Abstract
APOE ε4, the most significant genetic risk factor for Alzheimer disease (AD), may mask effects of other loci. We re-analyzed genome-wide association study (GWAS) data from the International Genomics of Alzheimer’s Project (IGAP) Consortium in APOE ε4+ (10,352 cases and 9,207 controls) and APOE ε4− (7,184 cases and 26,968 controls) subgroups as well as in the total sample testing for interaction between a SNP and APOE ε4 status. Suggestive associations (P<1x10−4) in stage 1 were evaluated in an independent sample (stage 2) containing 4,203 subjects (APOE ε4+: 1,250 cases and 536 controls; APOE ε4-: 718 cases and 1,699 controls). Among APOE ε4− subjects, novel genome-wide significant (GWS) association was observed with 17 SNPs (all between KANSL1 and LRRC37A on chromosome 17 near MAPT) in a meta-analysis of the stage 1 and stage 2 datasets (best SNP, rs2732703, P=5·8x10−9). Conditional analysis revealed that rs2732703 accounted for association signals in the entire 100 kilobase region that includes MAPT. Except for previously identified AD loci showing stronger association in APOE ε4+ subjects (CR1 and CLU) or APOE ε4− subjects (MS4A6A/MS4A4A/ MS4A6E), no other SNPs were significantly associated with AD in a specific APOE genotype subgroup. In addition, the finding in the stage 1 sample that AD risk is significantly influenced by the interaction of APOE with rs1595014 in TMEM106B (P=1·6x10−7) is noteworthy because TMEM106B variants have previously been associated with risk of frontotemporal dementia. Expression quantitative trait locus analysis revealed that rs113986870, one of the GWS SNPs near rs2732703, is significantly associated with four KANSL1 probes that target transcription of the first translated exon and an untranslated exon in hippocampus (P≤1.3x10−8), frontal cortex (P≤1.3x10−9), and temporal cortex (P≤1.2x10−11). Rs113986870 is also strongly associated with a MAPT probe that targets transcription of alternatively spliced exon 3 in frontal cortex (P=9.2x10−6) and temporal cortex (P=2.6x10−6). Our APOE-stratified GWAS is the first to show GWS association for AD with SNPs in the chromosome 17q21.31 region. Replication of this finding in independent samples is needed to verify that SNPs in this region have significantly stronger effects on AD risk in persons lacking APOE ε4 compared to persons carrying this allele, and if this is found to hold, further examination of this region and studies aimed at deciphering the mechanism(s) are warranted.
INTRODUCTION
The common late-onset form of Alzheimer disease (AD) has a strong genetic component,1 a portion of which is explained by APOE and several other genes identified by positional mapping, targeted gene analysis and genome-wide association studies (GWAS).2–4 Together, these loci account for less than one-half of the heritable component in AD susceptibility, of which 20%–25% is due to APOE.4,5 Because many of the known AD loci cluster in biological pathways, including those involved in inflammation, lipid metabolism and processing, and intracellular trafficking of Aβ, there are likely more AD risk loci that are difficult to detect because of very weak effect size, allelic heterogeneity, or rare variants. To examine yet another hypothesis, namely, that associations for some loci may be obscured by confounding or interaction with other loci, we conducted a two-stage GWAS in APOE genotype subgroups using the large resources of the International Genomics of Alzheimer’s Project (IGAP).
METHODS
Study Population
Details of the stage 1 sample from the International Genomics of Alzheimer’s Project (IGAP) Consortium including subject recruitment, genotyping, imputation, quality control, population substructure, and statistical methods for association analyses were previously described.4 In brief, phenotype and genotype data, including APOE genotypes, for a total of 53,711 subjects were assembled by IGAP from the Alzheimer’s Disease Genetic Consortium (ADGC), the Cohorts for Heart and Ageing Research in Genomic Epidemiology (CHARGE) consortium, the European Alzheimer’s Disease Initiative (EADI), and the Genetic and Environmental Risk in Alzheimer’s Disease (GERAD) consortium. Characteristics of this sample are in Supplementary Table S1.
The stage 2 dataset included GWAS and APOE genotype data for 4,203 subjects of European ancestry from the ADC4, ADC5, ADC6, MTV, Pfizer, and TARCC datasets in the ADGC. These individuals were recruited under protocols approved by the appropriate Institutional Review Boards. Details of the individual datasets are provided in the Supplementary Materials and summarized in Supplementary Table S1.
Procedures
QC, Imputation, and Population Substructure in Stage 2 Datasets
Quality control of the clinical and genotype data in these cohorts was performed using procedures described elsewhere.4 SNP genotypes in each stage 2 dataset were imputed with IMPUTE2 using reference haplotypes from the March 2012 release of 1000 Genomes. We compared imputation results for selected variants in the stage 1 datasets using the March 2012 release of 1000 Genomes and prior imputation on the December 2010 release, and found no significant difference in the distribution of genotype probabilities between old and new imputations for the same samples among the original ADGC datasets. We used actual APOE genotypes when available because previously we observed that imputation in this region using the 1000 Genomes reference panel is unreliable.5 Population substructure was evaluated within each dataset by principal components (PC) analysis using EIGENSTRAT (http://www.hsph.harvard.edu/alkes-price/software/) and a subset of 21,109 SNPs common to all genotyping platforms.
Statistical Analysis
Genome-wide Association Study
Within each stage 1 dataset, genome-wide association analyses were conducted separately in subgroups of subjects with and without the APOE ε4 allele using a logistic generalized linear model (GLM) in case-control datasets and a logistic generalized estimating equation (GEE) in family-based datasets. The potential independent effect of the APOE ε2 allele was not examined because of the paucity of carriers of this allele, thus rendering very small cell sizes particularly among AD cases and in smaller datasets. Cox-proportional hazards models were used to evaluate association with incident AD in three CHARGE cohorts. A quantitative estimate between 0 and 2 for the dose of the reference allele for a SNP was used to incorporate the uncertainty of the imputation estimates. Interaction between a SNP and APOE genotype was evaluated in the APOE genotype subgroups combined within each dataset using regression models including age, sex, the first three PCs, and terms for the SNP, APOE ε4 status, and interaction between the SNP and APOE ε4 status. Results for each model across datasets were combined by meta-analysis using the inverse variance method implemented in the software package METAL (http://www.sph.umich.edu/csg/abecasis/Metal/). Effect sizes were weighted by their inverse variance and a combined estimate was calculated by summing the weighted estimates and dividing by the summed weights. SNPs with a minor allele frequency >5% that were available in at least 50% of the datasets were included in the meta-analysis. The meta-analysis P-value for association was estimated by the summarized test statistic, after applying genomic control within each individual study.
Follow-up Analysis in Stage 2 Datasets
SNPs attaining a P-value <10−4 in the stage 1 GWAS were evaluated in each of the stage 2 GWAS datasets, containing a total of 1,786 APOE ε4+ and 2,417 APOE ε4− subjects (Supplementary Table S1), using the same approach described above.
Gene Expression Analysis
The effect of top-ranked SNPs on gene expression was evaluated using an open access database of control brain microarray data (BRAINEAC) made publically available by the UK Human Brain Expression Consortium (http://caprica.genetics.kcl.ac.uk/BRAINEAC). This dataset contains information generated by analysis of tissue samples obtained from 12 different central nervous system regions in 134 individuals. Details of the expression quantitative trait locus (eQTL) analysis are reported elsewhere.6 In this study, the experiment-wise significance threshold for association of a genetic marker with expression was determined to be 1.6x10−7 at the gene level and 1.8x10−6 for individual exons. Potential for functionality of the top-ranked SNPs was assessed using the Regulome database (http://www.regulomedb.org).
RESULTS
We conducted a genome-wide association study for AD using datasets stratified by APOE genotype assembled by IGAP which were from the ADGC, CHARGE consortium, EADI, and GERAD consortium. Meta-analyses were performed separately in APOE ε4+ (10,246 cases and 11,924 controls) and APOE ε4− (7,231 cases and 19,603 controls) subgroups, as well as the total sample using a model including a term for the interaction of the SNP with the APOE ε4 status. There was limited genomic inflation in the GWAS results in the APOE ε4+ (λ=1.05) and APOE ε4− (λ=1.06) groups, but not in the total sample (λ=0.98) testing the ε4 * SNP interaction (Supplementary Figure S1). Genome-wide significant (GWS) association (P<5x10−8) for AD was found in five distinct regions (CR1, BIN1, CLU, PICALM and APOE) in the APOE ε4+ subgroup (Supplementary Figure S2A, Supplementary Table S2) and four distinct regions (BIN1, HBEGF, MS4A6A/MS4A4A, SLC24A4, and APOE) in the APOE ε4− subgroup (Supplementary Figure S2B, Supplementary Table S2). No significant SNP*APOE interactions were found in the total group (Supplementary Figure S2C). Suggestive association (P<10−6) was observed with SNPs in five novel loci in the APOE ε4− subgroup (SOX14/CLDN18, ACSL6, FAM20C, MAPT region, and CDR2L; Supplementary Figure S2B, Supplementary Table S3) and with 21 TMEM106B SNPs (top result: rs1595014, P=1.6x10−7) (Supplementary Figure S2C, Supplementary Table S3).
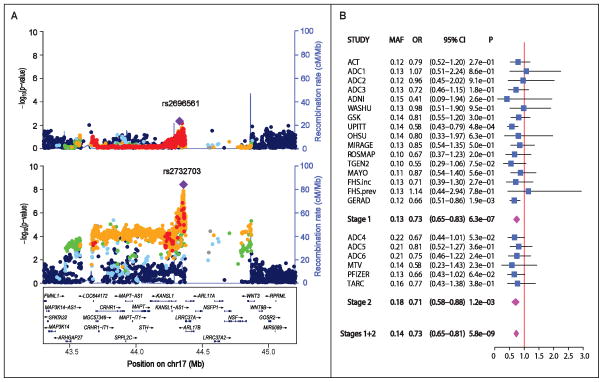
Approximately 1,130 SNPs from 38 regions (including seven previously established AD loci) were tested in Stage 2 (Supplementary Table S3). Follow-up analyses of the novel loci confirmed association with SNPs in CDC42SE2-ACSL6, KANSL1/LRRC37A, and CDR2L in the stage 2 sample (Table 1, Supplementary Table S2), but only SNPs near MAPT and between KANSL1 and LRRC37A (Figure 1A) were genome-wide significant after combining results from the stage 1 and stage 2 samples (best SNP: rs2732703, meta-analysis: P=5.8x10−9). The association was consistent in nearly all datasets which contained rs2732703 information (Figure 1B). To verify the reliability of the association with rs2732703, an imputed SNP, we compared rs2732703 allele dosages obtained directly by genotyping using a Taqman assay with those derived from imputation among 1,010 subjects from the ACT, ADC4, ADC5 and ADC6 datasets. The correlation of these values, 0.813 in the entire sample and 0.834 among APOE ε4− subjects, as well as a genotype misclassification rate of only 3.5% among subjects with imputed probability scores > 0.8 for a particular genotype, suggest that our association findings were not influenced substantially by imputation quality.
Table 1.
Association results (P<10−6) in novel AD loci among APOE ε4− subjects in the combined stage 1 and stage 2 samples.
| SNP | CH | Region or Closest Gene | MA | MAF | Stage 1
|
Stage 2
|
Stages 1 + 2
|
|||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | |||||
| rs16847609 | 3 | SOX14/CLDN18 | A | 0.09 | 1.21 (1.12–1.29) | 2.3x10−7 | 1.09 (0.87–1.37) | 0.47 | 1.19 (1.11–1.28) | 5.3x10−7 |
| rs382216 | 5 | CDC42SE2-ACSL6 | T | 0.36 | 0.88 (0.83–0.93) | 6.5x10−6 | 0.78 (0.67–0.91) | 0.002 | 0.87 (0.82–0.92) | 2.0x10−7 |
| rs11168036 | 5 | PFDN1/HBEGF | T | 0.50 | 1.14 (1.09–1.19) | 9.3x10−9 | 0.97 (0.85–1.11) | 0.64 | 1.12 (1.07–1.17) | 3.2x10−7 |
| rs2732703 | 17 | KANSL1/LRRC37A | G | 0.13 | 0.73 (0.65–0.83) | 6.4x10−7 | 0.71 (0.58–0.88) | 0.001 | 0.73 (0.65–0.81) | 5.8x10−9 |
| rs71380849 | 17 | CDR2L | A | 0.06 | 1.45 (1.24–1.70) | 3.8x10−6 | 1.59 (1.01–2.50) | 0.04 | 1.47 (1.26–1.71) | 9.1x10−7 |
Figure 1.
Association of AD with SNPs in chromosome 17q21.31 in the combined stage 1 and stage 2 samples. (A) Regional Manhattan plot in the APOE ε4+ (upper panel) and the APOE ε4− (lower panel) subgroups. SNPs with the lowest P-value are indicated with a purple diamond. Computed estimates of linkage disequilibrium (r2) of SNPs in this region with the most significant SNP are shown as red circles for r2 ≥ 0.8, orange circles for 0.6 ≤ r2 < 0.8, green circles for 0.4 ≤ r2 < 0.6, light blue circles for 0.2 ≤ r2 < 0.4, and blue circles for r2 < 0.2. Unannotated SNPs are shown as grey circles. (B) Forest plot of association results for rs2732703 in the Stage 1, Stage 2 and total samples among APOE ε4− subjects.
Further examination of this region in the total sample revealed an association peak spanning more than 1.25 Mb that contains 15 genes (Figure 1A). Within this region, 17 SNPs were GWS, have MAFs ranging from 0.13 to 0.17, and are located in a 10.2 kb segment upstream of both KANSL1 and LRRC37A (Supplementary Table S4). Nominally significant association was observed with only one of these SNPs among ε4+ subjects (rs2732703, P=0.02) (Supplementary Table S3). Although the odds ratios (OR) for effect of the effect of minor allele on AD risk were substantially lower for all of the GWS SNPs in the ε4− group (0.54< OR <0.86) than in the ε4+ group (0.76< β <1.04), there was no evidence of interaction with APOE genotype (Supplementary Table S3). The minor alleles of these SNPs reduced AD risk by 20%–37% in the ε4− group. The 350 kb gap in the broad association signal is punctuated at one end by a “cliff” adjacent to the MAPT-KANSL1-LRRC37A association peak (Figure 1). This gap is populated by relatively few SNPs and contains several copy number variation (CNV) polymorphisms.7,8 To explore the possibility that the association observed in the present analysis is explained by previously identified haplotypes H1/H2 in the MAPT region,8 we evaluated six models in the entire dataset conditioning on rs8070723 (an H1/H2 tagging SNP), rs2732703, or rs199533. Rs2732703 remained significant in models conditioning on rs8070723 (P=0.013) or rs199533 (P=0.0020), and rs8070723 was marginally significant in the model conditioning on rs199533 (P=0.043) (Supplementary Table S5, Supplementary Figure S3). These results suggest that KANSL1/LRRC37A is the only AD risk locus in this region.
We also examined the effect of APOE ε4 status on previously established AD loci (Supplementary Table S2). Four of these loci attained genome-wide significance in at least one of the APOE subgroups (Table 2), and the association signal in the MS4A cluster region was evident primarily in the APOE ε4− subgroup (Supplementary Figure S4). The association of AD with CR1, BIN1, and CLU was supported in both APOE subgroups.
Table 2.
Results (P<10−6) in previously known AD loci showing different pattern of association among APOE ε4+ and ε4− subjects in the combined datasets.
| SNP | CH | Region or Closest Gene | MA | MAF |
APOE ε4(+)
|
APOE ε4(−)
|
||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | |||||
| rs679515 | 1 | CR1 | T | 0.21 | 1.22 (1.14 –1.30) | 3.6x10−9 | 1.13 (1.07 – 1.19) | 1.6x10−5 |
| rs4663105 | 2 | BIN1 | C | 0.43 | 1.19 (1.12 – 1.25) | 2.5x10−9 | 1.19 (1.13 – 1.24) | 1.8x10−12 |
| rs9331896 | 8 | CLU | C | 0.38 | 0.84 (0.80 – 0.89) | 2.8x10−9 | 0.90 (0.86 – 0.94) | 9.6x10−6 |
| rs1582763 | 11 | MS4 region | A | 0.37 | 0.92 (0.87 – 0.97) | 0.003 | 0.87 (0.83 – 0.91) | 2.2x10−9 |
CH = chromosome; MA = minor allele; MAF = minor allele frequency.
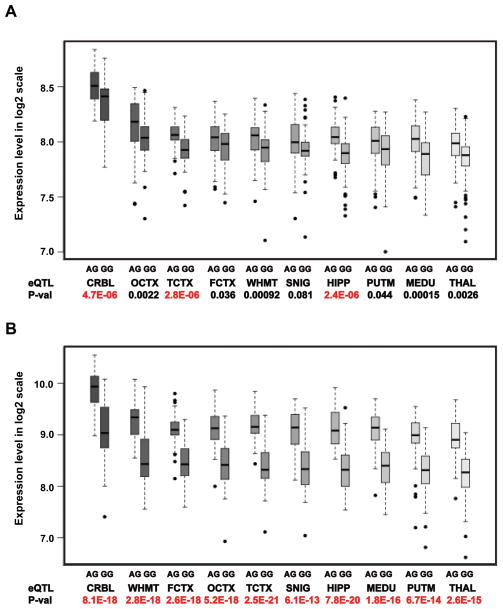
Next, we interrogated the BRAINEAC database to determine whether any of the 17 GWS SNPs located between KANSL1 and LRRC37A are cis-eQTLs. Data were available for only one of these SNPs (rs113986870) which is in high LD with and 2,461 base pairs away from rs2732703 (r2 and D′>0.9). Ten exon probes from four genes (KANSL1, LRRC37A4P, MAPT, and C17orf69) were significantly associated with rs113986870 when averaged across all brain regions (Table 3). Rs113986870 was significantly associated with gene-level expression (Figure 2A) as well as with exon-level expression (Figure 2B) in hippocampus, temporal cortex, and cerebellum. In these brain regions, rs113986870 was significantly associated with KANSL1 probes 3762011, 3762012 and 3762013 that measure expression of the first translated exon. Additionally, we observed that expression of probe 3760518 (Supplementary Figure S5A) present in all three transcripts (NM_001193466, NM_015443, and NM_001193465) and 3760219 in transcript variant 2 (NM_015443) was significantly associated with rs113986870 (Supplementary Figure S5B), while expression of probe 3760217 in transcript variant 1 ((NM_001193466) was not significant (Supplementary Figure S5C), indicating that alternative splicing may be a crucial mechanism for regulating KANSL1 expression. Rs113986870 was also strongly associated with MAPT transcription (Supplementary Figure S6A) and in particular with probe 3723712 that targets transcription of alternatively spliced exon 3 in frontal cortex (P≤9.2x10−6) and temporal cortex (P≤2.6x10−6) (Supplementary Figure S6B). The rs113986870 minor allele (A), which is associated with reduced risk of AD (Supplementary Table S4), increased expression of the target exons in KANSL1 and MAPT (Figure 2, Supplementary Figure S6, Supplementary Figure S7). The association with LRRC37A4P exon probe 3759898 was significant in all three AD-related brain regions (P≤3.6x10−9). The association of rs113986870 with exon probe 3723594 for C17orf69 was significant in hippocampus only (P=1.6x10−7). Five of the GWS SNPs including rs2732703 and rs113986870 are located within a transcription factor binding site or a DNase sensitivity peak and two of these five SNPs, including rs2668626 which is only 47 bp from rs2732703, have also been identified within an eQTL (Supplementary Table S4).
Table 3.
Exon probes covering the region between 43.5 and 45.0 Mb on chromosome 17 that reveal significant rs113986870 allelic expression differences averaged over of 10 brain areas
| Gene | ExprID | Start | End | AVGALL | FCTX | HIPP | TCTX |
|---|---|---|---|---|---|---|---|
| LRRC37A4P | 3759896 | 43583231 | 43583802 | 1.4x10−15 | 6.4x10−4 | 4.0x10−6 | 2.4x10−5 |
| LRRC37A4P | 3759898 | 43584264 | 43584884 | 1.7x10−20 | 8.0x10−11 | 5.3x10−10 | 3.6x10−9 |
| C17orf69 | 3723594 | 43716765 | 43716853 | 3.3x10−13 | 2.0x10−5 | 1.6x10−7 | 8.3x10−5 |
| C17orf69 | 3723604 | 43723359 | 43723556 | 4.9x10−10 | 0.004 | 9.8x10−4 | 1.3x10−5 |
| MAPT | 3723712 | 44051752 | 44051833 | 3.6x10−14 | 9.2x10−6 | 7.6x10−4 | 2.6x10−6 |
| KANSL1 | 3760158 | 44117069 | 44117161 | 9.8x10−14 | 2.8x10−5 | 0.008 | 6.2x10−5 |
| KANSL1 | 3760211 | 44247654 | 44247852 | 4.0x10−23 | 8.0x10−13 | 3.0x10−17 | 1.6x10−15 |
| KANSL1 | 3760212 | 44248224 | 44248977 | 1.4x10−24 | 2.6x10−18 | 7.8x10−20 | 2.5x10−21 |
| KANSL1 | 3760213 | 44249529 | 44249592 | 7.7x10−16 | 3.0x10−11 | 1.1x10−13 | 1.2x10−11 |
| KANSL1 | 3760219 | 44270189 | 44270252 | 4.3x10−13 | 1.3x10−9 | 1.3x10−8 | 1.3x10−11 |
Map position is based on 1000 Genomes database release GRCh37/hg19 assembly, February 2009. Significance threshold after multiple testing determined as 0.05/292,000 exon probes = 1.7x10−7; ExprID: exon-specific probeset ID. AVEALL: average expression levels across 10 regions including cerebellum (CRBL), frontal cortex (FCTX), hippocampus (HIPP), medulla (specifically inferior olivary nucleus, MEDU), occipital cortex (specifically primary visual cortex, OCTX), putamen (PUTM), substantia nigra (SNIG), thalamus (THAL), temporal cortex (TCTX), and intralobular white matter (WHMT).
Figure 2.
Genotype specific effect of the eQTL rs113986870 on expression of KANSL1. (A) Gene-level expression of KANSL1 transcript t3760137. Transcript-level expression represents the average across all KANSL1 exon probe sets. (B) Expression of exon probe 3760212. Probes 3760211, 3760212, and 3760213 measure expression of the first translated exon, are present in all three transcript variants, and were significantly associated with the eQTL. Expression profiles for probes 3760211 and 3760213 showed similar to those for probe 3760212 (Table 3). The distance from 3760212 to rs113986870 is 85,431 base pairs. Log2 scale of expression (Y-axis) is shown for 10 regions of cognitively normal human brains (X-axis) ordered by mean expression level. Rs113986870 genotype counts: AA=0, AG=56, and GG=76. Rs113986870 allele frequencies are 0.21 (A) and 0.79 (G). CRBL = cerebellum, FCTX = frontal cortex, HIPP = hippocampus, MEDU = medulla (specifically inferior olivary nucleus), OCTX = occipital cortex (specifically primary visual cortex), PUTM = putamen, SNIG = substantia nigra (SNIG), THAL = thalamus, TCTX = temporal cortex), WHMT = intralobular white matter.
DISCUSSION
This study was undertaken to identify loci whose effect on AD risk may be obscured by confounding or interaction with APOE genotype. Our APOE-stratified GWAS is the first to show GWS association for AD with SNPs in the chromosome 17q21.31 region including MAPT, KANSL1 and LRRC37A. Among the genes expected to emerge from GWAS but never seen before is MAPT which encodes the microtubule-associated protein tau (MAPT) found in AD neurofibrillary tangles. The association peak is located between KANSL1 and LRRC37A, approximately 200 kb downstream of MAPT, in a subset of subjects that do not possess the APOE ε4 allele. Although the association signal includes MAPT, conditional analysis suggests that the causal variant(s) are more likely located in a DNA segment between the 5′ end of KANSL1 and 5′ end of LRRC37A and not within MAPT or another gene distal to LRRC37A.
The nature of the AD-related functional variant could not be discerned from our genetic association findings. None of the GWS SNPs are within 42.1 kb of the KANSL1 start site or 16.8 kb of the LRRC37A start site, suggesting that the functional variant is not within the promoter region of either gene. KANSL1 is a widely expressed gene encoding a member of the nonspecific lethal (NSL) complex. The KANSL1 protein is an evolutionarily conserved regulator of the chromatin modifier KAT8, which influences gene expression through histone H4 lysine 16 (H4K16) acetylation.9 Notably, mutations in KANSL1 cause the 17q21.31 microdeletion syndrome which is associated with a wide range of abnormalities including intellectual disability and developmental delay, and is therefore thought to be involved in neuronal development.10,11 LRRC37A encodes a member of the leucine-rich repeat containing 37 family. Leucine-rich repeats (LRRs) are protein-ligand interaction motifs found in a large number of proteins with different structure, localization, and function.12 LRR motifs are important for intermolecular or intercellular interactions with exogenous factors in the immune system and/or with different cell types in the developing nervous system.12
However, expression analysis of exon array data in control brain tissue revealed that rs113986870, which is in high LD with the top-ranked SNP (rs2732703) in the GWAS, is an eQTL for expression of the first translated exon in KANSL1 and the alternatively spliced exon 3 in MAPT. Previous studies suggest that splicing of MAPT may be a crucial regulatory mechanism in the brain and tauopathies in particular,13 and that increased expression of exon 3 protects against neurodegeneration.14 Although rs113986870 is apparently not an eQTL for its adjacent gene LRRC37A, it was significantly associated with a closely related gene, LRRC37A4P, in all three AD-related brain regions. These results suggest that rs113986870 may have a potential function as a cis-acting regulatory element for multiple genes in this region. Another confounding feature of this region are copy number variations that in part overlap with the 5′ end of KANSL1 and possibly influence expression.7,8 Thus, it is possible that the exon probes targeting the first translated in KANSL1 may be tagging this duplication. In addition, interrogation of a database curating information about DNA features and regulatory regions revealed that five of the GWS SNPs, including rs2732703 and rs113986870, may have strong regulatory potential.
The association peak for AD on chromosome 17q21.31 is located in a well-recognized and perplexing genomic region containing a 900 kb inversion.8 Previous GWAS identified associations of variants within and at the edges of this inversion with Parkinson disease (PD)15 and progressive supranuclear palsy (PSP),16 but the most significant associations were not with SNPs between KANSL1 and LRRC37A (Supplementary Table S6). Multiple studies have identified more than 40 MAPT deletions, missense mutations, and splice site mutations that cause frontotemporal dementia (FTD).17 Although AD is only nominally associated with common variants in MAPT, previously we observed association of a rare MAPT variant (A152T) with increased risk for FTD and AD in a large sample,18 a finding which was supported by a subsequent smaller study.19 Ikram et al identified a GWS association peak with a KANSL1 SNP approximately 166 kb away from our most significant AD SNP (rs2732703) for a continuous measure of intracranial volume in a sample of nearly 10,000 community-dwelling elders (Supplementary Table S6).20 These two SNPs are moderately correlated (r2=0.71) which indicates that they may tag the same functional variant.
Other studies have focused on two divergent extended MAPT haplotypes, H1 and H2, which are in near complete LD with status of the inversion and contain independently derived partial duplications of KANSL1.8,16 The common H1 haplotype is associated with increased risk of FTD,21 PD,22 PSP,23 and corticobasal degeneration (CBD),23 while H2 is linked to recurrent deletion events associated with the 17q21.31 microdeletion syndrome.10 Among these non-AD forms of dementia, it is possible for FTD to masquerade clinically as AD and thereby cases of FTD could be present in our study group; however, any inadvertent inclusion of FTD cases is expected to be very small since the minimum age of dementia onset in our study group was 60 years and onset of dementia from FTD after age 69 years is relatively rare compared to AD that in most cases occurs after age 69.24 Furthermore, a recent review of almost 5000 autopsy brains from a subset of cases in the ADGC cohort failed to identify any case of FTD.25 Myers et al. reported association of AD with H1 and with common MAPT SNPs,26 but this association is controversial27 and did not reach genomewide significance in our study or previous GWAS. Another recent study showed that carriers of at least one H2 allele had a 5.4-fold increased risk of worsening hallucinations, but this result was marginally significant.28 Previously, we observed in a subset of the sample studied here that the H2-haplotype tagging rs8070723-G allele was associated with reduced risk of AD.29 However, this variant is no longer associated after conditioning on rs2732703 (Supplementary Table S5). In carriers of H2, the ancestral haplotype in both humans and chimpanzees,30 increased expression of exon 3 in MAPT has been associated with an eQTL located approximately 1,500 bp from rs113986870 which decreases aggregation of microtubules.31,32 These observations are consistent with our results showing that the rs113986870 minor allele is protective for AD and associated with elevated exon3 expression.
There is a large body of experimental evidence linking tau protein to AD pathogenesis,33 and some studies show evidence of association of AD with common MAPT SNPs.29,34 However, analysis of the MAPT coding sequence did not reveal disease-causing variants for early-onset AD 35 and other studies examining association of MAPT SNPs with late-onset AD were negative.27,36 Recently, Allen et al. reported that the rs8070723-G allele was associated with reduced MAPT expression in the cerebellum and temporal cortex of AD subjects.29 Robust genetic associations have also been identified for AD with several genes in cytoskeletal and axonal transport pathways including tau or leading to neurofibrillary tangles, most notably BIN1, EPHA1, RIN3, CASS4, and FERMT2.4
Based on the observation that overexpression of human ApoE4 in transgenic mouse neurons results in hyperphosphorylation of tau,37 it is possible that associations with AD-related loci in the chromosome 17q21.31 region are obscured by the much stronger effect of APOE ε4 on MAPT expression or function.38 This idea is consistent with lack of GWS association with 17q21.31 SNPs in the same dataset without stratification by APOE genotype,4 and no evidence for interaction between APOE and any SNPs in the MAPT-KANSL1-LRRC37A region in the current study. Another possible explanation for the significant association of 17q21.31 SNPs with AD only among subjects lacking APOE ε4 is genetic heterogeneity suggesting that variation at the chromosome 17q21.31 locus is associated with a distinct etiological subtype of AD where tau is the primary disease activator.39 Finally, the diagnosis of AD for most subjects in this dataset was established clinically suggesting the possibility of misdiagnosis or AD accompanied by other processes associated with other dementing illnesses. Further studies are needed to determine whether this subtype can be distinguished clinically or neuropathologically.
Our study also showed that the previously established association with the MS4A gene cluster is derived almost completely from subjects lacking APOE ε4, suggesting the contribution of the MS4A locus to AD may be mechanistically different than AD-related processes that are associated with APOE ε4. Members of the MS4A gene family encode membrane proteins, some of which have known roles in immune cell function,40 however, little is known about the function of MS4A6A, MS4A4A or MS4A6E in humans. Karch et al. showed that expression of MS4A6A was upregulated in AD brains of AD patients compared to brains of controls, and significantly correlated with AD status, AIF1 expression (a marker for microglia which is the immune cell of the brain), cognitive dementia rating score, and extent of AD neuropathologic change.41
The observed statistical interaction of genotypes for TMEM106B with APOE on AD risk in the stage 1 GWAS is noteworthy (rs1595014, P=1.6x10−7) even though it is not supported by results in the comparatively small stage 2 sample. TMEM106B is a glycoprotein predominantly localized at the lysosomal membrane where it might interact with intracellular progranulin (GRN).42,43 TMEM106B variants, particularly the p. T185S (rs3173615) mutation, are risk factors for FTD, especially among persons carrying a GRN mutation.44 TMEM106B variants are also associated with development of cognitive impairment in amyotrophic lateral sclerosis45 and implicated in the pathologic presentation of AD.46 Cruchaga et al observed association of the TMEM106B SNP rs1990622 risk allele with younger onset of the FTLD subtype with TAR DNA-binding protein inclusions (FTLD-TDP),47 a pattern reminiscent of the association of APOE ε4 with increased risk and younger onset of AD. The biological underpinning of the interaction of TMEM106B with APOE affecting AD risk is unclear.
Our top findings, including those that are genome-wide significant, should be confirmed in independent samples. Functional studies will be needed to understand the relationship between APOE and the causative variant(s) in 17q21.31 once they are identified, as well as with other loci showing much stronger association with AD in particular APOE genotype strata (e.g., MS4A6A/MS4A4A/ MS4A6E) or through interaction with APOE (e.g., TMEM106B). Our study provides a firm genetic connection of AD to several other pathologically distinct disorders in which dementia is a cardinal or common characteristic.
Supplementary Material
Acknowledgments
ADGC
The National Institutes of Health, National Institute on Aging (NIH-NIA) supported this work through the following grants: ADGC, U01 AG032984, RC2 AG036528; NACC, U01 AG016976; NCRAD, U24 AG021886; NIA LOAD, U24 AG026395, U24 AG026390; MIRAGE: R01 AG025259; Banner Sun Health Research Institute P30 AG019610; Boston University, P30 AG013846, U01 AG10483, R01 CA129769, R01 MH080295, R01 AG017173, R01AG33193; Columbia University, P50 AG008702, R37 AG015473; Duke University, P30 AG028377, AG05128; Emory University, AG025688; Group Health Research Institute, UO1 AG06781, UO1 HG004610; U01 HG006375; Indiana University, P30 AG10133; Johns Hopkins University, P50 AG005146, R01 AG020688; Massachusetts General Hospital, P50 AG005134; Mayo Clinic, P50 AG016574; Mount Sinai School of Medicine, P50 AG005138, P01 AG002219; New York University, P30 AG08051, MO1RR00096, and UL1 RR029893; Northwestern University, P30 AG013854; Oregon Health & Science University, P30 AG008017, R01 AG026916; Rush University, P30 AG010161, R01 AG019085, R01 AG15819, R01 AG17917, R01 AG30146; TGen, R01 NS059873; University of Alabama at Birmingham, P50 AG016582, UL1RR02777; University of Arizona, R01 AG031581; University of California, Davis, P30 AG010129; University of California, Irvine, P50 AG016573, P50, P50 AG016575, P50 AG016576, P50 AG016577; University of California, Los Angeles, P50 AG016570; University of California, San Diego, P50 AG005131; University of California, San Francisco, P50 AG023501, P01 AG019724; University of Kentucky, P30 AG028383; University of Michigan, P50 AG008671; University of Pennsylvania, P30 AG010124; University of Pittsburgh, P50 AG005133, AG030653; University of Southern California, P50 AG005142; University of Texas Southwestern, P30 AG012300; University of Miami, R01 AG027944, AG010491, AG027944, AG021547, AG019757; University of Washington, P50 AG005136; Vanderbilt University, R01 AG019085; and Washington University, P50 AG005681, P01 AG03991. The Kathleen Price Bryan Brain Bank at Duke University Medical Center is funded by NINDS grant # NS39764, NIMH MH60451 and by Glaxo Smith Kline. Genotyping of the TGEN2 cohort was supported by Kronos Science. The TGen series was also funded by NIA grant AG034504 to AJM, The Banner Alzheimer’s Foundation, The Johnnie B. Byrd Sr. Alzheimer’s Institute, the Medical Research Council, and the state of Arizona and also includes samples from the following sites: Newcastle Brain Tissue Resource (funding via the Medical Research Council, local NHS trusts and Newcastle University), MRC London Brain Bank for Neurodegenerative Diseases (funding via the Medical Research Council), SouthWest Dementia Brain Bank (funding via numerous sources including the Higher Education Funding Council for England (HEFCE), Alzheimer’s Research Trust (ART), BRACE as well as North Bristol NHS Trust Research and Innovation Department and DeNDRoN), The Netherlands Brain Bank (funding via numerous sources including Stichting MS Research, Brain Net Europe, Hersenstichting Nederland Breinbrekend Werk, International Parkinson Fonds, Internationale Stiching Alzheimer Onderzoek), Institut de Neuropatologia, Servei Anatomia Patologica, Universitat de Barcelona. Marcelle Morrison-Bogorad, PhD., Tony Phelps, PhD and Walter Kukull PhD are thanked for helping to co-ordinate this collection. ADNI Funding for ADNI is through the Northern California Institute for Research and Education by grants from Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson and Johnson, Eli Lilly and Co., Medpace, Inc., Merck and Co., Inc., Novartis AG, Pfizer Inc, F. Homan-La Roche, Schering-Plough, Synarc, Inc., Alzheimer’s Association, Alzheimer’s Drug Discovery Foundation, the Dana Foundation, and by the National Institute of Biomedical Imaging and Bioengineering and NIA grants U01 AG024904, RC2 AG036535, K01 AG030514. We thank Drs. D. Stephen Snyder and Marilyn Miller from NIA who are ex-officio ADGC members. Support was also from the Alzheimer’s Association (LAF, IIRG-08-89720; MP-V, IIRG-05-14147) and the US Department of Veterans Affairs Administration, Office of Research and Development, Biomedical Laboratory Research Program. P.S.G.-H. is supported by the Wellcome Trust, Howard Hughes Medical Institute, and Canadian Institute of Health
CHARGE
AGES: The AGES-Reykjavik Study is funded by NIH contract N01-AG-12100 (NIA with contributions from the NEI, NIDCD and NHLBI), the NIA Intramural Research Program, Hjartavernd (the Icelandic Heart Association), and the Althingi (the Icelandic Parliament).
ASPS/PRODEM: The Austrian Stroke Prevention Study and The Prospective Dementia Register of the Austrian Alzheimer Society was supported by The Austrian Science Fond (FWF) grant number P20545-P05 (H. Schmidt) and P13180; The Austrian Alzheimer Society; The Medical University of Graz.
Cardiovascular Health Study (CHS): This research was supported by NHLBI contracts HHSN268201200036C, HHSN268200800007C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086; and NHLBI grants U01HL080295, R01HL087652, R01HL105756, R01HL103612, and R01HL120393 with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through R01AG023629, R01AG15928, R01AG20098, R01AG027058 and R01AG033193 (Seshadri) from the National Institute on Aging (NIA). A full list of principal CHS investigators and institutions can be found at CHS-NHLBI.org. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center.
Framingham Heart Study (FHS): This work was supported by the National Heart, Lung and Blood Institute’s Framingham Heart Study (Contract No. N01-HC-25195) and its contract with Affymetrix, Inc for genotyping services (Contract No. N02-HL-6-4278). A portion of this research utilized the Linux Cluster for Genetic Analysis (LinGAII) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. This study as also supported by grants from the National Institute on Aging: AG08122 and AG033193 (Seshadri). Drs. Seshadri and DeStefano were also supported by additional grants from the National Institute on Aging: (R01 AG16495; AG031287, AG033040), the National Institute of Neurological Disorders and Stroke (R01 NS17950), and the National Heart, Lung and Blood Institute (U01 HL096917, HL093029 and K24HL038444, RC2-HL102419 and UC2 HL103010.
Fundació ACE would like to thank patients and controls who participated in this project. This work has been funded by the Fundación Alzheimur (Murcia), the Ministerio de Educación y Ciencia (PCT-010000-2007-18), (DEX-580000-2008-4), (Gobierno de España), Corporación Tecnológica de Andalucía (08/211) and Agencia IDEA (841318) (Consejería de Innovación, Junta de Andalucía). We thank to Ms. Trinitat Port-Carbó and her family for their generous support of Fundació ACE research programs.
Erasmus Rucphen Family Study: We thank the participants from the Genetic Research in Isolated Populations in the Erasmus Rucphen Family Study who made this work possible. This study is financially supported by the Netherlands Organisation for Scientific Research (NWO), the Internationale Stichting Alzheimer Onderzoek (ISAO), the Hersenstichting Nederland (HSN) and the Centre for Medical Systems Biology (CMSB1 and CMSB2) in the framework of the Netherlands Genomics Initiative (NGI).
The Rotterdam Study: The Rotterdam Study was funded by Erasmus Medical Center and Erasmus University, Rotterdam; the Netherlands Organization for Health Research and Development; the Research Institute for Diseases in the Elderly; the Ministry of Education, Culture and Science; the Ministry for Health, Welfare and Sports; the European Commission; and the Municipality of Rotterdam; by grants from the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), Internationale Stichting Alzheimer Onderzoek, Hersenstichting Nederland, the Netherlands Genomics Initiative–Netherlands Organization for Scientific Research (Center for Medical Systems Biology and the Netherlands Consortium for Healthy Aging), the Seventh Framework Program (FP7/2007-2013), the ENGAGE project (grant agreement HEALTH-F4-2007-201413), MRACE-grant from the Erasmus Medical Center, the Netherlands Organization for Health Research and Development (ZonMW Veni-grant no. 916.13.054).
ARIC: The Atherosclerosis Risk in Communities Study (ARIC) is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, N01-HC-55022 and grants R01-HL087641, RC2-HL102419 (Boerwinkle, CHARGE-S), UC2 HL103010, U01 HL096917 (Mosley) and R01-HL093029; NHGRI contract U01-HG004402; and NIH contract HHSN268200625226C and NIA: R01 AG033193 (Seshadri). Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research.
EADI
This work was supported by the National Foundation for Alzheimer’s disease and related disorders, the Institut Pasteur de Lille and the Centre National de Génotypage. The Three-City Study was performed as part of a collaboration between the Institut National de la Santé et de la Recherche Médicale (Inserm), the Victor Segalen Bordeaux II University and Sanofi-Synthélabo. The Fondation pour la Recherche Médicale funded the preparation and initiation of the study. The 3C Study was also funded by the Caisse Nationale Maladie des Travailleurs Salariés, Direction Générale de la Santé, MGEN, Institut de laLongévité, Agence Française de Sécurité Sanitaire des Produits de Santé, the Aquitaine and Bourgogne RegionalCouncils, Fondation de France and the joint French Ministry of Research/INSERM “Cohortes et collections de données biologiques” programme. Lille Génopôle received an unconditional grant from Eisai.
Belgium sample collection: Research at the Antwerp site is funded in part by the Interuniversity Attraction Poles program of the Belgian Science Policy Office, the Foundation for Alzheimer Research (SAO-FRA), a Methusalem Excellence Grant of the Flemish Government, the Research Foundation Flanders (FWO), the Special Research Fund of the University of Antwerp, Belgium. KB is a postdoctoral fellow of the FWO. The Antwerp site authors thank the personnel of the VIB Genetic Service Facility, the Biobank of the Institute Born-Bunge and the Departments of Neurology and Memory Clinics at the Hospital Network Antwerp and the University Hospitals Leuven.
Finish sample collection: Financial support for this project was provided by the Health Research Council of the Academy of Finland, EVO grant 5772708 of Kuopio University Hospital, and the Nordic Centre of Excellence in Neurodegeneration.
Italian sample collections: the Bologna site (FL) obtained funds from the Italian Ministry of research and University as well as Carimonte Foundation. The Florence site was supported by a grant from the Italian ministry of Health (RFPS-2006-7-334858) and grant RF-2010-2319722. The Milan site was supported by a grant from the “fondazione Monzino”. We thank the expert contribution of Mr. Carmelo Romano. The Roma site received financial support from Italian Ministry of Health, Grant RF07-08 and RC08-09-10-11-12. The Pisa site is grateful to Dr. Annalisa LoGerfo for her technical assistance in the DNA purification studies.
Spanish sample collection: the Madrid site (MB) was supported by grants of the Ministerio de Educación y Ciencia and the Ministerio de Sanidad y Consumo (Instituto de Salud Carlos III), and an institutional grant of the FundaciónRamón Areces to the CBMSO. We thank I. Sastre and Dr. A Martínez-García for the preparation and control of the DNA collection, and Drs. P. Gil and P. Coria for their cooperation in the cases/controls recruitment. We are grateful to the Asociación de Familiares de Alzheimer de Madrid (AFAL) for continuous encouragement and help. Swedish sample collection : Financially supported in part by the Swedish Brain Power network, the Marianne and Marcus Wallenberg Foundation, the Swedish Research Council (521-2010-3134), the King Gustaf V and Queen Victoria’s Foundation of Freemasons, the Regional Agreement on Medical Training and Clinical Research (ALF) between Stockholm County Council and the Karolinska Institutet, the Swedish Brain Foundation and the Swedish Alzheimer Foundation.
GERAD
Cardiff University was supported by the Wellcome Trust, Medical Research Council (MRC), Alzheimer’s Research UK (ARUK) and the Welsh Assembly Government. Cambridge University and Kings College London acknowledge support from the MRC. ARUK supported sample collections at the South West Dementia Bank and the Universities of Nottingham, Manchester and Belfast. The Belfast group acknowledges support from the Alzheimer’s Society, Ulster Garden Villages, N. Ireland R&D Office and the Royal College of Physicians/Dunhill Medical Trust. The MRC and Mercer’s Institute for Research on Ageing supported the Trinity College group. The South West Dementia Brain Bank acknowledges support from Bristol Research into Alzheimer’s and Care of the Elderly. The Charles Wolfson Charitable Trust supported the OPTIMA group. Washington University was funded by NIH grants, Barnes Jewish Foundation and the Charles and Joanne Knight Alzheimer’s Research Initiative. Patient recruitment for the MRC Prion Unit/UCL Department of Neurodegenerative Disease collection was supported by the UCLH/UCL Biomedical Centre and Queen Square Dementia Biomedical Research Unit. LASER-AD was funded by Lundbeck SA. The Bonn group was supported by the German Federal Ministry of Education and Research (BMBF), Competence Network Dementia and Competence Network Degenerative Dementia, and by the Alfried Krupp von Bohlen und Halbach-Stiftung. The GERAD1 Consortium also used samples ascertained by the NIMH AD Genetics Initiative.
International Genomics of Alzheimer’s Project Members
Alzheimer Disease Genetics Consortium (ADGC)
Perrie M. Adams, PhD; Marilyn S. Albert, PhD; Roger L. Albin, MD; Liana G. Apostolova, MD; Steven E. Arnold, MD; Sanjay Asthana, MD; Craig S. Atwood, PhD; Clinton T. Baldwin, PhD; Michjael M. Barmada, PhD; Lisa L. Barnes, PhD; Thomas G. Beach, MD PhD; James T. Becker, PhD; Eileen H. Bigio, MD; Thomas D. Bird, MD; Deborah Blacker, MD; Bradley F. Boeve, MD; James D. Bowen, MD; Adam Boxer, MD PhD, James R. Burke, MD PhD; Nigel J. Cairns, PhD FRCPath; Chuanhai Cao, PhD; Chris S. Carlson; PhD; Cynthia M. Carlsson, MD; Regina M. Carney, MD; Minerva M. Carrasquillo, PhD; Steven L. Carroll, MD PhD; Helena C. Chui, MD; David G. Clark, MD; Jason Corneveaux, BS; David H. Cribbs, PhD; Elizabeth A. Crocco, MD; Carlos Cruchaga, PhD; Philip L. De Jager, MD PhD; Charles DeCarli, MD; Steven T. DeKosky, MD; F. Yesim Demirci, MD; Malcolm Dick, PhD; Dennis W. Dickson, MD; Rachelle S. Doody, MD PhD; Ranjan Duara, MD; Nilufer Ertekin-Taner, MD PhD; Kelley M. Faber, MS; Thomas J. Fairchild, PhD; Kenneth B. Fallon, MD; Martin R. Farlow, MD; Steven Ferris, PhD; Matthew P. Frosch, MD PhD; Douglas R. Galasko, MD; Marla Gearing, PhD; Daniel H. Geschwind, MD PhD; Bernardino Ghetti, MD; John R. Gilbert PhD; Jonathan D. Glass, MD; Neill R. Graff-Radford, MD; Robert C. Green, MD MPH; John H. Growdon, MD; Hakon Hakonarson, MD PhD; Ronald L. Hamilton, MD; John Hardy, PhD; Lindy E. Harrell, MD PhD; Elizabeth Head, PhD; Lawrence S. Honig, MD PhD; Ryan M. Huebinger, PhD, Matthew J. Huentelman, PhD; Christine M. Hulette, MD; Bradley T. Hyman, MD PhD; Gail P. Jarvik, MD PhD; Gregory A. Jicha, MD PhD; Lee-Way Jin, MD PhD; Anna Karydas, BA; John S.K. Kauwe, PhD; Jeffrey A. Kaye, MD; Ronald Kim, MD; Edward H. Koo, MD; Neil W. Kowall, MD; Joel H. Kramer, PsyD; Frank M. LaFerla, PhD; James J. Lah, MD PhD; James B. Leverenz, MD; Allan I. Levey, MD PhD; Ge Li, MD PhD; Andrew P. Lieberman, MD PhD; Chiao-Feng Lin, PhD; Oscar L. Lopez, MD; Constantine G. Lyketsos, MD MHS; Wendy J. Mack, PhD; Daniel C. Marson, JD PhD; Frank Martiniuk, PhD; Deborah C. Mash, PhD; Eliezer Masliah, MD; Wayne C. McCormick, MD MPH; Susan M. McCurry, PhD; Andrew N. McDavid, BA; Ann C. McKee, MD; Marsel Mesulam, MD; Bruce L. Miller, MD; Carol A. Miller, MD; Joshua W. Miller, PhD; John C. Morris, MD; Shubhabrata Mukherjee, PhD; Jill R. Murrell, PhD, Amanda J. Myers, PhD; Sid O’Bryant, PhD; John M. Olichney, MD; Vernon S. Pankratz, PhD; Joseph E. Parisi, MD; Amanda Partch, MS; Henry L. Paulson, MD PhD; William Perry, MPH; Elaine Peskind, MD; Ronald C. Petersen, MD PhD; Aimee Pierce, MD; Wayne W. Poon, PhD; Huntington Potter, PhD; Joseph F. Quinn, MD; Ashok Raj, MD; Murray Raskind, MD; Barry Reisberg, MD; Joan S. Reisch, PhD; Christiane Reitz, MD PhD; John M. Ringman; MD; Erik D. Roberson, MD PhD; Ekaterina Rogaeva, PhD; Howard J. Rosen, MD; Roger N. Rosenberg, MD; Donald R. Royall, MD; Mark A. Sager, MD; Mary Sano, PhD; Andrew J. Saykin, PsyD; Julie A. Schneider, MD; Lon S. Schneider, MD; William W. Seeley, MD; Amanda G. Smith, MD; Joshua A. Sonnen, MD; Salvatore Spina, MD; Robert A. Stern, PhD; Rudolph E. Tanzi, PhD; Tricia A. Thornton-Wells, PhD; John Q. Trojanowski, MD PhD; Juan C. Troncoso, MD; Debby W. Tsuang, MD; Vivianna M. Van Deerlin, MD PhD; Linda J. Van Eldik, PhD; Badri N. Vardarajan, Ph.D.; Harry V. Vinters, MD; Jean Paul Vonsattel, MD; Sandra Weintraub, PhD; Kathleen A. Welsh-Bohmer, PhD; Jennifer Williamson, MS; Sarah Wishnek, MPH; Randall L. Woltjer, MD PhD; Clinton B. Wright, MD MS; Chuang-Kuo Wu, MD PhD; Chang-En Yu, PhD; Lei Yu, PhD
Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium
Rhoda Au, PhD; Philip A. Wolf, MD; Alexa Beiser, PhD; Claudia Satizabal, PhD; Andre G. Uitterlinden, PhD; Fernando Rivadeneira, MD, PhD; Peter J. Koudstaal, MD, PhD; William T. Longstreth, Jr, MD; James T. Becker, PhD; Lewis H. Kuller, MD; Thomas Lumley, PhD; Kenneth Rice, PhD; Tamara B. Harris, MD; Michael Nalls, PhD; Josef J.M. Marksteiner, MD; Peter Dal-Bianco, MD; Anna Maria Töglhofer, BSc; Paul Freudenberger, MSc; Gerhard Ransmayr, MD; Thomas Benke, MD; Anna M. Toeglhofer, MSc; Eric Boerwinkle, PhD; Jan Bressler, PhD; Myriam Fornage, PhD; Francisco J. Morón, PhD; Isabel Hernández, MD; Maitee Rosende Roca, MD; Ana Mauleón, MD; Montserrat Alegret, PhD; Reposo Ramírez-Lorca, PhD; Antonio González-Perez, MS
European Alzheimer’s Disease Initiative (EADI)
Annick, Alpérovitch, MD, PhD; Victoria Alvarez, PhD; Pascale Barberger-Gateau, MD; Karolien Karolien, PhD; Paola Bossù, MD; Alexis Brice, MD, PhD; Maria Bullido, MD; Paolo Caffara, MD; Jordi Clarimon, MD; Onofre Combarros, MD; Eliecer Coto, PhD; Maria del Zampo, PhD; Marc Delepine, PhD; Maria Candida Deniz Naranjo, PhD; Jacques Epelbaum, PhD; Laura Fratiglioni, MD, PhD; Daniela Galimberti, PhD; Caroline Graff, MD, PhD; Mikko Hiltunen, MD, PhD; Martin Ingelsson. MD, PhD; Lina Keller, MD; Lars Lannfelt, MD; Alberto Llèo, MD; Michelangelo Mancuso, MD; Ignacio Mateo, MD; Patrizia Mecocci, MD; Benedetta Nacmias, PhD; Francesco Panza, MD; Alberto Pilotto, MD; Florentino Sanchez Garcia, MD; Elio Scarpini, MD; Davide Seripa, PhD; Kristel Sleegers, PhD; Hlikka Soininen, MD; Sandro Sorbi, MD; Gianfranco Spalletta, MD; David Wallon, MD, PhD
Genetic and Environmental Risk in Alzheimer’s Disease (GERAD) Consortium
Charlene Thomas, BSc; Amy Gerrish, PhD; Jade Chapman, PhD; Alexandra Stretton, PhD; Angharad Morgan; Harriet Oldham, PhD; Michael J Owen, MD, PhD; Patrick G Kehoe, PhD; Christopher Medway, PhD; Kristelle Brown, PhD; Jenny Lord; James Turton; Nigel M Hooper; Emma Vardy; Jason D Warren; Jonathan M Schott; James Uphill; Paul Hollingworth, PhD, DClinPsy; Natalie Ryan; Martin Rossor, MD PhD; John Collinge, MD, PhD; Yoav Ben-Shlomo; Daniilidou Makrina; Olymbia Gkatzima; Michelle Lupton, PhD; Maria Koutroumani; Despoina Avramidou; Antonia Germanou; Frank Jessen, PhD; Steffi Riedel-Heller; Martin Dichgans, MD, PhD; Reiner Heun; Heike Kölsch; Britta Schürmann, PhD; Christine Herold; André Lacour; Dmitriy Drichel; Per Hoffmann; Johannes Kornhuber, MD, PhD; Wei Gu; Thomas Feulner; Manuel Mayhaus, PhD; Sabrina Pichler, PhD; Matthias Riemenschneider, MD; Hendrik van den Bussche, PhD; Brian Lawlor, MD, PhD; Aoibhinn Lynch, MD; David Mann, PhD; A. David Smith, DPhil; Donald Warden; Gordon Wilcock; Isabella Heuser, PhD; Jens Wiltfang, MD, PhD; Lutz Frölich, PhD; Michael Hüll, MD, PhD; Kevin Mayo, PhD; Gill Livingston, MD, PhD; Nicholas J Bass, MD, PhD; Hugh Gurling, MD, PhD; Andrew McQuillin, PhD; Rhian Gwilliam; Panagiotis Deloukas; Ammar Al-Chalabi, MB ChB; Christopher E Shaw, MD, PhD; Andrew B Singleton, PhD; Rita Guerreiro, PhD; Giancarlo Russo; Karl-Heinz Jöckel, PhD; Susanne Moebus, PhD; Norman Klopp, PhD; H-Erich Wichmann, MD, PhD; Dennis W Dickson; Neill R Graff-Radford; Li Ma; Gina Bisceglio; Elizabeth Fisher, PhD; Nick Warner MB ChB; Stuart Pickering-Brown, PhD; David Craig, MD; Janet A Johnston, PhD; Bernadette McGuinness, MD, PhD; Stephen Todd, MD, PhD; David C Rubinsztein, MB ChB, PhD; Simon Lovestone, PhD; Anthony Bayer, MD; John Gallacher, MD; Petroula Proitsi, PhD; Sara Ortega-Cubero, MD
References
- 1.Gatz M, Reynolds CA, Fratiglioni L, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63:168–174. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- 2.Farrer LA, Cupples LA, Haines JL, et al. Effects of age, gender and ethnicity on the association of apolipoprotein E genotype and Alzheimer disease. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 3.Sherva R, Farrer LA. Power and pitfalls of the genome wide association study approach to identify genes for Alzheimer disease. Cur Psych Rep. 2011;13:138–146. doi: 10.1007/s11920-011-0184-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambert J-C, Ibrahim-Verbaas CA, Harold D, et al. Extended meta-analysis of 74,538 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jun G, Vardarajan BN, Buros J, et al. A comprehensive search for Alzheimer disease susceptibility loci in the APOE region. Arch Neurol. 2012;69:1270–1279. doi: 10.1001/archneurol.2012.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trabzuni D1, Ryten M, Walker R, et al. Quality control parameters on a large dataset of regionally dissected human control brains for whole genome expression studies. J Neurochem. 2011;119:275–282. doi: 10.1111/j.1471-4159.2011.07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steinberg KM, Antonacci F, Sudmant PH, et al. Structural diversity and African origin of the 17q21. 31 inversion polymorphism. Nat Genet. 2012;44:872–880. doi: 10.1038/ng.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boettger LM, Handsaker RE, Zody MC, McCarroll SA. Structural haplotypes and recent evolution of the human 17q21. 31 region. Nat Genet. 2012;44:881–885. doi: 10.1038/ng.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X, Wu L, Corsa CA, Kunkel S, Dou Y. Two mammalian MOF complexes regulate transcription activation by distinct mechanisms. Mol Cell. 2009;36:290–300. doi: 10.1016/j.molcel.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koolen DA, Kramer JM, Neveling K, et al. Mutations in the chromatin modifier gene KANSL1 cause the 17q21. 31 microdeletion syndrome. Nat Genet. 2012;44:639–641. doi: 10.1038/ng.2262. [DOI] [PubMed] [Google Scholar]
- 11.Zollino M, Orteschi D, Murdolo M, et al. Mutations in KANSL1 cause the 17q21. 31 microdeletion syndrome phenotype. Nat Genet. 2012;44:636–638. doi: 10.1038/ng.2257. [DOI] [PubMed] [Google Scholar]
- 12.Giannuzzi G, Siswara P, Malig M, et al. Evolutionary dynamism of the primate LRRC37 gene family. Genome Res. 2013;23:46–59. doi: 10.1101/gr.138842.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trabzuni D, Wray S, Vandrovcova J, et al. MAPT expression and splicing is differentially regulated by brain region: relation to genotype and implication for tauopathies. Hum Mol Genet. 2012;21:4094–4103. doi: 10.1093/hmg/dds238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caffrey TM, Joachim C, Wade-Martins R. Haplotype-specific expression of the N-terminal exons 2 and 3 at the human MAPT locus. Neurobiol Aging. 2008;29:1923–1929. doi: 10.1016/j.neurobiolaging.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simón-Sánchez J, Schulte C, Bras JM, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet. 2009;41:1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Höglinger GU, Melhem NM, Dickson DW, et al. Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat Genet. 2011;43:699–705. doi: 10.1038/ng.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferrari R, Hardy J, Momeni P. Frontotemporal dementia: from Mendelian genetics towards genome wide association. J Mol Neurosci. 2011;45:500–515. doi: 10.1007/s12031-011-9635-y. [DOI] [PubMed] [Google Scholar]
- 18.Coppola G, Chinnathambi S, Lee JJ, et al. Evidence for a role of the rare p. A152T variant in MAPT in increasing the risk for FTD-spectrum and Alzheimer’s diseases. Hum Mol Genet. 2012;21:3500–3512. doi: 10.1093/hmg/dds161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee SE, Tartaglia MC, Yener G, et al. Neurodegenerative disease phenotypes in carriers of MAPT p. A152T, a risk factor for frontotemporal dementia spectrum disorders and Alzheimer disease. Alzheimer Dis Assoc Disord. 2013;27:302–309. doi: 10.1097/WAD.0b013e31828cc357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikram MA, Fornage M, Smith AV, et al. Common variants at 6q22 and 17q21 are associated with intracranial volume. Nat Genet. 2012;44:539–544. doi: 10.1038/ng.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verpillat P, Camuzat A, Hannequin D, et al. Association between the extended tau haplotype and frontotemporal dementia. Arch Neurol. 2002;59:935–939. doi: 10.1001/archneur.59.6.935. [DOI] [PubMed] [Google Scholar]
- 22.Zabetian CP, Hutter CM, Factor SA, et al. Association analysis of MAPT H1 haplotype and subhaplotypes in Parkinson’s disease. Ann Neurol. 2007;62:137–144. doi: 10.1002/ana.21157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pittman AM, Myers AJ, Abou-Sleiman P, et al. Linkage disequilibrium fine mapping and haplotype association analysis of the tau gene in progressive supranuclear palsy and corticobasal degeneration. J Med Genet. 2005;42:837–846. doi: 10.1136/jmg.2005.031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knopman DS1, Petersen RC, Edland SD, Cha RH, Rocca WA. The incidence of frontotemporal lobar degeneration in Rochester, Minnesota, 1990 through 1994. Neurology. 2004;62:506–508. doi: 10.1212/01.wnl.0000106827.39764.7e. [DOI] [PubMed] [Google Scholar]
- 25.Beecham GW, Hamilton K, Naj A, et al. Genome-wide association meta-analysis of neuropathologic features of Alzheimer’s disease and related dementias. PLoS Genet. 2014 doi: 10.1371/journal.pgen.1004606. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myers AJ, Kaleem M, Marlowe L, et al. The H1c Haplotype at the MAPT Locus is associated with Alzheimer’s disease. Hum Mol Genet. 2005;14:2399–2404. doi: 10.1093/hmg/ddi241. [DOI] [PubMed] [Google Scholar]
- 27.Abraham R, Sims R, Carroll L, et al. An association study of common variation at the MAPT locus with late-onset Alzheimer’s disease. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(8):1152–1155. doi: 10.1002/ajmg.b.30951. [DOI] [PubMed] [Google Scholar]
- 28.Creese B, Corbett A, Jones E, Fox C, Ballard C. Role of the Extended MAPT Haplotype in the Worsening of Psychotic Symptoms and Treatment Response in Alzheimer Disease. J Am Med Dir Assoc. 2014 doi: 10.1016/j.jamda.2014.08.011. In press. [DOI] [PubMed] [Google Scholar]
- 29.Allen M, Kachadoorian M, Quicksall Z, et al. Association of MAPT haplotypes with Alzheimer’s disease risk and MAPT brain gene expression levels. Alz Res Ther. 2014 doi: 10.1186/alzrt268. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zody MC, Jiang Z, Fung HC, et al. Evolutionary toggling of the MAPT 17q21. 31 inversion region. Nat Genet. 2008;40:1076–1083. doi: 10.1038/ng.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trabzuni D, Wray S, Vandrovcova J, et al. MAPT expression and splicing is differentially regulated by brain region: relation to genotype and implication for tauopathies. Hum Mol Genet. 2012;21:4094–4103. doi: 10.1093/hmg/dds238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong Q, Congdon EE, Nagaraja HN, Kuret J. Tau isoform composition influences the rate and extent of filament formation. J Biol Chem. 2012;287:20711–20719. doi: 10.1074/jbc.M112.364067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krstic D, Knuesel I. Deciphering the mechanism underlying late-onset Alzheimer disease. Nat Rev Neurol. 2013;9:25–34. doi: 10.1038/nrneurol.2012.236. [DOI] [PubMed] [Google Scholar]
- 34.Laws SM, Friedrich P, Diehl-Schmid J, et al. Fine mapping of the MAPT locus using quantitative trait analysis identifies possible causal variants in Alzheimer’s disease. Mol Psychiatry. 2007;12:510–517. doi: 10.1038/sj.mp.4001935. [DOI] [PubMed] [Google Scholar]
- 35.Roks G, Dermaut B, Heutink P, et al. Mutation screening of the tau gene in patients with early-onset Alzheimer’s disease. Neurosci Lett. 1999;277:137–139. doi: 10.1016/s0304-3940(99)00861-7. [DOI] [PubMed] [Google Scholar]
- 36.Cousin E, Macé S, Rocher C, et al. No replication of genetic association between candidate polymorphisms and Alzheimer’s disease. Neurobiol Aging. 2011;32:1443–1451. doi: 10.1016/j.neurobiolaging.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 37.Tesseur I, Van Dorpe J, Spittaels K, Van den Haute C, Moechars D, Van Leuven F. Expression of human apolipoprotein E4 in neurons causes hyperphosphorylation of protein tau in the brains of transgenic mice. Am J Pathol. 2000;156:951–964. doi: 10.1016/S0002-9440(10)64963-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cruchaga C, Kauwe JS, Harari O, et al. GWAS of cerebrospinal fluid tau levels identifies risk variants for Alzheimer’s disease. Neuron. 2013;72:256–268. doi: 10.1016/j.neuron.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morris GP, Clark IA, Vissel B. Inconsistencies and controversies surrounding the Amyloid Hypothesis of Alzheimer’s disease. Acta Neuropathol Commun. 2014;2:135. doi: 10.1186/s40478-014-0135-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuccolo J, Bau J, Childs SJ, et al. Phylogenetic analysis of the MS4A and TMEM176 gene families. PloS One. 2010;5:e9369. doi: 10.1371/journal.pone.0009369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karch CM, Jeng AT, Nowotny P, Cady J, Cruchaga C, Goate AM. Expression of novel Alzheimer’s disease risk genes in control and Alzheimer’s disease brains. PLoS One. 2012;7(11):e50976. doi: 10.1371/journal.pone.0050976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen-Plotkin AS, Unger TL, Gallagher MD, et al. TMEM106B, the risk gene for frontotemporal dementia, is regulated by the microRNA-132/212 cluster and affects progranulin pathways. J Neurosci. 2012;32:11213–11227. doi: 10.1523/JNEUROSCI.0521-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lang CM, Fellerer K, Schwenk BM, et al. Membrane orientation and subcellular localization of transmembrane protein 106B (TMEM106B), a major risk factor for frontotemporal lobar degeneration. J Biol Chem. 2012;287:19355–19365. doi: 10.1074/jbc.M112.365098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Deerlin VM, Sleiman PM, Martinez-Lage M, et al. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat Genet. 2010;42:234–239. doi: 10.1038/ng.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vass R, Ashbridge E, Geser F, et al. Risk genotypes at TMEM106B are associated with cognitive impairment in amyotrophic lateral sclerosis. Acta Neuropathol. 2011;121:373–380. doi: 10.1007/s00401-010-0782-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rutherford NJ, Carrasquillo MM, Li M, et al. TMEM106B risk variant is implicated in the pathologic presentation of Alzheimer disease. Neurology. 2012;79:717–718. doi: 10.1212/WNL.0b013e318264e3ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cruchaga C, Graff C, Chiang HH, et al. Association of TMEM106B gene polymorphism with age at onset in granulin mutation carriers and plasma granulin protein levels. Arch Neurol. 2011;68:581–586. doi: 10.1001/archneurol.2010.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.