ABSTRACT
Natural plasmid transformation of Escherichia coli is a complex process that occurs strictly on agar plates and requires the global stress response factor σS. Here, we showed that additional carbon sources could significantly enhance the transformability of E. coli. Inactivation of phosphotransferase system genes (ptsH, ptsG, and crr) caused an increase in the transformation frequency, and the addition of cyclic AMP (cAMP) neutralized the promotional effect of carbon sources. This implies a negative role of cAMP in natural transformation. Further study showed that crp and cyaA mutations conferred a higher transformation frequency, suggesting that the cAMP-cAMP receptor protein (CRP) complex has an inhibitory effect on transformation. Moreover, we observed that rpoS is negatively regulated by cAMP-CRP in early log phase and that both crp and cyaA mutants show no transformation superiority when rpoS is knocked out. Therefore, it can be concluded that both the crp and cyaA mutations derepress rpoS expression in early log phase, whereby they aid in the promotion of natural transformation ability. We also showed that the accumulation of RpoS during early log phase can account for the enhanced transformation aroused by additional carbon sources. Our results thus demonstrated that the presence of additional carbon sources promotes competence development and natural transformation by reducing cAMP-CRP and, thus, derepressing rpoS expression during log phase. This finding could contribute to a better understanding of the relationship between nutrition state and competence, as well as the mechanism of natural plasmid transformation in E. coli.
IMPORTANCE Escherichia coli, which is not usually considered to be naturally transformable, was found to spontaneously take up plasmid DNA on agar plates. Researching the mechanism of natural transformation is important for understanding the role of transformation in evolution, as well as in the transfer of pathogenicity and antibiotic resistance genes. In this work, we found that carbon sources significantly improve transformation by decreasing cAMP. Then, the low level of cAMP-CRP derepresses the general stress response regulator RpoS via a biphasic regulatory pattern, thereby contributing to transformation. Thus, we demonstrate the mechanism by which carbon sources affect natural transformation, which is important for revealing information about the interplay between nutrition state and competence development in E. coli.
INTRODUCTION
Horizontal gene transfer (HGT) is the transfer of genes between distantly related organisms. It is widely recognized that HGT contributes significantly to the evolution of bacterial genomes and the adaptation of bacteria to new environments (1, 2). In bacteria, the physical process of DNA transfer is accomplished by transduction, conjugation, and transformation. Natural transformation is characterized by the spontaneous uptake of free DNA from the environment by a competent cell, which then integrates said DNA into its chromosome or stabilizes the DNA extrachromosomally in plasmid form, leading to a new phenotype. So far, more than 80 bacterial species are known to be naturally transformable; they share a conserved DNA uptake and processing device but differ in their competence induction and regulatory mechanisms (3). Although Escherichia coli is not usually thought of as being capable of natural transformation, it has been found to develop natural competence and take up plasmid DNA under certain conditions (4–7). Our previous research showed that E. coli could develop natural competence and take up plasmid DNA on agar plates in the absence of either additional Ca2+ or heat shock (8). Furthermore, we found that DNA uptake gene orthologs required for other naturally transformable species were not involved in this transformation system (9), which implied that a different molecular mechanism of competence development and DNA uptake was being used. As reported by our group and others, the general stress response regulator RpoS (σS) was shown to mediate natural transformation (10), and transformation frequencies were apparently increased when rpoS was induced in the liquid culture stage but did not change significantly when rpoS was induced on solid medium (11). Moreover, OmpA, a membrane protein, has been reported to block DNA transfer, and the null mutation leads to a boost in transformation on agar plates (12).
Aside from investigation of the mechanisms of DNA uptake and processing, other issues in natural transformation relate to finding the relevant environmental cues that trigger competence induction and illustrating the relationship between competence and different ecological niches. The transcriptional dual regulator cyclic AMP receptor protein (CRP), signaled by cAMP, a global regulator of a large number of genes, is reported to be involved in natural competence induction. In Haemophilus influenzae and Vibrio cholerae, which are the best-characterized Gram-negative bacteria, natural competence is induced by cAMP-CRP, which couples the nutritional state to the induction of transformation (13). Spontaneous competence in H. influenzae is induced by deprivation of carbon, nitrogen, and cofactors when a rapidly growing cell shifts to a starvation state (14, 15). This effect of nutrition limitation on competence has been shown to be a consequence of intracellular cAMP elevation via the phosphotransferase system (PTS) (16, 17). Similarly, in V. cholerae, the level of cAMP plays a major role in natural transformation based on carbon catabolite repression (CCR) induced by chitin, a common environmental niche for this bacterium (18). To summarize, cAMP and CRP are induced by nutritional parameters and contribute to natural transformation in these bacteria. However, the correlations among carbon sources, cAMP-CRP, and natural transformation in E. coli remain undiscovered, which is intriguing in light of this relationship and the mechanism of competence development in E. coli.
As rpoS was reported previously to be a major player in natural transformation in E. coli, our attention focused on the pattern of its regulation by cAMP-CRP. rpoS encodes the alternative sigma factor σS, which acts as the regulator of the general stress response in E. coli. Regulation of σS is conducted in a complex manner throughout each stage of expression, and cAMP-CRP is reported to be the transcriptional regulator (19). There are two putative CRP-binding sites present upstream (centered at −61.5) and downstream (centered at +56.5) from the rpoS transcription start site. The former is similar to a classical activation site, and the latter is an inhibitory site. However, there are contradictory views regarding the regulation of rpoS expression by cAMP-CRP in E. coli. Hengge-Aronis and colleagues originally showed that cAMP-CRP is a negative regulator of rpoS transcription (20) and subsequently reported that cAMP-CRP positively controls rpoS transcription during entry into stationary phase (21). However, early work performed by McCann and colleagues demonstrated that rpoS transcription was regulated positively by the cyaA gene (22). Thus, the mechanism of cAMP-CRP modulation of rpoS has not yet been fully elucidated, and the effect of the regulation on transformation is unknown.
It is generally accepted that transformation confers potential evolutionary fitness and environmental adaptation advantages. However, the ability of microorganisms to transfer pathogenicity islands and antibiotic resistance genes among human-pathogenic bacteria presents a threat to human health (23, 24). Furthermore, versatile plasmids possessing a broad host range might increase the risk of gene spread prior to recombination. Understanding microbial natural transformation, including how bacteria acquire competence and take up DNA, will provide supportive evidence not only for the role of transformation in evolution but also for the influence of genetically engineered microorganisms on natural environments. In this study, we found that common intestinal carbon sources can increase the transformation frequency remarkably and that the inhibitory effect of carbon sources on cAMP is responsible. We then ascertained that cAMP-CRP mutants promote competence induction through derepression of rpoS in early log phase. Moreover, we observed a dual regulatory pattern of cAMP-CRP on rpoS, and both of the putative CRP-binding sites seem to be activation sites. Here, we demonstrate a relationship among the carbon source, cAMP-CRP, rpoS, and natural transformation, providing insight into the interplay between nutrition state and competence development in E. coli.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All of the strains and plasmids used in this study are listed in Table 1, and the primers are listed in Table 2. Cultures were grown in Luria-Bertani (LB) broth with gyratory shaking at 200 rpm or on LB agar plates. Cell growth was monitored spectrophotometrically at an optical density of 600 nm (OD600). All cultures were incubated at 37°C except those containing temperature-sensitive replicon plasmids, which were grown at 30°C. In order to evaluate the influence of the carbon source on transformation, cultures were grown in LB medium containing 0.2% glucose (Glc), fructose (Fru), trehalose (Tre), mannose (Man), mannitol (Mtl), N-acetylglucosamine (NAG), galactose (Gal), fucose (Fuc), maltose (Mal), glycerol (Gly), N-acetyl-neuraminic acid (NANA), lactose (Lac), arabinose (Ara), or N-acetyl-galactosamine (GalNAc). Antibiotics were supplemented as required at the following final concentrations: 25 μg/ml chloramphenicol, 100 μg/ml kanamycin, and 200 μg/ml ampicillin.
TABLE 1.
Bacterial strains and plasmids used in this study
| E. coli strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| BW25113 | F− Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ− rph-1 Δ(rhaD-rhaB)568 hsdR514 | CGSC |
| MG1655 | F− λ− rph-1 | Laboratory collection |
| JW5437 | BW25113 ΔrpoS::kan | CGSC |
| JW5702 | BW25113 Δcrp::kan | CGSC |
| JW3778 | BW25113 ΔcyaA::kan | CGSC |
| JW3000 | BW25113 ΔcpdA::kan | CGSC |
| JW2410 | BW25113 Δcrr::kan | CGSC |
| JW2408 | BW25113 ΔptsH::kan | CGSC |
| JW1087 | BW25113 ΔptsG::kan | CGSC |
| Δcrp ΔrpoS strain | BW25113 Δcrp ΔrpoS::kan | This study |
| ΔcyaA ΔrpoS strain | BW25113 ΔcyaA ΔrpoS::kan | This study |
| Plasmids | ||
| pDsRED | pUC19 carrying the red fluorescence gene, Ampr | 42 |
| pSU19 | p15A replication, Cmr | 43 |
| pSUcrp | pSU19 carrying a crp gene | This study |
| pRPS | pSU19 carrying an rpoS gene | This study |
| pRPS1 | pRPS containing mutated CRP-binding site I | This study |
| pRPS2 | pRPS containing mutated CRP-binding site II | This study |
| pRPS3 | pRPS containing mutated CRP-binding sites I and II | This study |
| pRPZ | pSU19 carrying the rpoS promoter region (−165 to +570) and lacZ gene fusion | This study |
| pRPZ1 | pRPZ containing mutated CRP-binding site I | This study |
| pRPZ2 | pRPZ containing mutated CRP-binding site II | This study |
| pRPZ3 | pRPZ containing mutated CRP-binding sites I and II | This study |
| pKD13 | Template for the kanamycin cassette, Ampr | 25 |
| pKD46 | Expresses λ Red recombinase, temperature-sensitive replicon, Ampr | 25 |
| pCP20 | Expresses FLP recombinase, temperature-sensitive replicon, Ampr | 25 |
| pARA | Modified from pSU19, containing the arabinose operon | This study |
| pARArpoS | pARA carrying an rpoS ORF | This study |
TABLE 2.
Oligonucleotides used in this study
| Purpose and primer | Sequencea |
|---|---|
| Construction of cloned genes in plasmids | |
| crp F | 5′-GGTCTAGACTTCACTCGCGCTTGCATTT-3′ |
| crp R | 5′-AGGGATCCCACTCCGACGGGATTAA-3′ |
| lacZ F | 5′-GGAAGCTTATGACCATGATTACGGAT-3′ |
| lacZ R | 5′-GATCTAGATTATTTTTGACACCAG-3′ |
| Construction of rpoS with site-directed mutagenesis | |
| rpoS−165 F | 5′-CGGGATCCGAACGTTGGTCAG-3′ |
| rpoS+1712 R | 5′-GCTCTAGACGACCATTCTCGG-3′ |
| rpoS+570 R | 5′-CCCAAGCTTCATAAGGTGGC-3′ |
| rpoS M1 R | 5′-TGCAGGCTTGATTATGACTCCTTGC-3′ |
| rpoS M2 R | 5′-GGCGCGTTTGCCGTGGCTGTGGTTGG-3′ |
| Construction of rpoS knockout mutant | |
| rpoS H1P1 | 5′-TGAGACTGGCCTTTCTGACAGATGCTTACTTACTCGCGGAACAGCGCTTCTGTAGGCTGGAGCTGCTTCG-3′ |
| rpoS H2P2 | 5′-TTTTTGACGAAAAGGCCTTAGTAGAATAGGAACCCAGTGATAACGATTTGATTCCGGGGATCCGTCGACC-3′ |
Modified nucleotides for site-directed mutagenesis are in boldface.
Natural transformation protocol.
Natural transformation of E. coli was carried out as described previously (8). All experiments were performed at 37°C. E. coli strains were grown overnight in LB broth and then inoculated at 1:100 (vol/vol) in 5 ml fresh LB broth with gyratory shaking at 200 rpm. When the culture's growth reached stationary phase, cells were transferred to an “open system” (a beaker covered by an air-permeable membrane) and incubated statically for 10 h. Each 50-μl aliquot of cultures and 2 μg of the plasmid pDsRED were mixed and spread on LB plates containing 5% agar and 200 μg/ml ampicillin. Meanwhile, cultures were diluted 106-fold and spread on agar plates for viable cell counts. Transformation frequency was calculated by dividing the number of transformants by the viable cell count.
Double-knockout mutant constructions.
A double-knockout mutant was constructed using the lambda Red recombination system, as described by Datsenko and Wanner (25). The first kanamycin cassette inserted into a Δcrp or ΔcyaA mutant was removed by using the temperature-sensitive plasmid pCP20, which carries the FLP recombinase gene. Primers used to construct rpoS deletion mutants containing 50-nucleotide homologue sequences of the rpoS gene and 20-nucleotide priming sequences of pKD13 were designed according to the E. coli MG1655 genomic sequence. A kanamycin resistance cassette from plasmid pKD13 was amplified and transformed into the Δcrp or ΔcyaA mutant carrying plasmid pKD46. Double-knockout mutants were selected on agar plates containing kanamycin and confirmed by PCR and DNA sequencing.
Measurement of β-galactosidase activity.
Cells were centrifuged and resuspended in an equal volume of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM 2-mercaptoethanol; pH 7.0) and then diluted with a suitable volume of Z buffer to bring the total volume to 1 ml. Next, 12.5 μl 0.1% sodium dodecyl sulfate (SDS) and 25 μl chloroform were added to permeabilize the cells. After vortexing for 5 s, cells were preheated at 30°C for 15 min, and β-galactosidase activity was determined according to the standard procedure described by Miller (26).
Site-directed mutagenesis of putative CRP-binding sites around the rpoS major promoter.
Two putative CRP-binding sites around the rpoS promoter were mutagenized using the PCR megaprimer method (27). The pRPZ plasmid containing an rpoS::lacZ fusion was constructed as a control, and site-directed mutagenesis was performed using pRPZ as the template directly. To amplify the mutant CRP-binding site I, three oligonucleotide primers (rpoS−165F, rpoS+570R, and rpoS M1 R) were utilized to perform two rounds of PCR. In the first round, rpoS−165F and rpoS M1 R were used to produce a short DNA fragment that served as the “megaprimer” for the second round of PCR with the other primer, rpoS+570R. The CRP-binding site II mutant product was amplified in the same manner as the CRP-binding site I mutant, using primers rpoS−165F, rpoS+570R, and rpoS M2 R. The double CRP-binding site I and CRP-binding site II mutant was constructed by digestion and ligation of the two mutant fragments, making use of the SnaBI restriction site located between the two binding sites. The mutagenized rpoS promoter DNA was cloned into the pMD18-T vector and confirmed by DNA sequencing. Afterwards, the mutagenized DNA fragments ligated to the lacZ open reading frame (ORF) were introduced into the pSU19 vector, resulting in the pRPZ1 (CRP-binding site I mutant), pRPZ2 (CRP-binding site II mutant), and pRPZ3 (double mutant) plasmid, respectively. The complete rpoS gene was cloned using primers rpoS−165F and rpoS+1712R and inserted into the pSU19 vector to generate the pRPS plasmid. pRPS1 (CRP-binding site I mutant), pRPS2 (CRP-binding site II mutant), and pRPS3 (double mutant) plasmids then were constructed using the PCR megaprimer method as mentioned above.
SDS-PAGE and Western blotting.
To measure protein levels in vitro, cells were harvested at various time points and resuspended in phosphate-buffered saline (PBS) buffer supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF). Cells were then sonicated for 150 s to completely break the cell walls and then centrifuged at 12,000 × g for 10 min at 4°C. The resulting supernatants were measured for microgram quantities of protein using the Bradford assay at an absorbance of 590 nm. Lysates were boiled for 10 min with loading buffer, followed by electrophoresis of equal protein quantities on a 10% SDS-PAGE gel. Proteins from the gel were transferred to a 0.22-μm nitrocellulose membrane, and Western blotting analyses were performed. The membrane was blocked with 5% (wt/vol) nonfat dry milk for 1 h. Monoclonal antibodies (Santa Cruz Biotechnology, Inc.) against RpoS or RpoB were used as the primary antibodies at dilutions of 1:1,000. Goat anti-mouse IgG conjugated to horseradish peroxidase (HRP; Protein Tech, China) was used as a secondary antibody at a dilution of 1:10,000. Blots were developed using a chemiluminescent HRP substrate (Millipore, Billerica, MA, USA).
cAMP assay.
Cultures were harvested during the exponential phase and adjusted to the same biomass according to OD600. cAMP concentrations in both supernatants and bacterial lysates were measured using a cAMP enzyme-linked immunosorbent assay (ELISA) kit (NewEast Biosciences, Malvern, PA, USA) in accordance with the manufacturer's instructions. Total cAMP concentrations (both intracellular and extracellular cAMP) were assayed and are presented as micromoles per liter of culture.
RESULTS
Effects of added carbon sources on natural transformation and cAMP levels in cells.
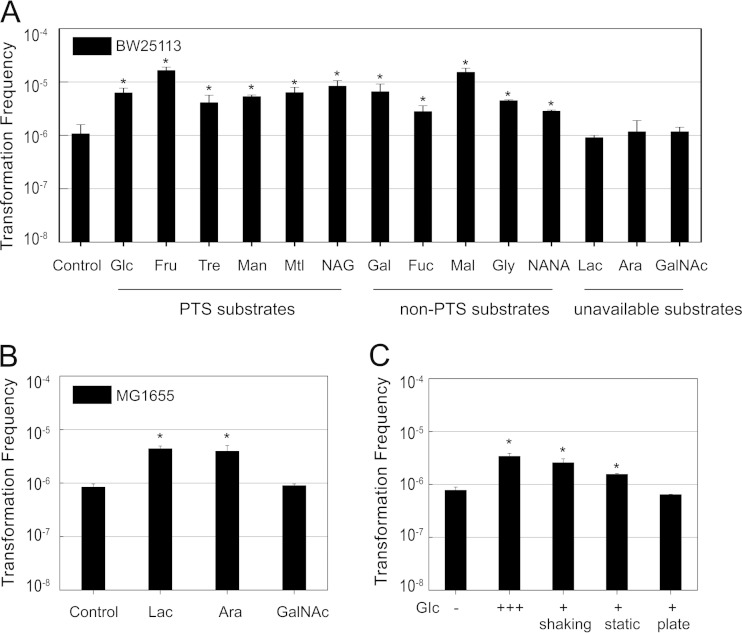
Based on our hypothesis that environmental cues and nutrition state may be involved in competence, we evaluated the impact of carbohydrates, including those commonly present in intestinal mucus (28), on transformation in E. coli strain BW25113. Following the addition of either PTS substrates (Glc, Fru, Tre, Man, Mtl, and NAG) or non-PTS substrates (Gal, Fuc, Mal, Gly, and NANA) to LB liquid medium, the transformation frequencies were increased markedly, to 3- to 15-fold that of the control, with the exceptions of lactose, arabinose, and GalNAc, which could not be used by BW25113 (Lac−, Ara−, and GalNAc−) (Fig. 1A). We selected another strain for the transformation test, MG1655, which is prototrophic for lactose and arabinose but auxotrophic for GalNAc. The results showed that lactose and arabinose but not GalNAc could still improve transformation (Fig. 1B). In general, carbon sources can facilitate transformation as a precondition to their metabolism. In addition, glucose, as the representative carbon source, was added at different stages of culture to identify the most effective stage (Fig. 1C). According to the results, the important stages were the shaking and static cultivation stages rather than growth on the agar plates.
FIG 1.
Effects of carbon sources on natural transformation. (A) Transformation frequencies of strain BW25113 cultivated in LB medium with additional carbon sources (0.2%) in shaking and static cultures. Natural transformation experiments were performed as described in Materials and Methods. (B) Transformation frequencies of MG1655 with additional carbon sources (0.2%) in shaking and static cultures. (C) Effect of glucose on transformation frequency during different culture stages. Glucose (0.2%) was added to LB medium at the shaking, static, and plating culture stages separately. “+++” indicates the addition of glucose at all three stages. Error bars signify standard deviations of the results for at least two independent experiments. Significant values compared with the results for controls as determined by Student's test are indicated by asterisks (P < 0.05).
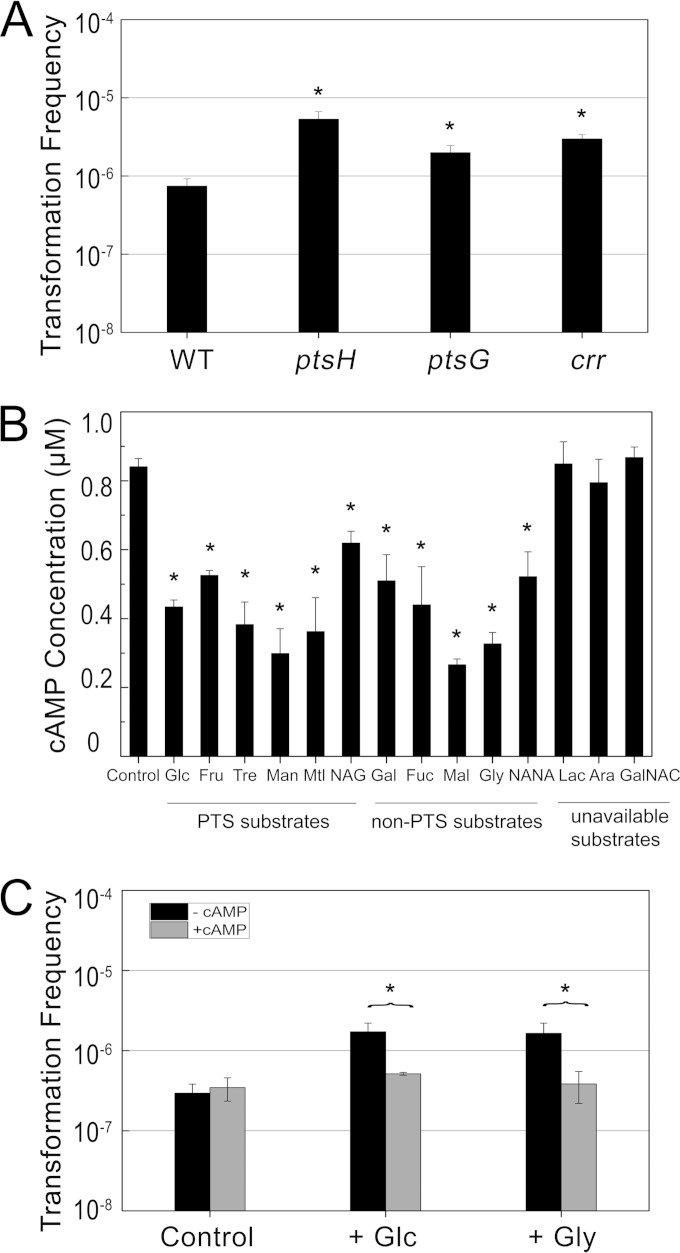
The PTS is recognized as a transport system that is involved in the uptake of numerous PTS carbohydrates and converts them into their respective phosphoesters. In contrast, non-PTS substrates are transported by specific permeases. Both PTS and non-PTS substrates can influence the phosphorylation of PTS through transport and metabolism, respectively (29). The phosphoryl group from phosphoenolpyruvate (PEP) is transferred through PTS protein, ultimately reaching the carbohydrate during its uptake. The phosphorylated and unphosphorylated forms of PTS protein are cyclic, and it is this cycle by which PTS-mediated regulation is carried out by the cell. The PTS mutants were subject to transformation tests; inactivation of HPr (ptsH), EIIBCGlc (ptsG), and EIIAGlc (crr) caused an increase in transformation frequency (Fig. 2A). Therefore, the effect of both PTS and non-PTS substrates on transformation is closely related to the activity of PTS and to the metabolic activity subsequently regulated by PTS. The model of PTS-mediated regulation of adenylate cyclase has been well characterized in studies of the relationship between carbon sources and cAMP levels. The phosphorylated EIIAGlc binds to and activates adenylate cyclase, which catalyzes ATP to cAMP. Substrate transport by PTS is thought to be the primary signal regulating adenylate cyclase activity. In addition, it has been demonstrated that both PTS and non-PTS substrates can decrease intracellular levels of cAMP by dephosphorylating EIIAGlc in the process of transport and reducing intracellular PEP/pyruvate ratios during high glycolytic fluxes, respectively (29). To confirm whether additional PTS and non-PTS substrates in LB broth could reduce the levels of cAMP, we measured the concentrations of cAMP in these cultures. It has been reported that over 99% of cAMP is excreted out of the cell via an unknown regulatory mechanism (30), and the rate of excretion is linearly correlated with intracellular cAMP concentrations (31). Therefore, the total content of cAMP can be precisely calculated by measuring both extracellular and intracellular cAMP levels. As expected, the results showed that the cultures treated with PTS and non-PTS carbon sources exhibited lower concentrations of cAMP than the control, while cultures treated with unavailable carbon sources displayed no significant variation (Fig. 2B).
FIG 2.
Relationship between carbon sources and cAMP and its effect on natural transformation. (A) Transformation frequencies of ΔptsH, ΔptsG, and Δcrr strains compared with that of wild-type strain BW25113. Transformation experiments were performed as described in Materials and Methods. (B) Total levels of cAMP in cultures with various carbon sources (0.2%). BW25113 cells were grown to log phase in LB broth with different carbon sources, and total cAMP concentrations (intracellular and extracellular) were assayed and are presented in micromoles per liter of culture. (C) Neutralizing effect of cAMP supplementation on increased transformation frequencies caused by carbon sources. External glucose or glycerol (0.2%) was added to wild-type strain culture with (+cAMP) or without (−cAMP) 5 mM cAMP added subsequently. The wild type cultured in LB broth without an external carbon source served as the control. Each bar indicates the average results from at least two independent replicates, and error bars indicate the standard deviations from the means. P values of <0.05 are indicated by asterisks.
To investigate whether cAMP is the main factor affecting the superiority of the transformation ability caused by the additional carbon sources, the test was performed using the wild-type strain by adding cAMP and glucose or glycerol, which serve as representative PTS and non-PTS substrates, respectively. The data showed that additional cAMP could neutralize the promotional effect of glucose or glycerol on transformation (Fig. 2C). Based on these results, we concluded that additional carbon sources most likely promote competence by diminishing the cAMP concentration.
Negative effects of cAMP-CRP on natural transformation.
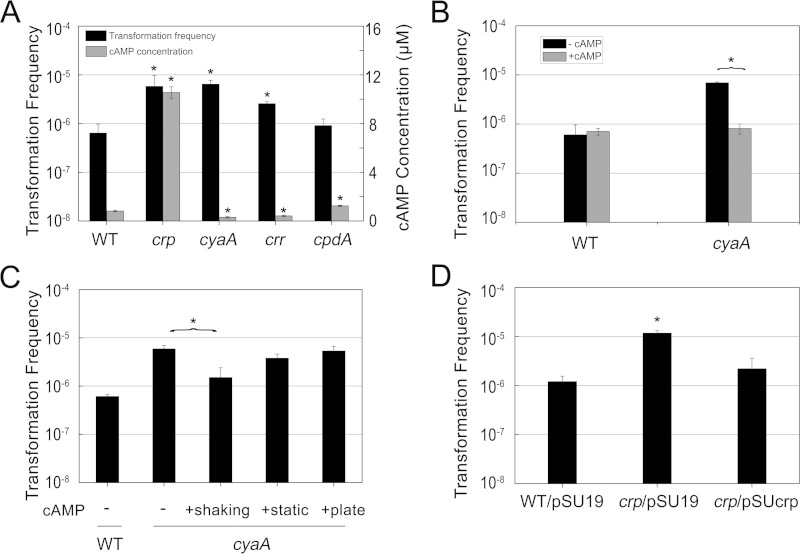
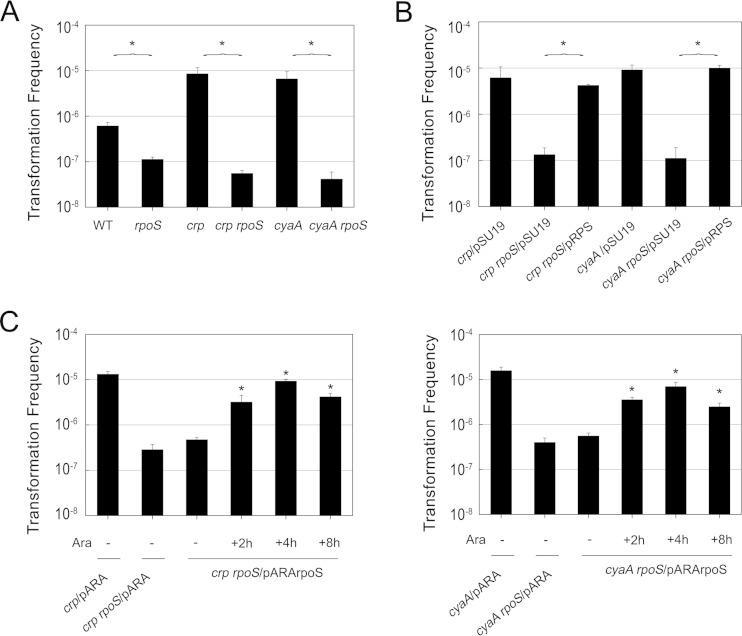
cAMP is an important signaling molecule in many biological processes. The formation of cAMP is catalyzed by adenylate cyclase, encoded by cyaA. The activity of adenylate cyclase is positively controlled by phosphorylated EIIAGlc, encoded by crr. cAMP phosphodiesterase encoded by cpdA can reduce cAMP levels by hydrolyzing cAMP. We measured the cAMP concentration and transformation frequency in these gene knockout mutants to confirm whether cAMP-CRP is involved in competence regulation. Although some mutant strains exhibited different growth stages and viable counts, the significant differences in transformation frequencies between the mutants and the wild type were not influenced (see Fig. S1 to S4 in the supplemental material). Δcrr and ΔcyaA mutants exhibited lower cAMP concentrations but approximately 4- and 10-fold higher transformation frequencies than the wild type, respectively. In contrast, the ΔcpdA mutant harbored a higher cAMP concentration than the wild type, but this mutation had no significant influence on transformation (Fig. 3A). Therefore, we speculated that relatively low levels of cAMP, but not constitutive or high levels, could facilitate transformation. To confirm this hypothesis, transformation experiments were performed by supplying the wild type and the ΔcyaA mutant with exogenous cAMP. The transformation frequency in the wild-type strain supplemented with exogenous cAMP did not exhibit a significant difference from that of the control. Nevertheless, supplementary cAMP could restore the transformation frequency of the ΔcyaA mutant to the wild-type level (Fig. 3B). Thus, we concluded that cAMP did repress the transformation frequency. In addition, adding cAMP during different culture phases seems to be an effective way to confirm the time point at which cAMP has the greatest effect. The results shed light on the importance of cAMP for transformation ability during the culture shaking stage rather than during growth on agar plates, in a similar manner to glucose (Fig. 3C).
FIG 3.
Effect of cAMP-CRP on natural transformation. (A) Transformation frequencies and cAMP concentrations in Δcrp, ΔcyaA, Δcrr, and ΔcpdA strains compared with those in wild-type strain BW25113. Cultures were harvested after incubation for 4 h, and total cAMP concentrations (intracellular and extracellular cAMP) were assayed as described in Materials and Methods. Transformation experiments were performed as described in Materials and Methods. (B) Transformation frequencies of BW25113 and the ΔcyaA mutant with 5 mM cAMP added during shaking and static stages. (C) Effects of supplementing cAMP during different culture stages on transformation frequency in the ΔcyaA mutant. “+” indicates 5 mM cAMP added during the shaking, static, and plating culture stages. (D) Results of a complementary experiment with the Δcrp mutant carrying the plasmid pSUcrp. BW25113 and the Δcrp mutant with plasmid pSU19 served as controls. Each strain was assayed in duplicate, and the average results and standard deviations are reported. P values of <0.05 are indicated by asterisks.
CRP, which is always coupled with cAMP, acts as a dual regulator controlling the transcription of many genes. In addition, CRP represses adenylate cyclase both at the transcriptional level, by binding to a site that overlaps the cyaA promoter, and at the posttranscriptional level, by reducing the level of phosphorylated EIIAGlc (32). Our research confirmed that the Δcrp mutation could result in overproduction of cAMP. Furthermore, it is worth noting that the Δcrp mutation significantly promoted the transformation frequency, in the same manner as the ΔcyaA mutation (Fig. 3A). Considering the synergistic effects of CRP and cAMP, we concluded that cAMP-CRP suppresses natural transformation. A plasmid complementation system for expression of the crp gene in the Δcrp mutant reduced the transformation frequency, which was in accordance with the results for the wild-type strain (Fig. 3D).
As described above, the results suggested that an imbalance in the cAMP pool can affect transformation ability and that cAMP-CRP has a negative effect on natural transformation. Based on these results, together with the promotional effect of carbon sources on transformation, it seems logical to conclude that additional carbon sources promote transformation by reducing cAMP levels and that the lower concentration of cAMP-CRP may participate in the regulation of downstream genes involved in natural transformation.
Regulation of rpoS expression by cAMP-CRP.
Since rpoS is important for natural transformation, as reported previously, and there are contradictory views on the regulation of rpoS expression by cAMP-CRP in E. coli, the relationship between rpoS and cAMP-CRP has been the subject of studies aimed at elucidating the mechanism of natural transformation suppression by cAMP-CRP.
(i) Effect of cAMP-CRP on rpoS expression.
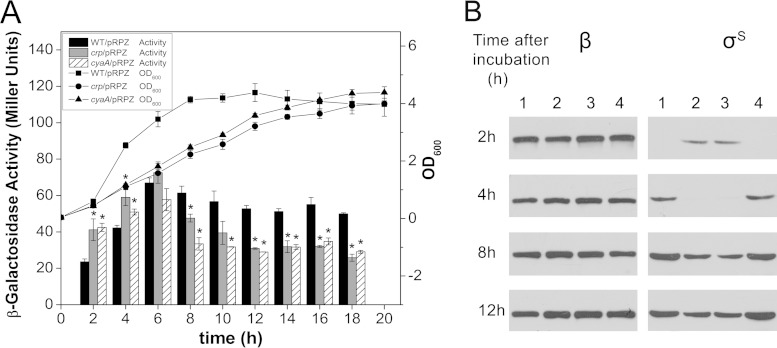
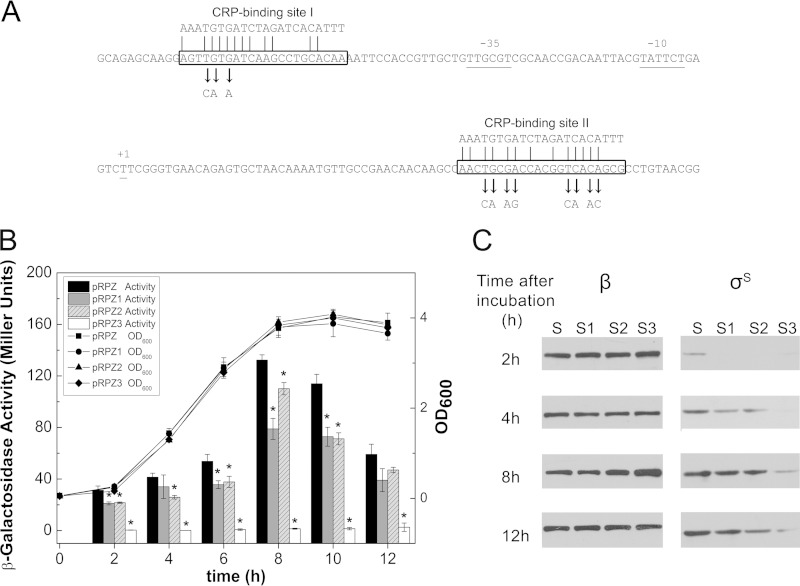
To investigate the effect of cAMP-CRP on rpoS, the rpoS::lacZ fusion-containing pRPZ plasmid was transferred into wild-type strain BW25113 and the Δcrp and ΔcyaA mutants. β-Galactosidase activity was measured along with the growth curve (Fig. 4A). In the Δcrp and ΔcyaA mutants, the increases in rpoS expression ranged from 20 to 80% relative to the expression in the wild-type strain in early log phase; however, the expression subsequently decreased to levels 20 to 40% lower than those of the wild-type strain. The results demonstrate that cAMP-CRP plays a biphasic role in rpoS expression during different phases of growth. Both the Δcrp and ΔcyaA mutants exhibited growth defects, so to avoid error caused by the different amounts of biomass, the β-galactosidase activity was divided by the OD600 value during calculation of the Miller units. The synthetic rates of β-galactosidase exhibited patterns similar to the results described above. In addition, there was no significant difference in the plasmid concentration among the three strains (data not shown).
FIG 4.
Effect of cAMP-CRP on rpoS expression at various growth stages. (A) Expression of rpoS::lacZ fusion plasmid pRPZ in wild-type BW25113 and Δcrp and ΔcyaA mutants. Cultures grown overnight were diluted 100-fold in fresh LB broth and cultivated at 37°C. The cultures were subjected periodically to OD600 readings and β-galactosidase activity assays. The symbols and lines represent typical growth curves at an optical density of 600 nm. The bars represent enzyme activity in Miller units. The data represent the average results of triplicate experiments, with standard deviations indicated. Significant values determined by Student's test (P < 0.05) are indicated with asterisks. (B) RpoS protein levels in BW25113 (lane 1) and the Δcrp (lane 2), ΔcyaA (lane 3), and ΔcyaA (lane 4) mutants grown in the presence of 5 mM exogenous cAMP. Cells were harvested at various time points and subjected to Western blotting using a monoclonal antibody against RpoS (right) and a monoclonal antibody against RpoB (left; β subunit of the core RNA polymerase) as an internal control. Incubation times of 2 h (OD600 of 0.9) and 4 h (OD600 of 3.0) represent the early and mid-log phases, respectively, in BW25113; 8 h (OD600 of 4.9) and 12 h (OD600 of 4.9) represent the stationary phase in BW25113. The results are from a single experiment and are representative of at least three independent experiments.
The cellular levels of σS in the Δcrp and ΔcyaA mutants were also compared with those in the wild-type strain using Western blotting. A monoclonal antibody against RpoS was used to assess the intracellular level of σS at various growth points. The level of the β subunit of the core RNA polymerase, which served as an internal control, was relatively constant across time points. The level of σS was tightly repressed in the log phase but induced during entry into the stationary phase. The RpoS levels in both the Δcrp and ΔcyaA mutants were found to increase in early log phase compared to the levels in the wild type. However, in contrast, the mutants showed lower RpoS levels than the wild type in the following incubation. The ΔcyaA mutant grown in the presence of additional cAMP exhibited an RpoS level comparable to that of the wild type (Fig. 4B).
The results described above suggest that cAMP-CRP plays a biphasic role in rpoS expression during different growth phases; in particular, cAMP-CRP has a negative effect on rpoS in the early log phase but a positive effect in the mid- and late log and stationary phases.
(ii) Effects of mutagenized putative CRP-binding sites on rpoS expression.
The two putative cAMP-CRP-binding sites around the rpoS main promoter are homologous to the consensus inverted repeat TGTGAN6TCACA. To determine whether both of these putative sites take part in biphasic regulation of rpoS, they were modified by site-directed mutagenesis as described in Materials and Methods (Fig. 5A). Additionally, according to current research on the regulator binding sites around the rpoS main promoter, CRP-binding site I overlaps with one of the ArcA binding sites (33). To avoid the influence that the mutant sites may have on the binding activity of the ArcA regulator, the nucleotides replaced by site-directed mutagenesis were not involved in the consensus sites for ArcA binding. Therefore, the binding activity of ArcA should not have been affected. Plasmids harboring the mutated DNA sequences and lacZ fusions, pRPZ1, pRPZ2, and pRPZ3, were used to measure the effects of these putative CRP-binding sites on rpoS expression. All of the rpoS mutant fusions exhibited lower levels of expression, especially the double mutants (Fig. 5B).
FIG 5.
Effects of mutagenized putative CRP-binding sites on rpoS expression. (A) Presence of the two putative CRP-binding sites around the major rpoS promoter. The promoter is flanked by two putative cAMP-CRP-binding sites, presented in the boxes matched to the CRP-binding site consensus sequences indicated above the DNA sequence. Nucleotides replaced by site-directed mutagenesis are indicated by arrows. (B) Expression plasmids pRPZ, pRPZ1, pRPZ2, and pRPZ3 in the ΔrpoS mutant. Strains were grown in LB medium and subjected to OD600 readings and β-galactosidase activity assays as described in the legend to Fig. 3. The symbols and lines represent typical growth curves at an optical density of 600 nm, and the bars represent enzymatic activity. Significant values determined by Student's test (P < 0.05) are indicated by asterisks. (C) RpoS protein levels derived from plasmids pRPS (S), pRPS1 (S1), pRPS2 (S2), and pRPS3 (S3) in the ΔrpoS strain. Strains were grown and analyzed by immunoblotting as described in the legend to Fig. 3. Incubation times of 2 h (OD600 of 0.35), 4 h (OD600 of 1.5), 8 h (OD600 of 4.2), and 12 h (OD600 of 4.2) represent the early, mid-log, and stationary (both 8 and 12 h) phase, respectively.
Moreover, Western blotting was performed to measure RpoS protein levels. Protein was extracted from cells harboring the pRPS1, pRPS2, and pRPS3 plasmids, all of which expressed the complete rpoS gene with mutated CRP-binding sites. These three mutants displayed lower levels of σS protein than those carrying the control plasmid pRPS, with the lowest level observed in the double mutant (Fig. 5C). The results were in general agreement with those of the β-galactosidase activity assay. In summary, we infer that rpoS expression is regulated positively by two putative CRP-binding sites in the rpoS promoter region. The details about the mechanism of cAMP-CRP modulation of rpoS will be discussed below.
A critical role for RpoS in transformation of Δcrp and ΔcyaA mutants.
As cAMP-CRP plays a biphasic role in rpoS expression during different growth phases, it raises the question of whether rpoS is related to the promotional effect of the Δcrp and ΔcyaA mutations on transformation. We constructed Δcrp ΔrpoS and ΔcyaA ΔrpoS double-knockout mutants, the transformation frequencies of which were compared to those of the single-knockout Δcrp, ΔcyaA, and ΔrpoS mutants and the wild-type strain. The results showed that the transformation frequencies of these double-knockout mutants showed a pronounced decrease to an even lower level than in the ΔrpoS mutant (Fig. 6A). This demonstrates the importance of rpoS in the promotional effect of the Δcrp and ΔcyaA mutations on transformation. Additionally, plasmid complementation of rpoS might compensate for the lower transformation ability of the double-knockout mutants and restore the transformation frequencies to the levels in the Δcrp and ΔcyaA mutants (Fig. 6B). So far, it has been conjectured that the Δcrp and ΔcyaA mutations promote transformation due to the derepression of rpoS in the early log phase. In order to further confirm this hypothesis, we induced rpoS expression in two double-knockout mutants at different time points using an arabinose-induced plasmid, pARArpoS. The transformation frequencies of the mutants induced in the different periods exhibited high levels similar to those of the Δcrp and ΔcyaA mutants (Fig. 6C). Based on the results, we conclude that rpoS is critical for the superior transformation ability of the Δcrp and ΔcyaA mutants and that the accumulation of RpoS during the early exponential phase plays a major role.
FIG 6.
Critical role of rpoS in the enhanced transformation ability of Δcrp and ΔcyaA mutants. (A) The transformation frequencies of double-knockout Δcrp ΔrpoS and ΔcyaA ΔrpoS mutants were compared with those of wild-type BW25113 and single-knockout ΔrpoS, Δcrp, and ΔcyaA mutants. (B) Results of a complementary experiment using Δcrp ΔrpoS and ΔcyaA ΔrpoS mutants with plasmid pRPS. Δcrp, ΔcyaA, Δcrp ΔrpoS, and ΔcyaA ΔrpoS mutants harboring empty plasmid pSU19 served as controls. (C) Effects on transformation frequency of inducing rpoS in Δcrp ΔrpoS (left) and ΔcyaA ΔrpoS (right) mutants at different time points. rpoS was induced without arabinose (−) or with 0.2% arabinose (+) added periodically after incubation. Δcrp, ΔcyaA, Δcrp ΔrpoS, and ΔcyaA ΔrpoS mutants harboring plasmid pARA served as controls. Significant values compared with the value for the Δcrp ΔrpoS/pARArpoS mutant or the ΔcyaA ΔrpoS/pARArpoS mutant without arabinose were determined by Student's test and are indicated with an asterisk (P < 0.05). Error bars signify standard deviations for the results of at least two independent experiments.
Effects of carbon sources on the transformation ability of the ΔrpoS mutant and intracellular σS levels.
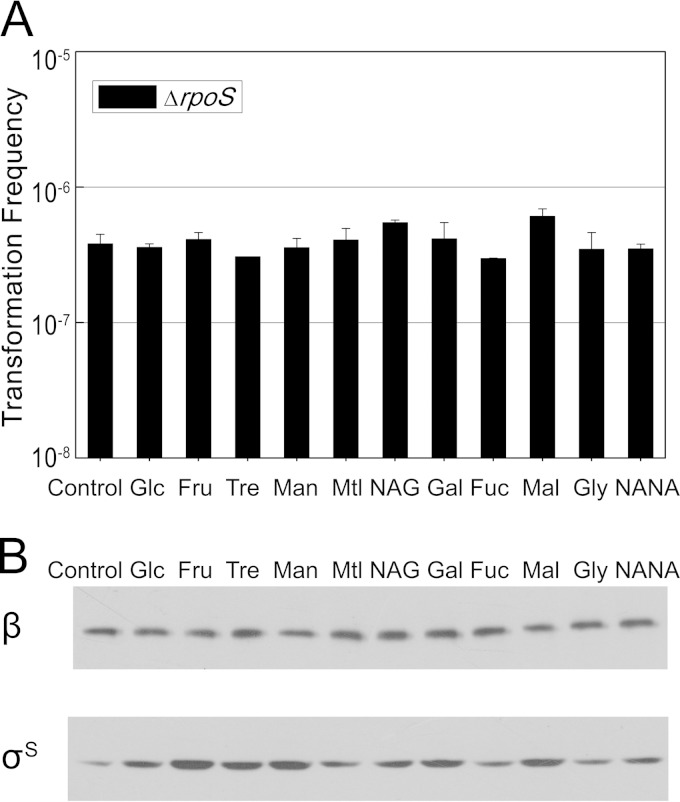
The metabolism of carbon sources leads to unbalanced cAMP synthesis. Furthermore, lower levels of cAMP-CRP can promote competence by derepressing σS in the early log phase. From the results described above, we inferred that carbon sources might promote rpoS expression by diminishing cAMP levels in cells, which in turn might contribute to a higher transformation frequency. To confirm this hypothesis, we tested the effects of the carbon source on transformation in the ΔrpoS mutant and on σS content in the wild-type strain during the log phase. The transformation frequency in the ΔrpoS mutant was not higher than that of the control even when carbon sources were supplied (Fig. 7A). This implies that rpoS is crucial for the transformation boost stimulated by additional carbon sources. The levels of σS during the log phase with additional carbon sources were also measured. All of the cultures supplied with a carbon source exhibited higher σS contents than the control, in accordance with a lower cAMP concentration that may cause derepression of rpoS in the early log phase (Fig. 7B). Taken together, these data provided support for our hypothesis that the carbon source contributes to a higher transformation frequency by diminishing cAMP levels in cells and removing the repressive action on σS in the log phase.
FIG 7.
Relationship between the carbon source and rpoS. (A) Effects of the carbon sources on the transformation ability of the ΔrpoS mutant. The ΔrpoS mutant was incubated in LB broth with various carbon sources (0.2%), and transformation experiments were performed as described in the legend to Fig. 1. (B) RpoS protein levels of BW25113 cultivated in LB medium with various carbon sources (0.2%) added. Strains were grown to log phase (OD600 of 2.0) and analyzed by immunoblotting as described in Materials and Methods.
DISCUSSION
Here, we present direct evidence to support the mechanism behind the impact of the carbon source on natural transformation of E. coli. Namely, carbon sources facilitate natural transformation by reducing cAMP-CRP, thereby derepressing rpoS expression.
Significance of the correlation between nutrition and natural transformation in E. coli.
In the investigation of the development of natural competence, it is important to identify the environmental factors that hint at relevant parameters potentially triggering competence induction. In some cases, competence induction is based on nutritional signals. Bacteria undergo a shift toward an unbalanced metabolic state to trigger competence induction. For example, in H. influenzae and Bacillus subtilis, nutrient limitation after transfer to minimal medium or upon reaching the stationary phase can trigger competence development (34). On the other hand, Streptococcus pneumoniae and Acinetobacter calcoaceticus develop competence in rich medium during log phase and in the circumstance of a nutrient upshift, respectively (35, 36). In this work, we found that various carbon sources potentially encountered by bacteria within a host could promote the natural transformation of E. coli, which implies that a carbon upshift can promote the development of competence. This phenomenon is consistent with competence induction by nutritional signaling or an unbalanced metabolic state. Thus, it seems that competence induction is more likely to occur in response to satiety than to starvation in E. coli, but further experimentation is required to confirm this. When cells were cultured in modified LB medium with tryptone or yeast extract concentration gradients, the transformation frequencies correlated positively only with a certain range of tryptone or yeast extract concentrations above the normal (1% tryptone or 0.5% yeast extract). Below the normal concentration, the transformation frequencies were not significantly different from those of controls (see Fig. S5 in the supplemental material). We observed that competence could be induced by nutrient upshifts in E. coli, which broadens our understanding of the relationship between nutrition state and competence development in bacteria. Moreover, combined with our previous finding that DNA uptake gene orthologs that participate in the uptake of DNA as nutrition are not involved in this transformation system (9, 37), a different molecular mechanism of competence induction and DNA uptake in E. coli is implied.
Molecular mechanism by which cAMP-CRP regulates rpoS.
The pattern of regulation of rpoS by cAMP-CRP in Salmonella enterica and Vibrio vulnificus has been reported (38, 39). In S. enterica, it has been demonstrated that Δcrp and ΔcyaA mutants both exhibit a 3-fold increase in rpoS transcription during the log phase and that cAMP-CRP functions as a repressor in the regulation of rpoS. Further investigation of this mechanism in V. vulnificus revealed that cAMP-CRP represses rpoS transcription by directly binding to its two promoter regions. In our work, we demonstrated the dual regulation of rpoS by cAMP-CRP in E. coli. Moreover, we found that both of the putative CRP-binding sites seem to be positive effectors, based on the results of site-directed mutagenesis. However, mutagenesis may change the secondary structure of the rpoS mRNA and so affect mRNA stability or small RNA-based control, which then influences rpoS translation. Therefore, the binding interaction between CRP protein and the two putative binding sites requires further confirmation. If the two putative CRP-binding sites are both actually activators, it can be speculated that the negative effect of cAMP-CRP on rpoS in the early log phase is indirect. The expression of rpoS is affected by the growth rate. One interpretation of the indirect regulatory mechanism may be linked to the growth deficiency exhibited by Δcrp and ΔcyaA mutants in the early log phase after inoculation. Another speculation is that CRP may influence certain factors involved in the transcriptional or translational regulation of rpoS. The posttranscriptional regulation of rpoS is complex and stringent. The RNA chaperone protein Hfq, coupled with three small RNAs (DsrA, RprA, and ArcZ), positively controls rpoS mRNA stability and stimulates translation by facilitating annealing between the small RNAs and the 5′ untranslated region (UTR) of the mRNA to open the hairpin (19). Previous research has shown that the expression of hfq is repressed by cAMP-CRP and that glucose is capable of increasing Hfq protein levels through reduction of cAMP levels (40). Therefore, it is justifiable to speculate that cAMP-CRP suppresses rpoS indirectly by repressing hfq expression and then interfering with rpoS mRNA stability. Moreover, the positive effect of carbon sources on rpoS expression is also well founded.
Target gene(s) downstream from rpoS.
σS is the master regulator of the general stress response to starvation, UV irradiation, oxidative stress, nonoptimal temperature, pH, osmolarity, and so on. It has been reported that up to 10% of the genes in E. coli are under the direct or indirect control of rpoS. Some of the genes are recognized as a common core set that changes in parallel with the σS level. Others are induced only under specific stress conditions, which can be attributed to extensive regulatory overlaps with other global regulons (e.g., cAMP-CRP) or the requirement for higher σS concentrations for transcription and competition with other sigma factors (19, 41). We previously reported the higher transformation ability of the wild-type strain with overexpression of rpoS (11), which suggests that higher levels of σS in crp and cyaA mutants contributed to transformation in the present study. To screen out the target genes downstream from rpoS, we detected the transformation frequencies of several single-knockout mutants. Unfortunately, the target gene(s) has not yet been identified. We speculate that (i) multiple target genes may collaboratively contribute to transformation and/or (ii) the target genes are induced under specific stress conditions (e.g., when transferred from liquid medium to solid medium) that have not been sufficiently studied. Further research on target genes is in progress.
Additionally, the double-knockout mutants exhibited lower transformation abilities than the ΔrpoS mutant. We infer that some rpoS-dependent target genes that are also regulated by cAMP-CRP have a role in transformation. Actually, cAMP-CRP plays a complex role in the regulation of rpoS itself and also controls some rpoS-dependent genes directly. It has been determined that 55% of rpoS-dependent genes have putative cAMP-CRP-binding sites in their regulatory regions, which indicates considerable overlap between the σS and cAMP-CRP regulons (41). The target gene(s) may be regulated by both RpoS and CRP, and the balance between the effects of RpoS and CRP determines the expression levels of the target gene(s). Therefore, the double-knockout mutants are insufficient to confer the expression levels of the target genes to the ΔrpoS mutant, which results in lower transformation frequencies in double-knockout mutants than in the ΔrpoS mutant. In addition, the fact that ΔrpoS cannot inhibit transformation completely suggests the involvement of a further regulatory pathway in the expression of target genes downstream from rpoS, which also gives us clues about the downstream regulation.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Hanna Oksanen for kindly providing the plasmid pSU19. The English in this document has been checked by at least two professional editors, both native speakers of English. For a certificate, please see http://www.textcheck.com/certificate/Rg7J9n.
This work was supported by the National Basic Research Program of China (973 Program, grant no. 2013CB933904), the National Natural Science Foundation of China (grants 30971573 and 21272182), the National Foundation for Fostering Talents of Basic Sciences (grant J1103513), and the Laboratory (Innovative) Research Fund of Wuhan University.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00291-15.
REFERENCES
- 1.Gogarten JP, Townsend JP. 2005. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol 3:679–687. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- 2.Cohan FM, Koeppel AF. 2008. The origins of ecological diversity in prokaryotes. Curr Biol 18:R1024–R1034. doi: 10.1016/j.cub.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 3.Johnston C, Martin B, Fichant G, Polard P, Claverys JP. 2014. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol 12:181–196. doi: 10.1038/nrmicro3199. [DOI] [PubMed] [Google Scholar]
- 4.Woegerbauer M, Jenni B, Thalhammer F, Graninger W, Burgmann H. 2002. Natural genetic transformation of clinical isolates of Escherichia coli in urine and water. Appl Environ Microbiol 68:440–443. doi: 10.1128/AEM.68.1.440-443.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsen S-D, Fang S-S, Chen M-J, Chien J-Y, Lee C-C, Tsen DH-L. 2002. Natural plasmid transformation in Escherichia coli. J Biomed Sci 9:246–252. doi: 10.1159/000059425. [DOI] [PubMed] [Google Scholar]
- 6.Maeda S, Ito M, Ando T, Ishimoto Y, Fujisawa Y, Takahashi H, Matsuda A, Sawamura A, Kato S. 2006. Horizontal transfer of nonconjugative plasmids in a colony biofilm of Escherichia coli. FEMS Microbiol Lett 255:115–120. doi: 10.1111/j.1574-6968.2005.00072.x. [DOI] [PubMed] [Google Scholar]
- 7.Etchuuya R, Ito M, Kitano S, Shigi F, Sobue R, Maeda S. 2011. Cell-to-cell transformation in Escherichia coli: a novel type of natural transformation involving cell-derived DNA and a putative promoting pheromone. PLoS One 6:e16355. doi: 10.1371/journal.pone.0016355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun D, Zhang Y, Mei Y, Jiang H, Xie Z, Liu H, Chen X, Shen P. 2006. Escherichia coli is naturally transformable in a novel transformation system. FEMS Microbiol Lett 265:249–255. doi: 10.1111/j.1574-6968.2006.00503.x. [DOI] [PubMed] [Google Scholar]
- 9.Sun D, Zhang X, Wang L, Prudhomme M, Xie Z, Martin B, Claverys JP. 2009. Transforming DNA uptake gene orthologs do not mediate spontaneous plasmid transformation in Escherichia coli. J Bacteriol 191:713–719. doi: 10.1128/JB.01130-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Shi C, Yu J, Ren J, Sun D. 2012. RpoS regulates a novel type of plasmid DNA transfer in Escherichia coli. PLoS One 7:e33514. doi: 10.1371/journal.pone.0033514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Guo M, Shen P, Xie Z. 2014. The stress response factor RpoS is required for the natural transformation of Escherichia coli. Chin Sci Bull 59:521–527. doi: 10.1007/s11434-013-0014-7. [DOI] [Google Scholar]
- 12.Sun D, Wang B, Zhu L, Chen M, Zhan L. 2013. Block and boost DNA transfer: opposite roles of OmpA in natural and artificial transformation of Escherichia coli. PLoS One 8:e59019. doi: 10.1371/journal.pone.0059019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seitz P, Blokesch M. 2013. Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microbiol Rev 37:336–363. doi: 10.1111/j.1574-6976.2012.00353.x. [DOI] [PubMed] [Google Scholar]
- 14.Redfield RJ. 1991. sxy-1, a Haemophilus influenzae mutation causing greatly enhanced spontaneous competence. J Bacteriol 173:5612–5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herriott RM, Meyer EM, Vogt M. 1970. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J Bacteriol 101:517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gwinn M, Yi D, Smith H, Tomb J. 1996. Role of the two-component signal transduction and the phosphoenolpyruvate: carbohydrate phosphotransferase systems in competence development of Haemophilus influenzae Rd. J Bacteriol 178:6366–6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macfadyen LP, Dorocicz IR, Reizer J, Saier MH Jr, Redfield RJ. 1996. Regulation of competence development and sugar utilization in Haemophilus influenzae Rd by a phosphoenolpyruvate:fructose phosphotransferase system. Mol Microbiol 21:941–952. doi: 10.1046/j.1365-2958.1996.441420.x. [DOI] [PubMed] [Google Scholar]
- 18.Lo Scrudato M, Blokesch M. 2012. The regulatory network of natural competence and transformation of Vibrio cholerae. PLoS Genet 8:e1002778. doi: 10.1371/journal.pgen.1002778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lange R, Hengge-Aronis R. 1994. The cellular concentration of the σS subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev 8:1600–1612. doi: 10.1101/gad.8.13.1600. [DOI] [PubMed] [Google Scholar]
- 21.Hengge-Aronis R. 2002. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol Mol Biol Rev 66:373–395. doi: 10.1128/MMBR.66.3.373-395.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCann MP, Fraley C, Matin A. 1993. The putative sigma factor KatF is regulated posttranscriptionally during carbon starvation. J Bacteriol 175:2143–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmed N, Dobrindt U, Hacker J, Hasnain SE. 2008. Genomic fluidity and pathogenic bacteria: applications in diagnostics, epidemiology and intervention. Nat Rev Microbiol 6:387–394. doi: 10.1038/nrmicro1889. [DOI] [PubMed] [Google Scholar]
- 24.Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J. 2010. Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol 8:251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 25.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 27.Sarkar G, Sommer SS. 1990. The “megaprimer” method of site-directed mutagenesis. Biotechniques 8:404–407. [PubMed] [Google Scholar]
- 28.Conway T, Cohen PS. 2007. Escherichia coli at the intestinal mucosal surface, p 175–196. In Brogden KA, Minion FC, Cornick N, Stanton TB, Zhang Q, Nolan LK, Wannemuehler MJ (ed), Virulence mechanisms of bacterial pathogens, 4th ed ASM Press, Washington, DC. [Google Scholar]
- 29.Görke B, Stülke J. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol 6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 30.Matin A, Matin MK. 1982. Cellular levels, excretion, and synthesis rates of cyclic AMP in Escherichia coli grown in continuous culture. J Bacteriol 149:801–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Epstein W, Rothman-Denes LB, Hesse J. 1975. cAMP as mediator of catabolite repression in Escherichia coli. Proc Natl Acad Sci U S A 72:2300–2303. doi: 10.1073/pnas.72.6.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi H, Inada T, Postma P, Aiba H. 1998. CRP down-regulates adenylate cyclase activity by reducing the level of phosphorylated IIAGlc, the glucose-specific phosphotransferase protein, in Escherichia coli. Mol Gen Genet 259:317–326. doi: 10.1007/s004380050818. [DOI] [PubMed] [Google Scholar]
- 33.Mika F, Hengge R. 2005. A two-component phosphotransfer network involving ArcB, ArcA, and RssB coordinates synthesis and proteolysis of σS (RpoS) in E coli. Genes Dev 19:2770–2781. doi: 10.1101/gad.353705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnsborg O, Eldholm V, Havarstein LS. 2007. Natural genetic transformation: prevalence, mechanisms and function. Res Microbiol 158:767–778. doi: 10.1016/j.resmic.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 35.Claverys JP, Havarstein LS. 2007. Cannibalism and fratricide: mechanisms and raisons d'être. Nat Rev Microbiol 5:219–229. doi: 10.1038/nrmicro1613. [DOI] [PubMed] [Google Scholar]
- 36.Palmen R, Hellingwerf KJ. 1997. Uptake and processing of DNA by Acinetobacter calcoaceticus: a review. Gene 192:179–190. doi: 10.1016/S0378-1119(97)00042-5. [DOI] [PubMed] [Google Scholar]
- 37.Palchevskiy V, Finkel SE. 2006. Escherichia coli competence gene homologs are essential for competitive fitness and the use of DNA as a nutrient. J Bacteriol 188:3902–3910. doi: 10.1128/JB.01974-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirsch M, Elliott T. 2005. Fis regulates transcriptional induction of RpoS in Salmonella enterica. J Bacteriol 187:1568–1580. doi: 10.1128/JB.187.5.1568-1580.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee HJ, Park SJ, Choi SH, Lee KH. 2008. Vibrio vulnificus rpoS expression is repressed by direct binding of cAMP-CRP receptor protein complex to its two promoter regions. J Biol Chem 283:30438–30450. doi: 10.1074/jbc.M802219200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin HH, Hsu CC, Yang CD, Ju YW, Chen YP, Tseng CP. 2011. Negative effect of glucose on ompA mRNA stability: a potential role of cyclic AMP in the repression of hfq in Escherichia coli. J Bacteriol 193:5833–5840. doi: 10.1128/JB.05359-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol 187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tolker-Nielsen T, Brinch UC, Ragas PC, Andersen JB, Jacobsen CS, Molin S. 2000. Development and dynamics of Pseudomonas sp. biofilms. J Bacteriol 182:6482–6489. doi: 10.1128/JB.182.22.6482-6489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bartolome B, Jubete Y, Martínez E, de la Cruz F. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75–78. doi: 10.1016/0378-1119(91)90541-I. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.