Abstract
Diagnosis of Roseolovirus infections mandates careful selection of patients, samples, and testing methods. We review advances in the field and highlight research priorities. Quantitative (q)PCR can accurately identify and distinguish between human herpesvirus 6 (HHV-6) species A and B. Whether screening of high-risk patients improves outcomes is unclear. Chromosomally integrated (ci)HHV-6 confounds test interpretation but can be ruled out with digital PCR. Reverse transcription qPCR may be a more specific and clinically applicable test for actively replicating Roseoloviruses, particularly among patients with ciHHV-6. Interpretation of Roseolovirus test results faces many challenges. However, careful application of refined and emerging diagnostic techniques will allow for increasingly accurate diagnosis of clinically significant infections and disease associations.
Introduction
The Roseolovirus genus of the betaherpesvirus subfamily is composed of three enveloped, double-stranded DNA viruses: human herpesvirus (HHV-) 6A, HHV-6B, and HHV-7 [1]. These viruses share many properties that include virion structure, genomic sequence, and epidemiology but have important molecular and biologic differences [2•]. Like other human herpesviruses, infection with Roseoloviruses occurs early in life, results in chronic viral latency in diverse cell types, and affects the population at large. These characteristics complicate diagnostic efforts to determine whether Roseoloviruses are causative in many implicated diseases. Additional confusion has developed due to the unique ability of HHV-6A and HHV-6B to integrate into chromosomal telomeres of infected cells [3] as reviewed in this issue by Kaufer et al. When this occurs in a germ cell, vertical transmission of inherited chromosomally integrated (ci)HHV-6 results in offspring with latent HHV-6 DNA in every nucleated cell of their body. To further complicate matters, there is evidence that biologically active HHV-6 can reactivate in individuals with inherited ciHHV-6 and cause disease [4,5••,6]. This review highlights important advances in the diagnosis of Roseolovirus infections and provides guidance for application of current and developing diagnostic methods.
Who to test
Roseoloviruses have been variably associated with many diseases in diverse patient groups. Primary HHV-6B infection occurs in the majority of children by two years of age and usually results in a typical presentation of exanthem subitum (roseola) with mild symptoms including fever and rash [7]. HHV-6A and HHV-7 primary infection have epidemiologic differences in comparison to HHV-6B but also appear to occur in childhood with similar presentations [8–10]. Serious complications are infrequent, although primary infection with Roseoloviruses leads to significant healthcare utilization [7], and HHV-6B or HHV-7 have been associated with approximately one-third of cases of febrile status epilepticus [11]. Although testing for Roseoloviruses in the setting of typical exanthem subitum is generally not indicated, quick and accurate diagnosis could play a role in stemming antimicrobial overuse, minimizing unnecessary hospitalization, informing potential utility of selective treatment, and advancing understanding of the clinical impact of primary infection (Table 1). Primary infections are reviewed in detail in this section by Tesini et al.
Table 1.
Summary of key diagnostic considerations for clinical testing of HHV-6Ba
| Patient selection | Comments | |
|---|---|---|
| • Primary infection | • Rarely results in significant morbidity, routine testing not indicated but may stem inappropriate use of healthcare resources | |
| • Reactivation after HCT | • Frequent finding with multiple associated complications, targeted testing indicated | |
| • Other | • Selective testing should be considered in other immunocompromised and immunocompetent patients with HHV-6B-associated complications | |
| Test selection | Strengths | Weaknesses |
| • Quantitative PCR | • Sensitive, quantitative, efficient, distinguishes species | • Not standardized, detects latent virus |
| • Digital PCR | • Better accuracy and precision, useful for detecting ciHHV-6 | • More expensive and labor intensive, detects latent virus |
| • Reverse transcription PCR | • Positive results represent active replication | • More expensive and labor intensive |
| Sample selection | Strengths | Weaknesses |
| • Whole blood, serum, plasma | • Easy to access and process | • May contain latent virus, not a perfect surrogate for end-organ disease |
| • Tissue | • Appropriate testing provides stronger evidence for causality | • May contain latent virus, difficult to obtain |
| • Other (e.g. CSF, BALF) | • Better surrogate for end-organ disease than blood fractions | • May contain latent virus, difficult to obtain |
HHV-6, human herpesvirus 6; HCT, hematopoietic cell transplantation; PCR, polymerase chain reaction; ciHHV-6, inherited chromosomally integrated HHV-6; CSF, cerebrospinal fluid; BALF, bronchoalveolar lavage fluid.
Testing for HHV-6A or HHV-7 should be considered on a case-by-case basis, as there is little evidence to support any definitive disease association for either virus.
The majority of known complications due to Roseoloviruses result from HHV-6B reactivation in immunocompromised patients, specifically those undergoing hematopoietic cell (HCT) or solid organ transplantation (SOT) as reviewed in this issue by Hill and Zerr [12]. Selective testing is important among these patients (Table 1). HHV-6B and HHV-7 reactivation after HCT or SOT occurs in 40–50% of patients, whereas HHV-6A reactivation is infrequent [13–15]. HHV-6A and HHV-7 do not appear to be important pathogens in these patients. However, HHV-6B has been associated with many complications in HCT recipients, most notably central nervous system (CNS) disease [13,16,17]. Accordingly, it is reasonable to test transplant recipients for HHV-6B in the setting of any end-organ disease and particularly those with encephalopathy. Although readily available antiviral medications can abrogate viral reactivation when used as a preventive measure, this has not resulted in statistically significant improvement in associated outcomes in a few small studies [18–20]. Whether routine monitoring for HHV-6 in transplant recipients can improve outcomes remains unclear [15].
Testing for Roseoloviruses in other patient groups with findings suggestive of herpesvirus pathogenicity and an otherwise negative workup should be considered (Table 1). Ultimately, testing should be ordered judiciously in all settings, and results must be interpreted in the context of the clinical scenario, sample source, and possibility of inherited ciHHV-6.
Clinical testing and specimen selection
We again underscore that test and specimen selection for Roseolovirus testing should be guided by the clinical context. Direct detection of Roseoloviruses by culture is considered the gold-standard test for active infection, but this method is labor intensive, slow, and unsuitable for routine clinical use [1]. Indirect methods to detect an immunological response have limited utility for clinical use [21]. Numerous serologic assays have been described, including indirect fluorescent-antibody and enzyme-linked immunosorbent assay. IgM testing is not useful for clinical diagnosis of primary infection [22], and most assays are unable to discriminate prior infections with HHV-6A from HHV-6B, although a recently described assay appears to enable variant-specific serologic testing [23]. Current antigenemia tests are inadequate for distinguishing low-level viral reactivation from clinically relevant infection [24,25]. Immunohistochemistry and in situ hybridization are rarely used clinically due to limited sensitivity and slow turn-around time. Selective application of DNA testing by polymerase chain reaction (PCR) assay, however, meets important criteria for clinical use: it is sensitive, quantitative, and precise; it can distinguish between species; and it can be efficiently performed [26•]. Accordingly, PCR for Roseolovirus DNA has become the mainstay of clinical diagnostics. We focus our discussion on diagnostic techniques for HHV-6 species (Table 1).
A variety of qPCR assays for measuring HHV-6 DNA viral load are in clinical use in laboratories across the world [26•,27,28]. Well-validated assays target conserved regions of the HHV-6 genome, and some are able to differentiate HHV-6A and HHV-6B. Early PCR assays that used qualitative, nested approaches had high sensitivity but were prone to false-positive results. Quantitative real-time PCR (qPCR) has emerged as the most sensitive and rapid method available for clinical diagnosis of Roseolovirus infection or reactivation. However, inter-lab quantitative agreement for HHV-6 viral load is poor [27,29], and there is currently no international standard available for HHV-6B or HHV-6A. These factors complicate implementation of commutable assays with clinically meaningful viral load thresholds to validate research findings and guide treatment decisions [30]. The development of an international standard, such as the one for CMV made available by the World Health Organization [31], would greatly improve inter-lab agreement to better evaluate the association of HHV-6 viral load with associated diseases (Table 2).
Table 2.
Research priorities
|
RT-qPCR, reverse transcription real-time polymerase chain reaction.
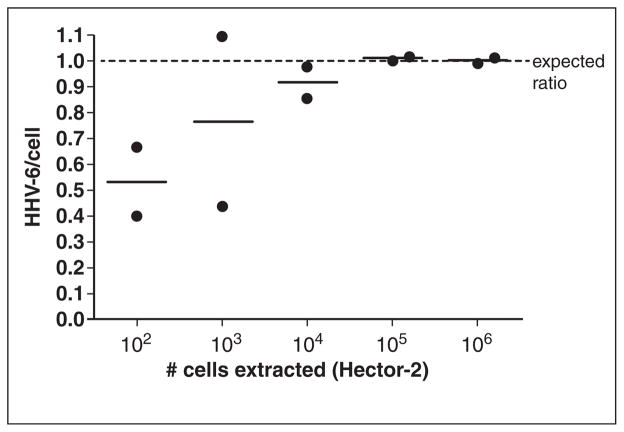
Digital PCR is another method that has recently been utilized for viral quantitation [32•,33•,34] (Table 1). Digital PCR uses the same chemistry as real-time qPCR, but this technique partitions the reaction into thousands of individual droplets, which are each read as positive or negative for DNA template. This allows for absolute quantitation of target DNA without the use of a standard curve [35]. Digital PCR is particularly well suited for the identification of inherited ciHHV-6 [36••,37•]. Previously, ciHHV-6 detection required fluorescence in situ hybridization, a labor-intensive procedure with limited availability, or HHV-6 PCR testing of hair follicle cells [38], an atypical sample type for many molecular diagnostics labs. Although HHV-6 DNA levels of >5.5 log10 copies/ml in whole blood samples is suggestive of inherited ciHHV-6, this can occur in the setting of primary infection or reactivation [3]. A digital PCR assay for inherited ciHHV-6 has been developed to concurrently amplify HHV-6 and human ribonuclease P (RPP30, a reference gene for cell count) DNA; inherited ciHHV-6 is ruled out if the ratio of HHV-6 DNA to cell genome equivalents (two RPP30/cell) falls outside a range of 1 ± 0.07 (Fig. 1) [36••]. This assay has high sensitivity and specificity when used with peripheral blood mononuclear cells (PBMCs) and other cellular samples, but it can also be utilized on study-banked plasma, sera, and other samples to aid in retrospective research, although with reduced specificity. Given mounting evidence to support in vitro and in vivo HHV-6 reactivation from inherited ciHHV-6 [4,5••,6], adapting this digital PCR method for high-throughput ciHHV-6 screening of immunocompromised individuals at high-risk for HHV-6 reactivation may be important.
Fig. 1.
Dilution series (10-fold) of Hector-2 ciHHV-6 cell line indicates that the droplet digital PCR assay provides a precise ratio of 1 HHV-6/cell with as few as 104 cells. Bars represent the mean of two replicate reactions (denoted by circles).
Source: Reprinted with permission from Clinical Chemistry, Vol. 60 no. 5, 765–772.
Limitations
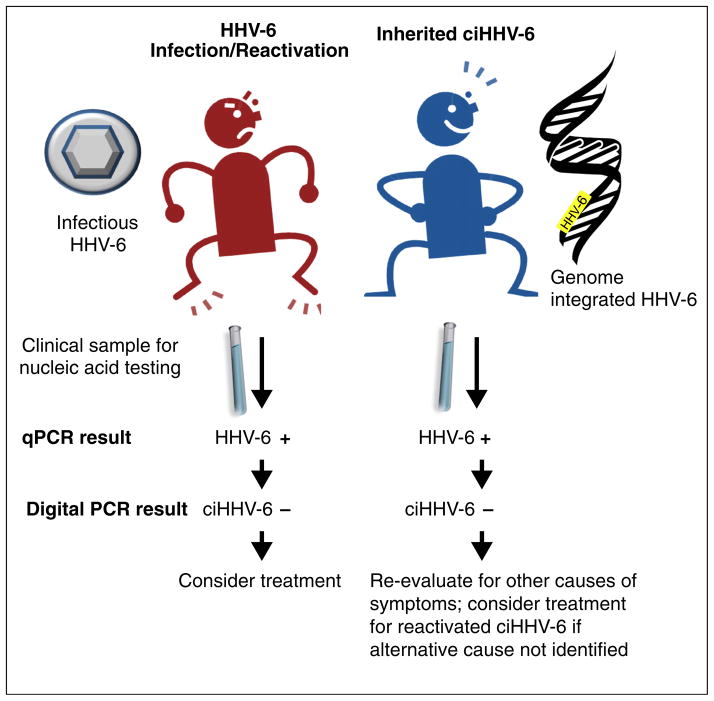
The use of qPCR to detect Roseolovirus DNA has important limitations (Table 1). Detection of HHV-6 DNA in serum or plasma appears to correlate well with indicators of active replication [39]. This may be misleading in some cases, however, as viral DNA may originate from latently infected cells that have lysed during sample preparation [40]. One study found the specificity of detecting HHV-6 DNA in plasma by qPCR to be 84% compared with viral culture [41]. PCR detection of HHV-6 DNA in plasma or serum is particularly problematic in patients with inherited ciHHV-6 (Fig. 2), who have a high burden of cell-associated latent HHV-6 DNA that can be released, especially if there is a delay in sample preparation and testing [38]. Detection of HHV-6 DNA in whole blood or PBMCs does not correlate as well with active viral replication, as the mononuclear cell is a site of latency [42]. Results of PCR testing of other cellular clinical specimens (e.g. tissue biopsies) can be difficult to interpret for the same reasons.
Fig. 2.
Flow diagram of test results and implications in patients with inherited ciHHV-6 versus HHV-6 primary infection or reactivation using quantitative and digital PCR assays for HHV-6 DNA detection.
Additional limitations to consider relate to the use of HHV-6 DNA detection in fluid samples (e.g. blood specimens, cerebrospinal fluid [CSF], bronchoalveolar lavage fluid) as a biomarker for end-organ dysfunction (Table 1). Physicians are increasingly reliant on easy-to-access surrogate markers of disease in an effort to minimize invasive procedures, such as a biopsy. However, qPCR for HHV-6 DNA is relatively insensitive for this purpose. Although HHV-6B DNA detection in blood and CSF specimens appears to occur concurrently with most cases of HHV-6B-associated CNS disease, viral detection and viral load thresholds do not strictly predict end-organ disease [43–45]. HHV-6 DNA in CSF and brain samples may also last longer than in blood samples [46,47]. In liver transplant patients with HHV-6-associated graft hepatitis, HHV-6 DNA was infrequently detected in serum [48]. Bronchoalveolar lavage fluid with detectable HHV-6 DNA also appears to be an imperfect surrogate for pulmonary disease in small studies [49]. Ultimately, PCR for HHV-6 DNA has not provided an ideal means of predicting or diagnosing clinically significant reactivation and pathogenicity. Until a better understanding of risk factors, clinical presentations, and other biomarkers of disease is developed, alternative diagnostic methods that include tissue-based and immunologic studies will be important for defining the role of HHV-6 in associated diseases (Table 2).
Research methods and future directions
While HHV-6 DNA detection with qPCR provides evidence to support active infection, we have reviewed multiple confounding factors that limit the sensitivity of viral DNA detection alone. Research-based methods of culture, serology, immunohistochemistry, and in situ hybridization are useful for identifying active infection and correlating with DNA viral load [50]. However, adaptation of these techniques to routine clinical diagnostics is limited by their complexity, long turn-around time, and variable sensitivity. Perhaps the most promising method for definitive clinical diagnosis of active HHV-6 infection is the molecular detection of viral transcripts via reverse transcription real-time quantitative PCR (RT-qPCR). This method of amplifying messenger (m)RNA from PBMCs or other infected cells could provide a better approach to distinguish active from latent infections [51•], and it may be particularly useful for identifying HHV-6 reactivation in patients with inherited ciHHV-6.
HHV-6 mRNA detection to identify active infection has been reported in a few studies to date. An early study that compared traditional viral culture with a nested RT-PCR assay for the U100 transcript, expressed during the late stages of viral replication, determined that the RT-PCR assay was 95% sensitive and 98.8% specific for actively replicating virus in PBMC samples [52]. Subsequent studies developed nested RT-PCR assays for genes in other stages of the viral replication cycle, including immediate early genes U16/17 and U89/90 [53], early gene U79/80 [54,55], late gene U60/66 [53], and latency-associated gene U94 [56••]. All of these studies were limited by the use of nested RT-PCR, a sensitive but qualitative molecular method historically prone to false-positive test results. Given these limitations, RT-qPCR assays that effectively quantitate viral transcript levels have been developed [51•,57••]. These assays have targeted immediate early (U90), early (U12), or late (U100) gene transcripts specifically from HHV-6B and show promising results regarding correlation of transcript levels with high-level viremia (>1000 copies/ml DNA) and viral culture in immunocompetent and immunocompromised patients. However, additional steps to optimize findings (e.g. specific processing and storage of clinical samples to augment RNA preservation) are required to further increase sensitivity and standardization. Large studies that correlate transcript detection with DNA detection and active disease will be critical to establish actionable DNA and mRNA transcript thresholds for treatment. Although additional work is needed to validate the utility and feasibility of RT-qPCR in the clinical setting (Table 2), this technique will likely play a bigger role in routine HHV-6 diagnostics, especially in the setting of inherited ciHHV-6.
Conclusions
The definitive establishment of Roseoloviruses as causative pathogens in their many associated diseases is challenging due to the ubiquity of infection, their latency in a variety of cell types, the ability of HHV-6A and HHV-6B to integrate into the human genome, lack of standardized testing metrics, and poor correlation of current diagnostic techniques with end-organ disease. While much work has been done to advance our understanding of the molecular virology, pathogenesis, and disease associations of these viruses, additional studies using immunologic and tissue-based diagnostics will be important to establish the role of Roseoloviruses in end-organ disease and inform clinically applicable testing methods. Ultimately, Roseolovirus detection does not necessarily imply causation, and interpretation of test results must account for the clinical context, sample type, and diagnostic technique in order to formulate valid clinical and scientific conclusions.
Acknowledgments
The authors thank the staff of the University of Washington Molecular Virology Laboratory for their outstanding service to our patients with roseolovirus infections. This review was presented in part at the NIAID-sponsored conference entitled “Roseoloviruses: Clinical Impact, Interventions, and Research Needs” on June 2, 2014, Natcher Center, NIH, Bethesda, MD.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Yamanishi K, Mori Y, Pellett PE. Human herpesvirus 6 and 7. In: Knipe D, Howley P, editors. In fields virology. Wolters Kluwer Health; 2013. p. 2058. [Google Scholar]
- 2•.Ablashi D, Agut H, Alvarez-Lafuente R, Clark DA, Dewhurst S, Diluca D, Flamand L, Frenkel N, Gallo R, Gompels UA, et al. Classification of HHV-6A and HHV-6B as distinct viruses. Arch Virol. 2014;159:863–870. doi: 10.1007/s00705-013-1902-5. A review of the distinctions between HHV-6A and HHV-6B to highlight the importance of diagnostic differentiation of these viruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pellett P, Ablashi D. Chromosomally integrated human herpesvirus 6: questions and answers. Rev Med Virol. 2012 doi: 10.1002/rmv.715. http://dx.doi.org/10.1002/rmv. [DOI] [PMC free article] [PubMed]
- 4.Arbuckle JH, Medveczky MM, Luka J, Hadley SH, Luegmayr A, Ablashi D, Lund TC, Tolar J, De Meirleir K, Montoya JG, et al. The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc Natl Acad Sci USA. 2010;107:5563–5568. doi: 10.1073/pnas.0913586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5••.Endo A, Watanabe K, Ohye T, Suzuki K, Matsubara T, Shimizu N, Kurahashi H, Yoshikawa T, Katano H, Inoue N, et al. Molecular and virological evidence of viral activation from chromosomally integrated HHV-6A in a patient with X-SCID. Clin Infect Dis. 2014 doi: 10.1093/cid/ciu323. http://dx.doi.org/10.1093/cid/ciu323A study utilizing multiple diagnostic methodologies to demonstrate HHV-6 reactivation from a patient with inherited ciHHV-6. [DOI] [PubMed]
- 6.Flamand L. Pathogenesis from the reactivation of chromosomally-integrated HHV-6: Facts rather than fiction. Clin Infect Dis. 2014 doi: 10.1093/cid/ciu326. http://dx.doi.org/10.1093/cid/ciu326. [DOI] [PubMed]
- 7.Zerr DM, Meier AS, Selke SS, Frenkel LM, Huang M-L, Wald A, Rhoads MP, Nguy L, Bornemann R, Morrow RA, et al. A population-based study of primary human herpesvirus 6 infection. N Engl J Med. 2005;352:768–776. doi: 10.1056/NEJMoa042207. [DOI] [PubMed] [Google Scholar]
- 8.De Bolle L, Naesens L, De Clercq E. Update on human herpesvirus 6 biology, clinical features, and therapy. Clin Microbiol Rev. 2005;18:217–245. doi: 10.1128/CMR.18.1.217-245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bates M, Monze M, Bima H, Kapambwe M, Clark D, Kasolo FC, Gompels UA. Predominant human herpesvirus 6 variant A infant infections in an HIV-1 endemic region of Sub-Saharan Africa. J Med Virol. 2009;81:779–789. doi: 10.1002/jmv.21455. [DOI] [PubMed] [Google Scholar]
- 10.Hall CB, Caserta MT, Schnabel KC, McDermott MP, Lofthus GK, Carnahan JA, Gilbert LM, Dewhurst S. Characteristics and acquisition of human herpesvirus (HHV) 7 infections in relation to infection with HHV-6. J Infect Dis. 2006;193:1063–1069. doi: 10.1086/503434. [DOI] [PubMed] [Google Scholar]
- 11.Epstein LG, Shinnar S, Hesdorffer DC, Nordli DR, Hamidullah A, Benn EKT, Pellock JM, Frank LM, Lewis DV, Moshe SL, et al. Human herpesvirus 6 and 7 in febrile status epilepticus: the FEBSTAT study. Epilepsia. 2012;53:1481–1488. doi: 10.1111/j.1528-1167.2012.03542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill JA, Zerr DM. Roseoloviruses in transplant recipients: clinical consequences and prospects for treatment and prevention trials. Curr Opin Virol. 2014 doi: 10.1016/j.coviro.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zerr DM, Boeckh M, Delaney C, Martin PJ, Xie H, Adler AL, Huang M-L, Corey L, Leisenring WM. HHV-6 reactivation and associated sequelae after hematopoietic cell transplant. Biol Blood Marrow Transplant. 2012;18:1700–1708. doi: 10.1016/j.bbmt.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Razonable RR. Human herpesviruses 6, 7 and 8 in solid organ transplant recipients. Am J Transplant. 2013;13:67–78. doi: 10.1111/ajt.12008. [DOI] [PubMed] [Google Scholar]
- 15.Olson AL, Dahi PB, Zheng J, Devlin SM, Lubin M, Gonzales AM, Giralt SA, Perales M-A, Papadopoulos EB, Ponce DM, et al. Frequent human herpesvirus-6 viremia but low incidence of encephalitis in double-unit cord blood recipients transplanted without antithymocyte globulin. Biol Blood Marrow Transplant. 2014 doi: 10.1016/j.bbmt.2014.02.010. http://dx.doi.org/10.1016/j.bbmt.2014.02.010. [DOI] [PMC free article] [PubMed]
- 16.Ogata M, Satou T, Kadota J-I, Saito N, Yoshida T, Okumura H, Ueki T, Nagafuji K, Kako S, Uoshima N, et al. Human herpesvirus 6 (HHV-6) reactivation and HHV-6 encephalitis after allogeneic hematopoietic cell transplantation: a multicenter, prospective study. Clin Infect Dis. 2013;57:671–681. doi: 10.1093/cid/cit358. [DOI] [PubMed] [Google Scholar]
- 17.Dulery R, Salleron J, Dewilde A, Rossignol J, Boyle EM, Gay J, de Berranger E, Coiteux V, Jouet J-P, Duhamel A, et al. Early human herpesvirus type 6 reactivation after allogeneic stem cell transplantation: a large-scale clinical study. Biol Blood Marrow Transplant. 2012;18:1080–1089. doi: 10.1016/j.bbmt.2011.12.579. [DOI] [PubMed] [Google Scholar]
- 18.Galarraga MC, Gomez E, de Oña M, Rodriguez A, Laures A, Boga JA, Melon S. Influence of ganciclovir prophylaxis on citomegalovirus, human herpesvirus 6, and human herpesvirus 7 viremia in renal transplant recipients. Transplant Proc. 2005;37:2124–2126. doi: 10.1016/j.transproceed.2005.03.123. [DOI] [PubMed] [Google Scholar]
- 19.Ogata M, Satou T, Inoue Y, Takano K, Ikebe T, Ando T, Ikewaki J, Kohno K, Nishida A, Saburi M, et al. Foscarnet against human herpesvirus (HHV)-6 reactivation after allo-SCT: breakthrough HHV-6 encephalitis following antiviral prophylaxis. Bone Marrow Transplant. 2012;48:257–264. doi: 10.1038/bmt.2012.121. [DOI] [PubMed] [Google Scholar]
- 20.Ishiyama K, Katagiri T, Hoshino T, Yoshida T, Yamaguchi M, Nakao S. Preemptive therapy of human herpesvirus-6 encephalitis with foscarnet sodium for high-risk patients after hematopoietic SCT. Bone Marrow Transplant. 2011;46:863–869. doi: 10.1038/bmt.2010.201. [DOI] [PubMed] [Google Scholar]
- 21.Ward KN. The natural history and laboratory diagnosis of human herpesviruses-6 and -7 infections in the immunocompetent. J Clin Virol. 2005;32:183–193. doi: 10.1016/j.jcv.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 22.De Oliveira Vianna RA, Siqueira MM, Camacho LAB, Setúbal S, Knowles W, Brown DW, de Oliveira SA. The accuracy of anti-human herpesvirus 6 IgM detection in children with recent primary infection. J Virol Methods. 2008;153:273–275. doi: 10.1016/j.jviromet.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Thäder-Voigt A, Jacobs E, Lehmann W, Bandt D. Development of a microwell adapted immunoblot system with recombinant antigens for distinguishing human herpesvirus (HHV)6A and HHV6B and detection of human cytomegalovirus. Clin Chem Lab Med. 2011;49:1891–1898. doi: 10.1515/CCLM.2011.666. [DOI] [PubMed] [Google Scholar]
- 24.Volin L, Lautenschlager I, Juvonen E, Nihtinen A, Anttila V-J, Ruutu T. Human herpesvirus 6 antigenaemia in allogeneic stem cell transplant recipients: impact on clinical course and association with other beta-herpesviruses. Br J Haematol. 2004;126:690–696. doi: 10.1111/j.1365-2141.2004.05101.x. [DOI] [PubMed] [Google Scholar]
- 25.Sampaio AM, Thomasini RL, Guardia AC, Stucchi RSB, Rossi CL, Costa SCB, Boin IFSF. Cytomegalovirus, human herpesvirus-6, and human herpesvirus-7 in adult liver transplant recipients: diagnosis based on antigenemia. Transplant Proc. 2011;43:1357–1359. doi: 10.1016/j.transproceed.2011.03.062. [DOI] [PubMed] [Google Scholar]
- 26•.Cassina G, Russo D, De Battista D, Broccolo F, Lusso P, Malnati MS. Calibrated real-time polymerase chain reaction for specific quantitation of HHV-6A and HHV-6B in clinical samples. J Virol Methods. 2013;189:172–179. doi: 10.1016/j.jviromet.2013.01.018. A description of the development of two sensitive qPCR assays for HHV-6A and HHV-6B DNA detection in clinical specimens. [DOI] [PubMed] [Google Scholar]
- 27.Flamand L, Gravel A, Boutolleau D, Alvarez-Lafuente R, Jacobson S, Malnati MS, Kohn D, Tang Y-W, Yoshikawa T, Ablashi D. Multicenter comparison of PCR assays for detection of human herpesvirus 6 DNA in serum. J Clin Microbiol. 2008;46:2700–2706. doi: 10.1128/JCM.00370-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karlsson T, Mannonen L, Loginov R, Lappalainen M, Höckerstedt K, Lautenschlager I. Development of a new quantitative real-time HHV-6-PCR and monitoring of HHV-6 DNAaemia after liver transplantation. J Virol Methods. 2012;181:25–36. doi: 10.1016/j.jviromet.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 29.De Pagter PJ, Schuurman R, de Vos NM, Mackay W, van Loon AM. Multicenter external quality assessment of molecular methods for detection of human herpesvirus 6. J Clin Microbiol. 2010;48:2536–2540. doi: 10.1128/JCM.01145-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agut H. Deciphering the clinical impact of acute human herpesvirus 6 (HHV-6) infections. J Clin Virol. 2011;52:164–171. doi: 10.1016/j.jcv.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 31.Hirsch HH, Lautenschlager I, Pinsky BA, Cardeñoso L, Aslam S, Cobb B, Vilchez RA, Valsamakis A. An international multicenter performance analysis of cytomegalovirus load tests. Clin Infect Dis. 2013;56:367–373. doi: 10.1093/cid/cis900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.Hayden RT, Gu Z, Ingersoll J, Abdul-Ali D, Shi L, Pounds S, Caliendo AM. Comparison of droplet digital pcr to real-time pcr for quantitative detection of cytomegalovirus. J Clin Microbiol. 2012 doi: 10.1128/JCM.02620-12. http://dx.doi.org/10.1128/JCM.02620-12A study detailing the direct comparison between qPCR and digital PCR in viral diagnostics. [DOI] [PMC free article] [PubMed]
- 33•.Sedlak RH, Cook L, Cheng A, Magaret A, Jerome KR. Clinical utility of droplet digital PCR for human cytomegalovirus. J Clin Microbiol. 2014;52:2844–2848. doi: 10.1128/JCM.00803-14. A review of the advantages of digital PCR for clinical virology diagnostics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hall Sedlak R, Jerome KR. The potential advantages of digital PCR for clinical virology diagnostics. Expert Rev Mol Diagn. 2014;14:501–507. doi: 10.1586/14737159.2014.910456. [DOI] [PubMed] [Google Scholar]
- 35.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36••.Sedlak RH, Cook L, Huang M-L, Magaret A, Zerr DM, Boeckh M, Jerome KR. Identification of chromosomally integrated human herpesvirus 6 by droplet digital PCR. Clin Chem. 2014;60:765–772. doi: 10.1373/clinchem.2013.217240. A study of the first clinically validated viral assay using digital PCR to identify patients with inherited ciHHV-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Leibovitch EC, Brunetto GS, Caruso B, Fenton K, Ohayon J, Reich DS, Jacobson S. Coinfection of human herpesviruses 6A (HHV-6A) and HHV-6B as demonstrated by novel digital droplet PCR assay. PLOS ONE. 2014;9:e92328. doi: 10.1371/journal.pone.0092328. A study of the use of digital PCR to identify HHV-6A and HHV-6B infection, as well as inherited ciHHV-6, in clinical samples from patients with multiple sclerosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward K, Leong H. Human herpesvirus 6 chromosomal integration in immunocompetent patients results in high levels of viral DNA in blood, sera, and hair follicles. J Clin Microbiol. 2006;44:1571–1574. doi: 10.1128/JCM.44.4.1571-1574.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boutolleau D, Fernandez C, André E, Imbert-Marcille B-M, Milpied N, Agut H, Gautheret-Dejean A. Human herpesvirus (HHV)-6 and HHV-7: two closely related viruses with different infection profiles in stem cell transplantation recipients. J Infect Dis. 2003;187:179–186. doi: 10.1086/367677. [DOI] [PubMed] [Google Scholar]
- 40.Achour A, Boutolleau D, Slim A, Agut H, Gautheret-Dejean A. Human herpesvirus-6 (HHV-6) DNA in plasma reflects the presence of infected blood cells rather than circulating viral particles. J Clin Virol. 2007;38:280–285. doi: 10.1016/j.jcv.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 41.Caserta MT, Hall CB, Schnabel K, Lofthus G, Marino A, Shelley L, Yoo C, Carnahan J, Anderson L, Wang H. Diagnostic assays for active infection with human herpesvirus 6 (HHV-6) J Clin Virol. 2010;48:55–57. doi: 10.1016/j.jcv.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kondo K, Kondo T, Okuno T, Takahashi M, Yamanishi K. Latent human herpesvirus 6 infection of human monocytes/macrophages. J Gen Virol. 1991;72(Pt 6):1401–1408. doi: 10.1099/0022-1317-72-6-1401. [DOI] [PubMed] [Google Scholar]
- 43.Hill JA, Boeckh MJ, Sedlak RH, Jerome KR, Zerr DM. Human herpesvirus 6 can be detected in cerebrospinal fluid without associated symptoms after allogeneic hematopoietic cell transplantation. J Clin Virol. 2014 doi: 10.1016/j.jcv.2014.07.001. http://dx.doi.org/10.1016/j.jcv.2014.07.001. [DOI] [PMC free article] [PubMed]
- 44.Hill JA, Koo S, Guzman Suarez BB, Ho VT, Cutler C, Koreth J, Armand P, Alyea EP, Baden LR, Antin JH, et al. Cord-blood hematopoietic stem cell transplant confers an increased risk for human herpesvirus-6-associated acute limbic encephalitis: a cohort analysis. Biol Blood Marrow Transplant. 2012;18:1638–1648. doi: 10.1016/j.bbmt.2012.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogata M, Kikuchi H, Satou T, Kawano R, Ikewaki J, Kohno K, Kashima K, Ohtsuka E, Kadota J-I. Human herpesvirus 6 DNA in plasma after allogeneic stem cell transplantation: incidence and clinical significance. J Infect Dis. 2006;193:68–79. doi: 10.1086/498531. [DOI] [PubMed] [Google Scholar]
- 46.Fotheringham J, Akhyani N, Vortmeyer A, Donati D, Williams E, Oh U, Bishop M, Barrett J, Gea-Banacloche J, Jacobson S. Detection of active human herpesvirus-6 infection in the brain: correlation with polymerase chain reaction detection in cerebrospinal fluid. J Infect Dis. 2007;195:450–454. doi: 10.1086/510757. [DOI] [PubMed] [Google Scholar]
- 47.Zerr DM, Gupta D, Huang M-L, Carter R, Corey L. Effect of antivirals on human herpesvirus 6 replication in hematopoietic stem cell transplant recipients. Clin Infect Dis. 2002;34:309–317. doi: 10.1086/338044. [DOI] [PubMed] [Google Scholar]
- 48.Pischke S, Gösling J, Engelmann I, Schlue J, Wölk B, Jäckel E, Meyer-Heithuis C, Lehmann U, Strassburg CP, Barg-Hock H, et al. High intrahepatic HHV-6 virus loads but neither CMV nor EBV are associated with decreased graft survival after diagnosis of graft hepatitis. J Hepatol. 2012;56:1063–1069. doi: 10.1016/j.jhep.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 49.Nagate, et al. Detection and quantification of human herpesvirus 6 genomes using bronchoalveolar lavage fluid in immunocompromised patients with interstitial pneumonia. Int J Mol Med. 2001;8:379. doi: 10.3892/ijmm.8.4.379. [DOI] [PubMed] [Google Scholar]
- 50.Crawford JR, Santi MR, Thorarinsdottir HK, Cornelison R, Rushing EJ, Zhang H, Yao K, Jacobson S, Macdonald TJ. Detection of human herpesvirus-6 variants in pediatric brain tumors: association of viral antigen in low grade gliomas. J Clin Virol. 2009;46:37–42. doi: 10.1016/j.jcv.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51•.Ihira M, Enomoto Y, Kawamura Y, Nakai H, Sugata K, Asano Y, Tsuzuki M, Emi N, Goto T, Miyamura K, et al. Development of quantitative RT-PCR assays for detection of three classes of HHV-6B gene transcripts. J Med Virol. 2012;84:1388–1395. doi: 10.1002/jmv.23350. A study demonstrating the utility of a RT-qPCR assay for identifying active HHV-6B infection in patients with primary infection and reactivation after HCT. [DOI] [PubMed] [Google Scholar]
- 52.Norton RA, Caserta MT, Hall CB, Schnabel K, Hocknell P, Dewhurst S. Detection of human herpesvirus 6 by reverse transcription-PCR. J Clin Microbiol. 1999;37:3672–3675. doi: 10.1128/jcm.37.11.3672-3675.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van den Bosch G, Locatelli G, Geerts L, Fagà G, Ieven M, Goossens H, Bottiger D, Oberg B, Lusso P, Berneman ZN. Development of reverse transcriptase PCR assays for detection of active human herpesvirus 6 infection. J Clin Microbiol. 2001;39:2308–2310. doi: 10.1128/JCM.39.6.2308-2310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kondo K, Kondo T, Shimada K, Amo K, Miyagawa H, Yamanishi K. Strong interaction between human herpesvirus 6 and peripheral blood monocytes/macrophages during acute infection. J Med Virol. 2002;67:364–369. doi: 10.1002/jmv.10082. [DOI] [PubMed] [Google Scholar]
- 55.Pradeau K, Bordessoule D, Szelag J-C, Rolle F, Ferrat P, Le Meur Y, Turlure P, Denis F, Ranger-Rogez S. A reverse transcription-nested PCR assay for HHV-6 mRNA early transcript detection after transplantation. J Virol Methods. 2006;134:41–47. doi: 10.1016/j.jviromet.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 56••.Strenger V, Caselli E, Lautenschlager I, Schwinger W, Aberle SW, Loginov R, Gentili V, Nacheva E, DiLuca D, Urban C. Detection of HHV-6-specific mRNA and antigens in PBMCs of individuals with chromosomally integrated HHV-6 (ciHHV-6) Clin Microbiol Infect. 2014 doi: 10.1111/1469-0691.12639. http://dx.doi.org/10.1111/1469-0691.12639A study using RT-PCR to demonstrate viral gene expression in four of eleven patients with inherited ciHHV-6. [DOI] [PubMed]
- 57••.Bressollette-Bodin C, Nguyen TVH, Illiaquer M, Besse B, Peltier C, Chevallier P, Imbert-Marcille B-M. Quantification of two viral transcripts by real time PCR to investigate human herpesvirus type 6 active infection. J Clin Virol. 2014;59:94–99. doi: 10.1016/j.jcv.2013.11.014. A study of the utility of RT-qPCR for monitoring HHV-6 transcription in HCT recipients. [DOI] [PubMed] [Google Scholar]