Abstract
Purpose
Circulating microRNAs (miRNAs) are emerging as promising diagnostic biomarkers for colorectal cancer (CRC), but their usefulness for detecting early colorectal neoplasms (CRNs) remains unclear. This study aimed to identify serum miRNA biomarkers for the identification of patients with early CRNs.
Experimental Design
A cohort of 237 serum samples from 160 patients with early CRNs (148 precancerous lesions and 12 cancers) and 77 healthy subjects was analyzed in a three-step approach that included: a comprehensive literature review for published biomarkers, a screening phase, and a validation phase. RNA was extracted from sera, and levels of miRNAs were examined by real-time RT-PCR.
Results
Nine miRNAs (miR-18a, miR-19a, miR-19b, miR-20a, miR-21, miR-24, miR-29a, miR-92 and miR-125b) were selected as candidate biomarkers for initial analysis. In the screening phase, serum levels of miR-21, miR-29a and miR-125b were significantly higher in patients with early CRN compared to healthy controls. Elevated levels of miR-21, miR-29a and miR-125b were confirmed in the validation phase using an independent set of subjects. Area under the curve (AUC) values for serum miR-21, miR-29a, miR-125b, and their combined score in discriminating early CRN patients from healthy controls were 0.706, 0.741, 0.806 and 0.827 respectively. Serum levels of miR-29a and miR-125b were significantly higher in patients who only had small CRNs (≤5mm) compared to healthy subjects.
Conclusions
Since serum levels of miR-21, miR-29a and miR-125b discriminated early CRN patients from healthy controls, our data highlight the potential clinical use of these molecular signatures for noninvasive screening of patients with colorectal neoplasia.
Keywords: Serum, microRNA, biomarker, early colorectal neoplasia, screening
Introduction
Colorectal cancer (CRC) is the third leading cause of cancer-related deaths in men and women, with more than 50,000 deaths annually in the United States (1). The 5-year survival for patients with localized disease is 89.8%, so CRC is a potentially curable disease if diagnosed early. However, only 39.6% of patients are diagnosed at this stage (2). Moreover, since most CRCs develop through a stepwise adenoma-carcinoma sequence or the serrated pathway, most patients would be cured if the disease were detected and resected at a precancerous stage. Therefore, detection of early colorectal neoplasms (CRNs) - including precancerous lesions and early CRCs - is essential in reducing mortality associated with CRC.
Current guidelines recommend colonoscopy and fecal occult blood tests for CRC screening (3–5). Although colonoscopy is regarded as the gold standard for detecting CRNs, this approach has several limitations. It is an invasive and expensive procedure, requires an unpleasant bowel preparation, its efficacy depends on the skill and experience of the endoscopist, and a significant percentage of adults prefer noninvasive options for CRC screening (4). Fecal occult blood testing is a commonly used non-invasive test for CRC screening, and has demonstrated a reduction in CRC mortality by 33% to 15%, if used annually or semi-annually respectively (4, 6). However, fecal tests are not recommended for detecting precancerous lesions, due to the limited sensitivity and specificity, which is further compromised by inappropriate handling of specimens or poor adherence to recommended guidelines (4, 7). Thus, alternative minimally invasive or noninvasive tests to detect early CRNs are urgently needed.
Imperiale et al. recently reported a multi-target stool DNA and hemoglobin test as an alternative screening method for CRC. Although this assay was able to detect CRCs, this approach has important limitations; the detection of precancerous lesions was moderate, and the sensitivity for proximal lesions was inferior to that for distal lesions (8). There is considerable room for improving the noninvasive approach to CRN screening.
MicroRNAs (miRNAs) are small non-coding RNA sequences of 19–25 nucleotides which function as post-transcriptional regulators of gene expression (9), and dysregulation of miRNAs has been implicated in human cancers (10). In colorectal carcinogenesis, it has been shown that many miRNAs are dysregulated during the progression from normal to precancerous and cancerous pathology (11–13). As some of these dysregulated miRNAs are secreted into blood, and circulating cell-free miRNAs can be detected in serum or plasma in highly stable form, circulating miRNAs have emerged as potential blood-based biomarkers for human cancer (9, 14, 15). To date, several miRNAs have been reported as promising diagnostic biomarkers for CRC (16–24), indicating the potential role of circulating miRNAs as minimally invasive biomarkers for CRC. Some evidence suggests that circulating miRNAs may be able to discriminate individuals with advanced adenomas - which represent a subset of precancerous lesions including large (≥10 mm) tubular adenomas (TA), adenoma with a villous component, and high-grade intraepithelial neoplasia (HGIN) - from healthy controls (17–20, 23). These results are important as they show the potential of circulating miRNAs for detecting individuals with precancerous CRNs, but their use in clinical medicine is limited due to the small number of studies, small sample sizes and lack of clinicopathological information on studied patients. Consequently, systematic studies are required to further elucidate the utility of circulating miRNAs as biomarkers for early detection of CRNs.
Herein, we have analyzed a large cohort of serum specimens from patients with early CRNs, the majority of whom had only precancerous lesions, and conducted a systematic investigation to identify serum miRNA(s) that can potentially serve as blood-based biomarkers for CRC screening. We aimed to identify and validate serum miRNA(s) that can discriminate early CRN patients from healthy controls, and to elucidate the relationship between validated biomarkers and clinicopathological factors of CRNs.
Materials and Methods
Study subjects
Study subjects were prospectively enrolled at participating hospitals in Japan between January 2012 and May 2014. Case subjects consisted of patients who underwent endoscopic resection of early CRNs. Early CRNs included tubular adenomas (TAs), tubulovillous adenomas (TVAs), serrated polyps (SPs), high grade intraepithelial neoplasms (HGINs), and invasive cancers. SPs included sessile serrated polyps/adenomas, traditional serrated adenomas and mixed serrated polyps. The majority of enrolled patients had only precancerous lesions, but some individuals with invasive cancers were enrolled because they underwent endoscopic resection based on an indefinite endoscopic diagnosis of intraepithelial neoplasia or invasive cancer, and pathological examination of the resected specimen yielded the diagnosis of invasive cancer. Most of these invasive cancers showed only slight invasion into the submucosa. Clinicopathological characteristics of the study subjects are summarized in Table 1. An advanced neoplasia (AN) was defined as either TA or SP 10 mm or larger, TVA, HGIN, or an invasive cancer (8). Enrolled patients were categorized according to the histology of the index lesion. In patient with multiple CRNs, the most advanced or largest lesion among equivalently advanced lesions was designated as the index lesion. Serum samples were drawn within 3 months prior to the endoscopic resection. Serum samples were also collected from asymptomatic healthy volunteers as controls. Volunteers were enrolled from employees, subjects for annual health checks without major abnormalities, and patients undergoing screening colonoscopy in the participating hospitals. Individuals with a personal history of colorectal cancer, malignant tumors in other organs, inflammatory bowel disease, familial adenomatous polyposis or Lynch syndrome were excluded from both groups. Clinicopathological information was obtained from medical charts and questionnaires filled out by the participants. Written informed consent was obtained from all subjects, and the study protocol was approved by the Institutional Review Board of participating institutions.
Table 1.
Clinicopathological characteristics of study subjects
| Screening set | Validation set | |||
|---|---|---|---|---|
|
| ||||
| HC (n=25) | CRN (n=24) | HC (n=52) | CRN (n=136) | |
| Age | ||||
| Median (range) | 34 (22–49) | 67 (37–84) | 58 (23–77) | 68 (37–86) |
| Gender | ||||
| Male (%) | 19 (76.0) | 13 (54.2) | 23 (44.2) | 93 (68.4) |
| Female (%) | 6 (24.0) | 11 (45.8) | 29 (55.8) | 43 (31.6) |
| Location* | ||||
| Right (%) | - | 13 (54.2) | - | 32 (23.7) |
| Left (%) | - | 7 (29.2) | - | 55 (40.7) |
| Both (%) | - | 4 (16.7) | - | 48 (35.6) |
| Morphology | ||||
| Protruded (%) | - | 3 (12.5) | - | 45 (33.1) |
| Superficial (%) | - | 13 (54.2) | - | 59 (43.4) |
| Both (%) | - | 8 (33.3) | - | 32 (23.5) |
| Size of the index lesion (mm) | ||||
| Median (range) | - | 8 (4–32) | - | 10 (3–55) |
| Number of neoplastic lesions | ||||
| Median (range) | - | 2 (1–9) | - | 1.5 (1–16) |
| Histology of the index lesion | ||||
| LGIN | - | 12 (50.0) | - | 94 (69.1) |
| Tubular adenoma | - | 12 (50.0) | - | 70 (51.5) |
| Tubulo-villous adenoma | - | 0 (0.0) | - | 15 (11.0) |
| Sessile serrated polyp/adenoma | - | 0 (0.0) | - | 5 (3.7) |
| Traditional serrated adenoma | - | 0 (0.0) | - | 2 (1.5) |
| Mixed serrated polyp | - | 0 (0.0) | - | 2 (1.5) |
| HGIN | - | 8 (33.3) | - | 34 (25.0) |
| Cancer | - | 4 (16.7) | - | 8 (5.9) |
Location of one case in the validation phase was unknown.
Abbreviations: HC, healthy control, CRN, colorectal neoplasia, LGIN, low-grade intraepithelial neoplasia, HGIN, high-grade intraepithelial neoplasia
Study design
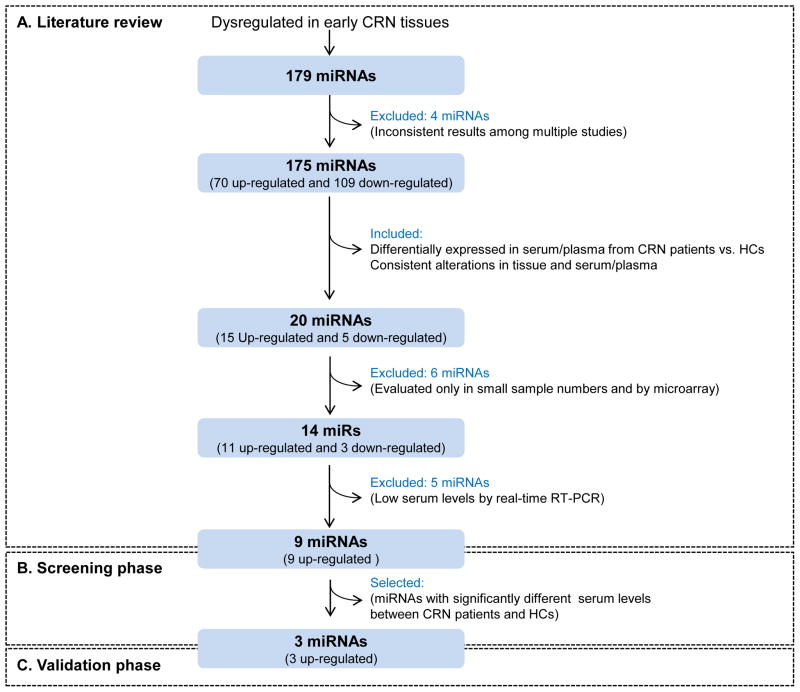
Schematic representation of this study is illustrated in Figure 1. Our study consisted of three parts: a systematic literature review for candidate miRNA biomarker selection, a screening phase, and a validation phase. The screening phase included patients with 24 early CRN and 25 healthy subjects, and a validation phase included 136 patients with early CRN and 52 healthy subjects. In the screening phase, serum levels of 9 miRNAs (miR-18a, miR-19a, miR-19b, miR-20a, miR-21, miR-24, miR-29a, miR-92a, and miR-125b) were investigated by quantitative real-time RT-PCR in sera from 24 early CRN patients and 25 healthy controls. In the validation phase, an independent cohort of 136 early CRN patients and 52 healthy controls were utilized to examine serum levels of miR-21, miR-29a and miR-125b.
Figure 1.
Schematic representation of the approach used for candidate miRNA biomarker selection in this study. HC, healthy control.
RNA extraction
RNA was extracted from serum samples using miRNeasy Serum/Plasma Kits (QIAGEN, Hilden, Germany, catalog number 217184) according to the manufacturer’s instruction. Briefly, 250 μL of serum was thawed on ice and centrifuged at 16,000g at 4°C for 10 minutes to remove cellular debris. Thereafter, 200 μL of supernatant was lysed in 1000 μL of QIAzol Lysis Reagent. After incubation for 5 minutes, 25 fmol of synthetic cel-miR-39 (Syn-cel-miR-39-3p miScript miRNA Mimic, QIAGEN, catalog number MSY0000010) was added to each sample as an external spiked-in control. Total RNA including small RNA was extracted and eluted in 30 μL of RNase-free water using a QIAcube devise (QIAGEN).
Real-time RT-PCR
Serum levels of candidate miRNA biomarkers and cel-miR-39 were examined by real-time RT-PCR using TaqMan MicroRNA Assays (Applied Biosystems, Foster City, CA, USA, catalog number 4427975). A fixed volume (2 μL) of total RNA was reverse transcribed using TaqMan MicroRNA Reverse Transcription Kits (Applied Biosystems, catalog number 4366597) in a total volume of 15μL with the following conditions: 16°C for 30 minutes, 42°C for 30 minutes, 85°C for 5 minutes and maintained at 4°C. Real-time PCR was conducted using MicroRNA Assay Kits and TaqMan Universal Master Mix II, no UNG (Applied Biosystems, catalog number 4440040), and performed in duplicate on the StepOne Plus system (Applied Biosystems) with the following cycling conditions: 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Cycle threshold (Ct) values were calculated using StepOne Software v2.3 (Applied Biosystems). Expression levels of miRNAs were normalized to those of cel-miR-39 and determined by the 2−ΔΔCt method. ΔCt was calculated as follows: ΔCt = Ct (miRNA of interest) – Ct (cel-miR-39). Then, ΔΔCt was calculated by using a sample from a healthy volunteer as a calibrator: ΔΔCt = ΔCt (tested sample) – ΔCt (calibrator).
Power calculations and statistical analysis
Power calculations to determine the required sample sizes in the validation phase were performed by G*Power 3 (25) based on the data from the screening phase. To compare the serum miRNA levels between two groups, the Mann-Whitney U test was conducted. The Steel-Dwass test was used to perform all-paired multiple comparisons among three or more groups. Correlation between the size of the index lesion and the serum levels of miR-21, miR-29a and miR-125b were analyzed by the Spearman’s rank correlation coefficient (ρ). All P-values were two-sided and a P-value of <0.05 was considered significant. Receiver operating characteristic (ROC) curves were generated and the area under the ROC curve (AUC) with 95% confidence intervals (CI) were computed to assess the discriminating performance of miR-21, miR-29a and miR-125b. All analyses were carried out using JMP® 10 (SAS institute Inc., Cary, NC, USA) and graphs were generated using Graphpad Prism 5.00 for Windows (Graphpad software, San Diego, CA, USA) except for the ROC curves, which were analyzed using Medcalc Statistical Software version 12.7.7 (Medcalc Software bvba, Ostend, Belgium).
Results
Candidate miRNA biomarker selection
Since one of the primary goals of our study was identification of diagnostic biomarkers for early CRNs, we first selected miRNAs that had been reported to be dysregulated in early CRN tissues (11, 13, 26–30). We found 175 miRNAs that have been reported to be dysregulated in early steps during colorectal carcinogenesis. To narrow our candidate list exclusively to secretory miRNAs that may be detectable in blood, we next selected miRNAs dysregulated in serum or plasma from CRC and/or colorectal adenoma patients. (16–24). We chose only miRNAs showing consistent dysregulation in tissues and serum or plasma among multiple studies, and identified 20 such miRNAs. We thereafter excluded 6 miRNAs that were discovered solely by microarrays in small sample numbers and not validated by subsequent quantitative methods such as real-time RT-PCR. Through this approach, 14 miRNAs remained as candidate biomarkers (Supplementary Table S1). Five miRNAs (miR-409-3p, miR-424, miR-575, miR-601 and miR-760) were further excluded because serum expression levels of these miRNAs were too low to be accurate quantifed by real-time RT-PCR in our preliminary experiment (data not shown). As a result, 9 miRNAs (miR-18a, miR-19a, miR-19b, miR-20a, miR-21, miR-24, miR-29a, miR-92 and miR-125b) were selected as candidate biomarkers for the early detection of CRNs.
The screening phase
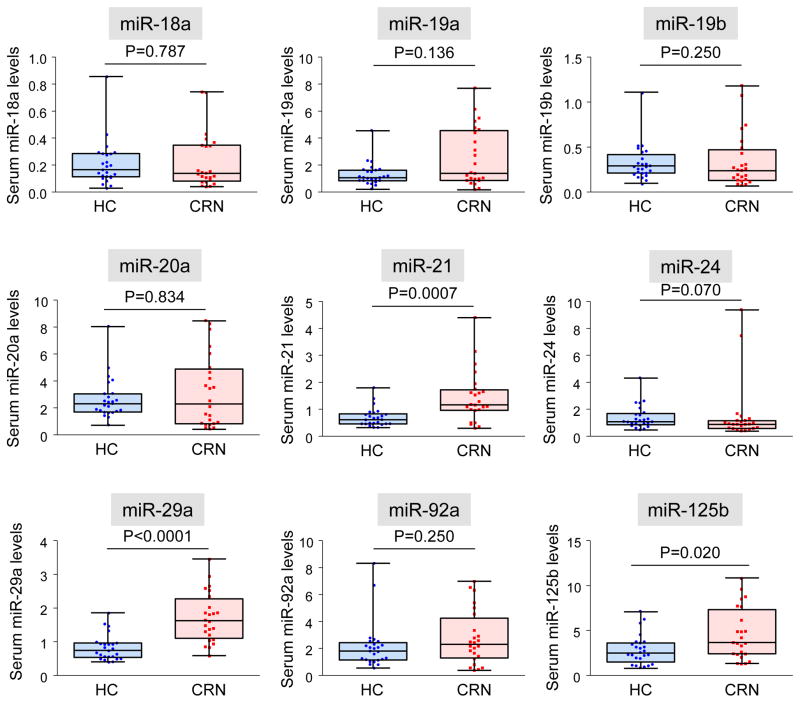
In the screening phase, expression levels of the above-mentioned 9 miRNAs in serum samples from 24 early CRN patients and 25 healthy controls were examined. As shown in Figure 2, serum levels of miR-21 (P=0.0007), miR-29a (P<0.0001) and miR-125b (P= 0.020) were significantly higher in early CRN patients than in healthy controls.
Figure 2.
Serum levels of candidate miRNA biomarkers in the screening phase. Levels of serum miR-18a, miR-19a, miR-19b, miR-20a, miR-21, miR-24, miR-29a, miR-92a and miR-125b were compared between healthy controls (HC) and early CRN patients (CRN). The Mann-Whitney U test was performed for comparisons between groups.
The validation phase
First, we performed power calculations by G*Power 3 (25) to determine the sample size required in the validation phase. Based on the miR-125b data in the screening phase, which showed the smallest effect size among the three candidate miRNAs, we estimated a minimal sample size of 122 early CRN patients and 30 healthy controls would be required to achieve 0.95 power.
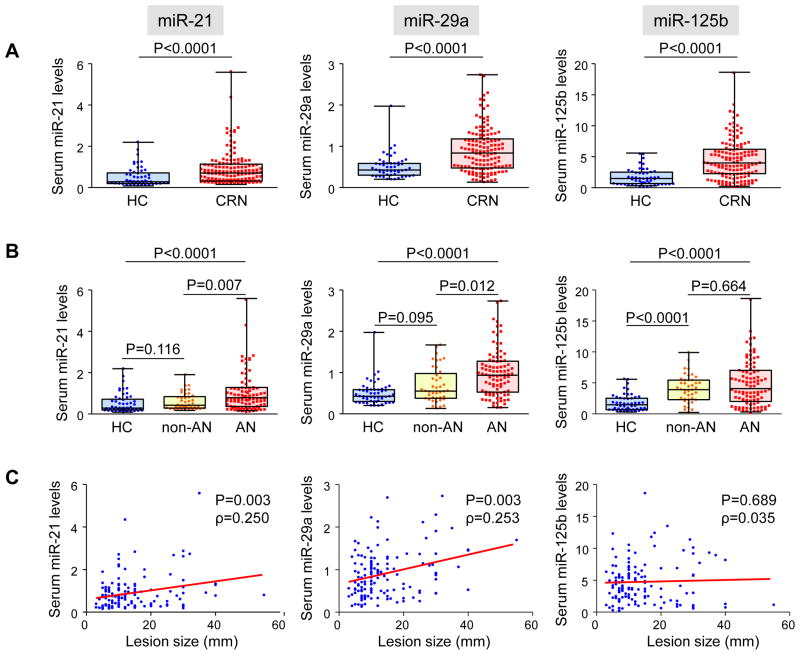
Finally, we utilized serum samples from 136 early CRN patients and 52 healthy controls (Table 1) to validate the discriminatory capability of miR-21, miR-29a and miR-125b. As shown in Figure 3A, serum levels of miR-21 (P<0.0001), miR-29a (P<0.0001) and miR-125b (P<0.0001) were significantly higher in early CRN patients than in healthy controls. As shown in Supplementary figure 1, increased levels of 3 miRNAs were significant even when we excluded 8 patients with CRCs and compared only patients with non-invasive CRNs to healthy subjects. When we divided CRN patients into either having ANs or not (non-ANs), expression levels of miR-21 and miR-29a were significantly elevated in AN patients compared to non-AN patients and healthy controls, whereas miR-125b levels increased in both non-AN and AN patients compared to healthy controls (Figure 3B). Serum levels of miR-21 and miR-29a significantly correlated with the size of the index lesions, while miR-125b showed no correlation (Figure 3C).
Figure 3.
Serum miR-21, miR-29a and miR-125b in the validation phase. A: Levels of miRNAs were compared between healthy controls (HC) and early colorectal neoplasia (CRN) patients. Statistical analyses were performed using the Mann-Whitney U test. B: Levels of miRNAs were compared among HCs, patients with non-AN CRNs (non-AN) and patients with ANs (AN). All-pairs multiple comparison was conducted by the Steel-Dwass test. C: Correlation between levels of serum miRNAs and the size of the index lesion. Spearman’s rank correlation coefficient (ρ) is shown.
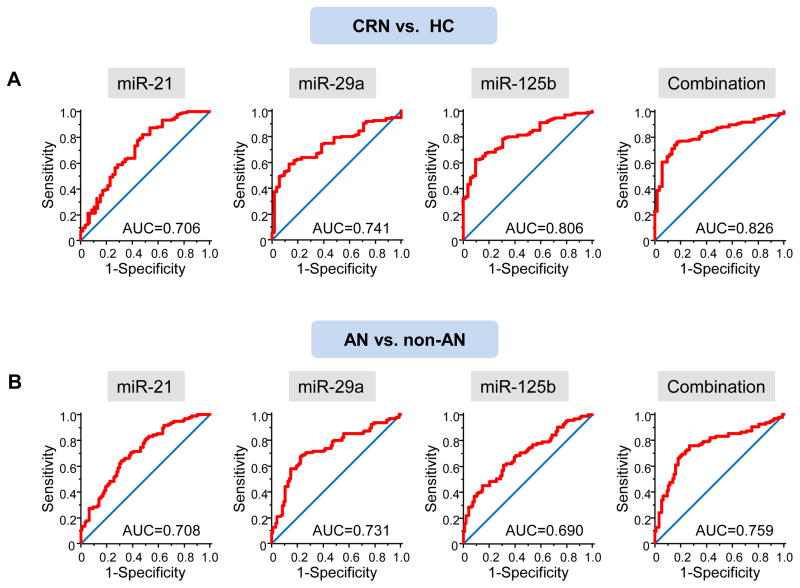
Next, we generated ROC curves to evaluate the performance of the three miRNAs as serum biomarkers for the detection of early CRNs. AUC values for serum miR-21, miR-29a and miR-125b in discriminating early CRN patients from healthy controls were 0.706 (95% CI, 0.635 to 0.770), 0.741 (95% CI, 0.673 to 0.802) and 0.806 (95% CI, 0.742 to 0.860), respectively. To discriminate AN patients from individuals without ANs (i.e. patients with non-AN CRNs plus healthy controls), AUC values for serum miR-21, miR-29a and miR-125b were 0.708 (95% CI, 0.638 to 0.772), 0.731 (95% CI, 0.662 to 0.793) and 0.690 (95% CI, 0.618 to 0.755), respectively. Combination of three miRNAs showed further improvement in AUC=0.826 (95% CI, 0.765 to 0.878) for all early CRNs, and AUC=0.759 (95% CI, 0.691 to 0.818) for ANs. (Figure 4) The results were almost identical when we excluded 8 patients with CRCs from the analyses. (Supplementary figure 2)
Figure 4.
ROC curves for miR-21, miR-29a, miR-125b and the combination of the three miRNAs in discriminating patients with early CRNs from healthy controls (HCs) (A), and in discriminating patients with ANs from individuals without ANs (B). AUC values are shown.
Serum miR-21, miR-29a and miR-125b levels and characteristics of early CRNs
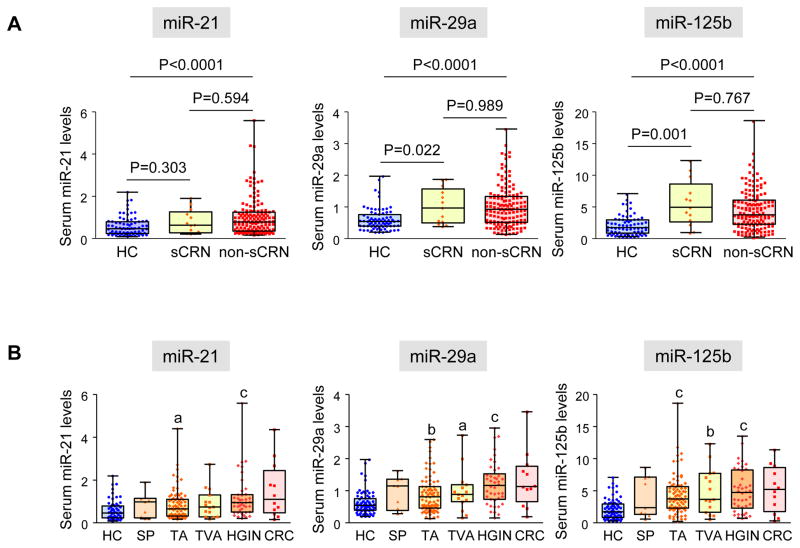
Given that serum miR-125b levels were significantly elevated in non-ANs, which consisted of TAs and SPs less than 10 mm in size, we determined whether the levels of miRNAs were elevated in patients with even smaller CRNs. We calculated the total diameters of all CRNs detected in an individual, and found that 11 of 136 patients had CRNs with total diameter of 5 mm or less. Serum expression levels of miR-29a and miR-125b were significantly higher in patients who only had small CRNs (total diameter of CRNs ≤5 mm) than in healthy controls. (P=0.022 and P=0.001, respectively, Figure 5A)
Figure 5.
A: Serum miR-21, miR-29a and miR-125b in patients with small CRNs. Levels of miRNAs were compared among HCs, patients with small CRNs (total diameter of CRNs in an individual ≤5 mm, sCRN) and with larger CRNs (total diameter of CRNs >5 mm, non-sCRN). All-pairs multiple comparison was conducted by the Steel-Dwass test. B: Serum miRNA levels according to various histological subtypes of CRNs. Statistical analysis was conducted by the Steel-Dwass test, and significant differences of miRNA levels compared to HCs are noted. (a; P<0.05, b; P<0.01, c; P<0.0001)
We analyzed serum miRNA expression levels according to histological subtypes of CRNs. Compared to healthy controls, patients with TAs and HGINs showed significantly higher serum levels of all three miRNAs, while patients with TVAs had significantly elevated levels of serum miR-29a and miR-125b. (Figure 5B)
Finally, we examined the association between serum levels of miR-21, miR-29a and miR-125b and clinicopathological factors. As shown in Supplementary Table S2, there were no significant association between serum levels of the three miRNAs and age, gender, the presence of concurrent disorders, location or morphology of the CRNs.
Discussion
In this study, we analyzed a cohort of 237 serum samples in independent screening and validation sets, and found that the levels of miR-21, miR-29a and miR125b in serum could discriminate patients with early CRNs from healthy controls. To our knowledge, this is the first study to systematically investigate circulating miRNA biomarkers focused on the identification of precancerous CRNs – the optimal target lesion for a CRC screening strategy.
Although genome-wide miRNA profiling approaches such as microarrays have been successfully utilized for the discovery of novel miRNA biomarkers for the early detection of CRNs, the outcomes have been largely inadequate for clinical decision making due to the limited number of studies and small numbers of samples analyzed. In fact, a few recent studies have found multiple miRNAs that could discriminate precancerous adenoma patients from healthy controls by using microarray; however, none of these studies identified common overlapping miRNA biomarkers, raising concerns about the discovery and validation methodologies used in these reports (18–20). In the current study, we started with a systematic literature review rather than genome-wide approaches for the selection of candidate miRNAs. This enabled us to choose miRNAs that have been found by independent researchers using different patient cohorts. Since reproducibility is one of the most critical components in the discovery of clinically relevant biomarkers, we believe our strategy has an advantage in finding more generally applicable markers.
The most remarkable findings of our study were that serum miR-21, miR-29a and miR-125b could discriminate early CRN patients from healthy controls. By performing a power calculation, we were able to analyze a sufficient number of subjects to reach this conclusion with appropriate statistical power. MiR-125b showed the best performance among the three miRNAs to discriminate early CRN patients from healthy subjects. Serum miR-125b levels were elevated in non-AN patients (i.e. TAs and/or SPs smaller than 10 mm) compared to healthy controls, and no significant increase was observed in AN patients. Surprisingly, elevated levels of miR-125b were seen even in patients whose total diameters of CRNs were 5 mm or less. Thus, the increase in serum miR-125b levels most likely reflects an earlier step during colorectal carcinogenesis. In contrast, serum levels of miR-21 and miR-29a were elevated significantly in patients with ANs compared to healthy controls and non-AN patients. Considering that the most relevant lesions in CRC screening would be precancerous lesions which possess higher risks of progressing to cancers, miR-21 and miR-29a might be better biomarkers to detect more risky lesions such as ANs.
Precancerous CRNs contain various histological subtypes including SPs, TAs, TVAs and HGINs. Since different types of CRNs possess different molecular alterations and possibly different miRNA signatures, it is important to consider the relevance of biomarker miRNAs in each subtype of precancerous lesion. Our data showed that serum miR-21, miR-29a and miR-125b had similar elevations in patients with TAs, TVAs and HGINs, suggesting their potential in detecting these precancerous CRNs. Although levels of miR-21, miR-29a and miR-125b were elevated in a subset of patients with SPs, another subset of precancerous lesion with different molecular characteristics (31), it was difficult to draw conclusions due to the small numbers of subjects with SPs in this study.
In addition to the performance of individual miRNA biomarkers, a biomarker panel created by combining the expression of three miRNAs demonstrated enhanced performance in detecting AN patients with an AUC value of 0.759, and was comparable to the multitarget stool testing (AUC = 0.73) reported by Imperiale and colleagues (8). Compared to stool DNA and hemoglobin testing, serum miRNA testing has several potential advantages as a screening method for early CRNs. First, our approach is simple, and requires only the quantification of the serum levels of three miRNAs by real-time RT-PCR, whereas the stool DNA testing requires transporting stool to a centralized lab, multiple assays of DNA mutation and methylation, and a hemoglobin immunoassay. Second, serum miRNAs in this study showed no significant difference according to the location of CRNs, suggesting their superiority in detecting CRNs regardless of their location. By contrast, stool DNA and hemoglobin testing were less sensitive for proximal lesions. Finally, since stool samples are collected by the subjects themselves, quality control may be easier using blood rather than stool. In addition, the use of blood samples may lead to better compliance than expecting patients to properly collect and ship stool.
There are some potential limitations of our study. First, the healthy volunteers in our study were younger than CRN patients. To minimize the bias caused by the age differences between the two groups, we assigned healthy volunteers of a similar age range with subjects undertaking CRC screening in the general population to the validation set. Second, hemolysis is known to affect levels of circulating biomarker miRNAs (32). As we saw some hemolyzed samples in our study subjects, hemolysis might confound our results. Impacts of confounding factors including hemolysis should be evaluated in the future study. Third, our study lacked an independent, large validation cohort, which must be considered in future investigations to further appreciate the clinical significance of the findings reported in this study.
In conclusion, we conducted a systematic investigation to identify circulating miRNA biomarkers for CRC screening, and found that serum miR-21, miR-29a and miR-125b levels could discriminate early CRN patients from healthy controls. Our data highlight the capability of serum miRNAs to detect precancerous CRNs, suggesting the potential clinical use of these molecular signatures for noninvasive screening of patients for CRC.
Supplementary Material
Translational relevance.
Circulating microRNAs are emerging as promising biomarkers for various human diseases including colorectal cancer. To reduce colorectal cancer-related mortality, diagnosis of the disease at curable stages such as precancerous lesions and early cancers is highly important. Although many studies have suggested the potential utility of circulating microRNAs in colorectal cancer detection, their potential for detecting precancerous lesions remains poorly understood. In this study, we analyzed a large cohort of serum samples from patients with early colorectal neoplasms, in which the majority only had precancerous lesions, and demonstrated that expression levels of miR-21, miR-29a and miR-125b could discriminate patients from healthy controls. Our findings of serum microRNAs as diagnostic biomarkers of precancerous colorectal lesions provide rationale for the further development of these molecular signatures as screening biomarkers for this fatal malignancy.
Acknowledgments
Financial support: The present work was supported by grant R01 CA72851 and CA 181572 from the National Cancer Institute, National Institutes of Health, and funds from the Baylor Research Institute to AG and CRB. This study was also supported by a pilot project grant from the Charles A Sammons Cancer Center, Baylor University Medical Center to AG.
The authors thank Dr. Yoshitaka Konda, Dr. Chiharu Kawanami, Dr. Shujiro Yazumi, Dr. Shinya Ohashi, Dr. Shigehiko Fujii, Ms. Hiromi Yoshida, Ms. Yukie Nakai, Ms. Mihoko Tsurumaki, Ms. Noriko Fujita, Ms. Mayumi Mizuguchi, Ms. Hoki Matsumoto and Ms. Eriko Ban for their supports in patients’ enrollment and sample collection.
Abbreviations
- CRC
colorectal cancer
- CRN
colorectal neoplasia
- miRNA
microRNA
- TA
tubular adenoma
- HGIN
high-grade intraepithelial neoplasia
- TVA
tubulo-villous adenoma
- SP
serrated polyp
- AN
advanced neoplasia
- LGN
low-grade neoplasia
- HGN
high-grade neoplasia
- ROC
Receiver operating characteristic
- AUC
area under the ROC curve
Footnotes
Conflict of interest: The authors have nothing to disclose.
Specific author contributions: Atsushi Yamada: study design, acquisition, analysis, and interpretation of data, drafting of the manuscript; Takahiro Horimatsu, Naoshi Nishida, Hajime Honjo, Hiroshi Ida, Tadayuki Kou, Toshihiro Kusaka, Yusuke Amanuma, Osamu Kikuchi, Yu Sasaki, Yagi Makato, Atsushi Nakajima, Takuma Higurashi, Norio Yukawa and Yoshiyuki Ueno: material support and acquisition of data; Yoshinaga Okugawa, Manabu Muto and Tsutomu Chiba: material support and critical revision of the manuscript for important intellectual content; C. Richard Boland: interpretation of data and study supervision and editing the manuscript; Ajay Goel: study concept and design, analysis and interpretation of data, drafting of the manuscript.
References
- 1.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64(2):104–17. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 2.Howlader N, NA, Krapcho M, Garshell J, Miller D, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA, editors. SEER Cancer Statistics Review, 1975–2011. National Cancer Institute; Bethesda, MD: 2014. [Google Scholar]
- 3.Rex DK, Johnson DA, Anderson JC, Schoenfeld PS, Burke CA, Inadomi JM. American College of Gastroenterology guidelines for colorectal cancer screening 2009 [corrected] Am J Gastroenterol. 2009;104(3):739–50. doi: 10.1038/ajg.2009.104. [DOI] [PubMed] [Google Scholar]
- 4.Levin B, Lieberman DA, McFarland B, Andrews KS, Brooks D, Bond J, et al. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. Gastroenterology. 2008;134(5):1570–95. doi: 10.1053/j.gastro.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Qaseem A, Denberg TD, Hopkins RH, Jr, Humphrey LL, Levine J, Sweet DE, et al. Screening for colorectal cancer: a guidance statement from the American College of Physicians. Ann Intern Med. 2012;156(5):378–86. doi: 10.7326/0003-4819-156-5-201203060-00010. [DOI] [PubMed] [Google Scholar]
- 6.Mandel JS, Bond JH, Church TR, Snover DC, Bradley GM, Schuman LM, et al. Reducing mortality from colorectal cancer by screening for fecal occult blood. Minnesota Colon Cancer Control Study. N Engl J Med. 1993;328(19):1365–71. doi: 10.1056/NEJM199305133281901. [DOI] [PubMed] [Google Scholar]
- 7.Quintero E, Castells A, Bujanda L, Cubiella J, Salas D, Lanas A, et al. Colonoscopy versus fecal immunochemical testing in colorectal-cancer screening. N Engl J Med. 2012;366(8):697–706. doi: 10.1056/NEJMoa1108895. [DOI] [PubMed] [Google Scholar]
- 8.Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370(14):1287–97. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- 9.Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11(3):145–56. doi: 10.1038/nrclinonc.2014.5. [DOI] [PubMed] [Google Scholar]
- 10.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartley AN, Yao H, Barkoh BA, Ivan C, Mishra BM, Rashid A, et al. Complex patterns of altered MicroRNA expression during the adenoma-adenocarcinoma sequence for microsatellite-stable colorectal cancer. Clin Cancer Res. 2011;17(23):7283–93. doi: 10.1158/1078-0432.CCR-11-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi X, Zhang Y, Cao B, Lu N, Feng L, Di X, et al. Genes involved in the transition from normal epithelium to intraepithelial neoplasia are associated with colorectal cancer patient survival. Biochem Biophys Res Commun. 2013;435(2):282–8. doi: 10.1016/j.bbrc.2013.04.063. [DOI] [PubMed] [Google Scholar]
- 13.Wang S, Wang L, Bayaxi N, Li J, Verhaegh W, Janevski A, et al. A microRNA panel to discriminate carcinomas from high-grade intraepithelial neoplasms in colonoscopy biopsy tissue. Gut. 2013;62(2):280–9. doi: 10.1136/gutjnl-2011-301554. [DOI] [PubMed] [Google Scholar]
- 14.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 16.Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58(10):1375–81. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 17.Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127(1):118–26. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 18.Wang Q, Huang Z, Ni S, Xiao X, Xu Q, Wang L, et al. Plasma miR-601 and miR-760 are novel biomarkers for the early detection of colorectal cancer. PLoS One. 2012;7(9):e44398. doi: 10.1371/journal.pone.0044398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanaan Z, Rai SN, Eichenberger MR, Roberts H, Keskey B, Pan J, et al. Plasma miR-21: a potential diagnostic marker of colorectal cancer. Ann Surg. 2012;256(3):544–51. doi: 10.1097/SLA.0b013e318265bd6f. [DOI] [PubMed] [Google Scholar]
- 20.Giraldez MD, Lozano JJ, Ramirez G, Hijona E, Bujanda L, Castells A, et al. Circulating microRNAs as biomarkers of colorectal cancer: results from a genome-wide profiling and validation study. Clin Gastroenterol Hepatol. 2013;11(6):681–8. e3. doi: 10.1016/j.cgh.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 21.Hofsli E, Sjursen W, Prestvik WS, Johansen J, Rye M, Trano G, et al. Identification of serum microRNA profiles in colon cancer. Br J Cancer. 2013;108(8):1712–9. doi: 10.1038/bjc.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo X, Stock C, Burwinkel B, Brenner H. Identification and evaluation of plasma microRNAs for early detection of colorectal cancer. PLoS One. 2013;8(5):e62880. doi: 10.1371/journal.pone.0062880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y, et al. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105(12):849–59. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang S, Xiang J, Li Z, Lu S, Hu J, Gao X, et al. A plasma microRNA panel for early detection of colorectal cancer. Int J Cancer. 2013 doi: 10.1002/ijc.28136. [DOI] [PubMed] [Google Scholar]
- 25.Faul F, Erdfelder E, Lang AG, Buchner A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods. 2007;39(2):175–91. doi: 10.3758/bf03193146. [DOI] [PubMed] [Google Scholar]
- 26.Diosdado B, van de Wiel MA, Terhaar Sive Droste JS, Mongera S, Postma C, Meijerink WJ, et al. MiR-17-92 cluster is associated with 13q gain and c-myc expression during colorectal adenoma to adenocarcinoma progression. Br J Cancer. 2009;101(4):707–14. doi: 10.1038/sj.bjc.6605037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamamichi N, Shimomura R, Inada K, Sakurai K, Haraguchi T, Ozaki Y, et al. Locked nucleic acid in situ hybridization analysis of miR-21 expression during colorectal cancer development. Clin Cancer Res. 2009;15(12):4009–16. doi: 10.1158/1078-0432.CCR-08-3257. [DOI] [PubMed] [Google Scholar]
- 28.Akao Y, Nakagawa Y, Hirata I, Iio A, Itoh T, Kojima K, et al. Role of anti-oncomirs miR-143 and -145 in human colorectal tumors. Cancer Gene Ther. 2010;17(6):398–408. doi: 10.1038/cgt.2009.88. [DOI] [PubMed] [Google Scholar]
- 29.Fassan M, Pizzi M, Giacomelli L, Mescoli C, Ludwig K, Pucciarelli S, et al. PDCD4 nuclear loss inversely correlates with miR-21 levels in colon carcinogenesis. Virchows Arch. 2011;458(4):413–9. doi: 10.1007/s00428-011-1046-5. [DOI] [PubMed] [Google Scholar]
- 30.Oberg AL, French AJ, Sarver AL, Subramanian S, Morlan BW, Riska SM, et al. miRNA expression in colon polyps provides evidence for a multihit model of colon cancer. PLoS One. 2011;6(6):e20465. doi: 10.1371/journal.pone.0020465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rex DK, Ahnen DJ, Baron JA, Batts KP, Burke CA, Burt RW, et al. Serrated lesions of the colorectum: review and recommendations from an expert panel. Am J Gastroenterol. 2012;107(9):1315–29. doi: 10.1038/ajg.2012.161. quiz 4, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirschner MB, Edelman JJ, Kao SC, Vallely MP, van Zandwijk N, Reid G. The Impact of Hemolysis on Cell-Free microRNA Biomarkers. Front Genet. 2013;4:94. doi: 10.3389/fgene.2013.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.