial tumor separating into two populations, and the recurrence forming a third (Figure 3F, top; Figure S3D and Table S3). We then performed exome sequencing of these same pieces of tissue to identify somatic mutations and constructed a phylogenetic tree (Figure 3F, bottom; Tables S2 and S5). This phylogenetic tree mirrored the evolutionary relationships defined from DNA methylation (Spearman's rho = 0.56). We further pursued this question with Patient49 who underwent a single resection for an initial tumor from which we profiled six spatially distinct pieces. Construction of a phyloepigenetic tree revealed that the six pieces separate into two groups, in agreement with the phylogenetic tree derived from exome sequencing of the same pieces of tissue (Spearman's rho = 0.64) (Figures 3G and S3E; Tables S2, S3 and S5). Thus, even in the absence of malignant progression to GBM, DNA methylation changes among tumor cell clones yielded a very similar evolutionary trajectory as was inferred from somatic mutations.
Enhanced model of tumor evolution derived from variation between phyloepigenetic and phylogenetic trees
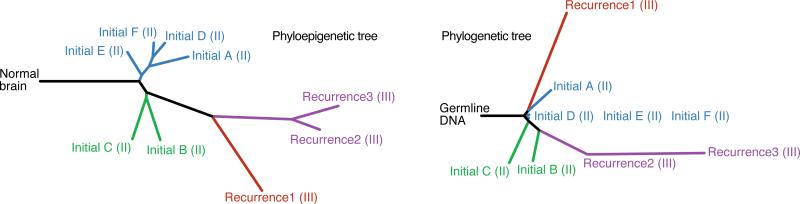
To further address phyloepigenetic relationships over time, we examined tumor samples from Patient04, who had four sequential surgical resections over five years. We profiled six spatially distinct pieces of tumor from the initial surgery, and one from each of the three subsequent surgeries for tumor recurrence. The phyloepigenetic tree reveals two distinct populations within the initial tumor and an evolutionary trajectory shared among the three recurrences, with a relatively closer relationship between recurrences 2 and 3 (Figure 4, left; Figure S4 and Table S3). The phylogenetic tree again reveals many similar clonal relationships, but also reveals differences that may be informative (Figure 4, right; Tables S2 and S5) (Spearman's rho = 0.78). Based on somatic mutations, the first recurrence shares evolutionary history with the initial tumor, while the second recurrence diverged earlier in the evolution of the tumor and therefore independently progressed to grade III (Johnson et al., 2014). Despite divergent genetic paths, methylation patterns are shared among the first recurrence and the second and third recurrences. This raises the possibility that the last common ancestor of the first and second recurrences was primed for progression with a set of DNA methylation changes required for progression to a higher grade. This case illustrates how differences in genetic and epigenetic phylogenies may bring to light an enhanced understanding of the evolution of a tumor.
Figure 4. Phyloepigenetic trees coupled with phylogenetic trees from a low-grade glioma patient with three recurrences reveal an enhanced understanding of evolutionary relationships.
Phyloepigenetic (left) and phylogenetic (right) trees of Patient04 present evolutionary relations across four surgical time points (Spearman's rho = 0.78). Tumor grade is provided in parentheses after each sample name.
See also Figure S4 and Tables S3, S5 and S6.
Gene-level genetic and epigenetic convergence
The common evolutionary histories defined from mutations and DNA methylation led us to examine if there was also convergence at the level of individual genes. We identified a small number of intra-patient single gene convergence events in which some samples from a patient had a mutation, while other samples which lack the mutation show differential methylation at the same gene (Table S6; see Supplemental Experimental Procedures). We also identified a small number of genes with inter-patient convergence. These are genes that are mutated in one patient but show methylation alteration in another patient (Table S6; see Supplemental Experimental Procedures). However, the vast majority of mutations and methylation changes occur in different sets of genes, consistent with our prior low-resolution analysis of gliomas (Zardo et al., 2002). In contrast to single genes, single pathways such as the cell cycle pathway are commonly altered by multiple genetic (Johnson et al., 2014) and epigenetic (Figure 2B) alterations within and across tumor samples.
DISCUSSION
DNA methylation patterns record a remarkable breadth of information about cells, including their chronological age, developmental history and differentiation potential. Here, we show that despite epigenome plasticity, chemotherapy, and the ubiquitous IDH1 mutation-driven G-CIMP pattern, patient-specific tumor phyloepigenetic analyses replicated and extended tumor phylogenetic analyses. From this striking result, we conclude that the precise chronological order of epigenetic changes, from initiating to late events, can be determined from intratumoral methylation patterns, thus surpassing prior binary categorization of epigenetic events as early or late. While our study is focused on methylation and somatic mutations in IDH1-mutant gliomas, a study of prostate cancer and prostate cancer metastasis showed a complementary unified model of evolution for DNA methylation and copy number alterations (Brocks et al., 2014). Thus, genomicepigenomic co-dependency may be a feature of multiple types of cancer, and may span somatic mutations, copy number, and DNA methylation.
The importance of epigenetic variation within individual human tumors is just beginning to be uncovered. Recent work in chronic lymphocytic leukemia suggests that stochastic changes in the methylome lead to increased heterogeneity, allowing for selection of more malignant epi-phenotypes coupled with an adverse clinical outcome (Landau et al., 2014). Somatic genetic events, like IDH1 mutations, have been directly linked to alterations in the methylome (Noushmehr et al., 2010; Turcan et al., 2012), while germline variants have been indirectly associated with specific DNA methylation patterns (Heyn et al., 2013; Kerkel et al., 2008; Shi et al., 2014). Consistent with these theories, the widespread correlation between somatic mutations and DNA methylation patterns suggests that in addition to IDH1 mutation and G-CIMP, other epigenetic patterns might be directly or indirectly induced by mutations, or vice versa. It will be of interest to determine the extent to which these findings hold for IDH1-wild-type low-grade gliomas and their recurrences.
We also discovered a convergence of genetic and epigenetic changes driving aberrant cell cycle function (Figure 5). We previously found that recurrent tumors that underwent malignant progression to GBM acquired somatic mutations in the RB pathway that inactivate the G1/S cell cycle checkpoint (Johnson et al., 2014). Here we identified a pattern of functional DNA hypomethylation specific to recurrence as GBM that alters cell cycle genes. This phenotypic convergence of genetic and epigenetic mechanisms on the same pathway underscores the importance of cell cycle deregulation on the process of malignant progression, while also raising questions about how these two processes might be connected. Of note, we identify hypomethylation at TP73 as a recurrent event. Transcription of TP73 is upregulated by E2F1 (Rufini et al., 2011), a transcription factor that itself activates cell cycle progression-related genes following inactivation of the RB pathway (Chen et al., 2009), which is deregulated by genetic mechanisms in these tumors. Further work will be required to deconvolute these relationships. By combining the information from somatic mutations, copy number alteration and DNA methylation patterns, we derived a comprehensive model of glioma evolution (Figure 5). Chronological ordering of IDH1, TP53, and ATRX mutations and copy number alterations was derived from our previous tumor phylogenetic analyses (Johnson et al, 2014), other studies (Lai et al., 2011; Watanabe et al., 2009), and additional data presented here. This model is derived from a total of 32 patients with paired initial and recurrent samples and includes 70 DNA methylation profiles, 26 mRNA expression profiles and 130 exome sequencing profiles. The model extends from the initiating genetic and epigenetic lesions and captures clinically divergent paths at recurrence, including an evolutionary path driven by treatment.
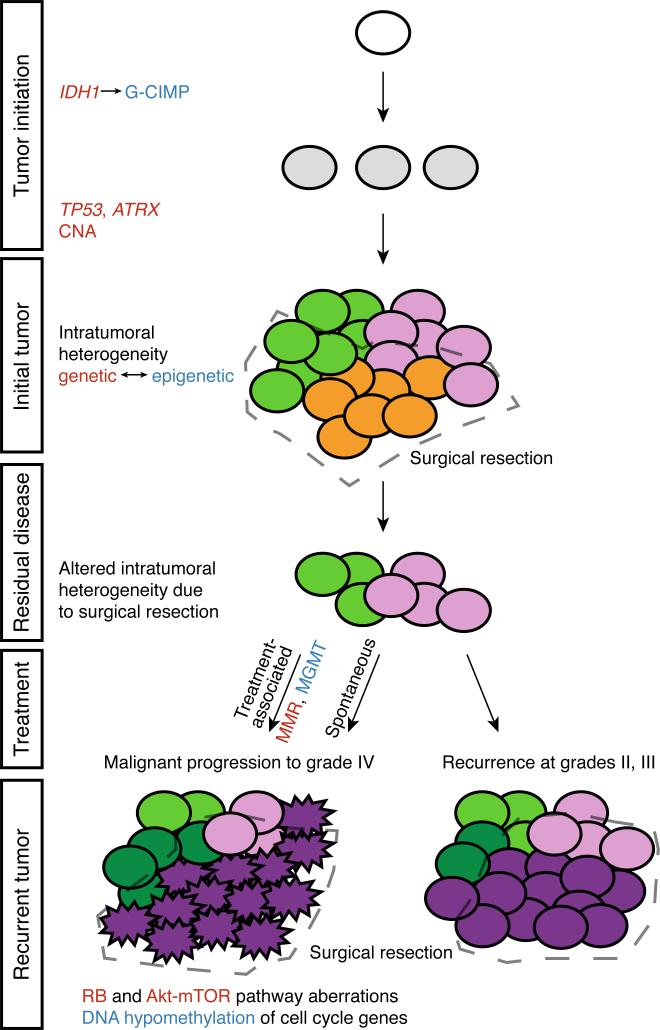
Figure 5. A genomic and epigenomic co-dependency model of clonal evolution.
Low-grade gliomas exhibit intratumoral heterogeneity at initial presentation, with subclones that share the initiating genetic (IDH1 followed by TP53 and ATRX and copy number alterations, CNA) and epigenetic (IDH1-associated glioma CpG island methylator phenotype, G-CIMP) alterations, but further develop distinct genetic and epigenetic characteristics. Following surgical resection, the outgrowth from residual disease may be grade II or III, while still continuing to evolve subclones with genetic and co-dependent epigenetic features that are distinct from the initial tumor. In other patients, residual disease may undergo malignant progression to GBM, either spontaneously or as a consequence of treatment-associated mutations, in either case acquiring genetic defects in the RB and Akt-mTOR pathways and promoter hypomethylation and activation of cell cycle genes. Treatment associated progression to GBM is uniquely associated with an increased epigenetic silencing of MGMT (van Thuijl, 2015) and acquisition of genetic defects in mismatch repair genes.
These findings underscore the power of integrated genetic and epigenetic analyses of tumors. Deregulated cell cycle control is among the essential phenotypes of cancer cells, and we demonstrate that this deregulation is encoded in both the genome and epigenome, raising the question of the extent to which this reflects a functional interaction between genetics and epigenetics. This finding also raises the possibility that other critical molecular phenotypes, such as genomic instability, angiogenesis or invasion may leave their imprint on DNA methylation patterns during tumor evolution.
EXPERIMENTAL PROCEDURES
Sample acquisition
Flash frozen tissue was acquired from patients undergoing surgical resection for glioma. Samples were obtained from the Neurosurgery Tissue Bank at the University of California San Francisco (UCSF). Sample use was approved by the Committee on Human Research at UCSF, and research was approved by the institutional review board (IRB) at UCSF. Additional samples were obtained from Erasmus Medical Center with the approval of the Medical Ethics Committee at Erasmus Medical Center Rotterdam and the OncoNeuroTheque tissue bank at Groupe Hospitalier Pitié-Salpêtrière with the approval of the Ethics Committee. All patients provided informed written consent. Genomic DNA was extracted, and where tissue availability was sufficient and high-quality RNA was obtained, strand-specific transcriptome sequencing was also performed. Complete details are provided in Supplemental Experimental Procedures.
DNA methylation analysis
Genomic DNA from 70 samples was bisulfite converted using the EZ DNA Methylation Kit (Zymo Research) and processed on Infinium HumanMethylation450 bead arrays (Illumina Inc.) according to the manufacturer's protocol. Probe-level signals for individual CpG sites were subject to both background and global dye-bias correction (Triche et al., 2013). Probes that map to regions with known germline polymorphisms, to multiple genomic loci (Price et al., 2013), or to either sex chromosome were filtered out. Descriptions of further analyses are provided in Supplemental Experimental Procedures.
Transcriptome sequencing analysis
Strand-specific transcriptome sequencing libraries were prepared as previously described (Johnson et al., 2014). All transcriptome sequencing data from initial and recurrent tumor pairs were aligned with TopHat (v2.0.12) (Trapnell et al., 2009) to the hg19 reference genome using a GENCODE V19 transcriptome-guided aligment. The aligned data were then processed through custom quality-control scripts to remove unmapped, improperly-matched, multi-mapping, and chimeric reads, as well as accumulation in non-assembled chromosomes. To estimate transcript abundance, aligned data were processed with the cuffnorm and cuffquant commands from the Cufflinks package (v2.2.1) (Trapnell et al., 2010). Further details on process and statistical analysis are provided in Supplemental Experimental Procedures.
Exome sequencing and mutation identification
Exome capture was performed using either Agilent or NimbleGen exome capture kits on 48 samples, 22 of which were previously published (Johnson et al., 2014; van Thuijl et al., 2015). All sequencing reported here acquired paired-end reads from Illumina HiSeq instrumentation. Exome alignment and mutation calling were performed as previously described (Johnson et al., 2014). To generate a list of only the highest quality variants for phylogenetic tree construction, further filtering was applied by excluding all SNVs that were not classified as “covered” by MuTect in all samples for that patient, SNVs with any variant reads detected in the patient-matched normal and all indels, unless validated by Sanger sequencing.
Construction of phylogenies
For the phylogeny analysis of both the genetic and epigenetic data, we employed an independent, but parallel, analysis of the methylation data and exome-seq mutations. For the exome-seq data, we used binary mutation calls to build a distance matrix for all samples from a patient using the Manhattan distance metric, including a normal tissue sample for which all mutations were absent. Similarly, for the methylation data, we used only the probes that had a beta value difference of at least 0.4 between any of the samples from a patient to build a Euclidean distance matrix. Using several other probe selection cut-offs produced similar results. A normal brain sample (adult insula tissue from a different individual) was not included in the probe selection, but was added to the distance matrix calculation to serve as the tree root. To compare the distance matrices from the mutation data and the methylation data, we calculated the Spearman's rho correlation. We then built the phylogeny trees using an ordinary least squares (OLS) minimum evolution (Desper and Gascuel, 2002) approach from the ape R package (Paradis et al., 2004) using the distance matrices from the genetic and epigenetic data independently.
Identifying discriminative methylation probes by Singular Value Decomposition
To identify the probes most responsible for a particular bifurcation on a phyloepigenetic tree (similar to identifying mutations that differ between two branches of a phylogenetic tree), we used Singular Value Decomposition (SVD) of the methylation data matrix to calculate the left and right singular vectors that form orthonormal bases of the subspaces spanned by the columns and rows of the data matrix, respectively. Projecting the columns, corresponding to samples, onto the two-dimensional subspace spanned by the first two left singular vectors (SV) reduces the data to the first two principal components that maximally separate the samples in the probe space. These projections are shown as arrows in Figure 3B, where the rows of the first two left SVs are plotted as scatter points representing probes. In this biplot, the probes that best separate samples have large absolute values in the SV1 direction. Additional details are provided in the Supplemental Experimental Procedures.
Supplementary Material
Significance.
Deciphering the evolutionary history of a tumor illuminates the sequence of events that occurred in tumorigenesis prior to diagnosis. The earliest events may provide ideal targets for precision therapeutic approaches, as these alterations are present in nearly all cells of a tumor. Here, we show that spatial and temporal patterns of either reversible DNA methylation or irreversible somatic mutations produce remarkably similar evolutionary histories. Phenotypically, mutations and promoter region DNA hypomethylation converge to deregulate the cell cycle as indolent low-grade tumors progress to high-grade malignancies. This study suggests strong interdependency of genetic and epigenetic alterations in these human brain tumors.
ACKNOWLEDGMENTS
The authors would like to acknowledge R. Nagarajan and S. Fouse for critical discussions; D. Johnson for assistance with mutation validation; N. Jabado and S. Papillon-Cavanagh for providing raw data for methylation analysis; K. Pollard and her lab for advice on phyloepigenetic evolution analyses. This project was generously supported by Accelerate Brain Cancer Cure and a gift from the Dabbiere family. Additional support by the National Institute Of General Medical Sciences T32GM008568 (T.M.), T32GM067547 (A.P.); the National Science Foundation 1144247 (A.P.); the National Institutes of Health 1T32CA15102201 (M.B.), R25NS070680 (M.B.), R01CA169316 (B.S.T and J.F.C.), and P50CA097257 (J.J.P., B.S.T., A.M.M., S.M.C., and J.F.C.); the National Cancer Institute P30CA82103 (A.B.O.), and R01CA163336 (J.S.S.); the Sontag Foundation (B.S.T., and J.S.S.); OncoNeuroThèque (A.I., and A.A.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
T.M. and J.F.C. designed the study and coordinated all aspects of the experiments, analyses and data interpretation. T.M., B.E.J., C.H., A.I., A.A., J.J.K., M.L.M.L., and S.M.C. coordinated sample acquisition and DNA and RNA isolation. T.M., B.E.J., C.H., and E.G.H. validated candidate mutations. T.M., A.P., B.E.J, C.H., R.J.A.B., I.V.S., B.S.T., and J.S.S. generated, analyzed, and interpreted sequencing data. T.M., A.P., and A.B.O. generated, analyzed, and interpreted Illumina 450K data. A.P., B.S.T., A.M.M., A.B.O., and J.S.S. contributed to statistical analyses and computation. G.F.R., J.J.P., M.B., and A.W.B. performed histopathology and immunohistochemistry analysis. B.S.T., A.M.M., A.B.O., S.M.C., and J.S.S. provided scientific advice. J.S.S provided oversight of statistical analyses. T.M., A.P., B.E.J., J.S.S, and J.F.C. wrote the manuscript.
ACCESSION NUMBERS
All sequencing and array data is deposited in the European Genome-Phenome Archive under accession codes EGAS00001000579, EGAS00001000685 and EGAS00001001255.
SUPPLEMENTAL INFORMATION
Supplemental information includes Supplemental Experimental Procedures, four figures and six tables and can be found with this article online.
The authors declare no competing financial interests.
REFERENCES
- Baeza N, Weller M, Yonekawa Y, Kleihues P, Ohgaki H. PTEN methylation and expression in glioblastomas. Acta Neuropathol. 2003;106:479–485. doi: 10.1007/s00401-003-0748-4. [DOI] [PubMed] [Google Scholar]
- Banine F, Bartlett C, Gunawardena R, Muchardt C, Yaniv M, Knudsen ES, Weissman BE, Sherman LS. SWI/SNF chromatin-remodeling factors induce changes in DNA methylation to promote transcriptional activation. Cancer research. 2005;65:3542–3547. doi: 10.1158/0008-5472.CAN-04-3554. [DOI] [PubMed] [Google Scholar]
- Brocks D, Assenov Y, Minner S, Bogatyrova O, Simon R, Koop C, Oakes C, Zucknick M, Lipka DB, Weischenfeldt J, et al. Intratumor DNA methylation heterogeneity reflects clonal evolution in aggressive prostate cancer. Cell reports. 2014;8:798–806. doi: 10.1016/j.celrep.2014.06.053. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9:785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello JF, Berger MS, Huang HS, Cavenee WK. Silencing of p16/CDKN2 expression in human gliomas by methylation and chromatin condensation. Cancer Res. 1996;56:2405–2410. [PubMed] [Google Scholar]
- Costello JF, Futscher BW, Kroes RA, Pieper RO. Methylation-related chromatin structure is associated with exclusion of transcription factors from and suppressed expression of the O-6-methylguanine DNA methyltransferase gene in human glioma cell lines. Mol Cell Biol. 1994a;14:6515–6521. doi: 10.1128/mcb.14.10.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello JF, Futscher BW, Tano K, Graunke DM, Pieper RO. Graded methylation in the promoter and body of the O6-methylguanine DNA methyltransferase (MGMT) gene correlates with MGMT expression in human glioma cells. J Biol Chem. 1994b;269:17228–17237. [PubMed] [Google Scholar]
- Desper R, Gascuel O. Fast and accurate phylogeny reconstruction algorithms based on the minimum-evolution principle. Journal of computational biology : a journal of computational molecular cell biology. 2002;9:687–705. doi: 10.1089/106652702761034136. [DOI] [PubMed] [Google Scholar]
- Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everhard S, Kaloshi G, Criniere E, Benouaich-Amiel A, Lejeune J, Marie Y, Sanson M, Kujas M, Mokhtari K, Hoang-Xuan K, et al. MGMT methylation: a marker of response to temozolomide in low-grade gliomas. Ann Neurol. 2006;60:740–743. doi: 10.1002/ana.21044. [DOI] [PubMed] [Google Scholar]
- Fouse SD, Costello JF. Epigenetics of neurological cancers. Future oncology (London, England) 2009;5:1615–1629. doi: 10.2217/fon.09.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, Fisher R, McGranahan N, Matthews N, Santos CR, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nature genetics. 2014 doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons RJ, McDowell TL, Raman S, O'Rourke DM, Garrick D, Ayyub H, Higgs DR. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nature Genetics. 2000;24:368–371. doi: 10.1038/74191. [DOI] [PubMed] [Google Scholar]
- Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegi ME, Diserens A-C, Gorlia T, Hamou M-F, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- Heyn H, Moran S, Hernando-Herraez I, Sayols S, Gomez A, Sandoval J, Monk D, Hata K, Marques-Bonet T, Wang L, Esteller M. DNA methylation contributes to natural human variation. Genome Res. 2013;23:1363–1372. doi: 10.1101/gr.154187.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill VK, Shinawi T, Ricketts CJ, Krex D, Schackert G, Bauer J, Wei W, Cruickshank G, Maher ER, Latif F. Stability of the CpG island methylator phenotype during glioma progression and identification of methylated loci in secondary glioblastomas. BMC cancer. 2014;14:506. doi: 10.1186/1471-2407-14-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C, Smith R, Cahill DP, Stephens P, Stevens C, Teague J, Greenman C, Edkins S, Bignell G, Davies H, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66:3987–3991. doi: 10.1158/0008-5472.CAN-06-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Killela PJ, Reitman ZJ, Rasheed AB, Heaphy CM, de Wilde RF, Rodriguez FJ, Rosemberg S, Oba-Shinjo SM, Nagahashi Marie SK, et al. Frequent ATRX, CIC, and FUBP1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3:709–722. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, Fouse SD, Yamamoto S, Ueda H, Tatsuno K, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkel K, Spadola A, Yuan E, Kosek J, Jiang L, Hod E, Li K, Murty VV, Schupf N, Vilain E, et al. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet. 2008;40:904–908. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- Kesari S, Schiff D, Drappatz J, LaFrankie D, Doherty L, Macklin EA, Muzikansky A, Santagata S, Ligon KL, Norden AD, et al. Phase II study of protracted daily temozolomide for low-grade gliomas in adults. Clin Cancer Res. 2009;15:330–337. doi: 10.1158/1078-0432.CCR-08-0888. [DOI] [PubMed] [Google Scholar]
- Kim TY, Zhong S, Fields CR, Kim JH, Robertson KD. Epigenomic profiling reveals novel and frequent targets of aberrant DNA methylation-mediated silencing in malignant glioma. Cancer Res. 2006;66:7490–7501. doi: 10.1158/0008-5472.CAN-05-4552. [DOI] [PubMed] [Google Scholar]
- Laffaire J, Everhard S, Idbaih A, Criniere E, Marie Y, de Reynies A, Schiappa R, Mokhtari K, Hoang-Xuan K, Sanson M, et al. Methylation profiling identifies 2 groups of gliomas according to their tumorigenesis. Neuro Oncol. 2011;13:84–98. doi: 10.1093/neuonc/noq110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A, Kharbanda S, Pope WB, Tran A, Solis OE, Peale F, Forrest WF, Pujara K, Carrillo JA, Pandita A, et al. Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J Clin Oncol. 2011;29:4482–4490. doi: 10.1200/JCO.2010.33.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau DA, Clement K, Ziller MJ, Boyle P, Fan J, Gu H, Stevenson K, Sougnez C, Wang L, Li S, et al. Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell. 2014;26:813–825. doi: 10.1016/j.ccell.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN. Molecular pathology of malignant gliomas. Annual review of pathology. 2006;1:97–117. doi: 10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan RP, Zhang B, Bell RJA, Johnson BE, Olshen AB, Sundaram V, Li D, Graham AE, Diaz A, Fouse SD, et al. Recurrent epimutations activate gene body promoters in primary glioblastoma. Genome Res. 2014 doi: 10.1101/gr.164707.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Yonekawa Y, Kleihues P, Ohgaki H. Promoter hypermethylation of the RB1 gene in glioblastomas. Lab Invest. 2001;81:77–82. doi: 10.1038/labinvest.3780213. [DOI] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell PC. The clonal evolution of tumor cell populations. Science (New York, NY) 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- Oakes CC, Claus R, Gu L, Assenov Y, Hüllein J, Zucknick M, Bieg M, Brocks D, Bogatyrova O, Schmidt CR, et al. Evolution of DNA methylation is linked to genetic aberrations in chronic lymphocytic leukemia. Cancer Discov. 2014;4:348–361. doi: 10.1158/2159-8290.CD-13-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okosun J, Bödör C, Wang J, Araf S, Yang C-Y, Pan C, Boller S, Cittaro D, Bozek M, Iqbal S, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nature genetics. 2014;46:176–181. doi: 10.1038/ng.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Price ME, Cotton AM, Lam LL, Farre P, Emberly E, Brown CJ, Robinson WP, Kobor MS. Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenetics Chromatin. 2013;6:4. doi: 10.1186/1756-8935-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufini A, Agostini M, Grespi F, Tomasini R, Sayan BS, Niklison-Chirou MV, Conforti F, Velletri T, Mastino A, Mak TW, et al. p73 in Cancer. Genes & cancer. 2011;2:491–502. doi: 10.1177/1947601911408890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanai N, Chang S, Berger MS. Low-grade gliomas in adults. J Neurosurg. 2011;115:948–965. doi: 10.3171/2011.7.JNS101238. [DOI] [PubMed] [Google Scholar]
- Shi J, Marconett CN, Duan J, Hyland PL, Li P, Wang Z, Wheeler W, Zhou B, Campan M, Lee DS, et al. Characterizing the genetic basis of methylome diversity in histologically normal human lung tissue. Nature communications. 2014;5:3365. doi: 10.1038/ncomms4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproul D, Kitchen RR, Nestor CE, Dixon JM, Sims AH, Harrison DJ, Ramsahoye BH, Meehan RR. Tissue of origin determines cancer-associated CpG island promoter hypermethylation patterns. Genome Biol. 2012;13:R84. doi: 10.1186/gb-2012-13-10-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taal W, Dubbink HJ, Zonnenberg CBL, Zonnenberg BA, Postma TJ, Gijtenbeek JMM, Boogerd W, Groenendijk FH, Kros JM, Kouwenhoven MCM, et al. First-line temozolomide chemotherapy in progressive low-grade astrocytomas after radiotherapy: molecular characteristics in relation to response. Neurooncology. 2011;13:235–241. doi: 10.1093/neuonc/noq177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams B. a., Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triche TJ, Jr., Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD. Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Res. 2013;41:e90. doi: 10.1093/nar/gkt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AWM, Lu C, Ward PS, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Thuijl HF, Mazor T, Johnson BE, Fouse SD, Aihara K, Hong C, Malmstrom A, Hallbeck M, Heimans JJ, Kloezeman JJ, et al. Evolution of DNA repair defects during malignant progression of low-grade gliomas after temozolomide treatment. Acta Neuropathol. 2015 doi: 10.1007/s00401-015-1403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. The American journal of pathology. 2009;174:1149–1153. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick W, Meisner C, Hentschel B, Platten M, Schilling A, Wiestler B, Sabel MC, Koeppen S, Ketter R, Weiler M, et al. Prognostic or predictive value of MGMT promoter methylation in gliomas depends on IDH1 mutation. Neurology. 2013;81:1515–1522. doi: 10.1212/WNL.0b013e3182a95680. [DOI] [PubMed] [Google Scholar]
- Wiencke JK, Zheng S, Jelluma N, Tihan T, VandenBerg S, Tamgüney T, Baumber R, Parsons R, Lamborn KR, Berger MS, et al. Methylation of the PTEN promoter defines low-grade gliomas and secondary glioblastoma. Neurooncology. 2007;9:271–279. doi: 10.1215/15228517-2007-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte T, Plass C, Gerhauser C. Pan-cancer patterns of DNA methylation. Genome medicine. 2014;6:66. doi: 10.1186/s13073-014-0066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Northcott PA, Dubuc A, Dupuy AJ, Shih DJH, Witt H, Croul S, Bouffet E, Fults DW, Eberhart CG, et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature. 2012;482:529–533. doi: 10.1038/nature10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zardo G, Tiirikainen MI, Hong C, Misra A, Feuerstein BG, Volik S, Collins CC, Lamborn KR, Bollen A, Pinkel D, et al. Integrated genomic and epigenomic analyses pinpoint biallelic gene inactivation in tumors. Nat Genet. 2002;32:453–458. doi: 10.1038/ng1007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.