Abstract
During prophase of meiosis I, homologous chromosomes interact and undergo recombination. Successful completion of these processes is required in order for the homologous chromosomes to mount the meiotic spindle as a pair. The organization of the chromosomes into pairs ensures orderly segregation to opposite poles of the dividing cell, such that each gamete receives one copy of each chromosome. Chiasmata, the cytological manifestation of crossover products of recombination, physically connect the homologs in pairs, providing a linkage that facilitates their segregation. Consequently, mutations that reduce the level of recombination are invariably associated with increased errors in meiotic chromosome segregation. In this review, we focus on recent biochemical and genetic advances in elucidating the mechanisms of meiotic DNA strand exchange catalyzed by the Dmc1 protein. We also discuss the mode by which two recombination mediators, Hop2 and Mnd1, facilitate rate-limiting steps of DNA strand exchange catalyzed by Dmc1.
Keywords: Dmc1, DNA repair, homologous chromosomes, homologous recombination, Hop2, meiosis, Mnd1, recombination mediators, strand exchange, zebrafish
Introduction
Homologous recombination (HR) is the only high-fidelity mechanism for the repair of DNA double-strand breaks (DSBs) generated during meiosis and mitosis. The repair of DSBs through HR promotes the proper transfer of genetic information from generation to generation without loss of crucial information. Meiotic HR also facilitates exchange between the alleles of maternal and paternal origin, which generates genetic diversity in gametes. Importantly, at the end of meiotic prophase, HR serves a third critical function by providing a physical link that holds homologous chromosome pairs together. These linkages are established by chiasmata, which are the cytological manifestation of the crossover product of HR, and, together with cohesin linkage between sister chromatids 1, ensure the orderly segregation of each chromosome in the pair to the opposite poles of the spindle and therefore are required to generate gametes with the correct number of chromosomes (Fig.1). This represents a seminal event in preparation of the genome for sexual reproduction, and is essential for species survival. At the heart of the HR pathway is formation of joint molecules by DNA strand exchange. These early DNA intermediates of recombination are later resolved to form the final genetic products of recombination, namely crossovers and non-crossovers. In meiosis, these intermediates have many unique features that distinguish them from mitotic recombination products. Several of these features are promoted by the recombinases and their ancillary proteins, which are critical for regulating homologous chromosome behavior during meiosis.
Figure 1.
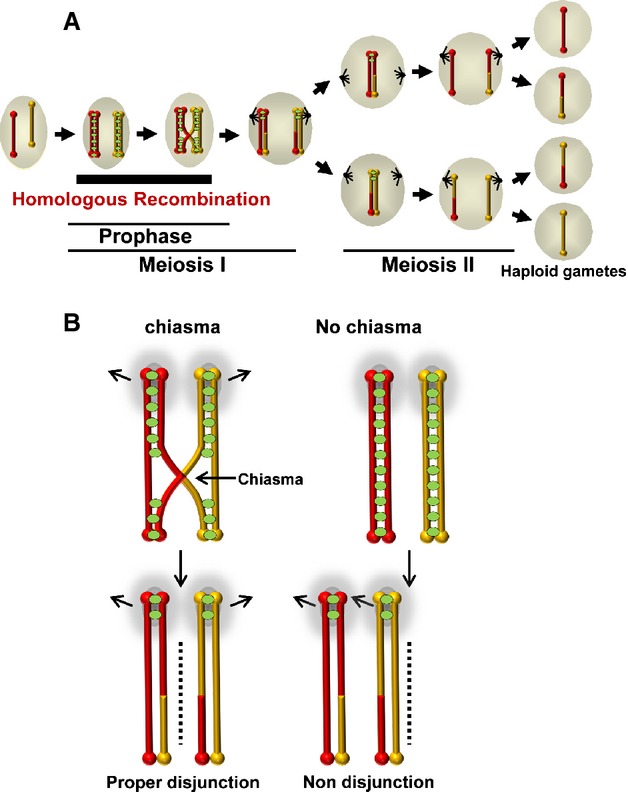
Homologous chromosome association and dysjunction. (A) Schematic of male mouse spermatogenesis. Note the period of prophase during which HR promotes exchange of genetic information and homologous chromosomes pairwise interactions. (B) The presence of chiasmata ensures that each chromosome of the homologous chromosome pair segregates to opposite poles of the spindle. The homologous chromosome pairs are represented in red and yellow. Each of the homologous chromosomes comprises two sister chromatids represented by two bars of the same color. The green ovals represent cohesins.
Meiotic recombination
The process of DSB repair in mammals appears to utilize pathways similar to those seen in lower eukaryotes, such as Saccharomyces cerevisiae (budding yeast) 2,3, and the final products are either crossovers, which involves exchange of flanking DNA markers between the homologs, or non-crossovers, in which the flanking DNA remains unchanged 2. In meiosis, the initial steps of HR involve introduction of DSBs at multiple chromosomal DNA sites catalyzed by the Spo11 protein 4 (Fig.2). This topoisomerase-like reaction cuts DNA to generate a covalent protein–DNA linkage to the 5′ DNA ends on either side of the break. After Spo11 is removed from the DNA ends, the process of HR involves exonuclease activity to generate 3′ single-stranded DNA (ssDNA) tails 5,6. After resection, two eukaryotic members of the RecA protein family, the ubiquitously expressed Rad51 DNA recombinase and the meiosis-specific Dmc1 DNA recombinase, bind the 3′ ssDNA tails to form helical nucleoprotein filaments, which perform a search for intact homologous double-stranded DNA (dsDNA) 7. Here we use the term homologous to describe DNA sequence similarity. It should be noted that this term is also often used with a different genetic meaning, i.e. homologous pairs of chromatids. Once the homologous sequence is found, the recombinases promote invasion of the ssDNA ends into the homologous duplex DNA (D-loops). After strand exchange, current models propose that HR intermediates are processed by one of two distinct pathways. The initial and relatively unstable strand invasion intermediates may be displaced from the invaded homolog and anneal to the second single-stranded end of the break. This leads to re-joining of the broken chromosome by synthesis-dependent strand annealing (SDSA) to generate non-crossovers (Fig.2, right branch) 2,8. In an optional pathway, they are processed by double-strand break repair (DSBR) 8,9, which includes DNA polymerase-dependent heteroduplex extension synthesis facilitated by Hfm1/Mer3, resulting in relatively more stable strand invasion 10,11. This alternative process is able to perform the second end capture, and leads to formation of double Holliday junctions. During and after the formation of joint molecules and DNA synthesis to restore sequences that were lost or damaged at the site of the original DSB lesion, joint molecules must be resolved to allow chromosome segregation and formation of chiasmata. The structure of the joint molecules dictates whether a DNA helicase, endonuclease, or a combination of both is required for resolution resulting in the formation of crossovers and non-crossovers 12. Whereas DSBR and SDSA occur both in cells that divide through mitosis and in cells that divide through meiosis, the major pathway for repair DSBs in mitosis appears to be the SDSA pathway, with DSBR primarily occurring in meiosis 13. During mitotic recombination, the recipient DNA duplex is generally a sister chromatid. In meiosis, however, the situation is more complex, as either the homolog chromatid or the sister chromatid may provide the template for repair (i.e. using either DSBR or SDSA). It has been suggested that the preferred meiotic inter-homolog recombination is promoted by meiosis-specific components that inhibit inter-sister chromatid recombination 14. Meiotic double Holliday junction intermediates (which are ultimately resolved as crossovers) are essential for the proper segregation of chromosomes. These crossovers also play an important role by shuffling parental genomes, generating genetic diversity.
Figure 2.
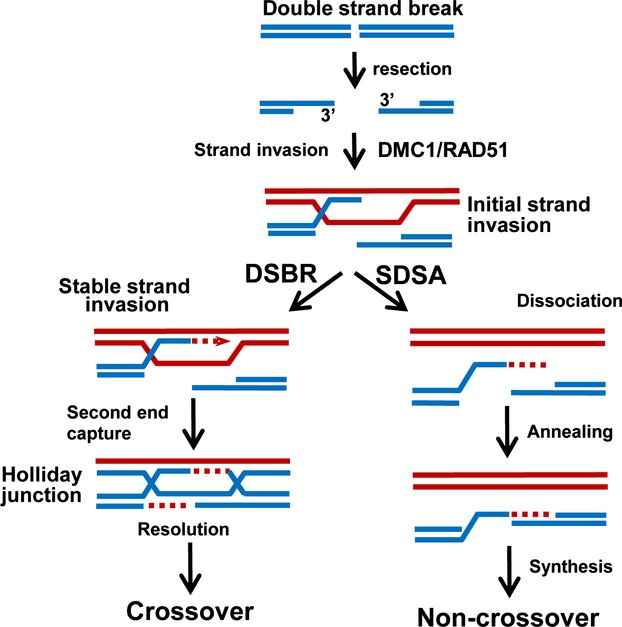
The pathway of meiotic recombination. Copies of homologous chromosomes are represented in red and blue. HR proceeds by two pathways: synthesis-dependent strand annealing (SDSA) and double-strand break repair (DSBR). While SDSA only produces non-crossovers, the second DSB end is captured during DSBR. After DNA synthesis and ligation, Holliday junctions are formed whose resolution results in formation of mostly crossovers and a small proportion of non-crossovers. Formation of both crossovers and non-crossovers between homolog chromatids is shown. Although use of the homologous chromatids is favored during meiotic recombination, final genetic products of recombination may also be generated using sister chromatids as DNA repair templates.
Dmc1 is at the center of meiotic recombination
DMC1 was first identified in a screen for genes specific to S. cerevisiae meiosis 15, and is present in almost all eukaryotes, including mice and humans 16. Deletion of DMC1 in budding yeast, plants and mice results in severe abnormalities that reflect an indispensable role of this protein in meiotic recombination 15,17,18. S. cerevisiae and mouse DMC1 mutants show a near-complete block of recombination 15,17, with the S. cerevisiae Dmc1-deficient strain showing no stable strand invasion or double Holliday junction formation 9,19, accompanied by defective synaptonemal complex formation. It has been suggested that Dmc1 promotes recombination almost exclusively between the homologous chromosomes 20, which is unique to meiosis 19. However, Dmc1 is not required in all organisms. For example, Drosophila melanogaster and Caenorhabditis elegans have Rad51, but lack Dmc1, and, in Schizosaccharomyces pombe (fission yeast), a mutation of DMC1 does not completely abolish meiotic recombination 21,22.
Dmc1-mediated strand invasion and DNA strand exchange
Whereas in vivo studies have revealed the indispensable role of the Dmc1 protein in meiotic recombination, the biochemical mechanism of action of Dmc1 is better studied using purified systems in vitro. Given its homology to RecA and Rad51, Dmc1 was predicted to exhibit the hallmarks of the reactions by which RecA family members promote the recognition of homology between ssDNA and a duplex DNA template to promote the formation of joint molecules (Fig.2) 23. Pioneering work, using purified human and mouse proteins, described essential structural characteristics 24 and enzymatic activities of Dmc1 25,26. Purified recombinant enzyme had a DNA-dependent ATPase activity, binds preferentially to ssDNA, and catalyzes formation of D-loops in super-helical DNA (strand invasion).
Pre-synaptic and synaptic events of recombination promoted by Dmc1: a biochemical view
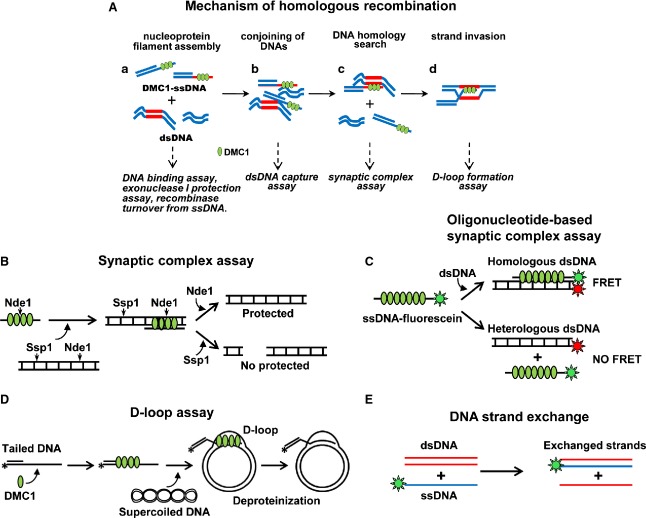
Figure3A shows the molecular events leading to strand exchange promoted by RecA-related proteins. It illustrates the obligatory phases and experimental approaches used to detect and characterize the molecular intermediates. In trying to understand the mechanism by which Dmc1 and other RecA-like recombinases promote strand exchange, it is useful to distinguish four consecutive steps 27–30. The first two are nucleoprotein filament assembly and conjoining of DNAs. Formation of these two intermediates requires no DNA homology, but they are a prerequisite to assemble the machinery promoting the subsequent steps of DNA homology search and strand exchange. During nucleoprotein filament assembly (the pre-synaptic phase), Dmc1 assembles on ssDNA, creating a helical nucleoprotein filament in which the DNA is stretched and under-wound relative to the B-form DNA. Nucleation of Dmc1 and Rad51 onto ssDNA is a slow process, which renders the pre-synaptic filament assembly prone to degradation by nucleases 15,19,31 or interference by other single-strand binding proteins, such as amounts of replication protein A (RPA) that are sufficient to saturate available ssDNA 32–34. Certain recombinase accessory factors facilitate the assembly of the Dmc1 pre-synaptic filament. As such, these recombination mediators are critical for the efficiency of homologous recombination. We expand on the mechanism of action of some of these recombination mediators below. Formation of a ternary complex of the Dmc1–ssDNA and Rad51–ssDNA filaments with dsDNA (conjoining of DNAs) initiates the synaptic phase. Here, we use the term ‘synapsis’ to describe the interaction of homologous DNA molecules. It should be noted that this term is also used with a different cytological meaning, which refers to the formation of the synaptonemal complex and consequently close juxtaposition of the homologous chromosomes. The synaptic phase has been proposed to be of critical importance in the reaction of homologous pairing catalyzed by RecA 35, Rad51 36 and Dmc1 30. Juxtaposition of three DNA strands within the synaptic filament permits rapid homology sampling through transient Watson–Crick base pairing between the ssDNA and the complementary strand of the duplex template (see below). In the third step, the search for DNA homology within a network of conjoined DNA molecules results in pairing of homologous DNA sequences or formation of stable DNA joints. In this reaction, a nucleoprotein filament complex of recombination proteins and ssDNA finds a homologous duplex DNA target, within otherwise heterologous DNA, to produce a synaptic complex (homologous alignment) (Fig.3A–C). Finally, the first stable intermediate product of homologous pairing and consequent strand exchange is revealed (D-loop and strand exchange in the in vitro assay; Fig.3D, E). During formation of this intermediate, incoming ssDNA forms a stable homoduplex with its complementary ssDNA strand in the targeted dsDNA.
Figure 3.
The mechanism of HR promoted by Dmc1. (A) Mechanism leading to formation of joint molecules and the experimental approaches used to detect and characterize such intermediates. Panel (a) shows the pre-synaptic polymerization of Dmc1 on ssDNA. Panel (b) shows homology-independent conjunction of ssDNA and dsDNA without homologous alignment. Panel (c) shows that the DNA homology search results in homologous DNA pairing. Panel (d) shows strand invasion and strand exchange. (B) Schematic of an enzyme-based synaptic complex assay. The duplex DNA containing the SspI and NdeI restriction endonuclease sites represents the duplex plasmid DNA substrate. (C) Schematic of an oligonucleotide-based synaptic complex assay. Fluorescein and rhodamine are represented in green and red, respectively. Note that this methodology may be used for real-time measurement of the reaction as it occurs in vitro. (D) Schematic of the D-loop formed by Dmc1. (E) Schematic of an oligonucleotide-based strand exchange reaction. dsDNA and ssDNA are represented in red and blue, respectively. The green label represents 3′ labeling by the fluorophore or radioisotope.
Several experimental approaches have been developed using purified systems that recapitulate all steps leading to strand exchange. For example, recombinase-dependent pairing of two homologous DNAs or synaptic complex formation (Fig.3B, C) 30,37,38 may be used to study the mechanism of homology search (see below). In the D-loop assay, a short radiolabeled oligonucleotide is used to detect invasion of ssDNA into negatively supercoiled duplex, displacing the non-complementary strand into a D-loop structure (Fig.3D) 39. The ssDNA in the D-loop assay represents the ssDNA tail of the resected DSBs, and the dsDNA plasmid with homology to the ssDNA represents the targeted sequence. In the oligonucleotide-based version of another common assay (DNA strand exchange), one strand of a linear double-stranded oligonucleotide is replaced by a labeled linear single-stranded nucleoprotein complex, creating a labeled duplex and a linear ssDNA (Fig.3E) 40,41. Methodologies have been also devised to evaluate less stable consecutive intermediates that lead to strand invasion: assembly and stability of nucleoprotein filaments formed by the recombinase and ssDNA may be studied by electron microscopy 24,26,42–44, DNA binding shift assay, surface plasmon resonance (for precise measurement of the kinetics of DNA binding) 30, exonuclease I protection 30,45,46 and recombinase turnover from ssDNA 36, while a dsDNA capture assay may be used to study homology-independent conjoining of DNA molecules 30,36.
Dmc1 mechanism of homology search
During a homology search, segments of the intact duplex DNA are bound by the Dmc1–ssDNA nucleoprotein filaments and tested reiteratively until homology is found. While it is still unclear how Dmc1 and other members of the RecA family of proteins perform genome-wide homology searches, several groups have shown that, upon identification of homology in the duplex DNA molecule, the pre-synaptic filament of RecA is able to form a stable synaptic complex consisting of three strands and the recombinase, in which strand exchange has already taken place 47,48. In this complex, the invading ssDNA is part of the new duplex, and the leaving strand has not yet been released. An important question here relates to the steps that lead to this exchange intermediate? Studies of RecA 48,49, Rad51 50,51 and Dmc1 38 have shown that formation of a synaptic complex by any of these recombinases includes the transition through several slower conformational changes, such as the later stages of homology recognition, which involve localized melting (base flipping) and annealing (switching) at A.T-rich regions. In these experiments, a fluorescein-labeled ssDNA oligonucleotide (negative strand) in combination with a rhodamine-labeled homologous dsDNA is used to measure fluorescence resonance energy transfer (Fig.3C). In synaptic complex formation, a ternary complex (three-strand) intermediate forms, bringing fluorescein and rhodamine into close proximity. In this state, the fluorescent dyes undergo fluorescence resonance energy transfer and fluorescein is quenched. This is detected as a reduction in the fluorescein-sensitized emission, and indicates formation of the first interactions between homologous DNA strands. In a synaptic complex formed with the recombinase (Dmc1, Rad51 or RecA), substitution of inosine for guanine (which destabilizes the duplex DNA that stimulates strand exchange) demonstrated a general effect of helix stability on recognition of homology, and A·T mismatches demonstrated a special role of A·T base pairs in recognition of homology 38. This suggests that the dynamic structure of the double helix (DNA ‘breathing’) significantly contributes to recognition of homology. In this case, it is possible that the extended conformation of the recombinase–ssDNA pre-synaptic filament allows rotational mobility of the bases, making them available to test interactions through collisions with transiently opened base pairs in the DNA duplex.
Dmc1 versus Rad51 in meiotic recombination
What is the functional relationship between Rad51 and Dmc1 during normal meiosis? From genetic experiments in budding yeast, it is known that, when Rad51 and Dmc1 work together, the repair of DSBs is preferentially directed to strand exchange between homologous chromosomes rather than sister chromatids. In addition, a Rad51 deletion leads to a notable reduction of inter-homolog recombination, and inter-sister chromatid repair prevails. In the absence of Dmc1, however, a dramatic reduction of both inter-sister and inter-homolog recombination is observed. In the latter case, the strand exchange activity of budding yeast Rad51 is inhibited by the Hed1 protein and the effector kinase Mek1. Removing this inhibition allows efficient recombination, although inter-homolog crossovers are reduced compared to wild-type. Two recent studies in S. cerevisiae reveal that the favored inter-homolog strand exchange activity mediated by Dmc1 requires inhibition of Rad51 strand exchange activity 52,53. These studies imply that the inhibitory action of Hed1 on Rad51 recombinational activity converts Rad51 from a recombination enzyme to a recombination mediator. In agreement with this idea, it has been shown that, in meiosis, the prominent strand exchange activity is exhibited by Dmc1, not Rad51 54. In a series of cleverly designed experiments, the authors used a separation-of-function mutant to show that the ability of Rad51 to interact with DNA and form nucleoprotein filaments, but not its strand exchange activity, is a prerequisite for normal meiotic recombination. In the same study, experiments with purified proteins showed that Rad51 is very efficient in stimulating the strand exchange activity of Dmc1.
What are the intrinsic biochemical properties that functionally distinguish Dmc1 from Rad51? Dmc1 and Rad51 share 54% amino acid identity in humans, 52% in mouse, and 45% in yeast. In vitro, both purified recombinant Rad51 and Dmc1 bind ssDNA to form helical nucleoprotein filaments and promote DNA strand exchange. Despite similarities in the general mechanism of recombination, Dmc1 and Rad51 show differences in a number of structural/biochemical properties. For example, DNA unstacking, and consequently reactivity toward chemical modification of thymines of Rad51/Dmc1 nucleoprotein complexes, are notably different 46. Differences have been also observed between Dmc1–ssDNA and Rad51–ssDNA nucleoprotein filaments, although the reported number of Dmc1 and Rad51 promoters per helical turn varies between reports 42,55,56. Dissimilarities in the structure of Rad51 versus Dmc1 nucleoprotein complexes may account for the increased resistance of native Dmc1 D-loops compared with Rad51 D-loops to dissociation by DNA translocases, such as bloom syndrome protein and Rad54 46. This difference in stability indicates a biochemical distinction between intermediates of recombination catalyzed by Rad51 and Dmc1. In addition to the differences stated above, the rate of ATP binding and hydrolysis, protein polymerization rate and the kinetics of DNA binding and dissociation are critical biochemical properties that have not yet been fully investigated in a comparative fashion and may account for the differences in Dmc1 and Rad51 functions. In an alternative view, unique meiotic functions for Rad51 and Dmc1 are more likely to result from the influence of distinct sets of accessory proteins than intrinsic differences in their biochemical properties.
Recombination mediators and Dmc1-promoted DNA strand exchange
Proper function of Dmc1 in vitro and in vivo requires interactions with several meiotic accessory proteins 7,57,58. These protein factors determine regulatory mechanisms that direct the choices of Dmc1 DNA repair pathways. Biochemically, they stimulate critical steps of the recombination mechanism catalyzed by Dmc1, such as the dynamics and targeting of filament assembly on ssDNA, the choice of homologous DNA for strand invasion, and protection of Dmc1 removal from DNA by factors such as helicases. Understanding HR requires a detailed understanding of the identities and activities of these accessory proteins. Here, we present an up-to-date overview of how one of these factors, the Hop2–Mnd1 complex, stimulates Dmc1.
Hop2 and Mnd1 have a dual role in recombinase enhancement
The MND1 gene was first described in a screen for genes with meiosis-specific expression in S. cerevisiae 59, and in the null mutant strain, cells initiate recombination, but do not form heteroduplex DNA and exhibit hyper-resected DSBs 60. This suggests that Mnd1 may be involved in strand exchange. Similarly, depletion of MCP7, the S. pombe ortholog of MND1, resulted in cell arrest in meiotic prophase, with a reduction in recombination rates 61. In higher eukaryotes, the MND1 gene is required for normal male and female fertility. For example, in mouse 41 and Arabidopsis thaliana 62,63, mutation of the MND1 gene results in normal recombination initiation, but meiotic DSBs are abnormally repaired, accompanied by aberrant chromosome synapsis. Similar to the MND1 mutant phenotype and consistent with a defect in a Hop2-dependent step during meiotic recombination, S. cerevisiae 64 and A. thaliana HOP2 deletion mutants exhibit a profound failure in meiosis, due to uniform arrest at meiosis I, with chromosomes engaged at synapsis with non-homologous partners 65. Analysis of mouse HOP2 knockout spermatocytes suggests that DSBs are created and processed, but their repair is abnormal as indicated by the accumulation of Dmc1 and Rad51 proteins at DNA repair sites 66. In sum, genetic and cellular analysis of deletion mutants in various species suggests that Hop2 and Mnd1 act in the same pathway of recombination, and the proteins have a conserved role in efficient DSB repair and normal homologous chromosome synapsis.
The functions of Hop2 and Mnd1 proteins when they act together as a complex have been studied more extensively. The functional interaction between Hop2 and Mnd1 was first suggested in studies of budding yeast, in which MND1 acts as a multi-copy suppressor of a HOP2 mutation defect in viable spore production, and these proteins co-immunoprecipitate from meiotic cell extracts 67. The cooperation between Hop2/Mnd1 and Dmc1/Rad51 is likely to be crucial in vivo. For example, in mice lacking Hop2 and/or Mnd1, progression of recombination is impaired immediately after Dmc1 and Rad51 are loaded onto the end of DSBs 41,66. Finally, in all organisms analyzed so far, HOP2 and MND1 only appear in those genomes that carry DMC1.
In vitro, Hop2 shows two distinctive activities. First, when it is incorporated into a Hop2–Mnd1 complex, it stimulates Dmc1/Rad51-promoted recombination. This appears to be a prominent activity of Hop2 that has so far been observed using purified recombinant proteins from mouse 36,68–71, budding yeast 72, fission yeast 73, and A. thaliana 74. Second, purified mouse Hop2 alone and independently of Dmc1 and Rad51 is capable of catalyzing strand invasion 41,68,69. Although this intrinsic recombinational activity of Hop2 shares mechanistic signatures characteristic of the mammalian RecA-like recombinases, it shows distinctive characteristics. For example, Hop2-mediated strand exchange does not require ATP, and, in contrast to Dmc1, joint molecules formed by Hop2 are more sensitive to mismatches and are efficiently dissociated by the branch migration protein Rad54 41. Recent work in mouse spermatocytes suggested that Hop2 may work alone as a recombinase 41. The authors reason that Dmc1 and Rad51 are inactive in the absence of the Hop2–Mnd1 complex. Therefore, deletion of Mnd1 in mouse spermatocytes leaves Hop2 as the only protein with recombinase activity. In agreement with this possibility, a proportion of Mnd1 knockout spermatocytes show a significantly high level of DSB repair (monitored by histone γ-H2AX and Dmc1/Rad51) and chromosome synapsis. Although these results indicate that DSB repair catalyzed solely by Hop2 may promote homologous chromosome pairing and synapsis, further evidence is required to demonstrate that Hop2 performs homology search and strand exchange in vivo in the absence of Dmc1 and Rad51 (i.e. analysis of Rad51−/−/Dmc1−/−/Mnd1−/− mutant spermatocytes is required).
In the context of Hop2 functions, a possible mode of action for the Mnd1 protein has been revealed, as interaction of Mnd1 with Hop2 down-regulates the D-loop formation activity of Hop2. Interestingly, Mnd1 inhibits the recombinase activity of Hop2, and, when incorporated into the Hop2–Mnd1 complex, promotes strand invasion mediated by Dmc1 and Rad51. It is proposed that Mnd1 works by producing changes in the biochemical properties and oligomerization state of Hop2. These changes result in a new molecular interface in the Hop2–Mnd1 complex that is responsible for Hop2-Mnd1 interaction and stimulation of the Dmc1 and Rad51 recombinases 68.
What is the molecular mechanism directing Hop2–Mnd1 complex stimulation of recombinase-mediated strand exchange? As stated above, efficient strand exchange promoted by Dmc1 and Rad51 requires formation of pre-synaptic ssDNA–Dmc1 nucleoprotein filaments and adjoining of homologous dsDNA and ssDNA. The Hop2–Mnd1 complex acts in these two critical stages of the recombination reaction 30,36. First, purified Hop2–Mnd1 binds and stabilizes the pre-synaptic filament formed by Dmc1/Rad51 on ssDNA. In a second reaction, Hop2–Mnd1 enhances the ability of the recombinase–ssDNA nucleoprotein filament to capture the duplex target DNA (Fig.4). Although Hop2–Mnd1 stimulation of the duplex DNA capture is homology-independent, this step is of paramount importance. Such capture is vital for promoting recombinase-mediated DNA pairing. In this reaction, it is proposed that unaligned DNA molecules transition to an aligned ternary complex that facilitates the homology search. Although strong in vitro evidence exists to support a bipartite action of Hop2 on both stabilization and the ability of a recombinase–ssDNA nucleoprotein filament to capture dsDNA, it remains to be determined whether these Hop2–Mnd1 activities contribute to the function of Dmc1 and Rad51 in vivo.
Figure 4.
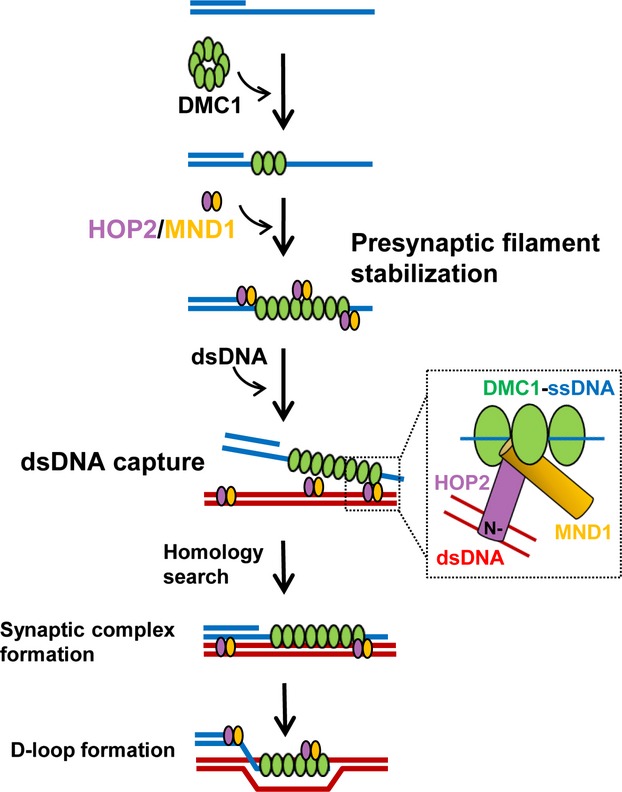
The Hop2–Mnd1 complex acts by enhancing two separate and critical stages of the DMC1-promoted recombination process. Hop2–Mnd1 first stabilizes Dmc1 filaments on the resected end of the DSBs, and then enhances the ability of the pre-synaptic filament to capture a dsDNA molecule.
The underlying molecular basis for this bipartite action of Hop2–Mnd1, i.e. in both stabilization of the Dmc1 pre-synaptic filament and assembly of the synaptic complex, has been further clarified in recent work. One of these reports utilized a combination of structural and biochemical approaches to show that the heterodimeric Hop2–Mnd1 complex is a V-shaped molecule, and that the dsDNA binding functions of the N-termini of Hop2 and Mnd1 work together to promote synaptic complex assembly, whereas the Hop2 C-terminus, which binds ssDNA, participates in stabilization of the Dmc1–ssDNA filament 71. The biochemical function of the Hop2 N-terminus is highlighted by the fact that this domain is a typical winged helix DNA-binding domain with specific amino acids involved in dsDNA coordination 75. The second report provides more mechanistic insights, showing that Hop2–Mnd1 has the ability to stimulate DNA strand exchange by modulating a range of Rad51 basic properties, particularly nucleotide and DNA binding 76. It has been shown that Hop2–Mnd1 enables Rad51 DNA strand exchange, even in the absence of divalent metal ions required for ATP binding. In addition, Hop2–Mnd1 acts in two steps of the Rad51-mediated recombination mechanism. First, during nucleoprotein formation, Hop2–Mnd1 helps to load Rad51 on ssDNA, restricting its dsDNA binding ability. Second, it promotes dsDNA binding during the homology search, by removing the inhibitory effect of ssDNA.
Hop2 as a potential tumor suppressor gene
To date, the number of ovarian and breast cancer susceptibility genes identified accounts for less than half of hereditary breast and ovarian cancers. The finding of germline mutations in BRCA2 and related genes, such as PALB2, RAD51C and RAD51D, suggests that DNA recombination repair pathways are linked to breast and ovarian cancer genes. The recent finding of a family tied to ovarian dysgenesis, carrying a deletion of Glu201 in the C-terminal acidic domain of Hop2, supports the critical role for this protein in ovarian development 77. Subsequent screening of germline mutations in familial and early-onset breast and ovarian cancers detected several mutations affecting Hop2 78,79. Interestingly, some of these mutations resulted in truncated versions of Hop2 with dominant-negative activity. Mutations in the HOP2 gene that de-regulated alternative splicing in cells derived from familial ovarian and breast cancer patients have also been found. Some of these splice variants act as dominant-negative mutants that abolish Rad51 foci formation during radiation-induced DNA damage. Together with results showing that constitutive expression of abnormal splice variants of HOP2 induces tumor growth in nude mice 79, these reports strongly suggest a role for inactivating HOP2 mutations in familial and early-onset breast and ovarian cancers.
Zebrafish, an emerging model for studies of recombination in meiosis
Current understanding of the genetic controls involved in meiotic recombination of eukaryotes primarily comes from studies of model organisms such as yeast. However, these studies have some limitations because many genes required for meiosis in higher eukaryotes have no orthologs in yeast, and there are clear meiotic differences between mouse and yeast 80–82. Alternatively, a mouse spermatocyte system, a model that more closely resembles humans, has proved an excellent model, advancing the field by allowing high-resolution observation of chromosome structures that is not possible in simple systems. A forward genetic screen in the mouse demonstrated that many vertebrate meiosis genes are yet to be found 82. By screening over 17 000 N-ethyl-N-nitrosourea-mutagenized mice, the Reproductive Genomics Group at Jackson Laboratory identified 44 mutant lines with reduced fertility. While many of the mutated genes from this screen have not yet been identified, this great effort revealed novel meiotic functions for six proteins 82. The discovery of the first meiosis gene from this screen, Mei1, exemplifies the advantage of screening for meiosis genes in vertebrates 83. Although it provided valuable new information and tools, the Jackson Laboratory screen also suggested that it will be very difficult to uncover all genes required for vertebrate meiosis through forward genetics studies in the mouse.
Reverse genetics studies in the mouse have been very effective in identifying factors necessary for vertebrate meiosis. Vertebrate meiosis gene candidates identified through genetic screens in lower organisms, gene expression analyses, or protein interaction studies have been functionally analyzed with great success using engineered mouse mutants 82. Although the mouse will probably continue to be the model-of-choice for vertebrates in the meiosis field, the development of state-of-the-art genetics tools such as engineered site-specific nucleases should make us reconsider whether other model organisms may be better suited to ‘reverse genetic’ screening for meiosis genes.
We propose that the zebrafish (Danio rerio) may serve as a powerful genetic screening platform for the meiosis field. A productive research community has grown around the use of zebrafish, but, without efficient targeted mutagenesis techniques, the use of zebrafish has predominantly been limited to studying embryonic development. The barrier to efficiently producing targeted mutations has been overcome by the development of engineered nucleases 84. While forward genetic screens for adult phenotypes in zebrafish requires more labor and space than most laboratories are able to commit, screening dozens or even hundreds of genes using engineered nucleases should be possible with the resources and capabilities of most small laboratories 84. This approach may also be taken with the mouse, but the zebrafish offers several advantages. First, adult zebrafish may be raised and maintained at higher densities and for less cost than mice. Second, the large clutch sizes, external development and accessibility of zebrafish embryos make it relatively easy for laboratories to quickly introduce nucleases by microinjection. Finally, homozygous gynogenetic diploid offspring may be generated from F1 fish or even highly mutagenized founders using the well-characterized early-pressure technique 85.
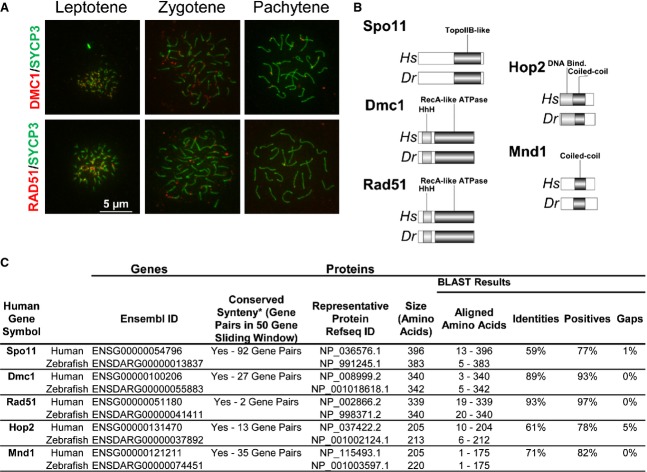
The use of immunofluorescence microscopy to visualize antibodies against components of the synaptic complex and cytological markers of recombination (i.e. Mlh1 and RPA) on whole-mount preparations of zebrafish spermatocytes has been shown to be an important resource for providing physical evidence of where and when recombination is occurring at the chromosomal level 86. Here, we present our results obtained with polyclonal antibodies that specifically detect Dmc1 and Rad51 on chromosome cores (revealed by Sycp3 immunostaining) of zebrafish spermatocyte nuclear spreads (Fig.5A). Initial studies on Dmc1 and Rad51 localization at recombination sites, in the context of meiotic progression, revealed that loading of both recombinases onto chromosomes is observed in leptotene and zygotene, and some remnant foci may still be detected at pachytene (Fig.5A).
Figure 5.
Zebrafish as a model for meiotic recombination studies. (A) Visualization of Dmc1 and Rad51 loading at chromosomal recombination sites. The methodologies used for chromosome surface spreading of zebrafish spermatocytes and immunolabeling are similar to those previously described for mouse spermatocytes 70,96. Sources and dilutions of primary antibodies used are as follows. Rabbit anti-Rad51 serum (H-92; Santa Cruz Biotechnology, Dallas, TX, USA), produced against an epitope corresponding to amino acids 1–92 of Rad51 of human origin (84% identity to the zebrafish Rad51 protein sequence), was used at a dilution of 1 : 100. Goat anti-Dmc1 serum (C-20; Santa Cruz Biotechnology), produced against an epitope corresponding to amino acids 290–340 of Dmc1 of human origin (90% identity to the zebrafish Dmc1 protein sequence), was used at a dilution of 1 : 100. Mouse anti-SYCP3 (Novus, Littleton, CO, USA) was used at a dilution of 1 : 400. Secondary antibodies were obtained from Jackson IR Laboratories (West Grove, PA, USA) and were used at a dilution of 1 : 300. Slides were counterstained using Vectashield mounting solution (Vector Laboratories, Burlingame, CA, USA) containing 2 μg·mL−1 4′,6-diamidino-2-phenylindole (DAPI). All images were acquired using a 40× objective oil immersion lens. We used an Axiovision SE 64 microscope (Zeiss, Thornwood, NY, USA) for imaging acquisition and processing. Scale bar = 5 μm for all images. (B) Structural conservation of proteins acting in homologous recombination in human and zebrafish. (C) Conservation of HR effectors between human and zebrafish.
Studies using N-ethyl-N-nitrosourea mutagenesis to generate zebrafish knockouts for proteins involved in meiotic prophase I 87, the successful use of reverse genetics 88–91, and outstanding imaging studies (this work and 86,87,92–94) illustrate the versatility of this model for studying the meiotic processes and defects. The meiotic segregation defects in zebrafish appear to have substantial similarities to those observed in mammals 88,90,95, and, importantly, may model mechanisms underlying human miscarriages. We found that homologs of important human meiosis genes are also present in zebrafish, and the protein sequences are highly conserved (Fig.5B, C). It is likely that additional vertebrate meiosis genes have yet to be discovered. The zebrafish promises to be a powerful model system for discovery and analysis of new vertebrate meiosis genes.
Perspective
Here we have reviewed advances regarding the critical activities in rate-limiting steps of early stages of recombination. We specifically focused in recent biochemical and genetic progress to elucidate the mechanisms of synaptic events in meiotic recombination. This information has revealed important mechanistic information regarding the molecular mode of action of recombinases and functional interaction of recombinases with ancillary proteins assisting in several steps of the DNA strand exchange process. However, these studies have also generated a number of hypotheses regarding the interplay between recombinases during the stand exchange reaction, and raised the question of whether certain ancillary proteins have a specific relationship with one of the two recombinases. Other important specific questions remain. What are the intrinsic structural and biochemical differences explaining the specific functions of Dmc1 and Rad51 in meiotic recombination? What is the 3D structure of DNA–protein complexes involved in strand exchange? What are the mechanisms by which ancillary factors channel recombination intermediates into various HR pathways? Future work is required to answer these questions. The answers may provide the information necessary to connect recombination events and chromosome interactions that ensure correct homologous chromosome distribution to create a balanced number of chromosomes in gametes.
Author contribution
CLS and RJP planned experiments, performed experiments, analyzed data, and wrote the paper.
Glossary
- DSB
double-strand break
- DSBR
double-strand break repair
- dsDNA
double-stranded DNA
- HR
homologous recombination
- SDSA
synthesis-dependent strand annealing
- ssDNA
single-stranded DNA
References
- Rankin S. Complex elaboration: making sense of meiotic cohesin dynamics. FEBS J. 2015 doi: 10.1111/febs.13301. doi: 10.1111/febs.13301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques F. Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Imai Y. de Massy B. Meiotic recombination in mammals: localization and regulation. Nat Rev Genet. 2013;14:794–806. doi: 10.1038/nrg3573. [DOI] [PubMed] [Google Scholar]
- Keeney S, Giroux CN. Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- Neale MJ, Pan J. Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Phelps SE, Gray S. Neale MJ. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature. 2011;479:241–244. doi: 10.1038/nature10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ. Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers T. Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- Hunter N. Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- Guiraldelli MF, Eyster C, Wilkerson JL, Dresser ME. Pezza RJ. Mouse HFM1/Mer3 is required for crossover formation and complete synapsis of homologous chromosomes during meiosis. PLoS Genet. 2013;9:e1003383. doi: 10.1371/journal.pgen.1003383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazina OM, Mazin AV, Nakagawa T, Kolodner RD. Kowalczykowski SC. Saccharomyces cerevisiae Mer3 helicase stimulates 3′-5′ heteroduplex extension by Rad51; implications for crossover control in meiotic recombination. Cell. 2004;117:47–56. doi: 10.1016/s0092-8674(04)00294-6. [DOI] [PubMed] [Google Scholar]
- Mimitou EP. Symington LS. Nucleases and helicases take center stage in homologous recombination. Trends Biochem Sci. 2009;34:264–272. doi: 10.1016/j.tibs.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Petronczki M, Siomos MF. Nasmyth K. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–440. doi: 10.1016/s0092-8674(03)00083-7. [DOI] [PubMed] [Google Scholar]
- Goldfarb T. Lichten M. Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLoS Biol. 2010;8:e1000520. doi: 10.1371/journal.pbio.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L. Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Masson JY. West SC. The Rad51 and Dmc1 recombinases: a non-identical twin relationship. Trends Biochem Sci. 2001;26:131–136. doi: 10.1016/s0968-0004(00)01742-4. [DOI] [PubMed] [Google Scholar]
- Pittman DL, Cobb J, Schimenti KJ, Wilson LA, Cooper DM, Brignull E, Handel MA. Schimenti JC. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol Cell. 1998;1:697–705. doi: 10.1016/s1097-2765(00)80069-6. [DOI] [PubMed] [Google Scholar]
- Couteau F, Belzile F, Horlow C, Grandjean O, Vezon D. Doutriaux MP. Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell. 1999;11:1623–1634. doi: 10.1105/tpc.11.9.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A. Kleckner N. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90:1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- Humphryes N. Hochwagen A. A non-sister act: recombination template choice during meiosis. Exp Cell Res. 2014;329:53–60. doi: 10.1016/j.yexcr.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, Sym M, Scherthan H. Roeder GS. Roles for two RecA homologs in promoting meiotic chromosome synapsis. Genes Dev. 1995;9:2684–2695. doi: 10.1101/gad.9.21.2684. [DOI] [PubMed] [Google Scholar]
- Young JA, Hyppa RW. Smith GR. Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics. 2004;167:593–605. doi: 10.1534/genetics.103.023762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A, Gasior S, Ogawa T, Kleckner N. Bishop DK. Saccharomyces cerevisiae recA homologues RAD51 and DMC1 have both distinct and overlapping roles in meiotic recombination. Genes Cells. 1997;2:615–629. doi: 10.1046/j.1365-2443.1997.1480347.x. [DOI] [PubMed] [Google Scholar]
- Sehorn MG, Sigurdsson S, Bussen W, Unger VM. Sung P. Human meiotic recombinase Dmc1 promotes ATP-dependent homologous DNA strand exchange. Nature. 2004;429:433–437. doi: 10.1038/nature02563. [DOI] [PubMed] [Google Scholar]
- Li Z, Golub EI, Gupta R. Radding CM. Recombination activities of HsDmc1 protein, the meiotic human homolog of RecA protein. Proc Natl Acad Sci USA. 1997;94:11221–11226. doi: 10.1073/pnas.94.21.11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson JY, Davies AA, Hajibagheri N, Van Dyck E, Benson FE, Stasiak AZ, Stasiak A. West SC. The meiosis-specific recombinase hDmc1 forms ring structures and interacts with hRad51. EMBO J. 1999;18:6552–6560. doi: 10.1093/emboj/18.22.6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camerini-Otero RD. Hsieh P. Homologous recombination proteins in prokaryotes and eukaryotes. Annu Rev Genet. 1995;29:509–552. doi: 10.1146/annurev.ge.29.120195.002453. [DOI] [PubMed] [Google Scholar]
- Bianco PR, Tracy RB. Kowalczykowski SC. DNA strand exchange proteins: a biochemical and physical comparison. Front Biosci. 1998;3:D570–D603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- Cox MM. The bacterial RecA protein as a motor protein. Annu Rev Microbiol. 2003;57:551–577. doi: 10.1146/annurev.micro.57.030502.090953. [DOI] [PubMed] [Google Scholar]
- Pezza RJ, Voloshin ON, Vanevski F. Camerini-Otero RD. Hop2/Mnd1 acts on two critical steps in Dmc1-promoted homologous pairing. Genes Dev. 2007;21:1758–1766. doi: 10.1101/gad.1562907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H. Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- New JH, Sugiyama T, Zaitseva E. Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- Haruta N, Kurokawa Y, Murayama Y, Akamatsu Y, Unzai S, Tsutsui Y. Iwasaki H. The Swi5-Sfr1 complex stimulates Rhp51/Rad51- and Dmc1-mediated DNA strand exchange in vitro. Nat Struct Mol Biol. 2006;13:823–830. doi: 10.1038/nsmb1136. [DOI] [PubMed] [Google Scholar]
- Flory SS, Tsang J, Muniyappa K, Bianchi M, Gonda D, Kahn R, Azhderian E, Egner C, Shaner S. Radding CM. Intermediates in homologous pairing promoted by RecA protein and correlations of recombination in vitro and in vivo. Cold Spring Harb Symp Quant Biol. 1984;49:513–523. doi: 10.1101/sqb.1984.049.01.058. [DOI] [PubMed] [Google Scholar]
- Chi P, San Filippo J, Sehorn MG, Petukhova GV. Sung P. Bipartite stimulatory action of the Hop2–Mnd1 complex on the Rad51 recombinase. Genes Dev. 2007;21:1747–1757. doi: 10.1101/gad.1563007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh P, Camerini-Otero CS. Camerini-Otero RD. The synapsis event in the homologous pairing of DNAs: RecA recognizes and pairs less than one helical repeat of DNA. Proc Natl Acad Sci USA. 1992;89:6492–6496. doi: 10.1073/pnas.89.14.6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RC, Golub E, Bi B. Radding CM. The synaptic activity of HsDmc1, a human recombination protein specific to meiosis. Proc Natl Acad Sci USA. 2001;98:8433–8439. doi: 10.1073/pnas.121005298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, DasGupta C, Cunningham RP. Radding CM. Purified Escherichia coli recA protein catalyzes homologous pairing of superhelical DNA and single-stranded fragments. Proc Natl Acad Sci USA. 1979;76:1638–1642. doi: 10.1073/pnas.76.4.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volodin AA, Bocharova TN, Smirnova EA. Camerini-Otero RD. Reversibility, equilibration, and fidelity of strand exchange reaction between short oligonucleotides promoted by RecA protein from Escherichia coli and human Rad51 and Dmc1 proteins. J Biol Chem. 2009;284:1495–1504. doi: 10.1074/jbc.M800612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezza RJ, Voloshin ON, Volodin AA, Boateng KA, Bellani MA, Mazin AV. Camerini-Otero RD. The dual role of HOP2 in mammalian meiotic homologous recombination. Nucleic Acids Res. 2014;42:2346–2357. doi: 10.1093/nar/gkt1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan SD, Yu X, Roth R, Heuser JE, Sehorn MG, Sung P, Egelman EH. Bishop DK. A comparative analysis of Dmc1 and Rad51 nucleoprotein filaments. Nucleic Acids Res. 2008;36:4057–4066. doi: 10.1093/nar/gkn352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Jacobs SA, West SC, Ogawa T. Egelman EH. Domain structure and dynamics in the helical filaments formed by RecA and Rad51 on DNA. Proc Natl Acad Sci USA. 2001;98:8419–8424. doi: 10.1073/pnas.111005398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passy SI, Yu X, Li Z, Radding CM, Masson JY, West SC. Egelman EH. Human Dmc1 protein binds DNA as an octameric ring. Proc Natl Acad Sci USA. 1999;96:10684–10688. doi: 10.1073/pnas.96.19.10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusetti SL, Voloshin ON, Inman RB, Camerini-Otero RD. Cox MM. The DinI protein stabilizes RecA protein filaments. J Biol Chem. 2004;279:30037–30046. doi: 10.1074/jbc.M403064200. [DOI] [PubMed] [Google Scholar]
- Bugreev DV, Pezza RJ, Mazina OM, Voloshin ON, Camerini-Otero RD. Mazin AV. The resistance of DMC1 D-loops to dissociation may account for the DMC1 requirement in meiosis. Nat Struct Mol Biol. 2011;18:56–60. doi: 10.1038/nsmb.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca AI. Cox MM. RecA protein: structure, function, and role in recombinational DNA repair. Prog Nucleic Acid Res Mol Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- Folta-Stogniew E, O’Malley S, Gupta R, Anderson KS. Radding CM. Exchange of DNA base pairs that coincides with recognition of homology promoted by E. coli RecA protein. Mol Cell. 2004;15:965–975. doi: 10.1016/j.molcel.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Bazemore LR, Takahashi M. Radding CM. Kinetic analysis of pairing and strand exchange catalyzed by RecA. Detection by fluorescence energy transfer. J Biol Chem. 1997;272:14672–14682. doi: 10.1074/jbc.272.23.14672. [DOI] [PubMed] [Google Scholar]
- Gupta RC, Folta-Stogniew E, O’Malley S, Takahashi M. Radding CM. Rapid exchange of A: T base pairs is essential for recognition of DNA homology by human Rad51 recombination protein. Mol Cell. 1999;4:705–714. doi: 10.1016/s1097-2765(00)80381-0. [DOI] [PubMed] [Google Scholar]
- Gupta RC, Folta-Stogniew E. Radding CM. Human Rad51 protein can form homologous joints in the absence of net strand exchange. J Biol Chem. 1999;274:1248–1256. doi: 10.1074/jbc.274.3.1248. [DOI] [PubMed] [Google Scholar]
- Lao JP, Cloud V, Huang CC, Grubb J, Thacker D, Lee CY, Dresser ME, Hunter N. Bishop DK. Meiotic crossover control by concerted action of Rad51-Dmc1 in homolog template bias and robust homeostatic regulation. PLoS Genet. 2013;9:e1003978. doi: 10.1371/journal.pgen.1003978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Gaines WA, Callender T, Busygina V, Oke A, Sung P, Fung JC. Hollingsworth NM. Down-regulation of Rad51 activity during meiosis in yeast prevents competition with Dmc1 for repair of double-strand breaks. PLoS Genet. 2014;10:e1004005. doi: 10.1371/journal.pgen.1004005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloud V, Chan YL, Grubb J, Budke B. Bishop DK. Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science. 2012;337:1222–1225. doi: 10.1126/science.1219379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okorokov AL, Chaban YL, Bugreev DV, Hodgkinson J, Mazin AV. Orlova EV. Structure of the hDmc1-ssDNA filament reveals the principles of its architecture. PLoS One. 2010;5:e8586. doi: 10.1371/journal.pone.0008586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X. Egelman EH. Helical filaments of human Dmc1 protein on single-stranded DNA: a cautionary tale. J Mol Biol. 2010;401:544–551. doi: 10.1016/j.jmb.2010.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J, Sung P. Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Kagawa W. Kurumizaka H. From meiosis to postmeiotic events: uncovering the molecular roles of the meiosis-specific recombinase Dmc1. FEBS J. 2010;277:590–598. doi: 10.1111/j.1742-4658.2009.07503.x. [DOI] [PubMed] [Google Scholar]
- Rabitsch KP, Toth A, Galova M, Schleiffer A, Schaffner G, Aigner E, Rupp C, Penkner AM, Moreno-Borchart AC, Primig M, et al. A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr Biol. 2001;11:1001–1009. doi: 10.1016/s0960-9822(01)00274-3. [DOI] [PubMed] [Google Scholar]
- Gerton JL. DeRisi JL. Mnd1p: an evolutionarily conserved protein required for meiotic recombination. Proc Natl Acad Sci USA. 2002;99:6895–6900. doi: 10.1073/pnas.102167899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito TT, Tougan T, Kasama T, Okuzaki D. Nojima H. Mcp7, a meiosis-specific coiled-coil protein of fission yeast, associates with Meu13 and is required for meiotic recombination. Nucleic Acids Res. 2004;32:3325–3339. doi: 10.1093/nar/gkh654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenichini S, Raynaud C, Ni DA, Henry Y. Bergounioux C. Atmnd1-delta1 is sensitive to gamma-irradiation and defective in meiotic DNA repair. DNA Repair (Amst) 2006;5:455–464. doi: 10.1016/j.dnarep.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Kerzendorfer C, Vignard J, Pedrosa-Harand A, Siwiec T, Akimcheva S, Jolivet S, Sablowski R, Armstrong S, Schweizer D, Mercier R, et al. The Arabidopsis thaliana MND1 homologue plays a key role in meiotic homologous pairing, synapsis and recombination. J Cell Sci. 2006;119:2486–2496. doi: 10.1242/jcs.02967. [DOI] [PubMed] [Google Scholar]
- Leu JY, Chua PR. Roeder GS. The meiosis-specific Hop2 protein of S. cerevisiae ensures synapsis between homologous chromosomes. Cell. 1998;94:375–386. doi: 10.1016/s0092-8674(00)81480-4. [DOI] [PubMed] [Google Scholar]
- Vignard J, Siwiec T, Chelysheva L, Vrielynck N, Gonord F, Armstrong SJ, Schlogelhofer P. Mercier R. The interplay of RecA-related proteins and the MND1-HOP2 complex during meiosis in Arabidopsis thaliana. PLoS Genet. 2007;3:1894–1906. doi: 10.1371/journal.pgen.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukhova GV, Romanienko PJ. Camerini-Otero RD. The Hop2 protein has a direct role in promoting interhomolog interactions during mouse meiosis. Dev Cell. 2003;5:927–936. doi: 10.1016/s1534-5807(03)00369-1. [DOI] [PubMed] [Google Scholar]
- Tsubouchi H. Roeder GS. The Mnd1 protein forms a complex with Hop2 to promote homologous chromosome pairing and meiotic double-strand break repair. Mol Cell Biol. 2002;22:3078–3088. doi: 10.1128/MCB.22.9.3078-3088.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezza RJ, Petukhova GV, Ghirlando R. Camerini-Otero RD. Molecular activities of meiosis-specific proteins Hop2, Mnd1, and the Hop2–Mnd1 complex. J Biol Chem. 2006;281:18426–18434. doi: 10.1074/jbc.M601073200. [DOI] [PubMed] [Google Scholar]
- Petukhova GV, Pezza RJ, Vanevski F, Ploquin M, Masson JY. Camerini-Otero RD. The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat Struct Mol Biol. 2005;12:449–453. doi: 10.1038/nsmb923. [DOI] [PubMed] [Google Scholar]
- Bannister LA, Pezza RJ, Donaldson JR, de Rooij DG, Schimenti KJ, Camerini-Otero RD. Schimenti JC. A dominant, recombination-defective allele of Dmc1 causing male-specific sterility. PLoS Biol. 2007;5:e105. doi: 10.1371/journal.pbio.0050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Saro D, Hammel M, Kwon Y, Xu Y, Rambo RP, Williams GJ, Chi P, Lu L, Pezza RJ, et al. Mechanistic insights into the role of Hop2–Mnd1 in meiotic homologous DNA pairing. Nucleic Acids Res. 2014;42:906–917. doi: 10.1093/nar/gkt924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YL, Brown MS, Qin D, Handa N. Bishop DK. The third exon of the budding yeast meiotic recombination gene HOP2 is required for calcium-dependent and recombinase Dmc1-specific stimulation of homologous strand assimilation. J Biol Chem. 2014;289:18076–18086. doi: 10.1074/jbc.M114.558601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploquin M, Petukhova GV, Morneau D, Dery U, Bransi A, Stasiak A, Camerini-Otero RD. Masson JY. Stimulation of fission yeast and mouse Hop2–Mnd1 of the Dmc1 and Rad51 recombinases. Nucleic Acids Res. 2007;35:2719–2733. doi: 10.1093/nar/gkm174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uanschou C, Ronceret A, Von Harder M, De Muyt A, Vezon D, Pereira L, Chelysheva L, Kobayashi W, Kurumizaka H, Schlogelhofer P, et al. Sufficient amounts of functional HOP2/MND1 complex promote interhomolog DNA repair but are dispensable for intersister DNA repair during meiosis in Arabidopsis. Plant Cell. 2013;25:4924–4940. doi: 10.1105/tpc.113.118521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moktan H, Guiraldelli MF, Eyster CA, Zhao W, Lee CY, Mather T, Camerini-Otero RD, Sung P, Zhou DH. Pezza RJ. Solution structure and DNA-binding properties of the winged helix domain of the meiotic recombination HOP2 protein. J Biol Chem. 2014;289:14682–14691. doi: 10.1074/jbc.M114.548180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev DV, Huang F, Mazina OM, Pezza RJ, Voloshin ON, Camerini-Otero RD. Mazin AV. HOP2–MND1 modulates RAD51 binding to nucleotides and DNA. Nat Commun. 2014;5:4198. doi: 10.1038/ncomms5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangen D, Kaufman Y, Zeligson S, Perlberg S, Fridman H, Kanaan M, Abdulhadi-Atwan M, Abu Libdeh A, Gussow A, Kisslov I, et al. XX ovarian dysgenesis is caused by a PSMC3IP/HOP2 mutation that abolishes coactivation of estrogen-driven transcription. Am J Hum Genet. 2011;89:572–579. doi: 10.1016/j.ajhg.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Bakker JL, Dicioccio RA, Gille JJ, Zhao H, Odunsi K, Sucheston L, Jaafar L, Mivechi NF, Waisfisz Q, et al. Inactivating mutations in GT198 in familial and early-onset breast and ovarian cancers. Genes Cancer. 2013;4:15–25. doi: 10.1177/1947601913486344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Yang Z, Zhang H, Jaafar L, Wang G, Liu M, Flores-Rozas H, Xu J, Mivechi NF. Ko L. GT198 splice variants display dominant-negative activities and are induced by inactivating mutations. Genes Cancer. 2013;4:26–38. doi: 10.1177/1947601913486345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel MA. Schimenti JC. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet. 2010;11:124–136. doi: 10.1038/nrg2723. [DOI] [PubMed] [Google Scholar]
- Hunter N. Synaptonemal complexities and commonalities. Mol Cell. 2003;12:533–535. doi: 10.1016/s1097-2765(03)00361-7. [DOI] [PubMed] [Google Scholar]
- Bolcun-Filas E. Schimenti JC. Genetics of meiosis and recombination in mice. Int Rev Cell Mol Biol. 2012;298:179–227. doi: 10.1016/B978-0-12-394309-5.00005-5. [DOI] [PubMed] [Google Scholar]
- Libby BJ, Reinholdt LG. Schimenti JC. Positional cloning and characterization of Mei1, a vertebrate-specific gene required for normal meiotic chromosome synapsis in mice. Proc Natl Acad Sci USA. 2003;100:15706–15711. doi: 10.1073/pnas.2432067100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao LE, Wente SR. Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci USA. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker C, Walsh GS. Moens C. Making gynogenetic diploid zebrafish by early pressure. J Vis Exp. 2009 doi: 10.3791/1396. doi: 10.3791/1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochakpour N. Immunofluorescent microscopic study of meiosis in zebrafish. Methods Mol Biol. 2009;558:251–260. doi: 10.1007/978-1-60761-103-5_15. [DOI] [PubMed] [Google Scholar]
- Saito K, Siegfried KR, Nusslein-Volhard C. Sakai N. Isolation and cytogenetic characterization of zebrafish meiotic prophase I mutants. Dev Dyn. 2011;240:1779–1792. doi: 10.1002/dvdy.22661. [DOI] [PubMed] [Google Scholar]
- Feitsma H, Leal MC, Moens PB, Cuppen E. Schulz RW. Mlh1 deficiency in zebrafish results in male sterility and aneuploid as well as triploid progeny in females. Genetics. 2007;175:1561–1569. doi: 10.1534/genetics.106.068171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal MC, Feitsma H, Cuppen E, Franca LR. Schulz RW. Completion of meiosis in male zebrafish (Danio rerio) despite lack of DNA mismatch repair gene mlh1. Cell Tissue Res. 2008;332:133–139. doi: 10.1007/s00441-007-0550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung O, Forbes MM. Marlow FL. Zebrafish vasa is required for germ-cell differentiation and maintenance. Mol Reprod Dev. 2014;81:946–961. doi: 10.1002/mrd.22414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Mari A. Postlethwait JH. The role of Fanconi anemia/BRCA genes in zebrafish sex determination. Methods Cell Biol. 2011;105:461–490. doi: 10.1016/B978-0-12-381320-6.00020-5. [DOI] [PubMed] [Google Scholar]
- Kochakpour N. Moens PB. Sex-specific crossover patterns in Zebrafish (Danio rerio. Heredity (Edinb) 2008;100:489–495. doi: 10.1038/sj.hdy.6801091. [DOI] [PubMed] [Google Scholar]
- Moens PB. Zebrafish: chiasmata and interference. Genome. 2006;49:205–208. doi: 10.1139/g06-021. [DOI] [PubMed] [Google Scholar]
- Iwai T, Yoshii A, Yokota T, Sakai C, Hori H, Kanamori A. Yamashita M. Structural components of the synaptonemal complex, SYCP1 and SYCP3, in the medaka fish Oryzias latipes. Exp Cell Res. 2006;312:2528–2537. doi: 10.1016/j.yexcr.2006.04.015. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Mari A, Wilson C, Titus TA, Canestro C, BreMiller RA, Yan YL, Nanda I, Johnston A, Kanki JP, Gray EM, et al. Roles of brca2 (fancd1) in oocyte nuclear architecture, gametogenesis, gonad tumors, and genome stability in zebrafish. PLoS Genet. 2011;7:e1001357. doi: 10.1371/journal.pgen.1001357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, Plug AW, van Vugt MJ. de Boer P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 1997;5:66–68. doi: 10.1023/a:1018445520117. [DOI] [PubMed] [Google Scholar]