Summary
We identified the minor allele (T) in SNP rs11676348 to have pleiotropic effect on risk of UC and CRC, particularly in tumors with an inflammatory component. Our findings offer the promise of risk stratification of UC patients for developing CRC.
Abstract
Although genome-wide association studies (GWAS) have separately identified many genetic susceptibility loci for ulcerative colitis (UC), Crohn’s disease (CD) and colorectal cancer (CRC), there has been no large-scale examination for pleiotropy, or shared genetic susceptibility, for these conditions. We used logistic regression modeling to examine the associations of 181 UC and CD susceptibility variants previously identified by GWAS with risk of CRC using data from the Genetics and Epidemiology of Colorectal Cancer Consortium and the Colon Cancer Family Registry. We also examined associations of significant variants with clinical and molecular characteristics in a subset of the studies. Among 11794 CRC cases and 14190 controls, rs11676348, the susceptibility single nucleotide polymorphism (SNP) for UC, was significantly associated with reduced risk of CRC (P = 7E−05). The multivariate-adjusted odds ratio of CRC with each copy of the T allele was 0.93 (95% CI 0.89–0.96). The association of the SNP with risk of CRC differed according to mucinous histological features (P heterogeneity = 0.008). In addition, the (T) allele was associated with lower risk of tumors with Crohn’s-like reaction but not tumors without such immune infiltrate (P heterogeneity = 0.02) and microsatellite instability-high (MSI-high) but not microsatellite stable or MSI-low tumors (P heterogeneity = 0.03). The minor allele (T) in SNP rs11676348, located downstream from CXCR2 that has been implicated in CRC progression, is associated with a lower risk of CRC, particularly tumors with a mucinous component, Crohn’s-like reaction and MSI-high. Our findings offer the promise of risk stratification of inflammatory bowel disease patients for complications such as CRC.
Introduction
Ulcerative colitis (UC) and Crohn’s disease (CD), collectively known as inflammatory bowel disease (IBD), are chronic inflammatory disorders of the gastrointestinal tract that are associated with an increased risk of colorectal cancer (CRC). The risk of CRC appears to increase with the duration, severity and anatomic extent of colonic inflammation (1–3). As a result, individuals with greater than a 8 to 12-year history of IBD are currently recommended to undergo annual colonoscopy with surveillance biopsies or chromoendoscopy with targeted biopsies to screen for precancerous and cancerous changes (4). Despite this compelling evidence for the association between IBD and CRC, there are no data on the potential for pleiotropy or shared genetic susceptibility between these conditions.
Genome-wide association (GWA) studies have identified more than 180 risk loci that are associated with risk of CD and UC (5–7). A number of these variants with potential biologic significance have shown to be associated with other chronic inflammatory disorders (5), and such studies previously led to identification of new and novel therapies for IBD (8–10). We therefore sought to establish whether known susceptibility variants for UC and CD are also associated with risk of CRC using data from the Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO) and the Colon Cancer Family Registry (CCFR) (11). In addition, we explored the potential functional significance of candidate variants with pleiotropic effect by examining their association with clinical and molecular characteristics of CRC using cases for which we had collected tissue for histologic and molecular analyses.
Methods
Study participants
Each study is described in detail in the Supplementary Material, available at Carcinogenesis Online and the number of cases and controls, as well as the age and sex distributions is listed in Supplementary Table 1, available at Carcinogenesis Online. In brief, CRC cases were defined as men and women diagnosed with colorectal adenocarcinoma and confirmed by medical records, pathologic reports, cancer registries or death certificates. All participants provided informed consent; each study was approved by their respective Institutional Review Boards.
Genotyping
GWAS in GECCO and CCFR
We conducted a meta-analysis of 181 previously identified IBD risk variants in 14 studies within the GECCO and the CCFR (11794 cases and 14190 controls). Details regarding genotyping and data quality control are described elsewhere (12). Owing to limited numbers, we did not analyze samples clustering with other HapMap populations (e.g. YRI), retaining those that clustered with the Utah Residents with Northern and Western European Ancestry (CEU) population and adjusting for population stratification using principle component analysis (See Statistical Analysis). Genotyped single nucleotide polymorphisms (SNPs) were excluded based on call rate (<98%), lack of Hardy Weinberg Equilibrium in controls (P < 1×10–4), and low minor allele frequency <5%. As imputation of genotypes is established as standard practice in the analysis of genotype data, we imputed the autosomal SNPs of all studies to the CEU population in HapMap II. Imputed SNPs were restricted based on minor allele frequency (≥1%) and imputation accuracy (R 2 > 0.3). In our detailed result table (Supplementary Table 2, available at Carcinogenesis Online), we list for each SNP the number of studies with directly genotyped or imputed data and the mean imputation R 2. These data show, as expected, that imputed SNPs tend to show very similar results to SNPs that were directly genotyped if the correlation is high between SNPs. The IBD SNPs were selected based on the most recent results of CD and UC genome-wide association studies (GWAS) (6,7,13). The risk allele is defined as the allele with reported association with CD or UC.
Pathological assessment of molecular characteristics
Beginning in 1997 in the Health Professional Follow up Study (HPFS) and 2001 in the Nurses’ Health Study (NHS), we began retrieving specimens from the pathology departments of treating hospitals for participants with confirmed diagnosis of CRC (14). Histological features including degree of differentiation, presence of mucinous, signet cell components, tumor infiltrating lymphocytes, or Crohn’s disease-like reaction and tumor growth pattern were collected by a pathologist (S.O.) as described previously (15,16). Real-time PCR (MethyLight) was used for quantitative DNA methylation to determine CpG island methylator phenotype (CIMP) status using DNA extracted from paraffin-embedded tissue (17). We quantified DNA methylation in eight CIMP-specific promoters as detailed elsewhere (18). CIMP-high was defined as ≥6/8 methylated markers using the eight-marker CIMP panel, and CIMP-low/negative was defined as 0/8 to 5/8 methylated markers, according to previously established criteria (19). Microsatellite instability (MSI) analysis was performed using 10 microsatellite markers (D2S123, D5S346, D17S250, BAT25, BAT26, BAT40, D18S55, D18S56, D18S67 and D18S487). MSI-high was defined as the presence of instability in ≥30% of the markers, and MSI-low/microsatellite stable (MSS) as instability in 0–29% of markers (20). We performed PCR and pyrosequencing targeted for KRAS (codons 12 and 13) and BRAF (codon 600) (20,21). TP53 and CDX2 expression were assessed by immunohistochemistry as detailed elsewhere (22,23).Tumor molecular phenotype was previously evaluated in the Diet, Activity, and Lifestyle Study (DALS) study for CIMP and MSI status, BRAF, TP53 and KRAS mutations using methods that have been described previously in detail (24–27).
Statistical analysis
GWAS in GECCO and CCFR
For each study, we estimated the association between SNPs and risk of CRC by calculating betas, odds ratios (ORs), standard errors, 95% confidence intervals (CIs) and P values using logistic regression models with log-additive genetic effects. Each directly genotyped SNP was coded as 0, 1 or 2 copies of the risk allele. For imputed SNPs, we used the expected number of copies of the risk allele (the ‘dosage’), which has been shown to give unbiased estimates in the association test for imputed SNPs (28). We adjusted for age, sex (when appropriate), study center (when appropriate), batch effects (ASTERISK only) and the first three principal components from EIGENSTRAT to account for population substructure. As CCFR Set 2 is a family-based study, we used logistic regression model stratified by family ID while adjusting for age and sex.
We conducted inverse-variance weighted, fixed-effects meta-analyses to combine beta estimates and standard errors across individual studies. In this approach, we weighted the beta estimate of each study by the inverse of its variance, and calculated a combined estimate by summing the weighted betas and dividing by the summed weights. For imputed SNPs, it has been shown that the inverse variance is approximately proportional to the imputation quality (28). Thus, the inverse variance weighting scheme automatically incorporates imputation quality in the meta-analysis for imputed SNPs. We calculated the heterogeneity P values based on Cochran’s Q statistic (29). Bonferroni-adjusted P value of 2.8E−04 (=0.05/181) was considered statistically significant. We used PLINK (30) and R to conduct the statistical analysis of the SNP and CRC associations across studies.
Characterization of variant shown to be associated with CRC (rs11676348)
Within the NHS and HPFS, we used logistic regression to estimate OR and corresponding 95% CI for the association of variant rs11676348 with CRC stratified by tumor phenotype (poorly differentiated tumors versus moderately well differentiated tumors), tumor histology (presence versus absence of mucinous, signet cell, Crohn’s-like reaction, tumor infiltrating lymphocytes or peritumor lymphocytic reaction, and expansible versus intermediate/infiltrative tumor growth pattern) and molecular characteristics (mutant versus wild-type KRAS; mutant versus wild-type BRAF; MSI-H versus MSI-L/MSS; CIMP-low/negative versus CIMP-high; presence versus absence of TP53 and CDX2 expressions). Because there was no heterogeneity for the association of SNP rs11676348 with CRC risk between women and men (P for heterogeneity > 0.40), we pooled data from the two studies of NHS and HPFS. To assess the effect of rs11676348 on CRC according to clinical phenotype or molecular characteristics in NHS/HPFS, we used a case–case logistic regression model comparing tumor subtypes. We used similar analyses to validate our findings with regards to the effect of variant rs11676348 on CRC according to molecular characteristics (CIMP and MSI status, BRAF, TP53 and KRAS mutations) in DALS. We used SAS V9.2 (SAS Institute, Cary, NC) for the analysis related to the characterization of variant rs11676348 and tumor subtypes in NHS/HPFS and DALS.
Results
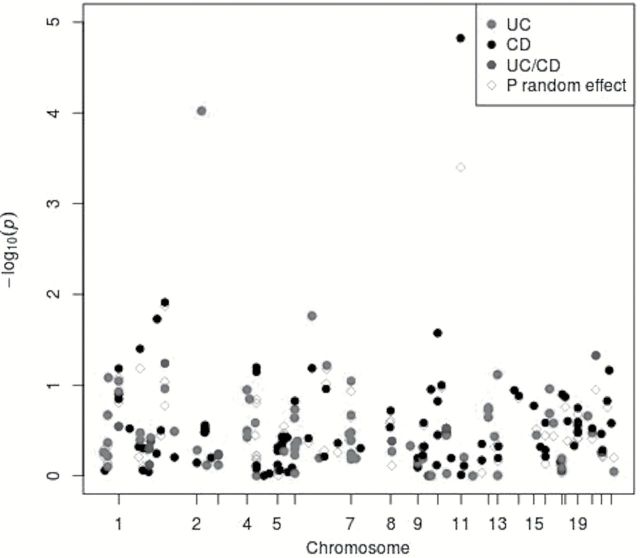
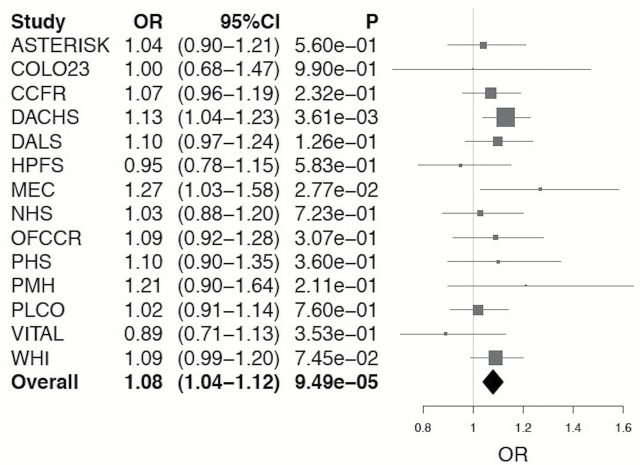
Among 11794 CRC cases and 14190 controls, we examined 181 SNPs previously associated with risk of UC and CD in relation to risk of CRC. Among these SNPs, we identified rs11676348, a susceptibility SNP for UC, as significantly inversely associated with risk of CRC (P = 7E−05) after Bonferroni correction for multiple comparisons (Figure 1). The mean allele frequency for the (T) allele was 0.50 with a range of 0.46–0.54 among the studies. The multivariate-adjusted odds ratio (OR) of CRC with each copy of minor allele (T) for rs11676348 was 0.93 (95% CI 0.89–0.96) (Figure 2). There was no statistically significant heterogeneity in the association among studies (P heterogeneity = 0.81). We also observed that rs102275, a susceptibility loci for CD located in TMEM258 gene on chromosome 11, was inversely associated with risk of CRC (OR = 0.92, 95% CI 0.88–0.96, P = 2E−05). However, because of heterogeneity among the studies (P heterogeneity = 0.09), the variant did not meet the Bonferroni-corrected significance using random-effect meta-analysis (P = 4E−04).
Figure 1.
Association between known IBD variants and risk of colorectal cancer. Manhattan plot of the meta-analysis association between risk variants of Crohn’ disease (CD) and ulcerative colitis (UC) and colorectal cancer. Each association is colored according to disease (CD or UC) for which the single nucleotide polymorphism was originally reported, and positioned on the x-axis according to its genomic position. Filled circles represent the results of fixed effect meta-analysis while blank circles reflect results of random effect meta-analysis.
Figure 2.
Meta-analysis of association of rs11676348 SNP with CRC.
To explore the possibility that the association between rs11676348 and risk of CRC is mediated by clinical diagnosis of UC or CD, we performed sensitivity analyses limiting our meta-analysis to the 11 studies with available data on reported diagnoses of UC and CD and excluded those with self-reported diagnosis of UC or CD (Supplementary Table 3, available at Carcinogenesis Online). Among 9963 cases and 12285 controls, we observed a consistent association of the (T) allele in rs11676348 with a lower risk of CRC (multivariate-adjusted OR = 0.93, P = 8.3E−04).
We explored the possibility that the inverse association of this UC susceptibility SNP with CRC may be limited to specific molecular subtypes of CRCs. Among 293 CRC cases with available molecular and histologic characteristics and 1172 controls in NHS/HPFS, the association of rs11676348 SNP with CRC appeared to vary according to MSI status (P heterogeneity = 0.03), presence of mucinous component (P heterogeneity = 0.008) and Crohn’s-like reaction within the tumor (P heterogeneity = 0.02) (Tables 1 and 2). The (T) allele at the UC susceptibility locus rs11676348 was associated with a lower risk of MSI-H CRC (multivariate OR 0.57; 95% CI 0.33–0.99) but not MSS/MSI-L CRC (multivariate-adjusted OR 1.05, 95% CI 0.86–1.29). Similarly, the (T) allele was associated with a lower risk of mucinous CRC (multivariate OR 0.77; 95% CI 0.58–1.02) but not CRC without a mucinous component (multivariate OR 1.21; 95% CI 0.96–1.52;). Finally, the (T) allele was associated with a lower risk of CRC with the presence of a Crohn’s-like reaction (multivariate OR 0.68; 95% CI 0.47–0.99) but not CRC without a Crohn’s-like reaction (multivariate OR 1.11; 95% CI 0.90–1.37). The association of the (T) allele with risk of CRC did not appear to differ according to other histologic or molecular characteristics including presence of a signet cell component or peritumoral lymphocytic reaction, tumor differential or growth, CIMP status, expression of CDX2 or TP53 or mutations in BRAF or KRAS.
Table 1.
Associations between rs11676348 variant and risk of CRC according to histologic subtypesa
| Controls | Cases | OR (95% CI) | Cases | OR (95% CI) | P heterogeneity b | |
|---|---|---|---|---|---|---|
| Mucinous component (−) | Mucinous component (+) | |||||
| CC | 287 | 47 | 1 | 42 | 1 | 0.008 |
| CT | 616 | 70 | 0.67 (0.45–1.00) | 51 | 0.56 (0.36–0.87) | |
| TT | 269 | 59 | 1.34 (0.91–2.12) | 24 | 0.66 (0.38–1.12) | |
| P trend | 0.11 | 0.07 | ||||
| T allele | 1.21 (0.96–1.52) | 0.77 (0.58–1.02) | ||||
| Crohn’s-like reaction (−) | Crohn’s-like reaction (+) | |||||
| CC | 287 | 60 | 1 | 27 | 1 | 0.02 |
| CT | 616 | 93 | 0.71 (0.49–1.01) | 28 | 0.46 (0.26–0.80) | |
| TT | 269 | 67 | 1.21 (0.82–1.79) | 13 | 0.55 (0.28–1.11) | |
| P trend | 0.33 | 0.04 | ||||
| T allele | 1.11 (0.90–1.37) | 0.68 (0.47–0.99) | ||||
| Tumor infiltrating lymphocytes (−) | Tumor infiltrating lymphocytes (+) | |||||
| CC | 287 | 57 | 1 | 30 | 1 | 0.05 |
| CT | 616 | 93 | 0.74 (0.51–1.06) | 27 | 0.40 (0.23–0.70) | |
| TT | 269 | 63 | 1.21 (0.81–1.81) | 18 | 0.67 (0.36–1.25) | |
| P trend | 0.35 | 0.11 | ||||
| T allele | 1.11 (0.89–1.37) | 0.75 (0.53–1.06) | ||||
| Peritumor lymphocytic reaction (−) | Peritumor lymphocytic reaction (+) | |||||
| CC | 287 | 12 | 1 | 76 | 1 | 0.55 |
| CT | 616 | 21 | 0.80 (0.39–1.66) | 98 | 0.58 (0.42–0.82) | |
| TT | 269 | 9 | 0.80 (0.33–1.95) | 72 | 1.05 (0.73–1.53) | |
| P trend | 0.61 | 0.87 | ||||
| T allele | 0.89 (0.57–1.40) | 1.02 (0.83–1.24) | ||||
| Well-moderate differentiated | Poorly differentiated | |||||
| CC | 287 | 86 | 1 | 6 | 1 | 0.54 |
| CT | 616 | 126 | 0.66 (0.49–0.91) | 7 | 0.51 (0.17–1.55) | |
| TT | 269 | 84 | 1.08 (0.76–1.53) | 4 | 0.76 (0.21–2.72) | |
| P trend | 0.74 | 0.59 | ||||
| T allele | 1.03 (0.86–1.24) | 0.82 (0.40–1.68) | ||||
| Tumor growth (expansible) | Tumor growth (intermediate-infiltrative) | |||||
| CC | 287 | 28 | 1 | 28 | 1 | 0.94 |
| CT | 616 | 31 | 0.49 (0.29–0.84) | 31 | 0.49 (0.29–0.84) | |
| TT | 269 | 26 | 1.05 (0.59–1.86) | 26 | 1.05 (0.59–1.86) | |
| P trend | 0.98 | 0.98 | ||||
| T allele | 1.01 (0.73–1.39) | 1.01 (0.73–1.39) | ||||
aModels were adjusted for age (years), cohort (NHS, HPFS) and the first three eigenvectors.
bCalculated using a case–case only logistic regression model comparing tumor subtypes.
Table 2.
Associations between rs11676348 variant and risk of CRC according to molecular subtypesa
| Controls | Cases | OR (95% CI) | Cases | OR (95% CI) | P heterogeneity b | |
|---|---|---|---|---|---|---|
| MSS/MSI-L | MSI-H | |||||
| CC | 287 | 68 | 1 | 14 | 1 | 0.03 |
| CT | 616 | 99 | 0.65 (0.46–0.92) | 12 | 0.40 (0.18–0.89) | |
| TT | 269 | 69 | 1.10 (0.75–1.62) | 5 | 0.42 (0.15–1.18) | |
| P trend | 0.65 | 0.05 | ||||
| T allele | 1.05 (0.86–1.29) | 0.57 (0.33–0.99) | ||||
| CIMP-low/negative | CIMP-high | |||||
| CC | 287 | 60 | 1 | 15 | 1 | 0.32 |
| CT | 616 | 96 | 0.72 (0.50–1.03) | 16 | 0.50 (0.24–1.04) | |
| TT | 269 | 62 | 1.12 (0.75–1.67) | 10 | 0.74 (0.33–1.70) | |
| P trend | 0.61 | 0.38 | ||||
| T allele | 1.06 (0.86–1.31) | 0.82 (0.52–1.29) | ||||
| BRAF mutation (−) | BRAF mutation (+) | |||||
| CC | 287 | 71 | 1 | 12 | 1 | 0.21 |
| CT | 616 | 100 | 0.66 (0.44–0.98) | 12 | 0.53 (0.33–0.86) | |
| TT | 269 | 69 | 0.94 (0.59–1.48) | 7 | 1.04 (0.63–1.74) | |
| P trend | 0.72 | 0.95 | ||||
| T allele | 0.96 (0.75–1.22) | 1.01 (0.76–1.34) | ||||
| KRAS mutation (−) | KRAS mutation (+) | |||||
| CC | 287 | 48 | 1 | 35 | 1 | 0.92 |
| CT | 616 | 72 | 0.63 (0.45–0.89) | 41 | 0.47 (0.21–1.06) | |
| TT | 269 | 42 | 1.05 (0.72–1.55) | 33 | 0.65 (0.25–1.67) | |
| P trend | 0.84 | 0.28 | ||||
| T allele | 1.02 (0.83–1.25) | 0.75 (0.44–1.26) | ||||
| CDX2 (+) | CDX2 (−) | |||||
| CC | 287 | 36 | 1 | 11 | 1 | 0.77 |
| CT | 616 | 47 | 0.58 (0.37–0.93) | 12 | 0.52 (0.22–1.19) | |
| TT | 269 | 32 | 0.96 (0.58–1.61) | 6 | 0.59 (0.22–1.63) | |
| P trend | 0.81 | 0.24 | ||||
| T allele | 0.97 (0.73–1.28) | 0.72 (0.42–1.24) | ||||
| TP53 expression (−) | TP53 expression (+) | |||||
| CC | 287 | 45 | 1 | 22 | 1 | 0.06 |
| CT | 616 | 54 | 0.55 (0.36–0.84) | 40 | 0.81 (0.47–1.40) | |
| TT | 269 | 34 | 0.81 (0.50–1.30) | 30 | 1.53 (0.85–2.73) | |
| P trend | 0.29 | 0.13 | ||||
| T allele | 0.87 (0.67–1.13) | 1.27 (0.93–1.74) | ||||
aModels were adjusted for age (years), cohort (NHS, HPFS) and the first three eigenvectors.
bCalculated using a case–case only logistic regression model comparing tumor subtypes.
We also confirmed the association between rs11676348 and risk of CRC according to molecular subtypes in a separate cohort of 548 cases and 728 controls in DALS. Similar to our findings in NHS and HPFS, the association of the SNP seemed to be modified by MSI status (P heterogeneity = 0.03). Compared to participants with TT genotype, the risk of developing MSI-H CRC among participants with CT and TT genotypes were 0.67 (95% CI 0.41–1.11) and 0.52 (95% CI 0.26–1.03), respectively (P trend = 0.04). Conversely, compared to participants with TT genotype, the risk of developing MSI-L/MSS CRC among participants with CT and TT genotypes were 1.14 (95% CI 0.86–1.50) and 1.16 (95% CI 0.83–1.63), respectively (P trend = 0.37). The association of the (T) allele with risk of CRC did not appear to differ according to CIMP status, expression of CDX2, or mutations in BRAF, TP53, or KRAS genes.
Discussion
In this large meta-analysis of individual-level data that included 11794 cases of CRC and 14190 controls, we examined the association of GWAS-identified risk variants for UC and CD on risk of CRC. We identified that the (T) allele in rs11676348, a UC susceptibility locus located on chromosome 2, to be inversely associated with risk of CRC. In addition, using a subgroup of CRC cases with known histologic and molecular data, we showed that rs11676348 was particularly associated with lower risk of CRC tumors with Crohn’s-like reaction, MSI-H status and mucinous components which are each characteristics of tumors with high inflammatory burden. We subsequently confirmed these findings in a separate population. Of note, we did not observe an association between rs11676348 and CRC cases with peritumoral lymphocytic reaction, which is typically associated with mucinous and MSI-H tumors. The lack of association is probably due to presence of at least mild lymphocytic reaction in most of our cases (>80%), consistent with previous reports (31,32).
Our findings are supported by biologically plausible mechanisms. rs11676348 was identified in GWAS of UC to be correlated with expression of CXCR2 gene (from whole blood samples) through analyses of expression quantitative trait loci (6). The CXCR2 gene, located 10 kB downstream from rs11676348, is a member of G-protein-coupled receptor family with high affinity for IL-8. CXCR2 primarily mediates migration of neutrophils to the site of inflammation and facilities the angiogenic effect of IL-8 in intestinal microvascular endothelial cells. More recently, CXCR2 has been implicated in development of CRCs associated with chronic colonic inflammation in experimental models. Katoh and colleagues (33) showed that loss of CXCR2 dramatically suppresses chronic colonic inflammation and colitis-associated tumorigenesis through inhibition of the infiltration of myeloid-derived suppressor cells into colonic mucosa and tumors in a mouse model of colitis-associated cancer. Similarly, using several mouse models of spontaneous and inflammation-driven neoplasia, Jamieson and colleagues showed that inhibition of CXCR2 profoundly suppresses inflammation-driven tumorigenesis (34).
Several studies have demonstrated a higher incidence of MSI-H in the cancers that develop in the setting of long-standing UC with severe inflammation (35–37). Specifically, compared with MSS stable tumors, MSI-H tumors are characterized by a higher inflammatory burden (35,38). Moreover, it has been demonstrated that chronic inflammation is associated with an imbalance in base excision-repair enzymes that contributes to microsatellite instability (39).
In addition to CXCR2, expression quantitative trait loci studies have linked rs11676348 to expression of MR1 gene (40). MR1 encodes an antigen-presenting molecule specialized in presenting microbial vitamin B6 metabolites (41,42). The protein is also involved in the development and expansion of a small population of T-cells expressing an invariant T-cell receptor alpha chain called mucosal-associated invariant T-cells (42,43). Mucosal-associated invariant T-cells lymphocytes are preferentially located in the gut lamina propria and therefore may be involved in monitoring commensal flora or serve as a distress signal (44,45). These findings may point to novel pathways associated with genetic variation that may potentially modify the effect of environmental factors (gut commensal bacterial and B vitamins) on risk of IBD and CRC (46–48).
The seemingly opposing effects of the (T) allele of rs11676348 with increased risk of UC but decreased risk of CRC may, at least at first, appear inconsistent. However, this SNP may still be a marker for shared pathways between UC and CRC that contribute to the initiation of mucosal inflammation and colitis but inhibit carcinogenesis. Similarly, we previously observed that a SNP associated with lower incident CRC risk was subsequently associated with poorer survival (49). Alternatively, genetic variation underlying the pathways associated with this SNP may predispose to distinctive phenotypic manifestations of a common mechanism (e.g. chronic inflammation versus cancer). The large number of subjects and the consistency of our observed associations across 14 separate, well-characterized studies add strength to our findings. In addition, we were able to deploy molecular pathological epidemiology approach to provide evidence for a role of the risk variant in specific pathogenic mechanisms (50). Indeed, the consistency of our associations with specific molecular and histological subtypes of CRC within two distinct studies provides additional evidence of a true association.
Limitations of our study include inclusion of only European ancestry participants. In addition, it is possible that a higher prevalence of IBD patients among the CRC cases may account for our observed associations. However, this is unlikely for several reasons. First, our sensitivity analyses excluding cases of IBD from our case–control set did not materially alter our effect estimates. Second, considering the low prevalence of IBD among cases and controls (<0.5%) together with the observation that other SNPs that are more strongly associated with risk of UC and CD were not found to be associated with risk of CRC, we believe that such oversampling is unlikely to have biased our results. Third, if the association between rs11676348 and CRC was mediated by its association with UC, we would expect that directionality of the association to be the same. In contrast, we observed that the T allele associated with UC was inversely associated with risk of CRC. Finally, as more recent GWAS have identified new IBD risk loci, these variants remain to be evaluated for their pleiotropic effects with CRC.
In conclusion, we identified a UC susceptibility locus, rs11676348, with pleotropic effect on risk of CRC. Our findings highlight the potential importance of common pathways underlying risk of IBD and CRC and have compelling biological plausibility as this SNP is in close proximity to CXCR2 and is more strongly associated with risk of specific subtypes of CRC (Crohn’s like-reaction, mucinous component or MSI-H status) that may have a distinct molecular pathogenesis associated with either CXCR2 function or chronic inflammation. Our finding further strengthens the potential importance of CXCR2-mediated pathways in inflammation and tumorigenesis. Whether these findings can be generalized to individuals with established IBD is unknown and yet to be determined. However, our finding provides proof-of-principle that genetic susceptibility loci could potentially be used to risk-stratify individuals with IBD for the development of CRC.
Supplementary material
Supplementary Tables 1–3 can be found at http://carcin.oxfordjournals.org/
Funding
Funded by R01 CA137178, R01 CA050385, R01 CA151993, P01 CA87969, P30 DK043351, P50 CA127003, UM1 CA 167552, UM1 CA186107, K23 DK099681, K24 [DK]098311, K07 CA190673 and K07 CA148894. Senior investigator grant from the Crohn’s and Colitis Foundation of America (CCFA) to A.T.C. A career development award from the American Gastroenterological Association (AGA) and by National Institute of Diabetes and Digestive and Kidney Diseases (K23 DK099681 to H.K.).
GECCO: National Cancer Institute, National Institutes of Health, U.S. Department of Health and Human Services (U01 CA137088; R01 CA059045).
ASTERISK: a Hospital Clinical Research Program (PHRC) and supported by the Regional Council of Pays de la Loire, the Groupement des Entreprises Françaises dans la Lutte contre le Cancer (GEFLUC), the Association Anne de Bretagne Génétique and the Ligue Régionale Contre le Cancer (LRCC).
COLO2&3: National Institutes of Health (R01 CA60987).
CCFR: National Institutes of Health (UM1 CA167551) and through cooperative agreements with members of the Colon Cancer Family Registry and P.I.s. This genome wide scan was supported by the National Cancer Institute, National Institutes of Health by U01 CA122839 and R01 CA143237 to G.C. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the CFRs, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government or the CFR. The following CFR centers contributed data to this manuscript and were supported by National Institutes of Health:
Australasian Colorectal Cancer Family Registry (U01/U24 CA097735), Familial Colorectal Neoplasia Collaborative Group (U01/U24 CA074799), Mayo Clinic Cooperative Family Registry for Colon Cancer Studies (U01/U24 CA074800), Ontario Registry for Studies of Familial Colorectal Cancer (U01/U24 CA074783), Seattle Colorectal Cancer Family Registry (U01/U24 CA074794) and the University of Hawaii Colorectal Cancer Family Registry (U01/U24 CA074806).
DACHS: German Research Council (Deutsche Forschungsgemeinschaft, BR 1704/6-1, BR 1704/6-3, BR 1704/6-4 and CH 117/1-1), and the German Federal Ministry of Education and Research (01KH0404 and 01ER0814).
DALS: National Institutes of Health (R01 CA48998 and CA61757 to M.L.S.);
OFCCR: National Institutes of Health, through funding allocated to the Ontario Registry for Studies of Familial Colorectal Cancer (U01 CA074783); see CCFR section. Additional funding toward genetic analyses of OFCCR includes the Ontario Research Fund, the Canadian Institutes of Health Research and the Ontario Institute for Cancer Research, through generous support from the Ontario Ministry of Research and Innovation.
PLCO: Intramural Research Program of the Division of Cancer Epidemiology and Genetics and supported by contracts from the Division of Cancer Prevention, National Cancer Institute, NIH, DHHS. Additionally, a subset of control samples were genotyped as part of the Cancer Genetic Markers of Susceptibility (CGEMS) Prostate Cancer GWAS (Yeager,M. et al. (2007) Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. , 39, 645–649), Colon CGEMS pancreatic cancer scan (PanScan) (Amundadottir,L et al. (2009) Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat. Genet., 41, 986–990, and Petersen,G.M. et al. (2010) A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat. Genet. , 42, 224–228), and the Lung Cancer and Smoking study (Landi,M.T. et al. (2009) A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am. J. Hum. Genet. , 85, 679–691). The prostate and PanScan study datasets were accessed with appropriate approval through the dbGaP online resource (http://cgems.cancer.gov/data/) accession numbers phs000207v.1p1 and phs000206.v3.p2, respectively, and the lung datasets were accessed from the dbGaP website (http://www.ncbi.nlm.nih.gov/gap) through accession number phs000093 v2.p2. Funding for the Lung Cancer and Smoking study was provided by National Institutes of Health (NIH), Genes, Environment and Health Initiative (GEI) Z01 CP 010200, NIH U01 HG004446 and NIH GEI U01 HG 004438. For the lung study, the GENEVA Coordinating Center provided assistance with genotype cleaning and general study coordination, and the Johns Hopkins University Center for Inherited Disease Research conducted genotyping.
PHS: National Institutes of Health (CA42182).
PMH: National Institutes of Health (R01 CA076366 to P.A.N.).
VITAL: National Institutes of Health (K05 CA154337).
WHI: The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201100046C, HHSN268201100001C, HHSN26820 1100002C, HHSN268201100003C, HHSN268201100004C and HHSN 271201100004C.
Supplementary Material
Acknowledgements
We would like to thank the participants and staff of ASTERISK, CCFR, COLO2&3, DACHS, DALS, HPFS, MEC, NHS, OFCCR, PHS, PLCO, PMH, WHI and VITAL for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- CD
Crohn’s disease
- CIMP
CpG island methylator phenotype
- CRC
colorectal cancer
- GWAS
genome-wide association studies
- HPFS
Health Professional Follow up Study
- IBD
inflammatory bowel disease
- MSI
microsatellite instability
- MSS
microsatellite stable
- NHS
Nurses’ Health Study
- SNP
single nucleotide polymorphism
- UC
ulcerative colitis
References
- 1. Ullman T.A., et al. (2011) Intestinal inflammation and cancer. Gastroenterology, 140, 1807–1816. [DOI] [PubMed] [Google Scholar]
- 2. Aust D.E., et al. (2002) The APC/beta-catenin pathway in ulcerative colitis-related colorectal carcinomas: a mutational analysis. Cancer, 94, 1421–1427. [DOI] [PubMed] [Google Scholar]
- 3. Bass A.J., et al. (2011) Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat. Genet., 43, 964–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Itzkowitz S.H., et al. (2005) Consensus conference: Colorectal cancer screening and surveillance in inflammatory bowel disease. Inflamm. Bowel Dis., 11, 314–321. [DOI] [PubMed] [Google Scholar]
- 5. Jostins L., et al. (2012) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature, 491, 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Anderson C.A., et al. (2011) Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet., 43, 246–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Franke A., et al. (2010) Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet., 42, 1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Elliott M., et al. (2009) Ustekinumab: lessons learned from targeting interleukin-12/23p40 in immune-mediated diseases. Ann. N. Y. Acad. Sci., 1182, 97–110. [DOI] [PubMed] [Google Scholar]
- 9. Sandborn W.J., et al. ; Ustekinumab Crohn’s Disease Study Group. (2008) A randomized trial of Ustekinumab, a human interleukin-12/23 monoclonal antibody, in patients with moderate-to-severe Crohn’s disease. Gastroenterology, 135, 1130–1141. [DOI] [PubMed] [Google Scholar]
- 10. Sandborn W.J., et al. ; CERTIFI Study Group. (2012) Ustekinumab induction and maintenance therapy in refractory Crohn’s disease. N. Engl. J. Med., 367, 1519–1528. [DOI] [PubMed] [Google Scholar]
- 11. Newcomb P.A., et al. ; Colon Cancer Family Registry. (2007) Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol. Biomarkers Prev., 16, 2331–2343. [DOI] [PubMed] [Google Scholar]
- 12. Peters U., et al. (2012) Meta-analysis of new genome-wide association studies of colorectal cancer risk. Hum. Genet., 131, 217–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McGovern D.P., et al. ; NIDDK IBD Genetics Consortium. (2010) Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat. Genet., 42, 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chan A.T., et al. (2009) Aspirin use and survival after diagnosis of colorectal cancer. JAMA, 302, 649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ogino S., et al. (2006) Distinct molecular features of colorectal carcinoma with signet ring cell component and colorectal carcinoma with mucinous component. Mod. Pathol., 19, 59–68. [DOI] [PubMed] [Google Scholar]
- 16. Ogino S., et al. (2009) Lymphocytic reaction to colorectal cancer is associated with longer survival, independent of lymph node count, microsatellite instability, and CpG island methylator phenotype. Clin. Cancer Res., 15, 6412–6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ogino S., et al. (2006) Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J. Mol. Diagn., 8, 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nosho K., et al. (2008) Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS One, 3, e3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ogino S., et al. (2007) Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J. Mol. Diagn., 9, 305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ogino S., et al. (2009) CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut, 58, 90–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ogino S., et al. (2005) Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J. Mol. Diagn., 7, 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morikawa T., et al. (2012) Tumor TP53 expression status, body mass index and prognosis in colorectal cancer. Int. J. Cancer, 131, 1169–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Baba Y., et al. (2009) Relationship of CDX2 loss with molecular features and prognosis in colorectal cancer. Clin. Cancer Res., 15, 4665–4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Samowitz W.S., et al. (2002) Prognostic significance of p53 mutations in colon cancer at the population level. Int. J. Cancer, 99, 597–602. [DOI] [PubMed] [Google Scholar]
- 25. Slattery M.L., et al. (2000) Associations between cigarette smoking, lifestyle factors, and microsatellite instability in colon tumors. J. Natl. Cancer Inst., 92, 1831–1836. [DOI] [PubMed] [Google Scholar]
- 26. Slattery M.L., et al. (2007) Diet and lifestyle factor associations with CpG island methylator phenotype and BRAF mutations in colon cancer. Int. J. Cancer, 120, 656–663. [DOI] [PubMed] [Google Scholar]
- 27. Samowitz W.S., et al. (2000) Relationship of Ki-ras mutations in colon cancers to tumor location, stage, and survival: a population-based study. Cancer Epidemiol. Biomarkers Prev., 9, 1193–1197. [PubMed] [Google Scholar]
- 28. Jiao S., et al. (2011) The use of imputed values in the meta-analysis of genome-wide association studies. Genet. Epidemiol., 35, 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ioannidis J.P., et al. (2007) Heterogeneity in meta-analyses of genome-wide association investigations. PLoS One, 2, e841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Purcell S., et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mogoanta SS, et al. (2014) Peritumoral inflammatory reaction in colon cancer. Histological and immunohistochemical study. Rom. J. Morphol. Embryol., 55, 1429–1435. [PubMed] [Google Scholar]
- 32. Morikawa T., et al. (2011) STAT3 expression, molecular features, inflammation patterns, and prognosis in a database of 724 colorectal cancers. Clin. Cancer Res., 17, 1452–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Katoh H., et al. (2013) CXCR2-expressing myeloid-derived suppressor cells are essential to promote colitis-associated tumorigenesis. Cancer Cell, 24, 631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jamieson T., et al. (2012) Inhibition of CXCR2 profoundly suppresses inflammation-driven and spontaneous tumorigenesis. J. Clin. Invest., 122, 3127–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ishitsuka T., et al. (2001) Microsatellite instability in inflamed and neoplastic epithelium in ulcerative colitis. J. Clin. Pathol., 54, 526–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cravo M.L., et al. (1998) Microsatellite instability in non-neoplastic mucosa of patients with ulcerative colitis: effect of folate supplementation. Am. J. Gastroenterol., 93, 2060–2064. [DOI] [PubMed] [Google Scholar]
- 37. Brentnall T.A., et al. (1996) Microsatellite instability in nonneoplastic mucosa from patients with chronic ulcerative colitis. Cancer Res., 56, 1237–1240. [PubMed] [Google Scholar]
- 38. Lyda M.H., et al. (1998) Multifocal neoplasia involving the colon and appendix in ulcerative colitis: pathological and molecular features. Gastroenterology, 115, 1566–1573. [DOI] [PubMed] [Google Scholar]
- 39. Hofseth L.J., et al. (2003) The adaptive imbalance in base excision-repair enzymes generates microsatellite instability in chronic inflammation. J. Clin. Invest., 112, 1887–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zeller T., et al. (2010) Genetics and beyond–the transcriptome of human monocytes and disease susceptibility. PLoS One, 5, e10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kjer-Nielsen L., et al. (2012) MR1 presents microbial vitamin B metabolites to MAIT cells. Nature, 491, 717–723. [DOI] [PubMed] [Google Scholar]
- 42. Reantragoon R., et al. (2012) Structural insight into MR1-mediated recognition of the mucosal associated invariant T cell receptor. J. Exp. Med., 209, 761–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lion J., et al. (2013) MR1B, a natural spliced isoform of the MHC-related 1 protein, is expressed as homodimers at the cell surface and activates MAIT cells. Eur. J. Immunol., 43, 1363–1373. [DOI] [PubMed] [Google Scholar]
- 44. Young M.H., et al. (2013) Mucosal associated invariant T cells: don’t forget your vitamins. Cell Res., 23, 460–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Martin E., et al. (2009) Stepwise development of MAIT cells in mouse and human. PLoS Biol., 7, e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sears C.L., et al. (2014) Microbes, microbiota, and colon cancer. Cell Host Microbe, 15, 317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chan AT, et al. 2010. et al. Primary prevention of colorectal cancer. Gastroenterology, 138, 2029–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kostic A.D., et al. (2014) The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology, 146, 1489–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Phipps AI, et al. (2012) Association between colorectal cancer susceptibility loci and survival time after diagnosis with colorectal cancer. Gastroenterology, 143, 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ogino S., et al. (2011) Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut, 60, 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.