Abstract
Autophagy is self-cleaning process that occurs at a constitutive basal level, and is upregulated in response to stress. Macroautophagy (hereafter autophagy) is the most robust type of autophagy, where cargo (specific or nonspecific) is engulfed within a double-membrane structure termed an autophagosome. This process needs to be tightly regulated to maintain normal cellular homeostasis and prevent dysfunction; therefore, a fuller knowledge of the mechanisms of autophagy regulation is crucial for understanding the entire pathway. The autophagy-related (Atg) proteins are the primary components that carry out autophagy. Many of these proteins are conserved from yeast to human. A number of significant discoveries with regard to protein functional domains, protein-protein interactions, or posttranslational modifications of proteins involved in autophagy have been reported in parallel with, or followed by, solving the NMR or crystal structures of autophagy proteins or their protein domains. In this work, we summarize structural insights gathered to date on the proteins of the autophagy machinery that are modulated by a posttranslational modification, specifically phosphorylation, acetylation, ubiquitination, and/or SUMOylation. For each protein, we link the reported results with information on the propensity of the corresponding amino acid sequence toward order/disorder. This integrative approach yields a comprehensive overview for each posttranslationally modified protein, and also reveals areas for further investigation.
Keywords: autophagy, intrinsically disordered region, posttranslational modification, protein structure, stress
Graphical Abstract
Introduction
Protein architecture is determined by the composition of amino acids in the primary sequence that may or may not be further modulated by one or more posttranslational modifications (PTMs). These modifications can occur at any stage of a protein’s existence. Modulation of protein structure via PTM can affect protein stability, cellular localization or protein engagement in protein-protein interactions, which in turn affects its biological function. The proteins of the autophagy machinery are phosphorylated, acetylated, ubiquitinated, SUMOylated, lipidated, O-GlcNAcylated, and/or modified on thiol groups [1]. The overall mechanism of nonselective and selective autophagy including autophagosome biogenesis has been described in several recent reviews [2–4]. In yeast, this pathway originates at a site called the phagophore assembly site (PAS). Posttranslationally modified proteins involved in nonspecific autophagy are described here in the order of hierarchy of their recruitment to the PAS, and are followed in description by receptors for selective autophagy.
PTM-modified structures in autophagy initiation
Upon starvation, a de novo-formed structure called the phagophore is initiated at the PAS. The phagophore is the initial sequestering compartment that subsequently matures into a completed autophagosome; as such, the phagophore is the dynamic intermediate organelle of autophagy. The current consensus is that the phagophore arises from small Atg9-containing vesicles not more than 20 nm in diameter.
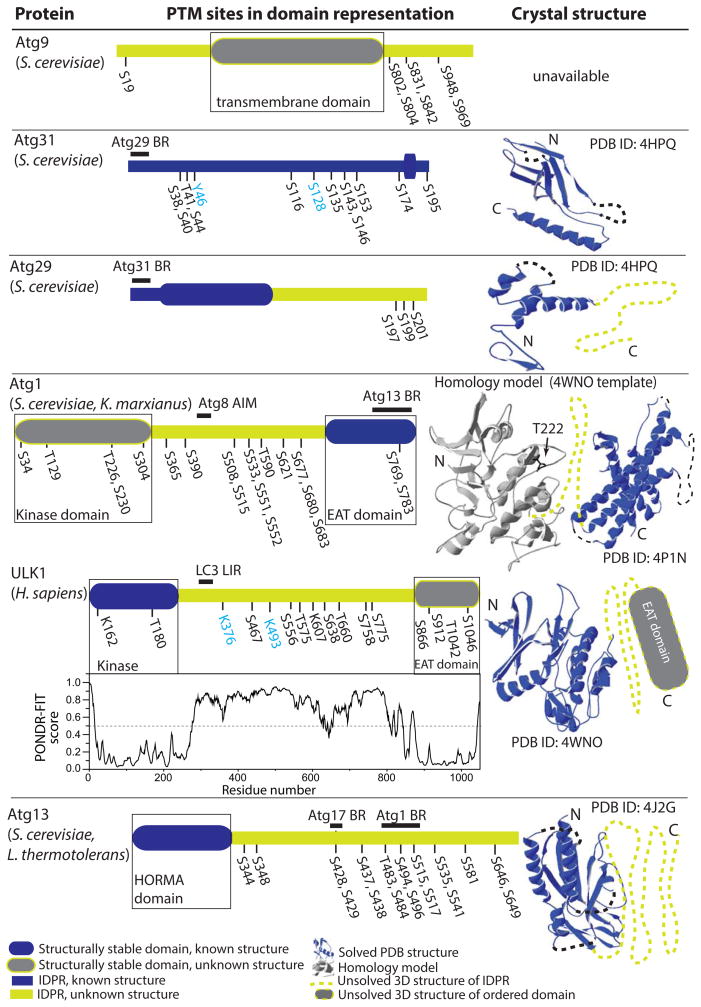
Atg9 is the only membrane protein in the core autophagy machinery. It consists of six predicted transmembrane α–helices, which orient the protein’s N and C termini to face the cytosol. The C terminus and especially the N terminus of this large 115-kDa protein has a high propensity for disorder (based on the PONDR-FIT algorithm in the DisProt database (http://www.disprot.org/pondr-fit.php; [5]) in contrast to the middle part of the sequence. In the yeast Saccharomyces cerevisiae, Atg9 is phosphorylated by the Atg1 kinase in early steps of autophagy on Ser19, Ser802, Thr804, Ser831, Ser842, Ser948, and Ser969 (Fig. 1). All of these phosphorylated residues are located in the disordered N and C termini of the protein, and this posttranslational modification is required for the recruitment of Atg8 and Atg18 to the PAS [6]. Atg9 is recruited relatively early to the PAS along with the trimeric Atg17–Atg31–Atg29 subcomplex. A crystal structure of the trimeric subcomplex shows that it dimerizes into a unique double crescent, of which the convex face harbors the molecular mass of Atg29 and Atg31, at least in nutrient-rich conditions [7, 8].
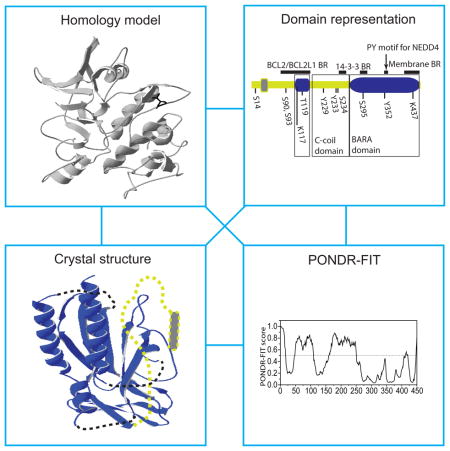
Figure 1.
Structure-related analysis of posttranslationally modified proteins related to Atg1 (Atg9, Atg31, Atg29, Atg1, ULK1, and Atg13). The PTM sites are denoted in the domain representation of the corresponding protein. The domain representation is the schematically depicted PONDR-FIT profile for each protein, where structurally stable domains are shown as round-cornered rectangles and disordered regions are bold lines. Domains that have been assigned to NMR or crystal structure are in blue, while unassigned structured or unstructured domains are gray or light green, respectively. For clarity of presentation, total lengths of protein domain representations are not comparable between each other. If known, the functionally important domains are marked by black-lined rectangles. Experimentally verified posttranslationally modified sites are denoted in black and predicted sites by ModPred are in light blue. PONDR-FIT scores for all Atg proteins were reported in ref. [9]. ULK1 was not included in that study, therefore, its PONDR-FIT profile is shown below the ULK1 domain representation. If the NMR or crystal structure of at least one domain is available for a particular protein, the corresponding putative 3D model is shown on the right side next to the domain representation. 3D structures recorded in the PDB are shown in blue, and black dashed lines show missing regions in those structures. The homology model based on the PDB template is in gray. Light-green dotted lines and gray rectangles in putative 3D models correspond to unsolved 3D structures of IDPRs and structurally stable domains, respectively. BR, binding region.
Atg29 and Atg31 are highly dynamic proteins, in fact the most flexible proteins of all Atg proteins known so far [9]. Their interacting N-termini undergo a disorder-to-order transition into a β sandwich [8]. The function of both proteins in the subcomplex still remains a mystery. Their dynamic properties, also confirmed by the difficulty to obtain crystallographic data (PDB ID:4HPQ, [8], would suggest that Atg29 and Atg31 function in signal sensing and/or regulation. It has been proposed that under growing conditions both proteins form a steric block that prevents the binding of Atg9 to Atg17 [8], and that upon stress conditions, this block needs to be removed to allow binding to occur. A stronger interaction of Atg9 with Atg17 upon rapamycin treatment relative to growing conditions has been demonstrated by immunoprecipitation [10], and could be interpreted in support of the hypothesis for removal of the steric block. The dynamic nature of Atg31 would also be consistent with the model where Atg31 pivots in its flexible linker between β–sheet 7 and α–helix 1 [8]. Phosphorylation of Atg31 has been demonstrated experimentally [11, 12]. Mass spec analysis identified 11 phosphorylation sites in Atg31 (Ser38, Ser40, Thr41, Ser44, Ser116, Ser135, Ser143, Ser146, Ser153, Ser174, and Ser195). Mutational analysis of these sites showed the strongest phenotype for Ser174. Further analysis revealed that phosphoserine 174 is important for formation of autophagosomes and the absence of this modification causes accumulation of Atg9-positive vesicles at the PAS [12]. Some of the experimentally determined phosphosites in Atg31 overlap with the outcome from the ModPred algorithm [13], which predicts with a medium level of confidence phosphorylation of Atg31 on Tyr46, Ser116, Ser128, Ser143, and Ser153 (Fig. 1). Under growing conditions, the highly mobile C terminus of Atg29 adopts an autoinhibitory conformation that regulates interaction with Atg11. Phosphorylation of 23 serine and threonine residues in the C-terminal half of Atg29 facilitates its binding to Atg11 [7]. Functionally the most important phosphoserines are Ser197, Ser199, and Ser201 right upstream of the C-terminal inhibitory peptide (Fig. 1).
Initiation of phagophore formation requires assembly of the trimeric Atg17-Atg31-Atg29 subcomplex with Atg13 and Atg1.
Atg1 is an essential kinase in autophagy that is highly regulated by phosphorylation. Atg1 in S. cerevisiae is phosphorylated at many sites [14, 15] (Fig. 1). Among these sites, phosphorylation on Ser508 and Ser515 is mediated by protein kinase A (PKA), whereas Thr226 and Ser390 are targets of autophosphorylation [15]. Ser34 phosporylation is inhibitory to kinase activity as well as to autophagy [15]. In contrast, phosphorylation of the conserved Thr226 and Ser230 is essential for kinase activity and for function of Atg1 in both autophagy and the autophagy-like cytoplasm-to-vacuole targeting pathways [14]. Regulation of Atg1 via phosphorylation is very complex [16] (further complicated by the fact that phosphoserines/phosphothreonines are located not only in the unstructured region of Atg1, but also in two structured functional domains, the N-terminal kinase domain and the C-terminal early autophagy targeting/tethering [EAT] domain; Fig. 1). The crystal structure of full-length Atg1 is not available, but the structure of the EAT domain from K. marxianus Atg1 has been solved (PDB ID: 4P1N; [17]). This domain forms an α–helical structure, where three N-terminal helices are stable and participate in dimerization of the EAT domain, whereas three C-terminal helices are highly dynamic and are stabilized by interaction with Atg13 [18]. Since the kinase domain of Atg1 from K. marxianus has not been crystalized, we used the SWISS model workspace [19–21] to build the homology model of this domain (Fig. 1) using the crystal structure of the human ULK1 kinase domain (PDB ID: 4WNO, [22]) as the template. In this model, the Atg1 target sequence covers 71% of the template. Autophosphorylated Thr180 of ULK1 in the activation loop maps in the model on Thr222 in K. marxianus Atg1, which is homologous to Thr226 in S. cerevisiae (Fig. 1).
ULK1 is the mammalian homolog of Atg1. The crystal structure of the human ULK1 kinase domain in a complex with a small-molecule inhibitor (PDB ID: 4WNO, [22]) is the only structure of the Atg1/ULK1 kinase domain currently available (Fig. 1). The ULK1-EAT domain has not been crystalized. We attempted to build the homology model of the ULK1-EAT domain based on the EAT domain of Atg1 from K. marxianus as the template (PDB ID: 4P1N, [17]), but the resulting model did not have a good coverage between the target and the template. The PONDR-FIT prediction from the amino acid sequence of human ULK1 shows, as expected, that the kinase and EAT domains are structurally stable protein regions. In contrast, the serine- and proline-rich spacer that connects them is a long intrinsically disordered protein region (IDPR) that contains many phosphorylation sites. Thr180 in the kinase domain is the autophosphorylation site necessary for kinase activity [22, 23].
AMPK-dependent phosphorylation of Ser556 and Thr660 (human protein numbering) facilitates interaction of ULK1 with the YWHAB/14-3-3 protein [23, 24]. Ser556 in ULK1 is also one of four AMPK targets (along with Ser467, Thr575, and Ser638). Phosphorylation of these sites by AMPK is required for mitochondrial homeostasis and cell survival during starvation [25]. Phosphorylation of Ser556, Ser638, and Thr660 in ULK1 is also important for correct localization of ATG9 to perinuclear clusters [24]. Interaction between ULK1 and AMPK takes place in nutrient-rich conditions and at early time points of starvation. After approximately 30 min of starvation, these two kinases dissociate due to dephosphorylation on Ser638 and Ser758 of ULK1. Ser638 and Ser758 in ULK1 are phosphorylated by the target of rapamycin complex 1 (TORC1) in nutrient-rich conditions, where phosphorylation on Ser638 is also maintained by AMPK; this latter PTM facilitates the interaction between AMPK and ULK1. Phosphorylated Ser638 facilitates proper phosphorylation of Ser758 by TORC1 and phosphoserine 758 is in turn required for association of ULK1 with AMPK [26].
TORC1 and AMPK are not the only kinases that can phosphorylate ULK1. AKT-mediated phosphorylation of ULK1 on Ser775 inhibits autophagy in response to insulin [23]. Some phosphorylation sites are also found in the EAT domain of ULK1. Ser1042 and Thr1046 in human ULK1 are homologous to Ser1043 and Ser1047 in the mouse protein, where autophosphorylation on Ser1047 promotes phosphorylation on Ser1043 [27]. Ser866 and Ser912 are homologous to phosphoserines in the EAT domain of mouse ULK1 [27].
Besides phosphorylation, ULK1 is also acetylated and ubiquitinated. There are two acetylation sites in ULK1. KAT5/TIP60 acetylase modifies K162 and K607 (human protein numbering), which promotes kinase activity and autophagy induction [28]. Ubiquitination of ULK1 by Lys63-linked chains promotes protein stability and function in autophagy [29]. The exact ubiquitination sites in ULK1 were not determined; however, ModPred predicts with high confidence ubiquitination of K376 and K493 in human ULK1 (Fig. 1).
Atg13 is a large highly dynamic protein. In S. cerevisiae, approximately 270 N-terminal residues of Atg13 fold into the HORMA domain, which is the only structured part of the protein, and is required for recruitment of Atg14 to the PAS (PDB ID: 4J2G, [30]). The remaining 470 residues of Atg13 make up an IDPR that is heavily regulated by phosphorylation and dephosphorylation [17, 31, 32] (Fig. 1). PKA mediates S. cerevisiae Atg13 phosphorylation on Ser344, Ser437, and Ser581. These phosphoserines negatively regulate localization of Atg13 to the PAS; specifically, these PTMs interfere with Atg13-Atg17 interaction, but not Atg13-Atg1 interaction [31]. TORC1 phosphorylates yeast Atg13 at least on Ser348, Ser496, Ser535, and Ser541, and possibly also on Ser437, Ser438, Ser646, and Ser649. Dephosphorylation of these residues directly induces autophagy, as it allows for association with Atg1 kinase [33]. Recent biochemical and mutagenesis experiments identified four serines (Ser494, Ser496, Ser515, and Ser517) in the MIM(C) region of Atg13 (492–521; S. cerevisiae numbering), dephosphorylation of which increases the binding affinity of Atg13 for Atg1 upon starvation. Thus, this region is regulatory, whereas the nearby MIM(N) region of Atg13 (460–491; S. cerevisiae numbering) is the basal binding site for Atg1, and is dephosphorylated at Thr483 and Ser484 to promote the interaction between these two proteins [17]. In the context of the Atg1-EAT crystal structure, Atg13-MIN(N) binds to the dynamic C-terminal subdomain of Atg1-EAT resulting in stabilization, whereas Atg13-MIN(C) binds to the N-terminal Atg1-EAT subdomain that dimerizes [17, 18]. Atg13 also binds Atg17 via a short region between residues 424 and 436. Phosphorylation of this region is again a major factor regulating Atg13-Atg17 interaction. Specifically, Ser428 and Ser429 of Atg13 are dephosphorylated upon starvation, which removes electrostatic repulsion forces with Asp427 in Atg17 and increases the binding affinity of Atg13 for Atg17 [17]. Atg13 appears to be tightly bound in the subcomplex with Atg1, and this subcomplex associates with the stable Atg17-Atg31-Atg29 subcomplex [18].
Along with the autophagy initiation complex, autophagosome formation requires the presence of the class III phosphatidylinositol 3-kinase (PtdIns3K) complex. This complex is responsible for production of PtdIns-3-phosphate (PtdIns3P) that is needed for recruitment of certain components of the autophagy machinery and phagophore expansion. The autophagy-promoting PtdIns3K complex in yeast consists of Vps34-Vps15-Vps30/Atg6-Atg14-Atg38; in humans there are multiple PtdIns3K complexes including two stimulating complexes consisting of PIK3C3/VPS34-PIK3R4/VPS15/p150-BECN1-and either ATG14 or UVRAG, where ATG14 and UVRAG are orthologs of yeast Atg14 and Atg38, respectively.
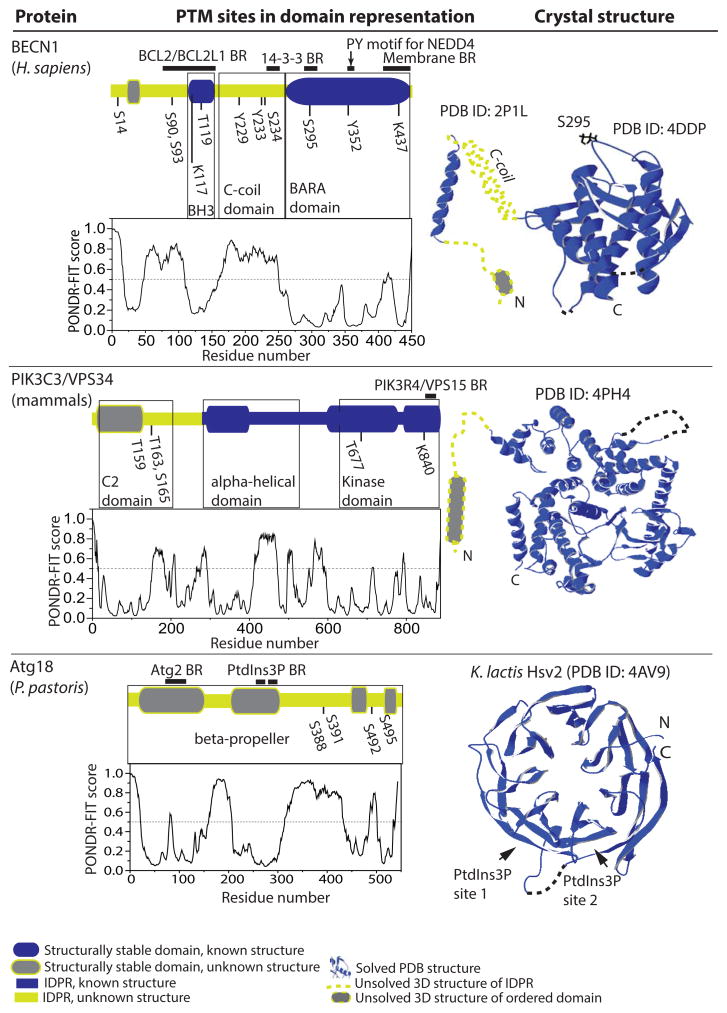
BECN1 in mammalian cells (Vps30/Atg6 in yeast) is the most studied subunit of the PtdIns3K complex, in part because it is a target of multiple regulatory signals. Both the mammalian and yeast protein contains coiled-coil and β–α repeated, autophagy-specific (BARA) domains. BECN1 also has a short BH3 domain preceding the coiled-coil domain. The BARA domain of S. cerevisiae Vps30/Atg6 (PDB ID:3VP7, [34]), the BARA and BH3 domains of human BECN1 (PDB ID: 4DDP, [35]; and PDBID: 2P1L, [36]) and the coiled-coil region of rat BECN1 (PDB ID: 3Q8T, [37]) have been crystalized. Analysis of the amino acid sequence of BECN1 by the PONDR-FIT algorithm shows that the BH3 and BARA domains are structurally stable protein regions. In contrast, the coiled-coil domain arises from an IDPR, and is the result of a disorder-to-order transition, probably induced by dimerization. BECN1 contains a number of PTM sites (Fig. 2). The interaction of BECN1 with negative regulators of autophagy (BCL2, KIAA0226/Rubicon) is increased via its phosphorylation on three tyrosine residues (Tyr229, Tyr233, and Tyr352). This phosphorylation, mediated by the EGFR (epidermal growth factor receptor) tyrosine kinase, promotes formation of homodimers that are unable to bind PIK3C3 and promote autophagy [38]. The BH3 domain is directly involved in binding of BCL2 or BCL2L1/Bcl-XL [36], which inhibits autophagy. Phosphorylation of BECN1 on Thr119 (located within the BH3 domain) by DAPK (death-associated protein kinase) promotes dissociation of BECN1 from BCL2 or BCL2L1, and induces autophagy [39]. The ULK1 kinase phosphorylates BECN1 on Ser14 [40], and this step is required for full autophagy induction.
Figure 2.
Structure-related analysis of posttranslationally modified proteins related to PtdIns3P synthesis or binding (BECN1, PIK3C3/VPS34, and Atg18). The PONDR-FIT profile of human BECN1, mammalian PIK3C3/VPS34, and P. pastoris Atg18 are included, as these proteins were not analyzed in ref. [9]. For other details, refer to the Figure 1 legend. C-coil, coiled-coil.
Finding that ULK1 mediates phosphorylation on Ser14 of BECN1 revealed a direct link between the autophagy initiation complex and the PdIns3K complex. Autophagy is also positively regulated by ATG14-controlled phosphorylation of BECN1 on Ser90 and Ser93 [41]. Conversely, AKT-mediated phosphorylation of BECN1 on Ser234 and Ser295 creates YWHAB/14-3-3 protein binding sites and inhibits autophagy via formation of a BECN1-YWHA-VIM/vimentin intermediate filament complex [42]. Phosphorylation is not the only posttranslational modification of BECN1. Ubiquitination of BECN1 on Lys117 by TRAF6 promotes autophagy by regulating BECN1 interaction with the negative regulator BCL2 [43]. AMBRA1-mediated ubiquitination on Lys437 increases activity of the PtdIns3K complex, and thus autophagy [44]. The E3 ubiquitin protein ligase NEDD4 ubiquitinates BECN1 by Lys11- and Lys63-linked chains via binding to the conserved PY motif (349LPLY in human BECN1), and thereby promotes its degradation [45].
PIK3C3/VPS34 is a kinase that is not categorized as an Atg protein. We included Vps34 in this minireview due to its importance for autophagy, the findings on its posttranslational modifications, and the recently reported insight into the structure of the human protein. PIK3C3 is an approximately 100-kDa protein that includes a kinase domain, an α-helical domain and the C2 domain involved in phospholipid binding (Fig. 2). Crystal structures of the kinase and helical domains of Pi3K59F/Vps34 from Drosophila melanogaster (PDB ID: 2X6H, [46]) and human PIK3C3 (PDB ID: 4PH4, [47]) were solved and reveal a similar overall topology. PIK3C3/VPS34 is directly phosphorylated on Thr159 by CDK1 (cyclin-dependent kinase 1) and CDK5, which inhibits its interaction with BECN1 [48]. In the absence of ATG14, PIK3C3 is phosphorylated by AMPK on Thr163 and Ser165. The presence of ATG14 prevents this phosphorylation and allows for phosphorylation of BECN1 by AMPK, thereby switching AMPK-mediated inhibition to activation [49]. Conversely, PRKD/protein kinase D activates PIK3C3 by phosphorylation in response to oxidative stress on multiple sites including Thr677 [50]. PIK3C3 is also SUMOylated on Lys840. This TRIM28/KAP1 protein-mediated modification increases the activity of PIK3C3 and promotes autophagosome formation [51].
In yeast, the PtdIns3P generated by PtdIns3K on the lipid membrane allows for recruitment of the Atg18-Atg2 subcomplex to the growing phagophore; however, the molecular function of these two proteins is still unknown, although they participate in regulating the localization of Atg9 [52].
Atg18 has two paralogs, Atg21 and Hsv2, in yeast. Hsv2 is a structured protein that was amenable to crystallization. The crystal structure of Kluyveromyces lactis Hsv2 (PDB ID:4AV9, [53]) reveals that this protein forms a seven-bladed β–propeller, where blades 5 and 6 create two PtdIns3P binding sites (Fig. 2). The hydrophobic loop of Hsv2 (partially missing from the crystal structure) provides an additional anchor for membrane binding [54]). The Atg18 amino acid sequence contains the conserved motif FRRG that separates two sequences homologous to those that create the PtdIns3P binding sites 1 and 2 in Hsv2. The Hsv2 β–propeller structure has been mapped onto Atg18 and mutational analysis confirms the presence of two PtdIns3P binding sites in S. cerevisiae Atg18 [54]. Another study showed that two sequences at the N terminus of S. cerevisiae Atg18 that map onto blade 2 of the β–propeller mediate its binding to Atg2 [55]. Atg18 from Pichia pastoris is phosphorylated on Ser388, Ser391, Ser492, and Ser495. This modification negatively regulates binding of the protein to the membrane [56].
PTM-modified structures promoting phagophore elongation
An essential family of proteins that arrives at the PAS after autophagy initiation is the Atg8/LC3/GABARAP family.
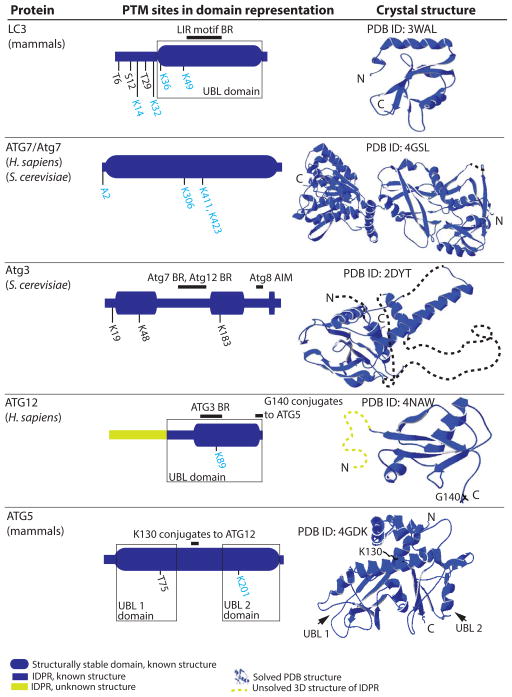
Atg8 in yeast (LC3/GABARAP in mammalian cells) contains a ubiquitin-like (UBL) domain packed against two α–helices at the N terminus. Figure 3 shows the crystal structure of human LC3A (PDB ID: 3WAL, [57]), which represents one of numerous crystal structures of the Atg8/LC3/GABARAP family that have been deposited in the Protein Data Bank (http://www.rcsb.org/pdb/home/home.do). The UBL domain in Atg8/LC3/GABARAP forms two hydrophobic pockets that bind all autophagy receptor proteins that carry the functional motif referred to as LIR/AIM (LC3-interacting region/Atg8-interacting motif) in their sequence [58]). In mammals, LC3 activity is regulated by phosphorylation and acetylation. PKA mediates phosphorylation of human LC3 on Ser12, which inhibits protein recruitment onto the phagophore membrane and suppresses autophagy [59]). Phosphorylation of rat LC3 on Thr6 and Thr29 by PRKC/protein kinase C has no impact on autophagy, and thus the role of these PTMs, if any, is unclear [60]. Acetylation of human LC3 by the acetyltransferase EP300/p300 has an inhibitory effect on autophagy [61], whereas deacetylation of human and mouse LC3 by SIRT1 stimulates autophagy [62]. Because the acetylation/deacetylation sites were not determined in these studies, we show the sites on human LC3A (K49) and LC3C (K14, K32, and K36) predicted by the ModPred predictor of posttranslational modification. These lysines were predicted with low confidence, and are the only predicted sites.
Figure 3.
Structure-related analysis of posttranslationally modified proteins related to Atg8/LC3 conjugation (LC3, ATG7/Atg7, Atg3, ATG12, and ATG5). LC3, ATG7, ATG12, and ATG5 have PONDR-FIT profiles very similar to their S. cerevisiae homologs [9]. For other details, please refer to the Figure 1 legend.
The Atg8/LC3/GABARAP proteins are conjugated to lipids to function in phagophore elongation. The covalent lipid-protein attachment requires activation of the Atg8/LC3/GABARAP proteins via the E1 enzyme, Atg7/ATG7, and then conjugation with a ubiquitin-conjugating enzyme (E2; Atg3/ATG3), which is facilitated by the E3 scaffold complex (Atg12–Atg5-Atg16/ATG12–ATG5-ATG16L1).
ATG7/Atg7 is the activating enzyme (E1) that is well-structured and hence amenable to crystallization. The crystal structure of the yeast protein in a complex with its interacting partners (PDB ID:4GSL, [63]) reveals that it forms a homodimer, where dimerizing C-terminal domains are each flexibly connected to the corresponding N-terminal domain. Figure 3 shows only one monomer of this dimer. A recent review summarizes many structural aspects of the autophagy conjugation system [64]. Briefly, for Atg7 the conserved shoulder groove in its N-terminal domain interacts with two E2 enzymes (Atg3 and Atg10) in a very different way. The flexible region of Atg3 dislocates and undergoes a disorder-to-order transition, forming an α-helix upon binding to Atg7. In contrast, Atg10 binds Atg7 via a β-hairpin. The function of the E1 enzyme appears to be modulated by PTM. Human ATG7 is acetylated by EP300 and deacetylated by SIRT1, with the same effects that have been observed for human LC3 [61, 62]. ModPred predicts with medium confidence that ATG7 is N-terminally acetylated on Ala2 and acetylated on three lysines (K306, K411, K423), two of which (K411, K423) are novel predictions.
Atg3 is one of two ubiquitin-conjugating enzymes (E2) required for autophagy to proceed. It is a relatively dynamic protein [9] containing a long flexible region (an IDPR) in the middle of the sequence. The solution structure of Atg3 has been solved (PDB ID:2DYT, [65]) (Fig. 3). Comparison of this structure with the crystal structure of Atg3 in the bound form (PDB ID: 4GSL, [63]) reveals that Atg3 undergoes a significant conformational change upon binding to Atg7. A molecular mechanism underlying functional reasons for this plasticity is still not well understood. It is possible that the IDPR of Atg3 serves as a scaffolding backbone for spatiotemporally regulated assembly in the complex with several partners (Atg8, Atg7, Atg12). In such a case, the presence of the Atg3-IDPR could prevent steric restrictions in the complex, and also allow for overlapping binding sites that Atg3 has for Atg7 and Atg12 [66]. These capabilities are typical features of intrinsically disordered proteins and protein regions, called assemblers [67]. Atg3 is posttranslationally modified by acetylation. In yeast, the histone acetyltransferase Esa1 acetylates Atg3 on K19 and K48, which promotes interaction between Atg3 and Atg8, and also on K183, which activates the enzyme in Atg8 lipidation [68]. K19 and K48 can conversely be deacetylated by the Rpd3 deacetylase, and this modification has an inhibitory effect on autophagy [68].
As mentioned above, the autophagy machinery involves also the E3 enzyme that consists of the Atg12–Atg5 conjugate bound to Atg16. The exact function of this complex is not fully understood, and it is not essential for lipidation of Atg8 [69]. In mammals, the ATG12–ATG5-ATG16L1 complex functions as a scaffold that promotes LC3 lipidation [70].
Mammalian ATG12 (Fig. 3) and its yeast homolog (Atg12) contain a ubiquitin-like domain, as does Atg8/LC3/GABARAP. This domain, however, does not form the entire architecture of the full-length protein. Both human ATG12 (Fig. 3) and yeast Atg12 have a long IDPR at the N terminus. This region is likely regulatory, because in yeast it is not required for conjugation with Atg5 or for autophagy [71]. In both the human and yeast E3 complex, ATG12/Atg12 functions as a binding module for ATG3/Atg3 (PDB ID: 4NAW, [72]; PDB ID: 3W1S, [73]). In the human E3 complex, the ATG12-ATG3 interface is on the opposite side of the ATG12 molecule than the ATG12–ATG5 interface. For binding of ATG3, ATG12 forms a hydrophobic pocket corresponding to the hydrophobic pocket of LC3 that binds a hydrophobic residue (L/I/V) in the LIR motif. The second, aromatic pocket seen in LC3 is not present on the surface of ATG12, so ATG12 cannot bind the LIR motif. The hydrophobic pocket on ATG12 interacts with the flexible region of ATG3, specifically with a fragment that undergoes disorder-to-order transition and forms a short β-sheet followed by an α-helix upon binding to ATG12 [72]. So far only two posttranslational modifications were found in ATG12. In human cells, autophagy is inhibited by EP300-mediated acetylation of ATG12, and is promoted by SIRT1-mediated deacetylation of the protein [61, 62]. ModPred predicts K89 as the medium-confidence acetylation site in ATG12 (Fig. 3). Given the long N-terminal IDPR in ATG12/Atg12 that escaped attention so far, it is possible that more PTMs of this protein will be revealed by future studies.
ATG5/Atg5 is a structured protein that functions as a scaffold, bringing together two more flexible partners, ATG12/Atg12 (see above) and the highly dynamic ATG16L1/Atg16. Besides noncovalent contacts, ATG5/Atg5 covalently binds ATG12/Atg12. Human ATG5 makes the covalent bond via Lys130 to Gly140 of ATG12, as revealed by the crystal structure of the human E3 complex (PDB ID: 4GDK, [74]). In yeast, Lys149 of Atg5 covalently binds Gly186 of Atg12 (PDB ID: 3W1S, [73]). The ATG5 fold includes two UBL domains with an α-helix-rich domain between them (Fig. 3). A similar architecture is observed in the yeast homolog [73]. Mammalian ATG5 is modified by phosphorylation and acetylation. Specifically, mouse ATG5 is phosphorylated on Thr75 by mitogen-activated protein kinase (MAPK)/p38, which inhibits starvation-induced autophagy [75]. Autophagy is also inhibited by acetylation of ATG5 by EP300 [61]. Conversely, deacetylation of both human and mouse ATG5 by SIRT1 promotes autophagy [62]. ModPred predicts with medium confidence that the acetylation site in the human form is K201.
PTM-modified structures involved in autophagy cargo recognition
A selective type of autophagy relies on the function of protein receptors that interact with the Atg11 scaffold and with the Atg8/LC3/GABARAP family of proteins, and thereby link the cargo to the autophagy machinery.
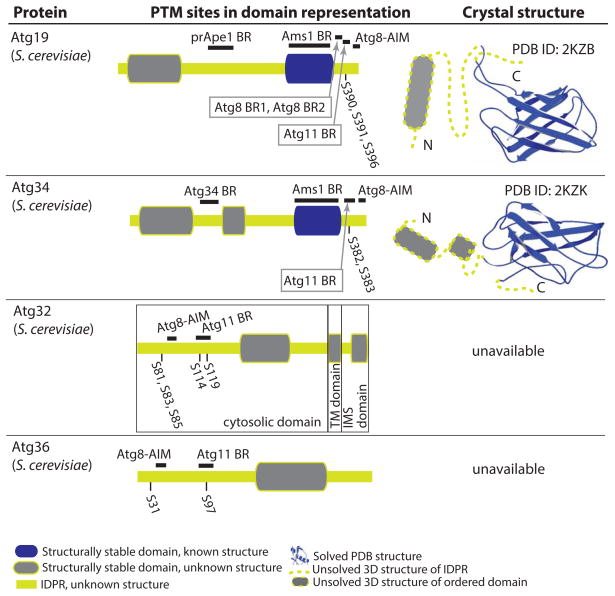
Atg19 is the receptor that functions in biosynthetic delivery of resident vacuolar hydrolases into the yeast vacuole [76]. The structure of the protein is not known, except for the Ams1 binding region (residues 254–367; S. cerevisiae numbering) that folds into a stable structure reminiscent of a β-barrel, as determined by NMR (PDB ID: 2KZB, [77]) (Fig. 4). The C terminus that follows the Ams1-binding region in Atg19 is very important for protein function (Fig. 4). This region contains several Atg8 binding sites [78], an Atg11 binding site, and three phosphorylation sites (S390, S391, S396) [79]. Direct phosphorylation of these serine residues by Hrr25 kinase in the Atg11 binding region modulates interaction of Atg19 with Atg11 [79]. PONDR-FIT analysis of the Atg19 amino acid sequence indicates that the C terminus along with the extreme N terminus and the middle domain (residues 124–253) consist of IDPRs. This is consistent with the functions reported for the Atg19 C terminus, because disordered protein regions carry short linear recognition motifs, that are often targets of posttranslational modifications, and/or contain adjacent/overlapping binding sites for multiple structured protein partners [67, 80–82].
Figure 4.
Structure-related analysis of posttranslationally modified autophagy receptors (Atg19, Atg34, Atg32, and Atg36). For other details, please refer to the Figure 1 legend. IMS, intermembrane space; TM, transmembrane.
Atg34 is paralog of Atg19 in yeast that also contains the Ams1 binding domain near the C terminus (residues 246–348; S. cerevisiae numbering) with a known NMR structure (PDB ID: 2KZK, [77]) (Fig. 4). As in Atg19, the disordered C terminus of Atg34 carries the Atg11 and Atg8 binding regions, and Hrr25-mediated phosphorylation on Ser382 and Ser383 regulates its binding to the Atg11 scaffold. Atg34 can self-dimerize via a somewhat flexible region between residues 130 and 157 [79] (Fig. 4).
Atg32 is the mitophagy receptor in the yeast S. cerevisiae. A crystal or NMR structure for this protein is not available. Prediction from amino acid sequence shows that the transmembrane and intermembrane space domains are structurally stable, in contrast to the largely disordered cytosolic domain (Fig. 4). The cytosolic domain binds Atg8 and Atg11, and is also regulated by phosphorylation. Phosphoserines on position 119 and especially 114 in S. cerevisiae Atg32 mediate its binding to Atg11 [83, 84], while phosphorylation of the same protein on Ser81, Ser83, and Ser85 upstream of the LIR motif facilitates its binding to Atg8 [84]. The MAPK Hog1 is involved in this phosphoryation, but does not phosphorylate Atg32 directly [83]; instead, this modification is carried out by Cka1/2 (casein kinase 2), and the role of Hog1 in this process is not known [52].
Atg36 is the pexophagy receptor in S. cerevisiae. It has one structurally stable region, whereas the rest of the protein consists of IDPRs; no crystal or NMR structure is available (Fig. 4). As in Atg32, binding of S. cerevisiae Atg36 to Atg8 and Atg11 is regulated by phosphorylation. In this case, phosphorylation on Ser31 facilitates binding of the Atg36 AIM motif to Atg8, and the same modification on Ser97 is required for interaction of Atg36 with Atg11 [84].
Conclusions
The structure-related analysis of posttranslational modifications of proteins presented here (Fig. 1–Fig. 4) shows that phosphorylation is the most frequent modification on autophagy proteins and that the majority of experimentally verified phosphoserines/phosphothreonines are located within disordered regions of these proteins. This is consistent with what has been observed for other phosphorylated proteins [85] and with the finding that kinases typically bind substrates with high specificity but weak affinity [80], which is the characteristic typical for intrinsically disordered proteins/IDPRs [67]. It is very likely that more posttranslational modifications of the autophagy proteins will emerge from future studies. New promising candidates certainly include Atg11, Atg14, Atg16, Atg20, Atg21, Atg23, and Atg33, as amino acid sequences of these proteins contain regions with a propensity to disorder [9].
Phosphorylation can have various consequences on protein structure or a protein region. For example, it can temporarily induce disorder-to-order transition or order-to-disorder transition in a protein [86, 87]. In transient functional unfolding, phosphorylation may act as an intramolecular modulator that shifts a protein conformation into a less ordered state, or it can prevent disorder-to-order transition of a molecular recognition sequence that in the unphosphorylated state folds into an inducible structured element, for example an α-helix. The latter mechanism was observed in EIF4E (eukaryotic translation initiation factor 4E) and its binding proteins [88], but it may also be present in autophagy, for example in the Atg1-Atg13 interaction. The low binding affinity of the Atg13-MIM(C) domain to the Atg1-MIT domain is due to phosphorylated serines in the IDPR of Atg13; dephosphorylation of these serines increases the binding affinity between the Atg13-MIM(C) and the Atg1-MIT [17]. Because the Atg13-MIM(C) undergoes disorder-to-order transition (i.e., it folds into an α-helix) during binding to the Atg1-MIT, the molecular mechanism explaining the change in affinity of binding during induction of autophagy could be that phosphoserines are the modulators that hold the Atg13-MIM(C) domain unfolded until it is dephosphorylated and can fold into an α-helix. Phosphorylation/dephosphorylation and posttranslational modification in general is an elegant molecular mechanism for a fast switch on or switch off that could be very useful in any regulatory pathway requiring a prompt response. It is reasonable to assume that the PTMs discovered so far in autophagy represent only the “tip of the iceberg”.
Acknowledgments
This work was supported by the National Institute of Health grant number R01GM053396 to D.J.K.
Abbreviations
- AMPK
AMP-activated protein kinase
- ATG
autophagy-related
- BARA
β–α repeated, autophagy-specific
- EAT
early autophagy targeting/tethering
- IDPR
intrinsically disordered protein region
- LIR
LC3-interacting region
- MAPK
mitogen-activated protein kinase
- PAS
phagophore assembly site
- PtdIns3K
phosphatidylinositol 3-kinase
- PtdIns3P
phosphatidylinositol 3-phopsphate
- PKA
protein kinase A
- PTM
posttranslational modification
- TORC
target of rapamycin complex
- UBL
ubiquitin-like
Footnotes
Author contributions
HP summarized the literature, created the initial figures and wrote the paper. DJK edited the paper, revised the figures and contributed to the writing.
References
- 1.Wani WY, Boyer-Guittaut M, Dodson M, Chatham J, Darley-Usmar V, Zhang J. Regulation of autophagy by protein post-translational modification. Laboratory investigation. 2015;95:14–25. doi: 10.1038/labinvest.2014.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reggiori F, Klionsky DJ. Autophagic Processes in Yeast: Mechanism, Machinery and Regulation. Genetics. 2013;194:341–361. doi: 10.1534/genetics.112.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng YC, He D, Yao ZY, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stanley RE, Ragusa MJ, Hurley JH. The beginning of the end: how scaffolds nucleate autophagosome biogenesis. Trends Cell Biol. 2014;24:73–81. doi: 10.1016/j.tcb.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, Szabo B, Tompa P, Chen J, Uversky VN, Obradovic Z, Dunker AK. DisProt: the database of disordered proteins. Nucleic Acids Res. 2007;35:D786–D793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papinski D, Schuschnig M, Reiter W, Wilhelm L, Barnes CA, Maiolica A, Hansmann I, Pfaffenwimmer T, Kijanska M, Stoffel I, Lee SS, Brezovich A, Lou JH, Turk BE, Aebersold R, Ammerer G, Peter M, Kraft C. Early Steps in Autophagy Depend on Direct Phosphorylation of Atg9 by the Atg1 Kinase (vol 53, pg 471, 2014) Mol Cell. 2014;53:515–515. doi: 10.1016/j.molcel.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mao K, Chew LH, Inoue-Aono Y, Cheong H, Nair U, Popelka H, Yip CK, Klionsky DJ. Atg29 phosphorylation regulates coordination of the Atg17-Atg31-Atg29 complex with the Atg11 scaffold during autophagy initiation. P Natl Acad Sci USA. 2013;110:E2875–E2884. doi: 10.1073/pnas.1300064110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 Complex as a Scaffold for Autophagosome Biogenesis. Cell. 2012;151:1501–1512. doi: 10.1016/j.cell.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Popelka H, Uversky VN, Klionsky DJ. Identification of Atg3 as an intrinsically disordered polypeptide yields insights into the molecular dynamics of autophagy-related proteins in yeast. Autophagy. 2014;10:1093–1104. doi: 10.4161/auto.28616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sekito T, Kawamata T, Ichikawa R, Suzuki K, Ohsumi Y. Atg17 recruits Atg9 to organize the pre-autophagosomal structure. Genes Cells. 2009;14:525–538. doi: 10.1111/j.1365-2443.2009.01299.x. [DOI] [PubMed] [Google Scholar]
- 11.Kabeya Y, Noda NN, Fujioka Y, Suzuki K, Inagaki F, Ohsumi Y. Characterization of the Atg17-Atg29-Atg31 complex specifically required for starvation-induced autophagy in Saccharomyces cerevisiae. Biochem Bioph Res Co. 2009;389:612–615. doi: 10.1016/j.bbrc.2009.09.034. [DOI] [PubMed] [Google Scholar]
- 12.Feng WZ, Wu T, Dan XY, Chen YL, Li L, Chen S, Miao D, Deng HT, Gong XQ, Yu L. Phosphorylation of Atg31 is required for autophagy. Protein Cell. 2015;6:288–296. doi: 10.1007/s13238-015-0138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pejaver V, Hsu WL, Xin FX, Dunker AK, Uversky VN, Radivojac P. The structural and functional signatures of proteins that undergo multiple events of post-translational modification. Protein Sci. 2014;23:1077–1093. doi: 10.1002/pro.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kijanska M, Dohnal I, Reiter W, Kaspar S, Stoffel I, Ammerer G, Kraft C, Peter M. Activation of Atg1 kinase in autophagy by regulated phosphorylation. Autophagy. 2010;6:1168–1178. doi: 10.4161/auto.6.8.13849. [DOI] [PubMed] [Google Scholar]
- 15.Yeh YY, Shah KH, Chou CC, Hsiao HH, Wrasman KM, Stephan JS, Stamatakos D, Khoo KH, Herman PK. The identification and analysis of phosphorylation sites on the Atg1 protein kinase. Autophagy. 2011;7:716–726. doi: 10.4161/auto.7.7.15155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie Y, Kang R, Sun X, Zhong M, Huang J, Klionsky DJ, Tang D. Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy. 2015;11:28–45. doi: 10.4161/15548627.2014.984267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujioka Y, Suzuki SW, Yamamoto H, Kondo-Kakuta C, Kimura Y, Hirano H, Akada R, Inagaki F, Ohsumi Y, Noda NN. Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat Struct Mol Biol. 2014;21:513–521. doi: 10.1038/nsmb.2822. [DOI] [PubMed] [Google Scholar]
- 18.Stjepanovic G, Davies CW, Stanley RE, Ragusa MJ, Kim DJ, Hurley JH. Assembly and dynamics of the autophagy-initiating Atg1 complex. P Natl Acad Sci USA. 2014;111:12793–12798. doi: 10.1073/pnas.1407214111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 20.Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc. 2009;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- 21.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazarus MB, Novotny CJ, Shokat KM. Structure of the Human Autophagy Initiating Kinase ULK1 in Complex with Potent Inhibitors. Acs Chem Biol. 2015;10:257–261. doi: 10.1021/cb500835z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bach M, Larance M, James DE, Ramm G. The serine/threonine kinase ULK1 is a target of multiple phosphorylation events. Biochem J. 2011;440:283–291. doi: 10.1042/BJ20101894. [DOI] [PubMed] [Google Scholar]
- 24.Mack HID, Zheng B, Asara JM, Thomas SM. AMPK-dependent phosphorylation of ULK1 regulates ATG9 localization. Autophagy. 2012;8:1197–1214. doi: 10.4161/auto.20586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ. Phosphorylation of ULK1 (hATG1) by AMP-Activated Protein Kinase Connects Energy Sensing to Mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shang LB, Chen S, Du FH, Li S, Zhao LP, Wang XD. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. P Natl Acad Sci USA. 2011;108:4788–4793. doi: 10.1073/pnas.1100844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dorsey FC, Rose KL, Coenen S, Prater SM, Cavett V, Cleveland JL, Caldwell-Busby J. Mapping the Phosphorylation Sites of Ulk1. J Proteome Res. 2009;8:5253–5263. doi: 10.1021/pr900583m. [DOI] [PubMed] [Google Scholar]
- 28.Lin SY, Li TY, Liu Q, Zhang CX, Li XT, Chen Y, Zhang SM, Lian GL, Liu Q, Ruan K, Wang Z, Zhang CS, Chien KY, Wu JW, Li QX, Han JH, Lin SC. GSK3-TIP60-ULK1 Signaling Pathway Links Growth Factor Deprivation to Autophagy. Science. 2012;336:477–481. doi: 10.1126/science.1217032. [DOI] [PubMed] [Google Scholar]
- 29.Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, Cecconi F. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol. 2013;15:406–+. doi: 10.1038/ncb2708. [DOI] [PubMed] [Google Scholar]
- 30.Jao CC, Ragusa MJ, Stanley RE, Hurley JH. A HORMA domain in Atg13 mediates PI 3-kinase recruitment in autophagy. P Natl Acad Sci USA. 2013;110:5486–5491. doi: 10.1073/pnas.1220306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. P Natl Acad Sci USA. 2009;106:17049–17054. doi: 10.1073/pnas.0903316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alers S, Wesselborg S, Stork B. ATG13 Just a companion, or an executor of the autophagic program? Autophagy. 2014;10:944–956. doi: 10.4161/auto.28987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y. Tor Directly Controls the Atg1 Kinase Complex To Regulate Autophagy. Mol Cell Biol. 2010;30:1049–1058. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noda NN, Kobayashi T, Adachi W, Fujioka Y, Ohsumi Y, Inagaki F. Structure of the Novel C-terminal Domain of Vacuolar Protein Sorting 30/Autophagy-related Protein 6 and Its Specific Role in Autophagy. J Biol Chem. 2012;287:16256–16266. doi: 10.1074/jbc.M112.348250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang WJ, Choi WY, Hu WQ, Mi N, Guo Q, Ma MS, Liu M, Tian Y, Lu PL, Wang FL, Deng HT, Liu L, Gao N, Yu L, Shi YG. Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein. Cell Res. 2012;22:473–489. doi: 10.1038/cr.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oberstein A, Jeffrey PD, Shi YG. Crystal structure of the Bcl-X-L-beclin 1 peptide complex - Beclin 1 is a novel BH3-only protein. J Biol Chem. 2007;282:13123–13132. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- 37.Li XH, He LQ, Che KH, Funderburk SF, Pan LF, Pan NN, Zhang MJ, Yue ZY, Zhao YX. Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat Commun. 2012:3. doi: 10.1038/ncomms1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei YJ, Zou ZJ, Becker N, Anderson M, Sumpter R, Xiao GH, Kinch L, Koduru P, Christudass CS, Veltri RW, Grishin NV, Peyton M, Minna J, Bhagat G, Levine B. EGFR-Mediated Beclin 1 Phosphorylation in Autophagy Suppression, Tumor Progression, and Tumor Chemoresistance. Cell. 2013;154:1269–1284. doi: 10.1016/j.cell.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-X-L and induction of autophagy. Embo Rep. 2009;10:285–292. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russell RC, Tian Y, Yuan HX, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–+. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fogel AI, Dlouhy BJ, Wang CX, Ryu SW, Neutzner A, Hasson SA, Sideris DP, Abeliovich H, Youle RJ. Role of Membrane Association and Atg14-Dependent Phosphorylation in Beclin-1-Mediated Autophagy. Mol Cell Biol. 2013;33:3675–3688. doi: 10.1128/MCB.00079-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang RC, Wei YJ, An ZY, Zou ZJ, Xiao GH, Bhagat G, White M, Reichelt J, Levine B. Akt-Mediated Regulation of Autophagy and Tumorigenesis Through Beclin 1 Phosphorylation. Science. 2012;338:956–959. doi: 10.1126/science.1225967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi CS, Kehrl JH. TRAF6 and A20 Regulate Lysine 63-Linked Ubiquitination of Beclin-1 to Control TLR4-Induced Autophagy. Sci Signal. 2010:3. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xia PY, Wang S, Du Y, Zhao ZN, Shi L, Sun L, Huang GL, Ye BQ, Li C, Dai ZH, Hou N, Cheng X, Sun QY, Li L, Yang X, Fan ZS. WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. Embo J. 2013;32:2685–2696. doi: 10.1038/emboj.2013.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Platta HW, Abrahamsen H, Thoresen SB, Stenmark H. Nedd4-dependent lysine-11-linked polyubiquitination of the tumour suppressor Beclin 1. Biochem J. 2012;441:399–406. doi: 10.1042/BJ20111424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller S, Tavshanjian B, Oleksy A, Perisic O, Houseman BT, Shokat KM, Williams RL. Shaping Development of Autophagy Inhibitors with the Structure of the Lipid Kinase Vps34. Science. 2010;327:1638–1642. doi: 10.1126/science.1184429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dowdle WE, Nyfeler B, Nagel J, Elling RA, Liu SM, Triantafellow E, Menon S, Wang ZC, Honda A, Pardee G, Cantwell J, Luu C, Cornella-Taracido I, Harrington E, Fekkes P, Lei H, Fang Q, Digan ME, Burdick D, Powers AF, Helliwell SB, D’Aquin S, Bastien J, Wang H, Wiederschain D, Kuerth J, Bergman P, Schwalb D, Thomas J, Ugwonali S, Harbinski F, Tallarico J, Wilson CJ, Myer VE, Porter JA, Bussiere DE, Finan PM, Labow MA, Mao XH, Hamann LG, Manning BD, Valdez RA, Nicholson T, Schirle M, Knapp MS, Keaney EP, Murphy LO. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat Cell Biol. 2014;16:1069–+. doi: 10.1038/ncb3053. [DOI] [PubMed] [Google Scholar]
- 48.Furuya T, Kim M, Lipinski M, Li JY, Kim D, Lu T, Shen Y, Rameh L, Yankner B, Tsai LH, Yuan JY. Negative Regulation of Vps34 by Cdk Mediated Phosphorylation. Mol Cell. 2010;38:500–511. doi: 10.1016/j.molcel.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, Guan KL. Differential Regulation of Distinct Vps34 Complexes by AMPK in Nutrient Stress and Autophagy. Cell. 2013;152:290–303. doi: 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eisenberg-Lerner A, Kimchi A. PKD is a kinase of Vps34 that mediates ROS-induced autophagy downstream of DAPk. Cell Death Differ. 2012;19:788–797. doi: 10.1038/cdd.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang YH, Fiskus W, Yong B, Atadja P, Takahashi Y, Pandita TK, Wang HG, Bhalla KN. Acetylated hsp70 and KAP1-mediated Vps34 SUMOylation is required for autophagosome creation in autophagy. P Natl Acad Sci USA. 2013;110:6841–6846. doi: 10.1073/pnas.1217692110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 53.Krick R, Busse RA, Scacioc A, Stephan M, Janshoff A, Thumm M, Kuhnel K. Structural and functional characterization of the two phosphoinositide binding sites of PROPPINs, a beta-propeller protein family. P Natl Acad Sci USA. 2012;109:E2042–E2049. doi: 10.1073/pnas.1205128109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baskaran S, Ragusa MJ, Boura E, Hurley JH. Two-Site Recognition of Phosphatidylinositol 3-Phosphate by PROPPINs in Autophagy. Mol Cell. 2012;47:339–348. doi: 10.1016/j.molcel.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe Y, Kobayashi T, Yamamoto H, Hoshida H, Akada R, Inagaki F, Ohsumi Y, Noda NN. Structure-based Analyses Reveal Distinct Binding Sites for Atg2 and Phosphoinositides in Atg18. J Biol Chem. 2012;287:31681–31690. doi: 10.1074/jbc.M112.397570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tamura N, Oku M, Ito M, Noda NN, Inagaki F, Sakai Y. Atg18 phosphoregulation controls organellar dynamics by modulating its phosphoinositide-binding activity. J Cell Biol. 2013;202:685–698. doi: 10.1083/jcb.201302067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suzuki H, Tabata K, Morita E, Kawasaki M, Kato R, Dobson RCJ, Yoshimori T, Wakatsuki S. Structural Basis of the Autophagy-Related LC3/Atg13 LIR Complex: Recognition and Interaction Mechanism. Structure. 2014;22:47–58. doi: 10.1016/j.str.2013.09.023. [DOI] [PubMed] [Google Scholar]
- 58.Rogov V, Dotsch V, Johansen T, Kirkin V. Interactions between Autophagy Receptors and Ubiquitin-like Proteins Form the Molecular Basis for Selective Autophagy. Mol Cell. 2014;53:167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 59.Cherra SJ, Kulich SM, Uechi G, Balasubramani M, Mountzouris J, Day BW, Chu CT. Regulation of the autophagy protein LC3 by phosphorylation. J Cell Biol. 2010;190:533–539. doi: 10.1083/jcb.201002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang HB, Cheng DM, Liu WH, Peng JM, Feng J. Protein kinase C inhibits autophagy and phosphorylates LC3. Biochem Bioph Res Co. 2010;395:471–476. doi: 10.1016/j.bbrc.2010.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee IH, Finkel T. Regulation of Autophagy by the p300 Acetyltransferase. J Biol Chem. 2009;284:6322–6328. doi: 10.1074/jbc.M807135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. P Natl Acad Sci USA. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaiser SE, Mao K, Taherbhoy AM, Yu SS, Olszewski JL, Duda DM, Kurinov I, Deng A, Fenn TD, Klionsky DJ, Schulman BA. Noncanonical E2 recruitment by the autophagy E1 revealed by Atg7-Atg3 and Atg7-Atg10 structures. Nat Struct Mol Biol. 2012;19:1242–+. doi: 10.1038/nsmb.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klionsky DJ, Schulman BA. Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nat Struct Mol Biol. 2014;21:336–345. doi: 10.1038/nsmb.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamada Y, Suzuki NN, Hanada T, Ichimura Y, Kumeta H, Fujioka Y, Ohsumi Y, Inagaki F. The crystal structure of Atg3, an autophagy-related ubiquitin carrier protein (E2) enzyme that mediates Atg8 lipidation. J Biol Chem. 2007;282:8036–8043. doi: 10.1074/jbc.M611473200. [DOI] [PubMed] [Google Scholar]
- 66.Qiu Y, Hofmann K, Coats JE, Schulman BA, Kaiser SE. Binding to E1 and E3 is mutually exclusive for the human autophagy E2 Atg3. Protein Sci. 2013;22:1691–1697. doi: 10.1002/pro.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van der Lee R, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, Dunker AK, Fuxreiter M, Gough J, Gsponer J, Jones DT, Kim PM, Kriwacki RW, Oldfield CJ, Pappu RV, Tompa P, Uversky VN, Wright PE, Babu MM. Classification of Intrinsically Disordered Regions and Proteins. Chem Rev. 2014;114:6589–6631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yi C, Ma MS, Ran LL, Zheng JX, Tong JJ, Zhu J, Ma CY, Sun YF, Zhang SJ, Feng WZ, Zhu LY, Le Y, Gong XQ, Yan XH, Hong B, Jiang FJ, Xie ZP, Miao D, Deng H, Yu L. Function and Molecular Mechanism of Acetylation in Autophagy Regulation. Science. 2012;336:474–477. doi: 10.1126/science.1216990. [DOI] [PubMed] [Google Scholar]
- 69.Cao Y, Cheong HS, Song H, Klionsky DJ. In vivo reconstitution of autophagy in Saccharomyces cerevisiae. J Cell Biol. 2008;182:703–713. doi: 10.1083/jcb.200801035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hanada T, Ohsumi Y. Structure-function relationship of Atg12, a ubiquitin-like modifier essential for autophagy. Autophagy. 2005;1:110–118. doi: 10.4161/auto.1.2.1858. [DOI] [PubMed] [Google Scholar]
- 72.Metlagel Z, Otomo C, Takaesu G, Otomo T. Structural basis of ATG3 recognition by the autophagic ubiquitin-like protein ATG12. P Natl Acad Sci USA. 2013;110:18844–18849. doi: 10.1073/pnas.1314755110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Noda NN, Fujioka Y, Hanada T, Ohsumi Y, Inagaki F. Structure of the Atg12-Atg5 conjugate reveals a platform for stimulating Atg8-PE conjugation. Embo Rep. 2013;14:206–211. doi: 10.1038/embor.2012.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Otomo C, Metlagel Z, Takaesu G, Otomo T. Structure of the human ATG12 similar to ATG5 conjugate required for LC3 lipidation in autophagy. Nat Struct Mol Biol. 2013;20:59–U79. doi: 10.1038/nsmb.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Keil E, Hocker R, Schuster M, Essmann F, Ueffing N, Hoffman B, Liebermann DA, Pfeffer K, Schulze-Osthoff K, Schmitz I. Phosphorylation of Atg5 by the Gadd45 beta-MEKK4-p38 pathway inhibits autophagy. Cell Death Differ. 2013;20:321–332. doi: 10.1038/cdd.2012.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shintani T, Huang WP, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Watanabe Y, Noda NN, Kumeta H, Suzuki K, Ohsumi Y, Inagaki F. Selective Transport of alpha-Mannosidase by Autophagic Pathways STRUCTURAL BASIS FOR CARGO RECOGNITION BY Atg19 AND Atg34. J Biol Chem. 2010;285:30026–30033. doi: 10.1074/jbc.M110.143545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sawa-Makarska J, Abert C, Romanov J, Zens B, Ibiricu I, Martens S. Cargo binding to Atg19 unmasks additional Atg8 binding sites to mediate membrane-cargo apposition during selective autophagy. Nat Cell Biol. 2014;16:425–U100. doi: 10.1038/ncb2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pfaffenwimmer T, Reiter W, Brach T, Nogellova V, Papinski D, Schuschnig M, Abert C, Ammerer G, Martens S, Kraft C. Hrr25 kinase promotes selective autophagy by phosphorylating the cargo receptor Atg19. Embo Rep. 2014;15:862–870. doi: 10.15252/embr.201438932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Uversky VN. Intrinsic Disorder-based Protein Interactions and their Modulators. Curr Pharm Design. 2013;19:4191–4213. doi: 10.2174/1381612811319230005. [DOI] [PubMed] [Google Scholar]
- 81.Van Roey K, Uyar B, Weatheritt RJ, Dinkel H, Seiler M, Budd A, Gibson TJ, Davey NE. Short Linear Motifs: Ubiquitous and Functionally Diverse Protein Interaction Modules Directing Cell Regulation. Chem Rev. 2014;114:6733–6778. doi: 10.1021/cr400585q. [DOI] [PubMed] [Google Scholar]
- 82.Tompa P. Multisteric Regulation by Structural Disorder in Modular Signaling Proteins: An Extension of the Concept of Allostery. Chem Rev. 2014;114:6715–6732. doi: 10.1021/cr4005082. [DOI] [PubMed] [Google Scholar]
- 83.Aoki Y, Kanki T, Hirota Y, Kurihara Y, Saigusa T, Uchiumi T, Kang DC. Phosphorylation of Serine 114 on Atg32 mediates mitophagy. Mol Biol Cell. 2011;22:3206–3217. doi: 10.1091/mbc.E11-02-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Farre JC, Burkenroad A, Burnett SF, Subramani S. Phosphorylation of mitophagy and pexophagy receptors coordinates their interaction with Atg8 and Atg11. Embo Rep. 2013;14:441–449. doi: 10.1038/embor.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Johnson LN, Lewis RJ. Structural basis for control by phosphorylation. Chem Rev. 2001;101:2209–2242. doi: 10.1021/cr000225s. [DOI] [PubMed] [Google Scholar]
- 87.Jakob U, Kriwacki R, Uversky VN. Conditionally and Transiently Disordered Proteins: Awakening Cryptic Disorder To Regulate Protein Function. Chem Rev. 2014;114:6779–6805. doi: 10.1021/cr400459c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tait S, Dutta K, Cowburn D, Warwicker J, Doig AJ, McCarthy JEG. Local control of a disorder-order transition in 4E-BP1 underpins regulation of translation via eIF4E. P Natl Acad Sci USA. 2010;107:17627–17632. doi: 10.1073/pnas.1008242107. [DOI] [PMC free article] [PubMed] [Google Scholar]