Abstract
The microbiota affects host health, and dysbiosis is involved in colitis. Sorghum bran influences butyrate concentrations during dextran sodium sulfate (DSS) colitis, suggesting microbiota changes. We aimed to characterize the microbiota during colitis, and ascertain if polyphenol-rich sorghum bran diets mitigate these effects. Rats (n = 80) were fed diets containing 6% fiber from cellulose, or Black (3-deoxyanthocyanins), Sumac (condensed tannins), or Hi Tannin black (both) sorghum bran. Inflammation was induced three times using 3% DSS for 48 h (40 rats, 2 week separation), and the microbiota characterized by pyrosequencing. The Firmicutes/Bacteroidetes ratio was higher in Cellulose DSS rats. Colonic injury negatively correlated with Firmicutes, Actinobacteria, Lactobacillales and Lactobacillus, and positively correlated with Unknown/Unclassified. Post DSS#2, richness was significantly lower in Sumac and Hi Tannin black. Post DSS#3 Bacteroidales, Bacteroides, Clostridiales, Lactobacillales and Lactobacillus were reduced, with no Clostridium identified. Diet significantly affected Bacteroidales, Bacteroides, Clostridiales and Lactobacillus post DSS#2 and #3. Post DSS#3 diet significantly affected all genus, including Bacteroides and Lactobacillus, and diversity and richness increased. Sumac and Hi Tannin black DSS had significantly higher richness compared to controls. Thus, these sorghum brans may protect against alterations observed during colitis including reduced microbial diversity and richness, and dysbiosis of Firmicutes/Bacteroidetes.
Keywords: short chain fatty acids, inflammatory bowel disease, Chao, Shannon–Weaver
The dysbiosis occurring during inflammatory bowel disease is improved by polyphenol-rich sorghum brans.
Graphical Abstract Figure.
The dysbiosis occurring during inflammatory bowel disease is improved by polyphenol-rich sorghum brans.
INTRODUCTION
Inflammatory bowel disease (IBD), which includes Crohn's disease (CD) and ulcerative colitis (UC), affects nearly 1.4 million people in the United States (Loftus 2004). Symptoms include abdominal cramping, constipation, abnormal bowel movements and patients are at an increased risk of colorectal cancer (Itzkowitz and Yio 2004). Although the etiology of IBDs is not fully known, dysbiosis of the native bacterial populations residing in the gastrointestinal (GI) tract have been identified as a contributing factor in the progression and severity of UC (Elson et al. 2005; Xavier and Podolsky 2007). Studies using gnotobiotic or knockout animals (e.g. TLR pathway) or experimental models that induce an inflammatory state in the bowel using dextran sodium sulfate (DSS) can help demonstrate the effect of the intestinal microbiota on health and disease states (Elson et al. 2005; Lee et al. 2010).
Previous studies have sought to elucidate alterations to the microbiota or identify which bacterial populations might be associated with the onset or recurrence of UC (Loftus 2004; Elson et al. 2005; Swidsinski et al. 2005; Xavier and Podolsky 2007). Some implicate an increase in pathogenic bacteria or a depletion of beneficial bacteria, such as lactic acid bacteria (Martin et al. 2004), yet it is becoming more apparent that no particular bacterial group can be implicated in the cause of UC. However, some studies report alterations in the ratio of the predominant bacterial phyla, Firmicutes and Bacteroidetes, in patients affected with both UC and CD compared to controls (Swidsinski et al. 2005; Sokol et al. 2006; Rajilić-Stojanović et al. 2011), and others note a reduction in bacterial diversity and species richness (Ott et al. 2004; Manichanh et al. 2006).
In recent years, diets containing bioactive compounds, such as polyphenols and tannins, have been identified as possible interventions for IBD due to their antimicrobial and antioxidant capacity (Larrosa et al. 2009). Bran isolated from some varieties of sorghum grain contain polyphenols, including 3-deoxyanthocyanins and condensed tannins, and have been characterized to have a higher antioxidant capacity compared to wheat bran, blueberries and pomegranates (Dykes and Rooney 2006; Burdette 2010). Diet impacts the colon luminal environment by affecting transit time and the production of microbial metabolites [short chain fatty acids (SCFA), e.g. butyrate] that alter luminal pH (Rastall et al. 2005; Walker et al. 2010). Bran from black and brown sorghum cultivars are known to alter rat fecal SCFA concentrations, which suggests possible changes in the intestinal microbiota (Turner et al. 2010). Furthermore, secondary plant metabolites similar to those found in sorghum bran are reported to differentially affect luminal bacterial populations (Okubo 1992; Chung, Lu and Chou 1998; Rastmanesh 2011).
In vitro studies have shown that tannins and other bioactive compounds can dramatically affect the survival of bacterial groups that typically reside in the GI tract (Ahn et al. 1998; Cueva et al. 2012). Moreover, numerous in vivo human and animal studies have also characterized how these compounds can promote the growth of certain health-promoting bacteria (i.e. Bifidobacterium and Lactobacillus spp.), while suppressing or eliminating other pathogenic bacteria such as Clostridium perfringens and C. difficile in the intestine (Okubo 1992; Smith and Mackie 2004). Hydrocaffeic acid, a polyphenol metabolite derived from colonic microbiota, produced anti-inflammatory effects including suppression of proinflammatory cytokine production (i.e. TNFα, IL-8, IL-1β) and reduction of oxidative DNA damage in rat colonic mucosa (Larrosa et al. 2009).
Based on the existing literature, we hypothesized that perturbations in the rat colonic environment occurring with DSS-induced colitis, including changes in microbiota, will be partially mitigated by feeding sorghum bran-containing diets. Therefore, the aim of this study was to determine the effect of diets containing sorghum bran with 3-deoxyanthocyanins, condensed tannins or both polyphenols on the intestinal microbiota. Furthermore, we aim to ascertain if sorghum bran diets can mitigate alterations to the microbiota during repeated inflammatory bouts produced by DSS challenge.
MATERIALS AND METHODS
Animals and diets
Eighty (40-day-old) male Sprague-Dawley rats (Harlan Sprague-Dawley, Houston, TX, USA) were stratified by body weight and assigned to one of four experimental diets (n = 20 per diet). Experimental diets were formulated from purified ingredients and contained 6% dietary fiber from cellulose (control diet) or 6% dietary fiber from bran isolated from sorghum grains that contain 3-deoxyanthocyanins and no condensed tannins (Black bran), high levels of both 3-deoxyanthocyanins and condensed tannins (Sumac bran), or intermediate levels of 3-deoxyanthocyanins and high levels of condensed tannins (Hi Tannin). A complete description of the diets including antioxidant capacity, total polyphenol content and tannin content is reported elsewhere (submitted work from Lauren E. Ritchie, Stella S. Taddeo, Brad R. Weeks, Raymond J. Carroll, Linda Dykes, Lloyd W. Rooney, Nancy D. Turner). The proportions of soluble (6.0, 9.5, 11.7 and 6.4%) and insoluble fiber (94.0, 90.5, 88.3 and 93.6%) were similar among the diets (Cellulose, Black bran, Sumac bran and Hi Tannin bran, respectively).
After 21 days of experimental diets, half of the rats were exposed to three sequential DSS (MP Biomedicals, Irvine, CA, USA) treatments in their drinking water [3% (w/v) DSS for 48 h], with 14 days between each DSS exposure. Between DSS exposures, water was supplied to treated animals, and the remaining half of the animals (non-DSS controls) received water throughout the course of the study.
Body weight and food intake were routinely monitored. On day 82, animals were euthanized by CO2 asphyxiation. A 1 cm segment was removed from the distal end of the colon and fixed in 70% ethanol solution prior to embedding in paraffin. The degree of inflammation and morphological injury caused by DSS exposure was assessed by hematoxylin and eosin staining as described previously (Jia et al. 2008).
Fecal sample collection and microbial DNA isolation
Fresh fecal samples were collected immediately upon defecation, placed in sterile cryotubes and then stored at −80°C. Samples collected after recovery from the second DSS treatment (day 47, n = 9 or 10 for Cellulose and Black, and/or Sumac and Hi Tannin DSS rats, respectively) and third DSS treatment (day 62, n = 5/diet for control rats and n = 10/diet for DSS rats) were used for microbial analyses. DNA was isolated using a FastDNA SPIN kit (MP Biomedicals, Solon, OH, USA) as described previously (Menon et al. 2013), and the purified DNA was stored at −80°C.
16S rRNA bacterial tag-encoded FLX amplicon pyrosequencing
16S rRNA amplicon sequencing was performed in the Microbiome Core Facility (University of North Carolina at Chapel Hill, NC, USA) as previously described (Devine et al. 2013). Briefly, initial amplification of the V1–V2 region of the bacterial 16S rRNA gene was performed on total DNA isolated from fecal samples. Master mixes for these reactions used the Qiagen Hotstar HiFidelity Polymerase Kit (Qiagen, Valencia, CA, USA) with a forward primer composed of the Roche Titanium Fusion Primer A (5′-CCATCTCATCCCTGCGTGTCTCCGACTCAG-3′), a 10-bp multiplex identifier (MID) sequence (Roche, Indianapolis, IN, USA) unique to each of the samples, and the universal primer for bacteria, 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) (Edwards et al. 1989). The reverse primer was composed of the Roche Titanium Primer B (5′-CCTATCCCCTGTGTGCCTTGGCAGTCTCAG-3′), the identical 10-bp MID sequence as the forward primer and the reverse bacteria primer 338R (5′-GCTGCCTCCCGTAGGAGT-3′) (Fierer et al. 2008), which span the V1–V2 hypervariable region of the bacterial 16S rRNA gene. Each sample was individually gel purified using the E-Gel Electrophoresis System (Life Technologies, Invitrogen). To ensure equal representation of each sample in the sequencing run, each barcoded sample was standardized by calculating equimolar amounts prior to pooling. Pooled samples of the 16S rDNA multiplexed amplicons were sequenced on a Roche 454 Genome Sequencer FLX Titanium instrument using the GS FLX Titanium XLR70 sequencing reagents and protocols.
Amplicon sequencing data analysis
Analysis of sequencing data was carried out using the QIIME pipeline (Caporaso et al. 2010). The combined raw sequencing data plus metadata describing the samples were de-multiplexed and filtered. Next, data were denoised using Denoiser software using standard parameters (Reeder and Knight 2010). Sequences were grouped into operational taxonomic units (OTUs) at a 97% level to approximate species-level phylotypes using Uclust (Edgar 2010). OTU sequences were aligned and OTU tables containing the counts of each OTU in each sample were used to calculate mean species diversity of each sample (alpha diversity) and the differentiation among samples (beta diversity). Alpha and beta diversity measures were used to calculate the Chao species richness estimate and Shannon–Weaver diversity index for each OTU. To evaluate the similarities between bacterial communities, a combination of Unifrac significance, principal coordinate analysis (PCoA) using Fast Unifrac (Lozupone, Hamady and Knight 2006) and network analysis (Ley et al. 2008) were performed to compare samples based on sample time and treatment.
Statistical analysis
Data were analyzed using two-way analysis of variance including variables of diet and DSS exposure in SAS 9.1 (SAS Institute, Inc.) considering a P-value of <0.05 as significant. On completion of all analyses, the uncorrected P-values were submitted to a single multiple-testing correction using the Benjamini and Hochberg false discovery rate method. Unadjusted P-values are presented in the tables and text with an asterisk indicating where results remained significant after adjusting for multiple comparisons. Relationships between microbial populations and colonic injury score were assessed by calculating Pearson's product moment correlation coefficient.
RESULTS
Body weight and experimental diet intake
Only minor differences in food intake or body weight were observed. Control rats fed Hi Tannin had higher body weights than control rats fed Cellulose at four time points (day 39, day 42, day 56 and day 60). The only difference in food intake occurred prior to the first DSS treatment (DSS#1) at day 39, where rats fed Cellulose DSS consumed less than rats fed Sumac DSS (P < 0.05).
Multivariate analysis of bacterial populations
Day 62 control rats (not exposed to DSS) feces enabled determination of the potential changes in microbiota that occurred from exposure to DSS within a diet group. The DSS-treated rat samples from day 47 and day 62 allowed the determination of how repeated bouts of inflammatory challenge impacted the microbiota, and to determine if there were differences in the response due to diet.
PCoA of weighted and unweighted UniFrac results (Hamady, Lozupone and Knight 2010) revealed distinct clustering of samples based on the experimental diet for both time points (Fig. 1A). Variation of data points along PC2 (10.3%) indicated clear clusters and therefore distinct differences in bacterial communities between rats fed the Cellulose, Black, Sumac and Hi Tannin black bran diets. The Hi Tannin black bran diet, which contains both condensed tannins and 3-deoxyanthocyanins, resulted in bacterial communities that clustered near both Black bran (contains 3-deoxyanthocyanins) and Sumac bran (contains condensed tannins) diets. Additionally, rats fed bran diets that contained condensed tannins (i.e. Sumac and Hi Tannin black bran) had bacterial communities that clustered together. Spatial relation of data points along PC3 (6.28%) revealed that rats consuming the Sumac bran diet had samples with the least amount of variation compared to other experimental diets. Furthermore, PCoA analysis revealed differences between samples collected after DSS#2 (day 47) and DSS#3 (day 62) (Fig. 1B), suggesting that the bacterial communities were sensitive to diet and the number of DSS exposures.
Figure 1.
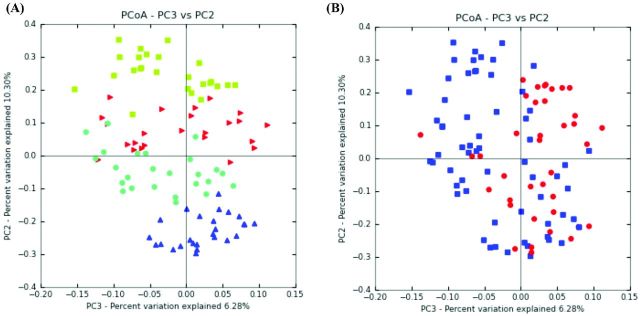
(A) PCoA plot of samples from both time points. Green squares (Cellulose), red triangles (Black bran), light blue circles (Hi Tannin black bran) and blue triangles (Sumac bran) illustrate differences in rat fecal bacterial populations due to experimental diets. (B) PCoA plot of amples post DSS#2 and DSS#3. Blue squares (post DSS#3) and red circles (post DSS#2) illustrate differences in rat fecal bacterial populations following DSS treatment over time.
Microbial taxonomic structure analysis
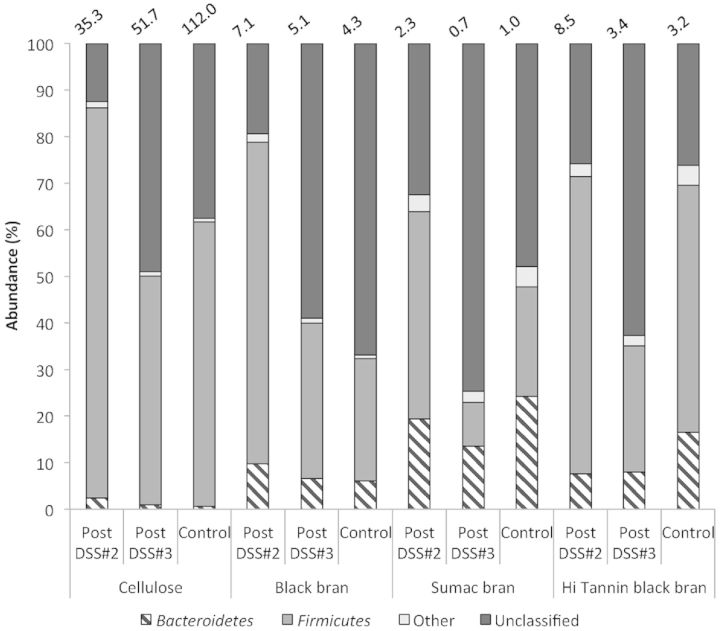
Phylogenetic classification of OTUs revealed two predominant phyla, Bacteroidetes and Firmicutes, for all experimental diets post DSS#2 and post DSS#3 (Fig. 2). The phyla Actinobacteria and Proteobacteria were also represented, but proportions did not exceed 5% for any experimental group at either time point. We observed a significant diet effect for all phyla at both time points (Table S1, Supporting Information). For each experimental diet at both time points, there were OTUs that did not match any known sequences in the RDP database, and the proportion of OTUs classified as ‘Unknown’ or ‘Unclassified’ was elevated in all experimental diets post DSS#3 compared to DSS#2 (≥2-fold increase).
Figure 2.
Phylogenetic classification of OTUs at the phylum level in fecal samples from control rats (day 62) and DSS-treated rats (on day 47—post DSS#2, and on day 62—DSS#3) for all diets. The Firmicutes/Bacteroidetes ratio is reported above each column.
We observed a significant effect of DSS on both the Firmicutes and Bacteroidetes phyla (P = 0.0006 and P < 0.0001, respectively) in samples collected post DSS#3. Thus, ratios of these phyla (% Firmicutes in a given experimental group /% Bacteroidetes in the same group) were calculated to characterize relative proportion of these predominant bacterial groups. The ratio of Firmicutes to Bacteroidetes was higher in Cellulose DSS rats post DSS#2 and post DSS#3 compared to Black, Sumac and Hi Tannin black bran DSS rats (Fig. 2).
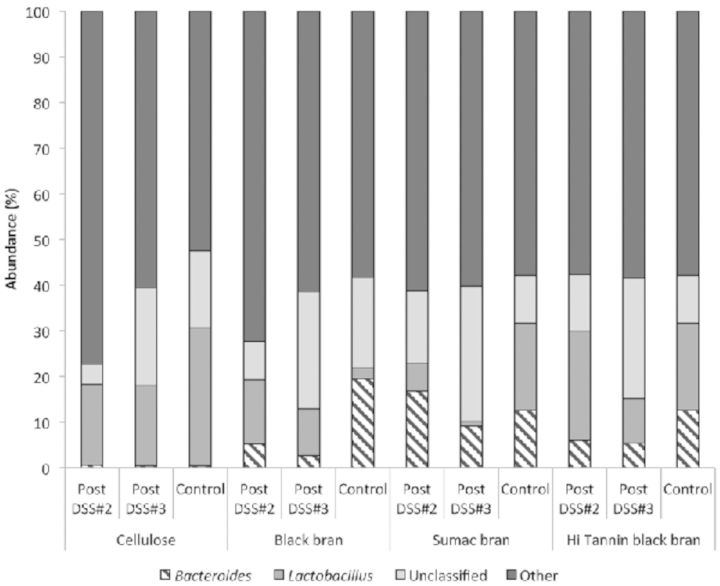
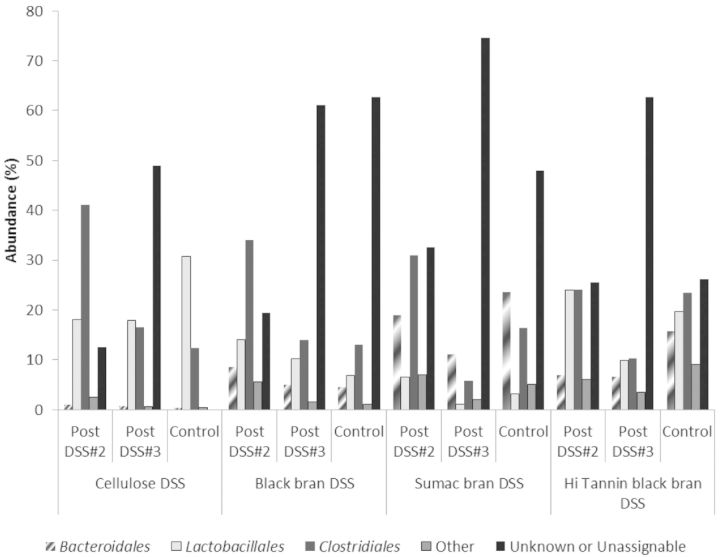
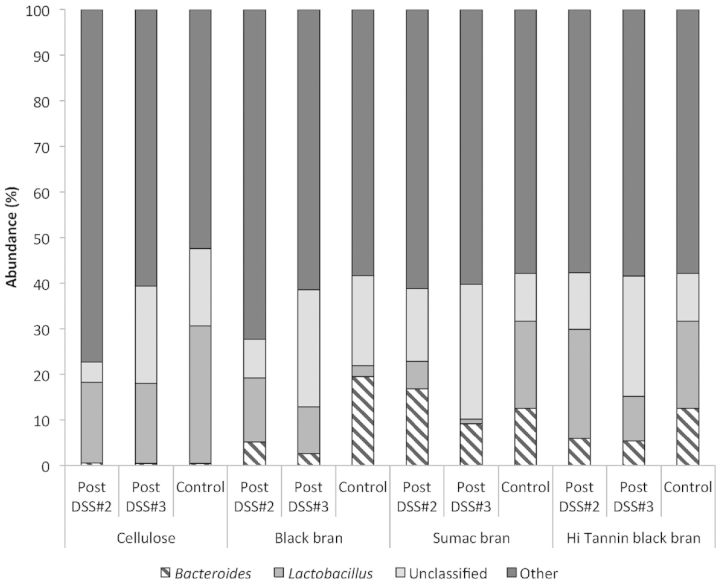
Further phylogenetic analysis of OTUs revealed three predominant bacterial orders (Bacteroidales, Clostridiales and Lactobacillales) and two genus (Bacteroides and Lactobacillus) in all collected samples (Figs 3 and 4, respectively). We observed significant diet effects post DSS#2 for bacteria in the Bacteroidales (P < 0.0001), Clostridiales (P = 0.0205) and Lactobacillales orders (P = 0.0417) (Table S2, Supporting Information), and for all characterized bacterial genus except Coprobacillus and Parasutterella (Table S3, Supporting Information). At the genus level, the confidence level for numerous OTUs was below 90% similarity to known sequences in the Ribosomal Database Project (RDP) database, and have been marked accordingly (Table S3, Supporting Information). Sumac DSS rats had significantly higher proportion of OTUs classified as Bacteroidales, and Hi Tannin black DSS rats had significantly higher proportions of Lactobacillales compared to the Sumac DSS rats, with the other two diets being intermediate (P < 0.05). These trends remained significant at the genus level as well (i.e. Bacteroides and Lactobacillus).
Figure 3.
Phylogenetic classification of OTUs at the order level in fecal samples from control rats (day 62) and DSS-treated rats (on day 47—post DSS#2, and on day 62—DSS#3) for all diets.
Figure 4.
Phylogenetic classification of OTUs at the genus level in fecal samples from control rats (day 62) and DSS-treated rats (on day 47—post DSS#2, and on day 62—DSS#3) for all diets.
Post DSS#3, proportions of OTUs classified in the predominant bacterial orders (i.e. Bacteroidales, Clostridiales and Lactobacillales) were reduced in all DSS-treated rats compared to post DSS#2 for all experimental diets (Fig. 3), with the largest reduction observed in the Clostridiales order (56–79% reduction). A similar trend was observed at the genus level for both Bacteroides and Lactobacillus, with proportions remaining the same or reduced following DSS#3 for all diets. Additionally, we observed significant diet, treatment and interactive effects for both Bacteroidales and Bacteroides (P < 0.0001, P < 0.0001, and P < 0.0005, respectively), Clostridiales (Diet P = 0.0005, and DSS P < 0.0001, respectively) and Turicibacter (Diet P = 0.0001) at this time point (Tables S2 and S3, Supporting Information), which all remained significant after adjusting for multiple comparisons. In Sumac control rats, we observed a significantly higher proportion of OTUs classified as Bacteroidales and Bacteroides, and a significantly higher proportion of Clostridiales in Hi Tannin control rats compared to all other diets (P < 0.05). We observed a significant effect of diet and treatment for OTUs classified in the Lactobacillales order (P < 0.001 and 0.0377, respectively), and a significant diet effect for all classified genus at this time point (i.e. Bacteroides, Lactobacillus, Turicibacter and Parasutterella), which remained significant after adjusting for multiple comparisons. Cellulose controls had a significantly higher proportion of OTUs classified as Lactobacillales (P < 0.05) compared to all other groups, with a similar trend at the genus level (Lactobacillus) as well. Additionally, DSS-treated rats fed Cellulose, Sumac and Hi Tannin black bran diets had lower proportions of both Lactobacillales and Lactobacillus compared to their diet-matched controls, a difference that was only significant for the Cellulose rats.
When samples from rats fed Sumac and Hi Tannin diets and treated with DSS were compared to their diet-matched controls, we observed a significant reduction in numerous bacterial orders following recovery from DSS#3 (Fig. 3). Sumac DSS rats had significantly lower proportions of Bacteroidales, Bacteroides, Clostridiales, Parasutterella and Burkholderiales, and Hi Tannin DSS showed lower proportions of Bacteroidales, Bacteroides, Clostridiales, Turicibacter and Erysipelotrichales compared to their diet-matched controls (P < 0.05) (Tables S2 and S3, Supporting Information). In contrast, Black bran DSS rats had higher abundance of Lactobacillales, Lactobacillus and Burkholderiales, and Cellulose DSS rats had higher abundance of Bacteroidales, Clostridiales and Erysipelotrichales compared to their diet-matched controls.
Diversity and species richness comparisons
To analyze the effects of diet and DSS-induced inflammation on bacterial species richness and diversity, sequences from each sample (approximately 1508) were used to perform alpha and beta diversity analyses and to calculate the Chao species richness estimate and the Shannon–Weaver diversity index. We observed a significant diet effect on both species richness and diversity post DSS#3 (P < 0.0001), with rats fed Black bran showing a significantly higher species richness (P < 0.05) compared to animals fed Sumac and Hi Tannin black bran diets, and a significantly higher diversity index (P < 0.05) compared to all other controls (Table 1). Sumac control rats had lower species richness and diversity indices compared to the Cellulose and Black bran fed rats. Following DSS#2, we observed no significant differences in bacterial diversity between Cellulose, Black, Sumac or Hi Tannin black bran DSS rats. Sumac bran DSS rats had a significantly lower species richness compared to Cellulose, Black and Hi Tannin black bran DSS rats (P < 0.05) at this time point. DSS-treated rats following DSS#2 had lower species richness and diversity indices compared to their diet-matched (non-DSS) controls, yet post DSS#3 there was a numerical increase in diversity (13.6–25%) and species richness (39–62%) compared to post DSS#2 for Cellulose and Sumac and Black, Sumac, and Hi Tannin black bran DSS rats (Table 1).
Table 1.
Chao and Shannon–Weaver indices for rat fecal microbial populations in samples from all treatment groups post DSS#2 and DSS#3.1
| Cellulose | Black bran | Sumac bran | Hi Tannin black bran | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DSS (n = 9) | DSS (n = 9) | DSS (n = 10) | DSS (n = 10) | Diet | ||||||||
| Post DSS#2 (day 47) | Chao | 117.33 ± 4.78b | 124.74 ± 6.23b | 92.76 ± 2.12a | 112.98 ± 5.91b | 0.0005 | ||||||
| Shannon–Weaver | 4.00 ± 0.18a | 4.35 ± 0.16a | 4.17 ± 0.08a | 4.17 ± 0.12a | ||||||||
| Control (n = 5) | DSS (n = 10) | Control (n = 5) | DSS (n = 9) | Control (n = 5) | DSS (n = 10) | Control (n = 5) | DSS (n = 9) | Diet | DSS | Diet* DSS | ||
| Post DSS#3 (day 62) | Chao | 207.32 ± 14.08d | 169.29 ± 5.45c | 222.91 ± 12.47d | 173.79 ± 14.27c | 118.27 ± 6.85a | 155.08 ± 6.87b,c | 133.45 ± 13.05a,b | 210.40 ± 9.17d | <0.0001 | <0.0001 | |
| Shannon–Weaver | 4.79 ± 0.33c,d | 4.86 ± 0.15c | 5.82 ± 0.04a | 5.13 ± 0.15b,c | 4.39 ± 0.14d | 4.78 ± 0.08c,d | 4.73 ± 0.16c,d | 5.50 ± 0.10a,b | <0.0001 | 0.0003 | ||
Data are LS mean ± SEM.
1Means in a row without a common superscript (a, b, c, d) differ (P < 0.05).
Post DSS#3, experimental diet and DSS treatment had an interactive effect on species richness and diversity (P < 0.0001 and P = 0.0003, respectively), in which Cellulose and Black bran DSS rats had a significantly lower Chao score compared to their diet-matched controls (P < 0.05). Sumac bran DSS rats had significantly higher richness (P = 0.0199) and numerically higher diversity index compared to Sumac controls, and Hi Tannin black bran DSS rats had significantly higher richness and diversity indices (P < 0.001 and P = 0.013, respectively) compared to Hi Tannin black bran controls at this time point (Table 1).
DSS-induced injury of distal colon
All DSS-treated rats had elevated colonic injury compared to their diet-matched controls, with significantly higher injury scores observed in Hi Tannin black bran DSS rats compared to Hi Tannin black control rats (P = 0.0072) (submitted work from Lauren E. Ritchie, Stella S. Taddeo, Brad R. Weeks, Raymond J. Carroll, Linda Dykes, Lloyd W. Rooney, Nancy D. Turner). DSS-treated rats fed bran diets had significantly higher injury scores compared to Cellulose DSS rats (P < 0.05). Cellulose controls had the lowest injury scores, which were significantly lower than Black and Sumac bran controls (P < 0.05). (submitted work from Lauren E. Ritchie, Stella S. Taddeo, Brad R. Weeks, Raymond J. Carroll, Linda Dykes, Lloyd W. Rooney, Nancy D. Turner).
In order to determine if there were relationships between the bacterial taxa characterized in fecal samples collected post DSS#3 and colon injury, we ran correlational analyses. In this UC model, significant correlations were observed for four phylogenetic groups (Table 2). At the phylum level, both Firmicutes and Actinobacteria were negatively correlated with colonic injury (P = 0.003 and P = 0.019). Within the Firmicutes phylum, Clostridiales, Erysipelotrichales, Lactobacillales and Lactobacillus negatively correlated with colonic injury, and the relationship reached significance for both Lactobacillales and Lactobacillus (P = 0.045 and P = 0.05). The abundance of OTUs classified as ‘Unknown’ or ‘Unclassified’ (OTUs that did not match any known sequences in the RDP database) were both positively correlated with colonic injury (both P = 0.003, Table 2).
Table 2.
Correlation of intestinal microbiota and colonic injury score in rats after three bouts of DSS-induced colonic injury.a
| Taxon | Pearson's correlation coefficient | ||
|---|---|---|---|
| Actinobacteria | −0.308a | ||
| Bacteroidetes | 0.076 | ||
| Bacteroidales | 0.049 | ||
| Bacteroides | 0.033 | ||
| Firmicutes | −0.387a | ||
| Clostridiales | −0.123 | ||
| Erysipelotrichales | −0.096 | ||
| Turicibacter | −0.088 | ||
| Lactobacillales | −0.264a | ||
| Lactobacillus | −0.258a | ||
| Proteobacteria | −0.012 | ||
| Burkholderiales | 0.080 | ||
| Parasutturella | 0.076 | ||
| Unknown | 0.382a | ||
| Unclassified | 0.387a | ||
aIndicates phylogenetic groups which proportion correlated significantly with injury score (P < 0.05).
DISCUSSION
The intestinal microbiota of mammals is composed of trillions of bacteria and over 400 individual species have been identified thus far (Rajilić-Stojanović, Smidt and De Vos 2007). These bacterial populations provide numerous benefits to the host, including immune system development, epithelial barrier maintenance, and providing metabolic substrate for colonocytes (Louis and Flint 2009; Wells et al. 2010). It is now understood that perturbations to the microbiota are involved in the initiation of intestinal inflammation and recurrence of inflammatory bouts, and is an important factor in the etiology of IBD such as UC (Manichanh et al. 2006; Frank et al. 2007; Sepehri et al. 2007). Another environmental input that has been shown to affect the microbiota is diet composition, and recent focus has been put on bioactive compounds and their possible role in mitigating the deleterious effects of intestinal inflammation (Gibson et al. 2004; Smith and Mackie 2004; Louis et al. 2007; Larrosa et al. 2009; Biasi et al. 2011; Possemiers et al. 2011). Brans utilized in this study contain concentrated levels of 3-deoxyanthocyanins (Black bran), condensed tannins (Sumac bran) or a combination of these compounds (Hi Tannin black bran) (Dykes and Rooney 2006; Dykes, Rooney and Rooney 2013). Our previous work demonstrated that these diets can alter fecal SCFA concentrations, a microbial metabolite, suggesting that these dietary ingredients alter the composition of the microbiota and/or microbial metabolism.
In the present study, PCoA analysis of microbial taxa from fecal samples suggests that there are distinct bacterial communities in animals fed experimental diets containing Cellulose or bran isolated from Black, Sumac or Hi Tannin sorghums. Additionally, samples collected at different time points (post DSS#2 and DSS#3) also grouped in different quadrants of the PCoA plot, suggesting, as expected, that DSS treatment and colonic inflammation also modify the composition of the gut microbial populations. Little research has been done to understand how the bioactive compounds found in sorghum (i.e. 3-deoxyanthocyanins and condensed tannins) can alter the luminal environment, particularly the intestinal bacterial populations. Therefore, the aim of this work was to characterize alterations to the microbiota during DSS-induced colitis and determine if polyphenol-rich sorghum diets have the ability to mitigate the dysbiosis (e.g. decreased bacterial diversity and richness; elevated Firmicutes/Bacteroidetes ratio) associated with UC.
Although research has been done to understand the role of the microbiota in UC, it is becoming more apparent that no specific microbiome component can be identified as an etiologic agent (Nagalingam and Lynch 2012). However, an overall dysbiosis in the ratio of predominant bacterial populations (i.e. increased Firmicutes to Bacteroidetes ratio) have been observed in both the feces and colon biopsies of IBD patients and experimental models of obesity and UC (Sokol et al. 2006; Nagalingam, Kao and Young 2011; Rajilić-Stojanović et al. 2011). In our study, the Firmicutes/Bacteroidetes ratio observed in Cellulose fed animals (both control and DSS) was at least 10-fold higher than that of bran fed animals. These results could be in part due to the presence of cellulose fermenting bacterial species within this phylum (e.g. Ruminococcus spp.) (Chassard, Gaillard-Martinie and Bernalier-Donadille 2005). Previous studies testing the effect of condensed tannins in vivo also reported inhibition of Gram-positive bacteria (i.e. Firmicutes phylum), specifically inhibition of the C. leptum group, while other enteric bacteria (i.e. Bacteroides fragilis and Bacteroides—Prevotella–Porphyromonas groups) increased significantly (Smith and Mackie 2004). Our observations are similar to these reports, as animals fed a bran diet containing tannins (i.e. Sumac and Hi Tannin black) had higher proportions of both Bacteroidales and Bacteroides compared to Cellulose and Black bran fed animals. Other dietary compounds such as tea polyphenols and other flavonoids have been reported to alter the composition of the intestinal microbiota in rats and human subjects, including decreased proportion of Clostridium spp. compared to those that did not consume these polyphenols (Okubo 1992; Hanske 2005).
A recent review documented the numerous changes observed in experimental models and patients with IBD, and no agreement has been reached on the association of specific microbial groups and UC (Nagalingam and Lynch 2012). Studies have documented elevated levels and enhanced epithelial adherence of pathogenic bacteria species (e.g. pathogenic Escherichia coli (E. coli)) following DSS-induced colitis (Heimesaat et al. 2007). In our study, we observed less than 5% of OTUs classified as Proteobacteria in control and DSS-treated animals at both time points. Plant polyphenols similar to those utilized in this study (i.e. gallic acid, methyl gallate, propyl gallate) have been observed to mitigate the effects of IBD and also been found to have inhibitory properties against potentially harmful bacterial species such as C. perfringens, C. paraputrificum, Eubacterium limosum, Bacteroides fragilis, Staphylococcus aureus and E. coli in culture (Ahn et al. 1998; Chung, Lu and Chou 1998). This inhibitory effect could be potentially beneficial and one reason for the reduced abundance of Proteobacteria in our study (≤4.29%), which harbors numerous pathogenic bacteria species such as Escherichia, Salmonella, Vibrio and Helicobacter.
Previous studies in experimental models and patients with UC have also documented a depletion in bacterial species that provide benefit to the host such as species of Bifidobacterium and Lactobacillus (Sokol et al. 2006; Frank et al. 2007; Heimesaat et al. 2007). These species produce antimicrobial substances, compete with pathogens for epithelial and mucin-binding sites (Ljungh and Wadstrom 2006), and have been shown to attenuate symptoms and maintain remission of UC (Garcia Vilela et al. 2008). During an active disease state (following DSS#2), we observed a significantly higher proportion of Lactobacillales and elevated proportion of Lactobacillus in Hi Tannin black DSS rats compared to Cellulose, Black and Sumac bran fed DSS rats. However, following DSS#3, we observed reduced abundance of Lactobacillales and Lactobacillus in DSS-treated rats fed Black, Sumac and Hi Tannin black bran diets, which is similar to other reports in patients and experimental models of UC that document a suppression of lactic acid bacteria (Heimesaat et al. 2007). Previous studies have reported that hydrolysable tannins have minimal effect on growth of lactic acid bacteria, specifically Bifidobacterium infantis and Lactobacillus acidophilus (Ahn et al. 1998; Chung, Lu and Chou 1998), which parallels animals fed Hi Tannin black and treated with DSS having a higher abundance of both Lactobacillales and Lactobacillus post DSS#2. In this study, we observed virtually undetectable levels of OTUs classified in the Actinobacteria phylum (≤0.80%), and Bifidobacterium in particular, which may suggest that this group is not a major constituent of the microbiota in any of our experimental groups. However, the low abundance of Bifidobacterium observed in our study could be also due to the facts that no Bifidobacterium-specific primers were used, which have been shown essential to quantify abundance changes in this group (Ritchie et al. 2010; Davis et al. 2011).
Previous studies and our results indicate that the effects of polyphenols on microbiota vary substantially and may be dependent upon the polyphenolic structure of these compounds. Very little is known regarding microbial metabolism of the 3-deoxyanthocyanins and condensed tannins found in sorghum bran. In general, polyphenols are poorly absorbed and therefore available to be catabolized by the colonic microbiota. The microbial metabolism of polyphenols is extremely complex, and numerous microbial catalytic and hydrolytic enzymes have been identified (van Duynhoven et al. 2011); therefore, a complete description is beyond the scope of this paper. However, unlike other flavonoids, many anthocyanins do not undergo extensive metabolism and their structure is stabilized in acidic conditions (Crozier, Del Rio and Clifford 2010). Furthermore, condensed tannins such as those described in this study are large non-hydrolysable proanthocyanindins, with three or more polymerizations, that are not easily fractionated by water and tannases (Ree 2001). One study demonstrated that condensed tannins isolated from sorghum bran were only isolated in excrement and not absorbed in chickens (Jimenez-Ramsey et al. 1994). Furthermore, both animal and human studies have shown that anthocyanins from berries (i.e. bilberry and raspberry) are poorly absorbed in the small intestine and typically <0.1% of the quantities ingested are detected in urine within 24 h of consumption (Borges et al. 2007; Sakakibara et al. 2009). This implies that compounds similar to those found in this study may not be absorbed or metabolized and therefore concentrated in the intestinal lumen where they are able to impact the microbiota and/or be metabolized by them. Metagenome and metabolomics analyses may help elucidate the relationship of the microbiota and the bioactive compounds found in sorghum; however, due to the large number of genes and metabolites identified in these analyses it could be quite difficult to elucidate the biomolecular mechanisms involved in the observed beneficial effects.
Alterations in other bacterial populations that are prevalent constituents of the commensal intestinal flora have been reported in experimental models and patients with UC, and both an elevation and suppression have been observed in species of the genera Bacteroides and Clostridium in feces and colonic tissue (Noor et al. 2010; Andoh et al. 2011). To understand how the microbiota was fluctuating during different stages of inflammation, we compared DSS-treated animals post DSS#2 to post DSS#3 and observed decreased proportions of Bacteroidales, Clostridiales and Lactobacillales in DSS animals for all experimental diets following DSS#3, with the highest reduction observed in the Clostridiales order (56–79%). At the genus level, we see similar reductions in Bacteroides and Lactobacillus, with minimal OTUs classified within the Clostridium genus (<3% post DSS #2 and none post DSS#3). The Clostridiales and Clostridium taxa are of particular importance, as they harbor bacterial species that have the ability to produce important bacterial metabolites such as butyrate (Wiegel, Kuk and Kohring 1989). Although this order also harbors opportunistic pathogens, alterations to this particular population could be detrimental to colonic health as butyrate is not only the predominant fuel for colonocytes, but suppressed availability and uptake of this metabolite have been implicated in the etiology of IBD (Thibault et al. 2007; Vicky De et al. 2010). Observed differences could be due, in part, to the progression of inflammation or severity of epithelial barrier injury, as it has been previously reported that inflamed tissue harbors different bacterial populations than non-inflamed tissues from the same individual (Bibiloni et al. 2006; Frank et al. 2007; Sepehri et al. 2007). Additionally, previous studies have shown correlations between bacterial populations and disease severity, which can further elucidate the relationship between the microbiota and UC. In the present study, we report a negative correlation between colonic injury and numerous phylogenetic groups, including significant correlations to Firmicutes, Actinobacteria, and both Lactobacillales and Lactobacillus. Similarly, a previous study reported that the abundance of certain taxa in the Firmicutes phylum (i.e. Clostridium clusters IV and IX) was negatively correlated with disease symptom score (e.g. abdominal pain, distention) in patients with irritable bowel syndrome (Rajilić-Stojanović et al. 2011). In contrast to our results, another study reported a positive correlation between the abundance of cecal E. coli and histologic colon score in HLA-B27 transgenic rats (Onderdonk et al. 1998). Our results could differ from previous studies due to the experimental model utilized, tissue analyzed (i.e. feces versus cecal content and/or colonic tissue) and phylogenetic characterization techniques (e.g. FISH, T-RFLP, DGGE, microbiological culture). Further analysis, such as identifying closest neighbors to unclassified and unidentified OTUs, will further elucidate the significance of these bacterial groups and their relation to colonic injury in our experimental groups.
Although we observed a decrease in the predominant bacterial orders post DSS#3, there was no reduction in bacterial diversity and species richness at this time point, which has been previously reported in both patients with UC and DSS-induced colitis animal models (Ott et al. 2004; Heimesaat et al. 2007; Andoh et al. 2011; Nagalingam, Kao and Young 2011). Following the second DSS treatment, we observed a decrease in both species richness and diversity compared to diet-matched controls; however, the values of these indices were elevated following recovery from DSS#3. Furthermore, following all DSS exposures diseased animals fed brans containing condensed tannins (i.e. Sumac and Hi Tannin black bran) showed bacterial richness and diversity indexes higher than their diet-matched controls. This result parallels a previous study using denaturing gel electrophoresis (DGGE) that reported tannins derived from the Acacia angustissima shrub significantly increased murine fecal bacterial diversity (Smith and Mackie 2004). A reduction in bacterial diversity and species richness could be detrimental due to decreased colonization resistance, imbalances in microbial-host signaling through pattern recognition receptors such as Toll-like receptors (TLR), as well as allowing for pathogenic bacteria to thrive (Croswell 2009). These data suggest that even though the bacterial populations may be affected during repeated DSS exposure (i.e. a more active disease state), feeding bran-based diets, particularly those containing condensed tannins, may be useful to restore bacterial species richness and diversity.
CONCLUSIONS
To our knowledge, there are no studies that describe the effect of sorghum bran on the microbiota. We demonstrated that sorghum bran diets may be able to prevent an overall dysbiosis of predominant bacterial populations and decreased microbial diversity commonly associated with UC. Furthermore, we observed distinct differences among experimental diets suggesting that the presence of bioactive compounds like 3-deoxyanthocianins and condensed tannins may be a factor in these diets ability to alter the luminal environment and microbial populations. Additional analyses, such as those employing species-specific bacterial primers, could elucidate additional population changes not captured in this study. Future studies elucidating mechanisms by which these bioactive compounds affect the intestinal microbiota during a healthy and diseased state are warranted.
SUPPLEMENTARY DATA
Supplementary data is available at FEMSEC online.
Acknowledgments
The authors appreciate the assistance with sample collection provided by Sujitta Raungrusmee and Adam Brum.
FUNDING
Research support from United Sorghum Checkoff Program (Roo31A-09 and HVM006-12), and the NIH/National Institute of Diabetes and Digestive and Kidney Diseases grant P30 DK34987. The work was also supported through a fellowship to Lauren Ritchie from the National Space Biomedical Research Institute through NCC 9-58 (EO01001) and Whole Systems Genomics at Texas A&M University.
Conflict of interest. None declared.
REFERENCES
- Ahn YJ, Lee CO, Kweon JH, et al. Growth-inhibitory effects of Galla Rhois-derived tannins on intestinal bacteria. J Appl Microbiol. 1998;84:439–43. doi: 10.1046/j.1365-2672.1998.00363.x. [DOI] [PubMed] [Google Scholar]
- Andoh A, Imaeda H, Aomatsu T, et al. Comparison of the fecal microbiota profiles between ulcerative colitis and Crohn's disease using terminal restriction fragment length polymorphism analysis. J Gastroenterol. 2011;46:479–86. doi: 10.1007/s00535-010-0368-4. [DOI] [PubMed] [Google Scholar]
- Biasi F, Astegiano M, Maina M, et al. Polyphenol supplementation as a complementary medicinal approach to treating inflammatory bowel disease. Curr Med Chem. 2011;18:4851–65. doi: 10.2174/092986711797535263. [DOI] [PubMed] [Google Scholar]
- Bibiloni R, Mangold M, Madsen KL, et al. The bacteriology of biopsies differs between newly diagnosed, untreated, Crohn's disease and ulcerative colitis patients. J Med Microbiol. 2006;55:1141–9. doi: 10.1099/jmm.0.46498-0. [DOI] [PubMed] [Google Scholar]
- Borges G, Roowi S, Rouanet JM, et al. The bioavailability of raspberry anthocyanins and ellagitannins in rats. Mol Nutr Food Res. 2007;51:714–25. doi: 10.1002/mnfr.200700024. [DOI] [PubMed] [Google Scholar]
- Burdette A. Anti-inflammatory activity of select sorghum (Sorghum bicolor) brans. J Med Food. 2010;13:879. doi: 10.1089/jmf.2009.0147. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassard C, Gaillard-Martinie B, Bernalier-Donadille A. Interaction between H2-producing and non-H2-producing cellulolytic bacteria from the human colon. FEMS Microbiol Lett. 2005;242:339–44. doi: 10.1016/j.femsle.2004.11.029. [DOI] [PubMed] [Google Scholar]
- Chung KT, Lu Z, Chou MW. Mechanism of inhibition of tannic acid and related compounds on the growth of intestinal bacteria. Food Chem Toxicol. 1998;36:1053–60. doi: 10.1016/s0278-6915(98)00086-6. [DOI] [PubMed] [Google Scholar]
- Croswell A. Prolonged impact of antibiotics on intestinal microbial ecology and susceptibility to enteric Salmonella infection. Infect Immun. 2009;77:2741–53. doi: 10.1128/IAI.00006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozier A, Del Rio D, Clifford MN. Bioavailability of dietary flavonoids and phenolic compounds. Mol Aspects Med. 2010;31:446–67. doi: 10.1016/j.mam.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Cueva C, Sánchez-Patán F, Monagas M, et al. In vitro fermentation of grape seed flavan-3-ol fractions by human faecal microbiota: changes in microbial groups and phenolic metabolites. FEMS Microbiol Ecol. 2012;83:792–805. doi: 10.1111/1574-6941.12037. [DOI] [PubMed] [Google Scholar]
- Davis LMG, Davis Is, MartÃnez J, et al. Barcoded pyrosequencing reveals that consumption of galactooligosaccharides results in a highly specific bifidogenic response in humans. PLoS One. 2011;6:e25200. doi: 10.1371/journal.pone.0025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine AA, Gonzalez A, Speck KE, et al. Impact of ileocecal resection and concomitant antibiotics on the microbiome of the murine jejunum and colon. PLoS One. 2013;8:e73140. doi: 10.1371/journal.pone.0073140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykes L, Rooney LW. Sorghum and millet phenols and antioxidants. J Cereal Sci. 2006;44:236–51. [Google Scholar]
- Dykes L, Rooney WL, Rooney LW. Evaluation of phenolics and antioxidant activity of black sorghum hybrids. J Cereal Sci. 2013;58:278–83. [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Edwards U, Rogall T, Blocker H, et al. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–53. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson CO, Cong Y, McCracken VJ, et al. Experimental models of inflammatory bowel disease reveal innate, adaptive, and regulatory mechanisms of host dialogue with the microbiota. Immunol Rev. 2005;206:260–76. doi: 10.1111/j.0105-2896.2005.00291.x. [DOI] [PubMed] [Google Scholar]
- Fierer N, Hamady M, Lauber CL, et al. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. P Natl Acad Sci USA. 2008;105:17994–9. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. P Natl Acad Sci USA. 2007;104:13780–5. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Vilela E, De Lourdes De Abreu Ferrari M, Oswaldo Da Gama Torres H, et al. Influence of Saccharomyces boulardii on the intestinal permeability of patients with Crohn's disease in remission. Scand J Gastroentero. 2008;43:842–8. doi: 10.1080/00365520801943354. [DOI] [PubMed] [Google Scholar]
- Gibson GR, Probert HM, Loo JV, et al. Dietary modulation of the human colonic microbiota: updating the concept of prebiotics. Nutr Res Rev. 2004;17:259–75. doi: 10.1079/NRR200479. [DOI] [PubMed] [Google Scholar]
- Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanske L. Xanthohumol does not affect the composition of rat intestinal microbiota. Mol Nutr Food Res. 2005;49:868. doi: 10.1002/mnfr.200500048. [DOI] [PubMed] [Google Scholar]
- Heimesaat MM, Fischer A, Siegmund B, et al. Shift towards pro-inflammatory intestinal bacteria aggravates acute murine colitis via Toll-like receptors 2 and 4. PLoS One. 2007;2:e662. doi: 10.1371/journal.pone.0000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzkowitz SH, Yio X. Inflammation and Cancer IV. Colorectal cancer in inflammatory bowel disease: the role of inflammation. Am J Physiol-Gastr L. 2004;287:G7–17. doi: 10.1152/ajpgi.00079.2004. [DOI] [PubMed] [Google Scholar]
- Jia Q, Lupton JR, Smith R, et al. Reduced colitis-associated colon cancer in Fat-1 (n-3 fatty acid desaturase) transgenic mice. Cancer Res. 2008;68:3985–91. doi: 10.1158/0008-5472.CAN-07-6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Ramsey LM, Rogler JC, Housley TL, et al. Absorption and distribution of 14C-labeled condensed tannins and related sorghum phenolics in chickens. J Agr Food Chem. 1994;42:963–7. [Google Scholar]
- Larrosa M, Luceri C, Vivoli E, et al. Polyphenol metabolites from colonic microbiota exert anti-inflammatory activity on different inflammation models. Mol Nutr Food Res. 2009;53:1044–54. doi: 10.1002/mnfr.200800446. [DOI] [PubMed] [Google Scholar]
- Larrosa M, Yañéz-Gascón MaJ, Selma MaV, et al. Effect of a low dose of dietary resveratrol on colon microbiota, inflammation and tissue damage in a DSS-induced colitis rat model. J Agr Food Chem. 2009;57:2211–20. doi: 10.1021/jf803638d. [DOI] [PubMed] [Google Scholar]
- Lee I-A, Bae E-A, Hyun Y-J, et al. Dextran sulfate sodium and 2,4,6-trinitrobenzene sulfonic acid induce lipid peroxidation by the proliferation of intestinal gram-negative bacteria in mice. J Inflamm. 2010;7:7. doi: 10.1186/1476-9255-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–51. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungh A, Wadstrom T. Lactic acid bacteria as probiotics. Curr Issues Intest Microbiol. 2006;7:73–90. [PubMed] [Google Scholar]
- Loftus EV., Jr Clinical epidemiology of inflammatory bowel disease: incidence, prevalence, and environmental influences. Gastroenterology. 2004;126:1504–17. doi: 10.1053/j.gastro.2004.01.063. [DOI] [PubMed] [Google Scholar]
- Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- Louis P, Scott KP, Duncan SH, et al. Understanding the effects of diet on bacterial metabolism in the large intestine. J Appl Microbiol. 2007;102:1197–208. doi: 10.1111/j.1365-2672.2007.03322.x. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Hamady M, Knight R. UniFrac–an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichanh C, Rigottier-Gois L, Bonnaud E, et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–11. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin HM, Campbell BJ, Hart CA, et al. Enhanced Escherichia coli adherence and invasion in Crohn's disease and colon cancer. Gastroenterology. 2004;127:80–93. doi: 10.1053/j.gastro.2004.03.054. [DOI] [PubMed] [Google Scholar]
- Menon R, Watson SE, Thomas LN, et al. Diet complexity and estrogen receptor β status affect the composition of the murine intestinal microbiota. Appl Environ Microb. 2013;79:5763–73. doi: 10.1128/AEM.01182-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalingam NA, Kao JY, Young VB. Microbial ecology of the murine gut associated with the development of dextran sodium sulfate-induced colitis. Inflamm Bowel Dis. 2011;17:917–26. doi: 10.1002/ibd.21462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalingam NA, Lynch SV. Role of the microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2012;18:968–84. doi: 10.1002/ibd.21866. [DOI] [PubMed] [Google Scholar]
- Noor S, Ridgway K, Scovell L, et al. Ulcerative colitis and irritable bowel patients exhibit distinct abnormalities of the gut microbiota. BMC Gastroenterol. 2010;10:134. doi: 10.1186/1471-230X-10-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubo T. In vivo effects of tea polyphenol intake on human intestinal microflora and metabolism. Biosci Biotech Bioch. 1992;56:588. doi: 10.1271/bbb.56.588. [DOI] [PubMed] [Google Scholar]
- Onderdonk AB, Richardson JA, Hammer RE, et al. Correlation of cecal microflora of HLA-B27 transgenic rats with inflammatory bowel disease. Infect Immun. 1998;66:6022–3. doi: 10.1128/iai.66.12.6022-6023.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott SJ, Musfeldt M, Wenderoth DF, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–93. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possemiers S, Bolca S, Verstraete W, et al. The intestinal microbiome: a separate organ inside the body with the metabolic potential to influence the bioactivity of botanicals. Fitoterapia. 2011;82:53–66. doi: 10.1016/j.fitote.2010.07.012. [DOI] [PubMed] [Google Scholar]
- Rajilić-Stojanović M, Biagi E, Heilig HGHJ, et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141:1792–801. doi: 10.1053/j.gastro.2011.07.043. [DOI] [PubMed] [Google Scholar]
- Rajilić-Stojanović M, Smidt H, De Vos WM. Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol. 2007;9:2125–36. doi: 10.1111/j.1462-2920.2007.01369.x. [DOI] [PubMed] [Google Scholar]
- Rastall RA, Gibson GR, Gill HS, et al. Modulation of the microbial ecology of the human colon by probiotics, prebiotics and synbiotics to enhance human health: an overview of enabling science and potential applications. FEMS Microbiol Ecol. 2005;52:145–52. doi: 10.1016/j.femsec.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Rastmanesh R. High polyphenol, low probiotic diet for weight loss because of intestinal microbiota interaction. Chem Biol Interact. 2011;189:1–8. doi: 10.1016/j.cbi.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Ree T. Tannins: classification and definition. Nat Prod Rep. 2001;18:641–9. doi: 10.1039/b101061l. [DOI] [PubMed] [Google Scholar]
- Reeder J, Knight R. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Methods. 2010;7:668–9. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie LE, Burke KF, Garcia-Mazcorro JF, et al. Characterization of fecal microbiota in cats using universal 16S rRNA gene and group-specific primers for Lactobacillus and Bifidobacterium spp. Vet Microbiol. 2010;144:140–6. doi: 10.1016/j.vetmic.2009.12.045. [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Ogawa T, Koyanagi A, et al. Distribution and excretion of bilberry anthocyanins in mice. J Agr Food Chem. 2009;57:7681–6. doi: 10.1021/jf901341b. [DOI] [PubMed] [Google Scholar]
- Sepehri S, Kotlowski R, Bernstein CN, et al. Microbial diversity of inflamed and noninflamed gut biopsy tissues in inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:675–83. doi: 10.1002/ibd.20101. [DOI] [PubMed] [Google Scholar]
- Smith AH, Mackie RI. Effect of condensed tannins on bacterial diversity and metabolic activity in the rat gastrointestinal tract. Appl Environ Microb. 2004;70:1104–15. doi: 10.1128/AEM.70.2.1104-1115.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Seksik P, Rigottier-Gois L, et al. Specificities of the fecal microbiota in inflammatory bowel disease. Inflamm Bowel Dis. 2006;12:106–11. doi: 10.1097/01.MIB.0000200323.38139.c6. [DOI] [PubMed] [Google Scholar]
- Swidsinski A, Weber J, Loening-Baucke V, et al. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol. 2005;43:3380–9. doi: 10.1128/JCM.43.7.3380-3389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault R, Thibault P, De Coppet K, et al. Down-regulation of the monocarboxylate transporter 1 is involved in butyrate deficiency during intestinal inflammation. Gastroenterology. 2007;133:1916–27. doi: 10.1053/j.gastro.2007.08.041. [DOI] [PubMed] [Google Scholar]
- Turner N, Taddeo SS, McDonough CM, et al. Polyphenol-rich sorghum brans promote fecal water retention and alter short chain fatty acids in Sprague Dawley rats. Cereal Foods World. 2010;55(4 Suppl.):A72–3. [Google Scholar]
- van Duynhoven J, Vaughan EE, Jacobs DM, et al. Metabolic fate of polyphenols in the human superorganism. P Natl Acad Sci USA. 2011;108:4531–8. doi: 10.1073/pnas.1000098107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicky De P, Greet V, Paul JR, et al. T1795 Impaired butyrate oxidation in ulcerative colitis is due to decreased butyrate uptake and a defect in the oxidation pathway. Gastroenterology. 2010;138:S-580. [Google Scholar]
- Walker AW, Ince J, Duncan SH, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2010;5:220–30. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JM, Rossi O, Meijerink M, et al. Epithelial crosstalk at the microbiota–mucosal interface. P Natl Acad Sci USA. 2010;108(Suppl. 1):4607–14. doi: 10.1073/pnas.1000092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegel J, Kuk S-U, Kohring GW. Clostridium thermobutyricum sp. nov., a moderate thermophile isolated from a cellulolytic culture, that produces butyrate as the major product. Int J Syst Bacteriol. 1989;39:199–204. [Google Scholar]
- Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–34. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.