ABSTRACT
Current knowledge regarding the mechanism that governs flagellar motor rotation in response to environmental stimuli stems mainly from the study of monotrichous and peritrichous bacteria. Little is known about how two polar flagella, one at each cell pole of the so-called amphitrichous bacterium, are coordinated to steer the swimming. Here we fluorescently labeled the flagella of Magnetospirillum magneticum AMB-1 cells and took advantage of the magnetically controllable swimming of this bacterium to investigate flagellar rotation in moving cells. We identified three motility behaviors (runs, tumbles, and reversals) and two characteristic fluorescence patterns likely corresponding to flagella rotating in opposite directions. Each AMB-1 locomotion mode was systematically associated with particular flagellar patterns at the poles which led us to conclude that, while cell runs are allowed by the asymmetrical rotation of flagellar motors, their symmetrical rotation triggers cell tumbling. Our observations point toward a precise coordination of the two flagellar motors which can be temporarily unsynchronized during tumbling.
IMPORTANCE Motility is essential for bacteria to search for optimal niches and survive. Many bacteria use one or several flagella to explore their environment. The mechanism by which bipolarly flagellated cells coordinate flagellar rotation is poorly understood. We took advantage of the genetic amenability and magnetically controlled swimming of the spirillum-shaped magnetotactic bacterium Magnetospirillum magneticum AMB-1 to correlate cell motion with flagellar rotation. We found that asymmetric rotation of the flagella (counterclockwise at the lagging pole and clockwise at the leading pole) enables cell runs whereas symmetric rotation triggers cell tumbling. Taking into consideration similar observations in spirochetes, bacteria possessing bipolar ribbons of periplasmic flagella, we propose a conserved motility paradigm for spirillum-shaped bipolarly flagellated bacteria.
INTRODUCTION
Mobile bacteria have developed strategies to efficiently explore their environment, in aqueous media as well as on solid surfaces (1, 2). In most cases, their movements are ensured by a highly efficient proteinaceous nanomachine, the flagellum. The flagellar apparatus comprises three main parts: the motor, the hook, and the flagellar filament. The flagellar motor, anchored in the plasma membrane, uses the proton motive force or the sodium ion gradient to power the rotation of the flagellar filament, which is connected to it through the structure called the hook (3, 4). The rotation of the motor determines the direction of flagellum rotation and, consequently, the swimming direction of the bacterium. Using that principle, chemotactic bacteria directly regulate motor rotation so as to swim toward an attractant or away from a repellent, which involves signal detection via chemoreceptors. The signal is then transmitted from the chemoreceptor to the flagellar motor through a phosphorylation-dephosphorylation cascade of dedicated chemotaxis proteins (Che proteins) (5).
While chemotaxis proteins are well conserved in phylogenetically and morphologically diverse bacteria, the mechanisms by which they govern flagellar propulsion are diverse. In fact, flagellar number, position, and regulation differ between microorganisms. In peritrichously flagellated bacterial species, such as Escherichia coli or Bacillus subtilis, the counterclockwise (CCW) rotation of all flagella (viewed from the flagellar tip toward its base) results in the formation of a bundle of flagella that propels the cell toward attractants. When the cell senses a repellent, a phosphorylation cascade leads to a change in the phosphorylation status of a response regulator, CheY. Activated CheY directly interacts with the motor switch proteins and causes the flagella to rotate in the opposite direction (clockwise [CW]). As a consequence, the bundle is disrupted, the flagella are spread around the cell, and their uncoordinated rotation triggers cell tumbling. The movements caused by Brownian motion help the cell to randomly reorient until an attractant is detected and a bundle is reassembled to cause the cell to resume swimming in a randomly selected direction (2). In monotrichous bacteria, which possess only one polar flagellum, several mechanisms allow cells to change direction. In Vibrio spp., the CCW rotation of the flagellum propels the cells forward while its CW rotation pulls the bacterium backward (6). In the case of Rhodobacter sphaeroides, the rotation speed of the flagellum can be modified, affecting its conformation and, in turn, cell velocity (4).
Compared to these very well described flagellar propulsion mechanisms, little is known about motility control in amphitrichously flagellated bacteria such as Campylobacter jejuni, Rhodospirillum rubrum, and Magnetospirillum spp., which possess one flagellum at each cell pole (7). Recently, Popp and colleagues studied Magnetospirillum gryphiswaldense motility and showed that swimming polarity is controlled by aerotaxis in this magnetotactic bacterium (MTB) (8). Two simple models can explain how a symmetrical cell can swim in an oriented manner, and both imply that the two flagella are operated differently. In one model, each flagellum would be able to assume cell movement in only one direction (in a monotrichous manner), whereas, in the second one, the two flagella would simultaneously rotate but must rotate in opposite directions. Motility control has been studied in spirochetes, bacteria which swim thanks to internal structures that are analogous to the polar flagella of amphitrichous bacteria. In fact, spirochetes move thanks to two polar bundles of periplasmic flagella, and it has been shown that oriented swimming of the cells is a consequence of the rotation of these bundles in opposite directions (9). However, direct observation of flagella during swimming in bacteria possessing single polar flagella has been limited due to flagellum size and the lack of molecular tools allowing their visualization without interfering with motility. The challenge here resides in being able to directly observe flagellar rotation during cell movement and decipher the molecular mechanisms ensuring coordination of flagella.
To get insights into the mechanism underlying oriented swimming in amphitrichously flagellated bacteria possessing only one flagellum at each cell pole, we took advantage of the magnetic properties of the MTB Magnetospirillum magneticum AMB-1. MTB synthesize intracellular magnetic nanocrystals within specialized membrane-bounded organelles termed magnetosomes that are arranged in a chain and confer a magnetic moment to the cells (10). Thanks to this internal compass, MTB can align passively and swim along the geomagnetic field lines, which was proposed to facilitate their search for an oxic-anoxic transition zone (OATZ), their most favorable living habitat (11). Once located on the more reduced side of the OATZ, they should swim upward by inverting the direction of swimming. In this study, cell motility analysis was simplified by applying an external magnetic field, which imposes an overall direction to the cells and limits their trajectories to one-dimensional runs. M. magneticum AMB-1 was picked for this study because it is one of the few MTBs for which molecular engineering is available (12, 13).
We investigated polar coordination of flagella during motility in this bacterium by directly visualizing the rotation of fluorescently labeled flagella during cell movement. We qualitatively and quantitatively described motility behaviors of AMB-1 and correlated them with a particular rotation direction of each flagellum. We show that cells are propelled by the counterclockwise rotation of the lagging flagellum. We inferred from the pattern of fluorescence observed at the leading pole that the leading flagellum is likely to rotate in the clockwise rotation during oriented swimming. We showed that cell reversals are caused by the simultaneous changes of rotation direction of both flagella and identified a “tumbling-like” motion caused by the rotation of both flagella in the same direction. We propose that the opposite rotations of the motors during cell runs might result from the assembly of distinct flagellar motors at the opposite poles or differential control of their rotation directions or both.
MATERIALS AND METHODS
General microbiology and molecular biology.
A motile strain of Magnetospirillum magneticum AMB-1 was used for all the experiments and was grown as described in reference 14. The strain was grown in 10 ml MG medium (14) in 12-ml polystyrene plastic tubes at 30°C without shaking for 24 to 72 h. Antibiotics were used as follows: kanamycin at 7 to 10 μg/ml in liquid media and at 15 μg/ml on agar plates and ampicillin at 20 to 30 μg/ml in liquid media as well as on agar plates. Escherichia coli DH5λPIR was used for cloning purposes, and E. coli WM3064 was used as a donor strain for conjugation experiments as described by Murat et al. (14). Gene deletions and mutant phenotype complementations were performed according to previously described strategies (14). Strains, plasmids, and oligonucleotides used in this work are listed in Table S1 in the supplemental material. DNA sequences were amplified using GoTaq Green Mix (Promega) and purified AMB-1 genomic DNA. Cloning was done either by restriction and ligation using NEB restriction enzymes and a Fast ligation kit from TaKaRa or by the use of an In-Fusion kit (ClonTech). Flagellin site-directed mutagenesis was performed using Pfu Turbo DNA polymerase (Invitrogen) and DpnI from NEB. After verification of the constructs by restriction and PCR, all plasmids were sequenced (Beckman Coulter Genomics). RNA extraction and reverse transcription were done with an RNAlater kit (Invitrogen) according to the manufacturer's instructions. RNAs were treated twice with Turbo DNA-free DNase (Ambion).
Flagellin purification and polyclonal antibody rising.
AMB-1 cells (in reaction volumes of 2 to 4 liters) were grown in individual 1-liter bottles to an optical density at 400 nm (OD400) of 0.2 to 0.3. Cells were collected by centrifugation and resuspended in 1/10 of the culture volume in 12 mM HEPES buffer (pH 7.2). Flagella were sheared from the cell bodies by subjecting the cell suspension to vortex mixing for 1 min followed by 20 passages through a needle (0.8 by 40 mm). Sheared cells were centrifuged at 20,000 × g for 5 min, and the supernatants were pooled and concentrated by ultracentrifugation (1 h at 40,000 × g). The flagellin concentration was determined using the DC protein assay from Bio-Rad. A total of 40 liters of culture was necessary to obtain a solution enriched with flagellin (2.2 mg in total). Flagellin purity was estimated by SDS-PAGE followed by Coomassie staining. One rabbit (Charles River Laboratories, Inc., Wilmington, MA, USA) was immunized using three flagellin injections, and after a 90-day protocol, an antiflagellin serum was collected and used for immunodetection of flagellin by Western blot analyses (1/20,000 dilution). SDS-PAGE electrophoresis and Western blot analyses were performed as described in reference 16.
Flagellum labeling.
AMB-1 cells (AMB-1 or Δfla strain) carrying the plasmid allowing the expression of the modified flagellin were grown in the presence of antibiotics for 48 h (AMB-1 strain) to 72 h (Δfla strain). Cell labeling was performed according to Turner et al. (17, 18) with slight modifications. Ten milliliters of AMB-1 cells was centrifuged for 10 min at 1,000 × g in 15-ml Falcon tubes. As the pellet was not visible at that stage, 8 ml of supernatant was cautiously discarded by pipetting. The remaining 2 ml of cell suspension was transferred to a 2-ml tube and centrifuged for 4 min at 1,000 × g to avoid breaking of flagellar filaments. After the supernatant was removed, cells were washed three times in 1 ml of motility buffer (MB) (100 μM EDTA, 10 mM potassium phosphate [pH 7], 67 mM NaCl, 0.0001% Triton). After the third centrifugation, cells were resuspended in 300 μl of motility buffer to which 30 μl of Alexa Fluor 488 C5 maleimide (Life Technologies) was added. The tube was wrapped with aluminum foil and placed on a horizontal rocking platform set at 18 oscillations per min for 1 h at room temperature. Excess dye was removed by three successive washes in 1 ml of MB. Finally, cells were resuspended in 500 μl of MB.
AMB-1 motility analysis.
AMB-1 motility behavior was analyzed using phase-contrast microscopy and a Zeiss inverted microscope equipped with custom-built electromagnetic coils (Magnetodrome) as previously described (22). In addition, swimming velocity and magnetic moments were determined using a custom-made plugin for Fiji-ImageJ.
The motion of fluorescence-labeled AMB-1 cells, placed on a glass slide or in μ-Slide VI0.4 poly-l-lysine-coated microchambers (Ibidi, Germany), was analyzed using an inverted Nikon TiE-PFS microscope and a Hammatsu Orca R2 camera. For observations on glass slides, 4.5 μl of labeled cells was observed for no longer than 30 min using a 100× phase numerical-aperture (NA) 1.3 immersion objective. Otherwise, 300 μl of cell suspension were placed in a microchamber and observed with a long-working-distance 60× NA 1.4 immersion objective. Every 30 min or so, the canal was washed with MB and fresh cells were injected to prevent AMB-1 cells from sticking to the bottom of the canal.
For tracking and motility analysis of fluorescently labeled AMB-1 cells, the video sequences were rotated to align the movement horizontally. Fluorescent objects were identified by the intensity threshold and automatically detected with a Fiji/ImageJ plugin (17). Data sheets corresponding to the positions of the centers of mass of the objects along the x axis (horizontal movement) were analyzed with R software (19) for each track, and several descriptors were calculated: (i) a linear regression model (r2 coefficient and slope), (ii) the average velocity along the track, (iii) the maximum amplitude of the displacement, and (iv) the number of contiguous points of instantaneous speed above or below a certain threshold. These descriptors were used to automatically sort tracks. The results were controlled by hand, and some tracks were reclassified according to the images on the original video.
RESULTS
AMB-1 cells swim as a right-handed helix.
M. magneticum AMB-1 is a spirillum-shaped amphitrichous bacterium producing a chain of intracellular single-domain magnetite nanocrystals (Fig. 1A). While its flagella appear to be curved in electron microscopy, at this point, we do not know whether they are helical. AMB-1 performs axial magnetotaxis, which means that even though most cells swim toward the magnetic north (north-seeking behavior), they can also perform southbound runs while maintaining a perfect alignment with the magnetic field lines (11). Such motility is observed when cells are placed in microchambers (Ibidi, Germany) (150-μm depth) and subjected to a local magnetic field. Under these conditions, AMB-1 cells frequently reverse swimming direction while remaining parallel to the applied magnetic field.
FIG 1.
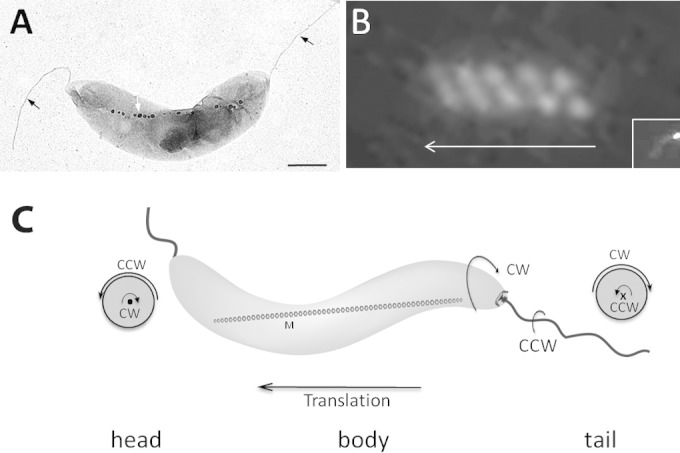
Rotation direction of AMB-1 cell body during runs. (A) Electron micrograph of a spirillum-shaped AMB-1 cell showing its two polar flagella (black arrows) and its magnetosome chain (white arrow). Bar, 500 nm. (B) Swimming trajectory reflected by single polar fluorescence labeling (inset) of MamU-GFP recorded during cell translation with a 100-ms exposure. The arrow indicates the direction of cell movement. (C) Schematic representation of an AMB-1 cell showing cell body and flagellar rotation directions. • and × show views of the flagella out-off and into the image, respectively.
To infer the rotation direction of AMB-1 flagella, we analyzed the motion of cell bodies during cell movement, considering that, to balance the force and torque generated by the rotation of the flagellum, the cell body would rotate in the direction opposite the direction employed by the flagellum. First, we used a polarly localized magnetosome protein labeled with green fluorescent protein (GFP) (MamU-GFP) to follow the rotation of AMB-1 cell body during movement. Since the position of MamU-GFP is fixed in the cell (Fig. 1B, inset), the trajectory of the fluorescent focus during cell movement is indicative of the trajectory of the cell body. A representative helical pattern of MamU-GFP fluorescence during AMB-1 translation is shown in Fig. 1B. Physicists define a helical object as right-handed when it moves away from an observer along such a helix and rotates in the clockwise direction. According to this definition, the fluorescent tracks of MamU-GFP show a distinctive right-handed helix, indicating that during movement, the AMB-1 cell body rotates in the clockwise direction.
Second, we took advantage of AMB-1 spiral morphology to determine cell body rotation direction in phase-contrast microscopy. As shown in Movie S1 in the supplemental material, the cell body rotates in the clockwise direction when it swims from right to left, and, after a pause lasting about 200 ms, the cell starts swimming from left to right and cell body rotation is inverted. As a consequence, cell movement is consistently associated with the clockwise rotation of the cell body. Importantly, this observation confirms the right-handed translational motility mode of AMB-1 cells and implies that the lagging flagellum rotates in the counterclockwise (CCW) direction (projected from the end of the flagellum toward to the basal body) (Fig. 1C).
AMB-1 flagella present two patterns of fluorescence.
To shed light on flagellar operation in amphitrichous bacteria, we needed to visualize the flagellar filaments during cell movement. We chose to label AMB-1 flagella with the fluorescent moieties that have been successfully used to label E. coli and B. subtilis flagellins (17, 18). Briefly, the Alexa 488 C5 maleimide fluorophore (Life Technologies) is linked to the flagellin in vitro through a covalent bond formed with the thiol group of cysteine residues. As described by Turner et al. and Blair et al., this strategy does not impede flagellar assembly or affect swimming in E. coli or B. subtilis (17, 18). Since AMB-1's only flagellin (Amb0684 was annotated as FliC) does not contain any cysteine, we introduced one (substituted to replace a threonine) at position 207 (FliCT207C) or 210 (FliCT210C) using site-directed mutagenesis. The modified allele was expressed under the control of its own promoter from a low-copy-number plasmid in AMB-1 cells. Flagellum labeling was performed according to the protocol of Turner et al. with slight modifications (see Materials and Methods). The same results were obtained with both altered fluorescent flagellins, but only those obtained with FliCT210C are shown here. We verified that the modified flagellin could complement the AMB-1 ΔfliC strain nonmotile phenotype and that it did not significantly affect wild-type AMB-1 swimming velocity (25.9 ± 5.2 and 23.4 ± 5.2 μm/s for AMB-1 and AMB-1-FliCT210C, respectively). Consistently, about two-thirds of the cells presented two 2- to 3-μm-long fluorescent flagella (Fig. 2A). The remaining third presented either only one labeled flagellum or none, which was most likely the consequence of flagella breaking off the cell surface, since many isolated flagella could be observed and AMB-1 flagella are particularly fragile.
FIG 2.
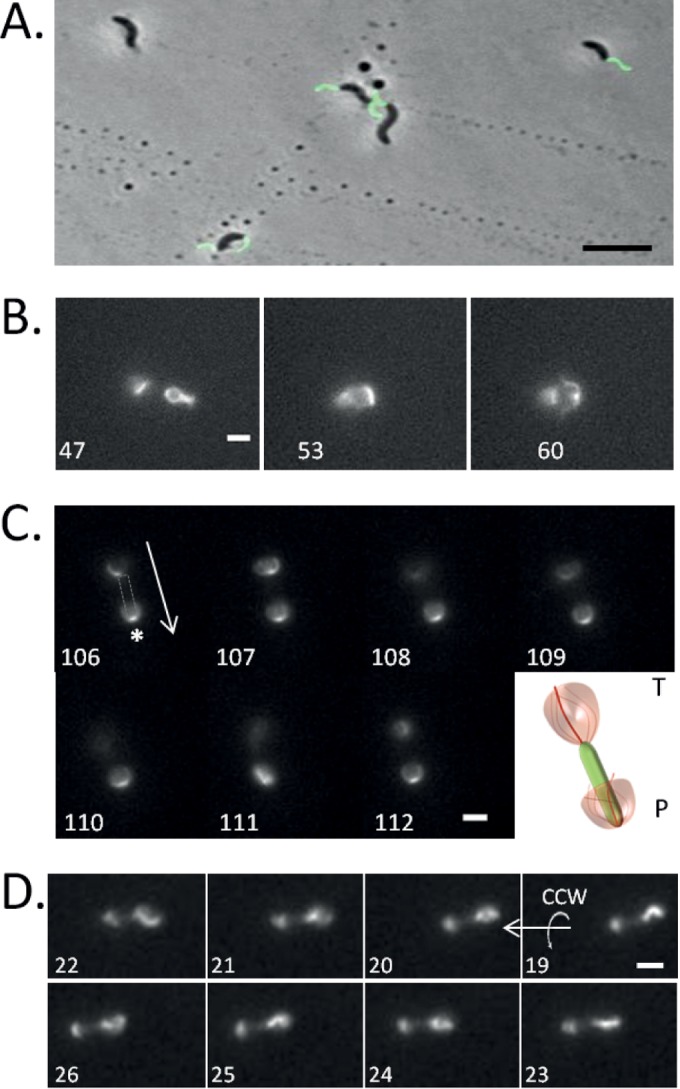
Rotation of AMB-1 fluorescently labeled flagella. (A) Superimposed images of phase-contrast (gray-scale) and fluorescence (green) microscopy of AMB-1 flagella labeled with Alexa 488 C5 maleimide. Bar, 5 μm. (B) Frames extracted from Movie S2 in the supplemental material. Exposure time, 100 ms. Bar, 2 μm. (C) Time-lapse images extracted from Movie S4 showing the fluorescently labeled flagella of an AMB-1 cell swimming downward. In frame 106, the gray shape indicates the position of the unlabeled cell body. The arrow indicates the direction of swimming. P, parachute; T, tuft. The asterisk indicates the position of the flagellum anchoring point at the pole. The pictogram on the right is a model representing flagellar rotation at each cell pole. Exposure time, 80 ms. Bar, 2 μm. (D) AMB-1 lagging flagellum rotates in the CCW direction. Frames 19 to 26 (from right to left) were extracted from Movie S5. The arrows indicate the translation and rotation direction. In all panels, the number at the bottom left corner corresponds to the frame number in the movie. Bar, 3 μm.
In moving cells, we observed two patterns of flagellar fluorescence, which we named “tuft” and “parachute” as shown in Fig. 2B and Movie S2 in the supplemental material and which correspond to two different positions of the flagellum with respect to the cell body. In the movie, an AMB-1 cell is shown immobilized on a glass slide and the right flagellum goes from being deployed outward (tuft, frame 47) to rotating about the cell body (parachute, frame 60). Because the cell body is immobilized and there is no flow in the microchamber, the change of the position of the flagella is likely caused by a change in the rotation direction of the flagellum. The capacity of flagellar motors to alternate between CW and CCW rotation directions was confirmed by attaching AMB-1 cells through their flagella to anti-FliC-coated glass slide. During these observations, we clearly saw cells which alternated between a CW rotation and a CCW rotation, again indicating the capacity of the motor to rapidly and spontaneously change rotation direction (see Movie S3). We use these two patterns of flagellar fluorescence as references to infer changes in flagellar rotation direction.
We found that AMB-1 cells could swim with different velocities. In faster cells, with our recording settings (100-ms exposure), a rapidly rotating flagellum appears as a blurry zone of fluorescence (Fig. 2C, frame 107), while the position of its anchoring point at the cell pole can be made out in each frame, as its position is constant (indicated by an asterisk in Fig. 2C, frame 106). The moving cell displays the two characteristic patterns of flagellar fluorescence: a parachute at the leading pole and a tuft at the lagging pole (Fig. 2C; see also Movies S4 and S7 in the supplemental material). The analysis of hundreds of cells (n > 300) in oriented runs showed that the parachute is systematically associated with the leading pole and the tuft with the lagging pole.
In slower cells, the position of the flagella can be precisely located in almost every recorded frame (Fig. 2C, frame 106). We took advantage of these cells to determine the rotation direction of AMB-1 flagella. Figure 2D and Movie S5 in the supplemental material show a representative “slower” AMB-1 cell swimming from right to left (frame 19 to 26). Frame-by-frame inspection clearly indicates that the lagging flagellum rotates in the CCW direction, which is fully consistent with the right-handed rotation of the AMB-1 cell body depicted in Fig. 1. Incidentally, the tuft pattern at the lagging pole most likely illustrates the counterclockwise rotation of the flagellum. Considering the consistent pattern of fluorescence observed at the leading pole, we propose that the parachute would correspond to the rotation of the flagellum in a CW direction.
M. magneticum AMB-1 performs runs, pauses, and reversals.
To study AMB-1 motility quantitatively, we developed a plugin for ImageJ to automatically identify and track fluorescently labeled cells in video recordings (see Materials and Methods and reference 20). After flagellin labeling, cells were placed in microchambers and a magnetic field was applied such that the magnetic north points to the left on all images and the majority of AMB-1 cells swim toward it. A total of 586 cell trajectories were obtained from 6 independent experiments (see Fig. S1 in the supplemental material). The trajectories obtained depict the variation of the horizontal coordinate (the vertical coordinate is constant) as a function of time. Negative slopes correspond to northbound runs (cells swim toward the left) and positive slopes to southbound runs (cells swim to the right). We identified three motility behaviors defined by characteristic trajectories: uninterrupted runs (motility group 1) (Fig. 3A), runs interrupted by pauses (motility group 2) (Fig. 3B), and runs interrupted by at least one reversal (motility group 3) (Fig. 3C). Motility group 1 exclusively consists of cells that performed linear runs during the course of the recording. When a single cell performed a pause and a reversal during the recording, it was classified in group 3 to compensate for the fact that the cells performing reversals were most often lost by our tracking device.
FIG 3.
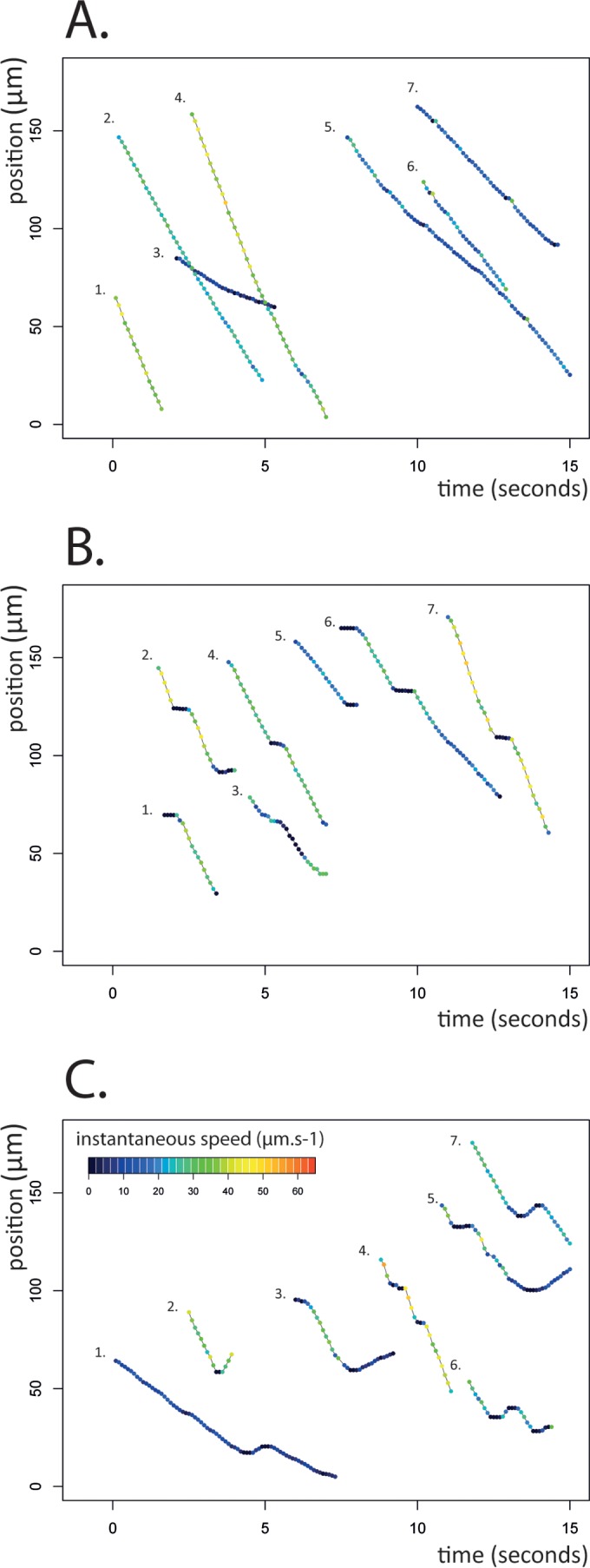
Three distinct motility behaviors of AMB-1 cells. The traces describe the position of a cell (y axis, distance in micrometers) as a function of time (seconds). Each color-coded dot corresponds to the instantaneous speed of the cell. Shown are seven examples of motility groups 1, 2, and 3, consisting of uninterrupted runs (A), runs interrupted by pauses (B), and runs interrupted by reversals (C), which represent 586 tracks obtained from six independent experiments (see the text and Materials and Methods for details).
Of the 586 trajectories we analyzed, about 74% performed uninterrupted runs (motility group 1) across the field of view (144 by 110 μm) with an unchanged velocity or a slight variation of instantaneous velocity (as indicated by the colors of the dots in Fig. 3A). The distribution of instantaneous speeds was rather wide (from 1.9 to 55.2 μm/s), with an average speed of 24.3 μm/s (standard deviation [SD] = 11.6 μm/s). The maximal speed of AMB-1 is close to 30 body lengths per second (65 μm/s), compared to 10 to 20 for E. coli and 60 for Vibrio cholerae (6). About 14% of the tracks (motility group 2) were characterized by at least one short pause illustrated by a plateau in the trace at the beginning (traces 1 and 6), in the middle (traces 2, 3, 4, 6, and 7), or at the end (traces 2 and 5) of the recorded run (Fig. 3B). The trajectories assigned to this group (see Materials and Methods) were individually verified so as to avoid potential tracking errors. While most cells performed one pause during the recording, some cells performed as many as five successive pauses in less than 10 s (three successive pauses are shown in trace 4) (Fig. 3B). The cell speed after a pause or between two successive pauses was determined and found not to be significantly different from the initial speed. Finally, results from 7% of the analyzed tracks indicate cells reversing their swimming direction during the run (motility group 3 is shown in Fig. 3C). Most cells in this group actually performed two successive reversals which allowed them to rapidly resume their initial northbound run. The southbound runs were systematically shorter than the northbound ones and for most of them lasted less than 400 ms. Yet we found that AMB-1 was able to reach similar maximal speeds when performing north- and southbound runs, suggesting that the two flagella can generate similar levels of torque and, consequently, similar cell speeds in the two directions.
A change in swimming direction is triggered by changes in the rotation direction of both flagella.
AMB-1 can perform short consecutive reversals while keeping its alignment with the magnetic field lines. A typical reversal is illustrated in Fig. 4 (see Movie S7 in the supplemental material). The time-lapse images show a cell reversing twice: north to south and, rapidly thereafter, south to north. Initially (frames 1 to 3), the cell swims toward the north: a parachute is visible at the leading pole (left) and a tuft at the lagging pole (right). In frame 16, the cell rapidly changes swimming direction and the fluorescence patterns of the two flagella are simultaneously inverted: the parachute becomes a tuft at the new lagging pole, and the tuft becomes a parachute at the new leading pole. Before going through a second reversal and resuming its northbound run, the cell shortly stalls as both its flagella are deployed outward (frames 29 and 30). Finally, the tuft on the left bends backward to rotate about the cell body and becomes the new leading pole (frames 47 and 48). This representative recording shows that a reversal is accompanied by simultaneous changes in both the fluorescence and the position of the flagella with respect to the AMB-1 cell body. This change of pattern is likely to correspond to a rapid change in flagellar rotation direction, implying the precise coordination of the two polar flagellar motors.
FIG 4.
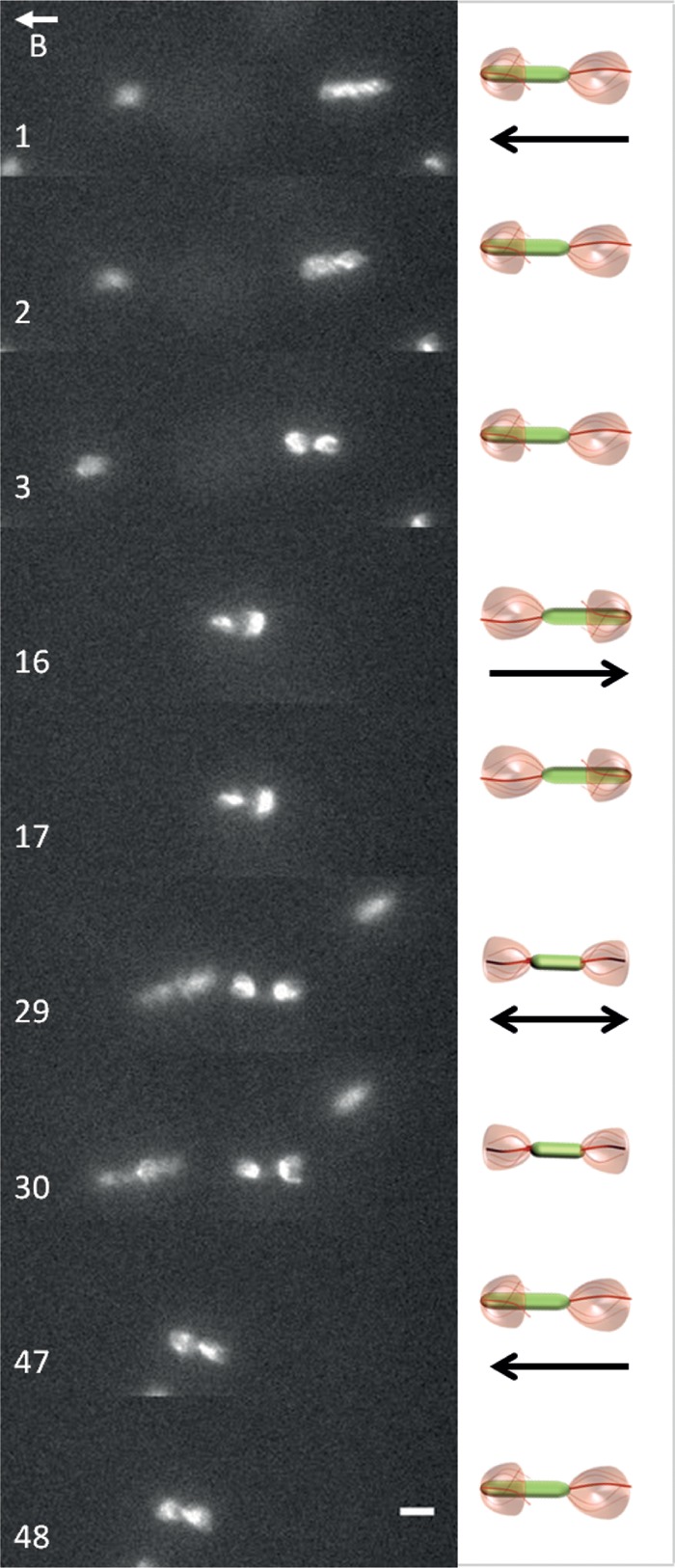
Cell reversals are triggered by simultaneous changes of the rotation direction of both flagella. Time-lapse images were extracted from Movie S7 in the supplemental material. Exposure time, 80 ms. Bar, 4 μm. The letter B and the vector above it indicate the orientation of the local magnetic field.
Cells tumble when both flagella are rotating in the same direction.
We found that AMB-1 cells can temporarily stall during an oriented run. Figure 5A and corresponding Movie S8 in the supplemental material show an AMB-1 cell from motility group 2 (trace 4 in Fig. 3B) swimming from top to bottom (frames 6, 8, 10, 16, and 18) and pausing in the middle of its run (frames 12 and 14). In the first three frames, the two flagella appear as one contiguous blur of fluorescence due to the fact that the cell was swimming too fast for the recording to capture. However, in frames 12 and 14, when the cell pauses, the two flagella appear as two sharp fluorescent curved lines which correspond to slowed-down or static flagella. After this brief interruption, the cell resumes swimming (frames 16 and 18) before pausing again (frame 20). This suggests that AMB-1 cells can control flagellar rotation speed such that they can either drastically slow down or temporarily stop swimming.
FIG 5.
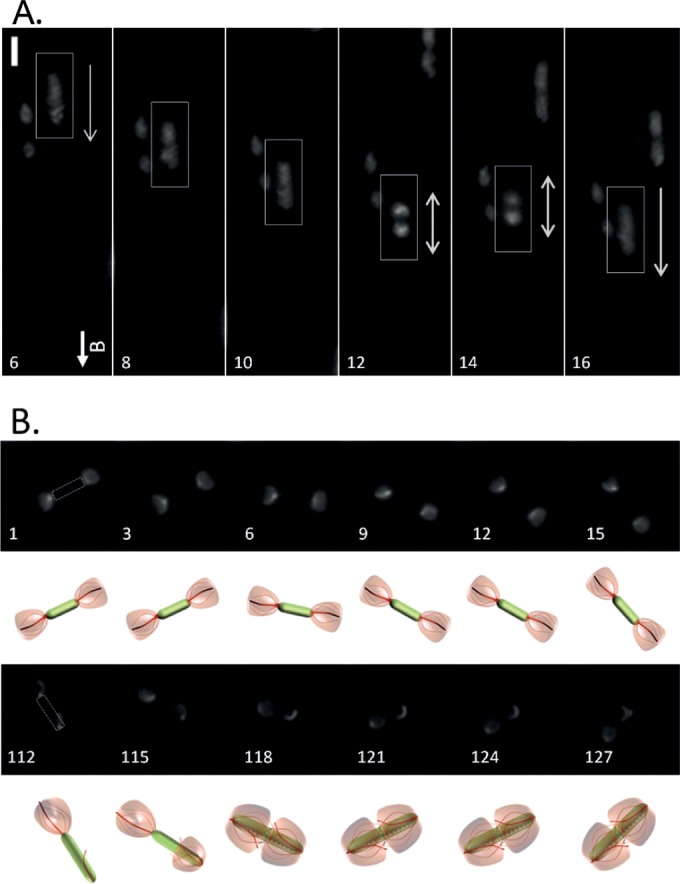
AMB-1 runs are interrupted by short pauses and tumbles. (A) Time-lapse images extracted from Movie S5 in the supplemental material showing an AMB-1 cell pausing in the midst of a run (frames 6 to 20). Exposure time, 100 ms. Bar, 4 μm. The letter B and the vector above it indicate the orientation of the local magnetic field. (B) Tumbles are caused by two flagella showing the same fluorescent pattern. Time-lapse images were extracted from Movie S8. The pictogram under each frame represents an interpretation of the flagellar motion depicted in the image above it. Exposure time, 80 ms. Bar, 2 μm.
In addition, some AMB-1 cells were found rotating in place for an extended period of time without any oriented movement, which is reminiscent of E. coli tumbling motion and illustrated in Fig. 5B and Movie S8 in the supplemental material. At the beginning of the recording, both flagella form tufts and flare away from the cell body. The cell is slightly rotating in place (frames 1 to 21). Then, one flagellum folds back on the cell body (frame 112) and stops rotating; the cell stays still. Soon after, the second flagellum (frame 118) folds back toward the cell body and keeps rotating around it. Finally, as the first flagellum resumes its rotation about the cell body (at frame 124), the cell resumes tumbling. Since all cells going through this tumbling motion show two parachutes or two tufts, this finding means that cell tumbling occurs when flagella rotate symmetrically, triggering opposite pushing or pulling forces.
DISCUSSION
While rotation of flagella in opposite directions in bipolarly flagellated bacteria has long been postulated, here we provide the first experimental evidence supporting this model by using fluorescence labeling of M. magneticum AMB-1 single polar flagella. Our results indicate that the motions of this apparently symmetrical helix-shaped bacterium rely on the asymmetrical rotation of its polar flagella. We observed three distinct motility behaviors (runs, reversals, and pauses), each of which was correlated with a flagellar rotation pattern. All of these observations are reminiscent of the findings in spirochetes that were reported by Charon and colleagues. An extensive analysis of spirochete swimming behavior combined with mathematical modeling led to a widely accepted model in which cell translation is permitted by the asymmetrical rotation of the flagella at opposite poles (9). During runs, the posterior flagellar ribbon would rotate in the CCW direction while the anterior ribbon would rotate in the CW direction. Cell reversals are thought to occur when both flagellar ribbons change their direction of rotation. As we also observed in AMB-1, Borrelia burgdorferi has a nontranslational motility mode which is thought to be provoked by the symmetrical rotation of the anterior and posterior flagella relative to one another. As the flagella are hidden under the outer membrane, testing these models in spirochetes is limited by obvious technical difficulties (9).
We showed that AMB-1 cell bodies rotate in a clockwise direction, and single-image analysis of fluorescently labeled flagella confirmed that the flagellum at the lagging pole rotates in a counterclockwise direction, as expected. Even though we have not been able to directly visualize the rotation direction of the leading flagellum during cell movement, the simplest model is one where the leading and lagging flagella rotate in opposite directions to allow swimming, as previously proposed in spirochetes. Therefore, it is likely that during runs, the flagellum associated with the leading cell pole rotates in the clockwise direction.
We also show that both flagella can alternate between the two positions we describe, which indicates that both flagellar motors are bidirectional. AMB-1 cells are able to scan the milieu by reversing their swimming direction. Such reversals result from seemingly simultaneous changes of rotation direction of the two flagella, while the cell body remains perfectly aligned in the magnetic field. This observation indicates that both flagellar motors must be placed under the control of at least one common factor that ensures the coordination of the flagella.
Statistical analysis of AMB-1 motility behavior was performed in microchambers, that is, in a chemically homogenous space without any conspicuous gradients. As a consequence, the frequency of pauses and reversals measured likely illustrates stochastic events and would be modulated by chemotaxis circuits in response to variations in the environment. Taking into consideration the microaerobic lifestyle of M. magneticum and the work by Popp and colleagues (8), we predict that these behaviors should be directly influenced by oxygen concentration. This work and the tools we developed are essential to study how oxygen concentration affects magnetoaerotaxis.
Two kinds of pausing behaviors were identified. First, pauses were identified that might have been the consequence of a temporary stop of the rotation of both flagella, reflected by sharp images of fluorescent flagellar filaments in video recordings. This behavior suggests the existence of a clutch-like mechanism like that described in B. subtilis (18). Second, AMB-1 cells displayed a tumbling-like motion which was triggered by the rotation of both flagella in the same direction (appearing as two parachutes or two tufts). Since the flagella were pushing or pulling the cell in opposite directions, the cell was unable to swim in any direction, which is reminiscent of the E. coli tumbling motion caused by uncoordinated peritrichous flagella (2). Interestingly, this tumbling motion was more frequently observed (and for extended periods of time) when AMB-1 cells were placed between a glass slide and a coverslip than when they were being maintained in microchambers. The altered behavior could have been due to a biological reaction to external stresses such as oxygen exposure or dehydration or to a hydrodynamic border effect (15), although such forces are less likely to occur in the context of a microchamber. The rapid switch between oriented runs and tumbling depends on the fact that the coordinated flagella can be temporarily decoupled to trigger tumbling.
Lastly, we noted that cell velocity could vary in the course of a run. This suggests that the overall flagellar torque can vary, which would likely be the consequence of slowed rotation of both flagella. It is possible that the presence of accessory proteins in the flagellar apparatus could help tune cell speed.
To date, several models have been proposed to explain how the two flagella are coordinated in amphitrichous bacteria. On the one hand, one motor could be active at a time, each flagellum being dedicated to moving the cell in only one direction. In that case, the rotation of the lagging flagellum would cause the cell body to rotate in the opposite direction, which would in turn trigger the passive rotation of the leading flagellum with the cell body. On the other hand, the two flagellar motors could be active simultaneously. The fact that both flagella can rotate in the same direction during cell tumbling provides unambiguous evidence in support of the latter model.
Asymmetrical rotation of the polar motors points to the asymmetrical distribution of soluble entities in the cell to or structural differences between the molecular motors assembled at the north and south poles. In fact, many cellular processes depend on the asymmetrical localization of proteins, secondary messengers, DNA, and even lipids, and several mechanisms by which these factors localize to the cell poles have been elucidated (21). For example, asymmetrically distributed proteins have already been shown to be essential for controlling cell movement, and for cell reversals in particular, in the gliding bacterium Myxococcus xanthus (1). One possibility would be that one or several chemotaxis proteins would be asymmetrically distributed or phosphorylated in AMB-1 cells, therefore allowing the opposite states of the flagellar motors at the poles during runs. The AMB-1 genome is particularly rich in predicted chemotactic proteins (i.e., 2 putative histidine kinases [CheA], 4 putative signal transducing proteins [CheW], 3 putative phosphatases [CheZ], and no fewer than 32 putative response regulators, which could correspond to as many CheY proteins) which could potentially be responsible for this phenomenon. Yet one cannot rule out the possibility that the two flagellar machineries assembled at the cell poles present slight differences, especially considering the fact that AMB-1 encodes two paralogs of stator proteins MotA and MotB as well as two hook proteins. However, in comparing AMB-1 gene content to that of other amphitrichously flagellated bacteria, we found only one set of stator proteins in the C. jejuni genome or the R. rubrum genome, indicating that the presence of two stators is not a common trait among amphitrichous bacteria. It rather suggests that motility control in these bacteria would also depend on the differential regulation of identical flagellar motors at the poles.
We are providing here accumulating evidence pointing toward a common motility paradigm for spirillum-shaped bipolarly flagellated bacteria, regardless of their phylogeny. This motility mode might be physically the most efficient one for these morphologically similar microorganisms and would have imposed this particular evolution of flagellar operation mechanism across phyla. Considering this, it is likely that this paradigm is also applicable to other amphitrichously flagellated microbes, including other species of magnetotactic bacteria (Magnetospirillum spp. and Magnetospira spp.), pathogenic species (Campylobacter jejuni), or photosynthetic bacteria (Rhodospirillum rubrum).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by l'Agence Nationale de la Recherche (ANR-2010-BLAN-1320-01 to L.-F.W.). D.M. and the Microscopy Core Facility were supported by grants from the Fondation pour la Recherche Médicale (SPF20110421349 and DGE 20101221257, respectively).
We thank Antonin Serrano for creating the AMB-1 pictograms shown in Fig. 2C, 4, and 5B. We thank C. Santini, T. Doan, E. Mauriello, and T. Mignot for critical reading of the manuscript and stimulating discussions.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00172-15.
REFERENCES
- 1.Mauriello EM, Mignot T, Yang Z, Zusman DR. 2010. Gliding motility revisited: how do the myxobacteria move without flagella? Microbiol Mol Biol Rev 74:229–249. doi: 10.1128/MMBR.00043-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wadhams GH, Armitage JP. 2004. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol 5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 3.Samatey FA, Matsunami H, Imada K, Nagashima S, Shaikh TR, Thomas DR, Chen JZ, Derosier DJ, Kitao A, Namba K. 2004. Structure of the bacterial flagellar hook and implication for the molecular universal joint mechanism. Nature 431:1062–1068. doi: 10.1038/nature02997. [DOI] [PubMed] [Google Scholar]
- 4.Armitage JP. 1999. Bacterial tactic responses. Adv Microb Physiol 41:229–289. doi: 10.1016/S0065-2911(08)60168-X. [DOI] [PubMed] [Google Scholar]
- 5.Porter SL, Wadhams GH, Armitage JP. 2011. Signal processing in complex chemotaxis pathways. Nat Rev Microbiol 9:153–165. doi: 10.1038/nrmicro2505. [DOI] [PubMed] [Google Scholar]
- 6.McCarter LL. 2001. Polar flagellar motility of the Vibrionaceae. Microbiol Mol Biol Rev 65:445–462, table of contents. doi: 10.1128/MMBR.65.3.445-462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lertsethtakarn P, Ottemann KM, Hendrixson DR. 2011. Motility and chemotaxis in Campylobacter and Helicobacter. Annu Rev Microbiol 65:389–410. doi: 10.1146/annurev-micro-090110-102908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Popp F, Armitage JP, Schueler D. 14 November 2014, posting date. Polarity of bacterial magnetotaxis is controlled by aerotaxis through a common sensory pathway. Nat Commun doi: 10.1038/ncomms6398. [DOI] [PubMed] [Google Scholar]
- 9.Charon NW, Cockburn A, Li C, Liu J, Miller KA, Miller MR, Motaleb MA, Wolgemuth CW. 2012. The unique paradigm of spirochete motility and chemotaxis. Annu Rev Microbiol 66:349–370. doi: 10.1146/annurev-micro-092611-150145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murat D. 2013. Magnetosomes: how do they stay in shape? J Mol Microbiol Biotechnol 23:81–94. doi: 10.1159/000346655. [DOI] [PubMed] [Google Scholar]
- 11.Bazylinski DA, Frankel RB. 2004. Magnetosome formation in prokaryotes. Nat Rev Microbiol 2:217–230. doi: 10.1038/nrmicro842. [DOI] [PubMed] [Google Scholar]
- 12.Murat D, Byrne M, Komeili A. 2010. Cell biology of prokaryotic organelles. Cold Spring Harb Perspect Biol 2:a000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quinlan A, Murat D, Vali H, Komeili A. 2011. The HtrA/DegP family protease MamE is a bifunctional protein with roles in magnetosome protein localization and magnetite biomineralization. Mol Microbiol 80:1075–1087. doi: 10.1111/j.1365-2958.2011.07631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murat D, Quinlan A, Vali H, Komeili A. 2010. Comprehensive genetic dissection of the magnetosome gene island reveals the step-wise assembly of a prokaryotic organelle. Proc Natl Acad Sci U S A 107:5593–5598. doi: 10.1073/pnas.0914439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lauga E, DiLuzio WR, Whitesides GM, Stone HA. 2006. Swimming in circles: motion of bacteria near solid boundaries. Biophys J 90:400–412. doi: 10.1529/biophysj.105.069401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang WJ, Santini CL, Bernadac A, Ruan J, Zhang SD, Kato T, Li Y, Namba K, Wu LF. 2012. Complex spatial organization and flagellin composition of flagellar propeller from marine magnetotactic ovoid strain MO-1. J Mol Biol 416:558–570. doi: 10.1016/j.jmb.2011.12.065. [DOI] [PubMed] [Google Scholar]
- 17.Turner L, Ryu WS, Berg HC. 2000. Real-time imaging of fluorescent flagellar filaments. J Bacteriol 182:2793–2801. doi: 10.1128/JB.182.10.2793-2801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. 2008. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science 320:1636–1638. doi: 10.1126/science.1157877. [DOI] [PubMed] [Google Scholar]
- 19.R Development Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- 20.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Treuner-Lange A, Sogaard-Andersen L. 2014. Regulation of cell polarity in bacteria. J Cell Biol 206:7–17. doi: 10.1083/jcb.201403136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang S-D, Petersen N, Zhang W-J, Cargou S, Ruan J, Murat D, Santini C-L, Song T, Kato T, Notareschi P, Li Y, Namba K, Gué A-M, Wu L-F. 2014. Swimming behaviour and magnetotaxis function of the marine bacterium strain MO-1. Environ Microbiol Rep 6:14–20. doi: 10.1111/1758-2229.12102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


