Abstract
Objective
Sepsis remains a predominant cause of mortality in the ICU, yet strategies to increase survival have proved largely unsuccessful. This study aimed to identify proteins linked to sepsis outcomes using a glycoproteomic approach to target extracellular proteins that trigger downstream pathways and direct patient outcomes.
Design
Plasma was obtained from the LacTATEs cohort. N-linked plasma glycopeptides were quantified by solid-phase extraction coupled with mass spectrometry. Glycopeptides were assigned to proteins using RefSeq and visualized in a heat map. Protein differences were validated by immunoblotting, and proteins were mapped for biological processes using Database for Annotation, Visualization and Integrated Discovery and for functional pathways using Kyoto Encyclopedia of Genes and Genomes databases.
Setting
Hospitalized care.
Measurements and Main Results
A total of 501 glycopeptides corresponding to 234 proteins were identified. Of these, 66 glycopeptides were unique to the survivor group and corresponded to 54 proteins, 60 were unique to the nonsurvivor group and corresponded to 43 proteins, and 375 were common responses between groups and corresponded to 137 proteins. Immunoblotting showed that nonsurvivors had increased total kininogen; decreased total cathepsin-L1, vascular cell adhesion molecule, periostin, and neutrophil gelatinase–associated lipocalin; and a two-fold decrease in glycosylated clusterin (all p < 0.05). Kyoto Encyclopedia of Genes and Genomes analysis identified six enriched pathways. Interestingly, survivors relied on the extrinsic pathway of the complement and coagulation cascade, whereas nonsurvivors relied on the intrinsic pathway.
Conclusion
This study identifies proteins linked to patient outcomes and provides insight into unexplored mechanisms that can be investigated for the identification of novel therapeutic targets. (Crit Care Med 2015; XX:00–00)
Keywords: blood coagulation factors, complement system proteins, computational biology, glycoproteins, proteomics, sepsis
Severe sepsis is a clinical syndrome in which organ dysfunction or hypoperfusion occurs as a result of a robust immune response to infection (1). If sepsis progresses to septic shock, blood pressure decreases dramatically and multiple organ failure may occur. Septic shock is a leading cause of death in patients who are admitted to the ICU, with a mortality rate approximating 30–50%, which results in about 750,000 annual deaths (1, 2). Treatment strategies aim to maintain blood pressure using IV fluids, sustain adequate oxygen levels by administration of supplemental oxygen, and target the bacterial load using broad-range antibiotics, all improve overall patient outcome (3). However, mortality remains high, and therapeutic strategies that further improve survival rates have yet to be elucidated. Thus, additional molecular targets for improved prognosis and therapeutics are needed for resolution of sepsis in the early phase, before the patient progresses to septic shock.
A high-throughput proteomic evaluation of plasma proteins can provide mechanistic insight and identify biomarkers for improved diagnostics (2, 4–6). A current limitation of well-established proteomic techniques, such as 2D gel electrophoresis, is that albumin, a high-abundance plasma protein, limits identification and measurement of changes in low-abundance plasma proteins (7). Albumin depletion protocols have been developed, and currently, several commercial kits are available; however, low-abundance proteins can generate complexes with albumin and be removed through this process, making depletion protocols problematic. Additionally, depletion protocols can remove other proteins, resulting in a low extraction yield (8). Because albumin is not glycosylated, a plasma glycoproteomic evaluation allows for analysis of low-abundance plasma proteins, without requiring the potentially problematic process of albumin depletion.
N-linked glycosylation is the most common glycan modification on human proteins (9). Extracellular proteins, including cell membrane proteins and secreted proteins, are accessible drug targets that hold high potential for diagnosis and therapeutics. Extracellular proteins are more often modified with carbohydrates than intracellular proteins, allowing N-linked glycoproteomic analysis to greatly enrich for the extracellular proteome (10, 11). Because plasma is composed of extracellular proteins, a glycoproteomics approach also lends itself to plasma analysis.
The goal of this study, accordingly, was to identify early changes in N-linked plasma glycoproteins that predict later mortality in patients with sepsis. Plasma samples from sepsis survivors and nonsurvivors were analyzed by solid-phase extraction of glycopeptides using hydrazide chemistry, followed by mass spectrometry (MS) analysis. Computational tools were used to map glycoprotein expression to cellular functions, and the pathways involved with these glycoproteins were mapped.
MATERIALS AND METHODS
Patient Cohort
The LactATEs cohort was a randomized controlled noninferiority trial comparing resuscitation protocols for treating early septic shock (12, 13). Patients admitted to the emergency department were enrolled in the study when the diagnosis of sepsis was made, within 6 hours of presentation. Inclusion criteria have been reported previously (12). Blood samples were collected into EDTA tubes and placed immediately on ice. Samples were centrifuged within 30 minutes of blood collection, the plasma layer was removed to cryovials, and plasma samples were stored at −80°C prior to use. Excessive freeze-thawing of the samples was avoided to maintain plasma integrity. The study was approved by the institutional review board at each institution and performed in accordance with Guidelines for Good Clinical Practice. Informed consent was obtained from all patients. From this cohort, plasma samples from 20 patients (10 survivors and 10 matched nonsurvivors) were obtained. Patients in each group were matched 1:1 for sex, race, age (± 10 yr), and severity of illness (as measured by Sequential Organ Failure Assessment score) (14).
Solid-Phase Extraction of Glycopeptides and
MS Analysis
The glycopeptides were isolated using a label-free method as described previously, with 96% specificity for glycopep-tide isolation (15–17). Briefly, plasma proteins were reduced, alkylated, and trypsin-digested into peptides. The glycopeptides were oxidized and conjugated to a solid support using hydrazide chemistry, and glycopeptides that were previously N-linked were released by PNGase F. Peptides were cleaned using Sep-Pak Vac C18 cartridge (Waters, Milford, MA) and analyzed label-free by liquid chromatography-tandem mass spectrometry using a Q Exactive (ThermoFisher, Waltham, MA) coupled with a 15 cm × 75 μm C18 column (5 μm particles with 100 Å pore size). A nano UPLC at 300 nL/min with a 90-minute linear acetonitrile gradient (from 5% to 35% B over 90 min; A = 0.2% formic acid in water, B = 0.2% formic acid in 90% acetonitrile) was used. A top 20 data-dependent tandem mass spectrometry (MS/MS) with exclusion for 25 seconds was set. The samples were run with higher-energy collisional dissociation fragmentation at normalized collision energy of 30 and an isolation width of 2 m/z. A lock mass of the polysiloxane peak at 371.10123 was used to correct the mass in MS and MS/MS. Target values in MS were 1e6 ions at a resolution setting of 70,000 and in MS2 1e5 ions at a resolution setting of 17,500.
MS/MS spectra were searched with SEQUEST using Proteome Discoverer (version 1.4; Thermo Fisher) against the human RefSeq database (November 3, 2013) containing 53,918 sequences. For this database search, the precursor mass tolerance and fragment mass tolerance were set at 10 ppm and 0.02 Da, respectively. Trypsin was specified as the protease. The fixed modification was set as carbamidomethylation (C), and other modifications were set as flexible modification as follows: deamidation (N) and oxidation (M). Full-tryptic end and two missed cleavage sites were allowed. A decoy version of the human RefSeq database was used to estimate peptide and protein false discovery rate (FDR). The FDR was set at 0.01 to eliminate low-probability protein identifications.
The identified peptides were quantified using spectral counting. The total spectral counts of each sample were used for normalization. Missing values were replaced with a small value (0.01) in order to calculate the ratio and p value.
Proteomic Data Repository
The MS proteomics data have been converted using PRIDE Converter 2 and deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (http://www.ebi.ac.uk/pride/) with the dataset identifier PXD001425 (15–17).
Protein Quantitation Using Immunoblotting
Total protein immunoblotting was performed to validate selected glycopeptide levels measured by MS, as previously described (18). A total of 22 proteins were selected for immunoblotting and included both those differentially expressed at the glycopeptide level and several that showed no change but had previously been associated with sepsis (19–24). Primary antibodies are listed in Supplemental Digital Content 1 (http://links.lww.com/CCM/B334).
Equal total protein concentrations (5 μg) or equal volumes (100 nL) of plasma samples were run on 26-well, 4–12% Criterion XT Bis–Tris gels (Bio-Rad, Cat. # 345-0125) and transferred onto nitrocellulose membranes (Bio-Rad). A reversible protein staining kit (Thermo Scientific) was used to confirm transfer efficiency and calculate total protein levels per sample. The membrane was cut at approximately 50 kDa and the upper section discarded prior to addition of the primary antibody, to eliminate nonspecific binding of primary antibodies to albumin. Membranes were blocked with 5% blotting grade buffer (Bio-Rad, Cat. # 170–6404) and incubated with primary antibodies (1:1,000 dilution) overnight at 4°C. Antibodies used for immunoblotting are listed in Supplemental Digital Content 1 (http://links.lww.com/CCM/B334). Membranes were incubated with a peroxidase-labeled goat-anti-rabbit secondary antibody (Vector Laboratories, Cat. # PI-1000) for 1 hour at room temperature, and chemiluminescence was detected using Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare, Waukesha, WI Cat. # RPN2236) (18). Densitometry was measured using the IQ-TL image analysis software (GE Healthcare). The densitometric units were normalized to that of the total protein stain for the entire lane for each sample.
Lectin precipitation coupled with immunoblotting was used to determine if glycosylated levels of four selected proteins matched glycopeptide levels (25). Agarose-bound concanavalin A (ConA) (Vector Laboratories, Cat. # AL-1003) was washed five times with 500 μL of binding buffer (10 mM HEPES, 0.15 M NaCl, 0.1 mM CaCl, 0.01 mM MnCl2). Plasma (2 μL) was diluted in 100 μL of binding buffer, and the diluted plasma was incubated with Con A-lectin for 1 hour at room temperature with gentle shaking. The samples were centrifuged to collect lectin-bound and unbound fractions, and immunoblotting was performed to measure differences in glycosylated proteins, as described above.
Bioinformatics Analysis
Glycopeptide levels in plasma of patients with sepsis were visualized using a heat map. Pairwise distances between peptide sequences in both survivor and nonsurvivor groups were evaluated using Euclidean metric on the log10-transformed peptide expression values. The peptide sequences were clustered based on pairwise distances. Because of the low absolute expression values with large variances, we used the log10 transformation of expression values to focus on the significantly differentially expressed peptide sequences among survivor and nonsurvivor groups. Because we want to focus on the differences between survivor and nonsurvivor groups among all peptides, we did not perform principle component analysis to reduce the data dimension at this step. The heat map was generated using Matlab R2012. Proteins corresponding to the glycopeptides identified in patient plasma were aligned with their official names based on the UniProt database. Gene Ontology (GO) was used to identify intracellular and extracellular proteins. Proteins that had been reviewed in UniProt were analyzed by GO enrichment to identify enriched Gene Ontology Biological Processes (GOBPs) and to create a list of annotated proteins. The Database for Annotation, Visualization and Integrated Discovery for involved GOBPs was used to generate a list of annotated proteins and construct a functional network (threshold set at p < 0.05). Differentially expressed proteins were analyzed by Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis to determine specific pathways involved in the sepsis response.
Statistical Analysis
All analyses were performed blinded to groups. Patient characteristics described by binary variables were assessed using Fisher exact test, whereas continuous variables were first assessed for normality. Differences between groups were assessed using Student t test or Mann-Whitney U test, as appropriate. The 501 glycopeptides identified by MS were analyzed by Student t test. Because multiple testing analysis increases the chance of type II errors, due to the large number of peptides identified relative to sample size, we did not perform correction for multiple testing. There were 126 peptides identified only in survivors (a total of 66 peptides) or nonsurvivors (a total of 60 peptides). These peptides were tested for normality and none passed. For this reason, a Wilcoxon signed-rank test was used to determine significance, setting the comparison to 0. Arbitrary units of normalized values for immunoblots and lectin immunoblots were analyzed by one-sample t test, setting the survivor group equal to 1.0 for proteins increased only in the nonsurvivor group. Graphpad Prism was used for all analyses, and p value less than 0.05 was set as the significance level.
RESULTS
Patient Characteristics
The survivor and nonsurvivor groups did not differ significantly in regard to age, sex, race, days in the ICU or hospital, Sequential Organ Failure Assessment score, IV fluids over the first 6 hours, history of diabetes, WBC and platelet counts, or hemoglobin, creatinine, and bilirubin levels (Table 1). Thus, patient outcomes were not predicted by these factors individually.
TABLE 1.
Patient Characteristics
| Survivors (n = 10) | Nonsurvivors (n = 10) | p | |
|---|---|---|---|
| Age (yr) | 66 ± 4 | 65 ± 3 | 0.83 |
| Sex (female/male) | 6/4 | 6/4 | 1.00 |
| Race (African American/Caucasian) | 4/6 | 4/6 | 1.00 |
| ICU days | 4.6 ± 1.2 | 8.6 ± 2.6 | 0.18 |
| Hospital days | 13.0 ± 2.5 | 11.8 ± 2.9 | 0.76 |
| Sequential Organ Failure Assessment total | 9.3 ± 1.4 | 9.3 ± 1.4 | 1.00 |
| IV fluids over first 6 hr | 5.3 ± 0.6 | 4.9 ± 0.6 | 0.68 |
| History of diabetes | 8/10 | 6/10 | 0.48 |
| WBCsa | 24.3 ± 6.9 | 16.1 ± 3.4 | 0.30 |
| Plateletsa | 273 ± 47 | 217 ± 54 | 0.44 |
| Hemoglobina | 10.7 ± 0.6 | 11.5 ± 0.8 | 0.38 |
| Creatininea | 2.8 ± 0.8 | 3.5 ± 1.0 | 0.62 |
| Bilirubina | 2.6 ± 1.0 | 1.1 ± 0.3 | 0.20 |
Initial values.
Glycoproteomic Analysis
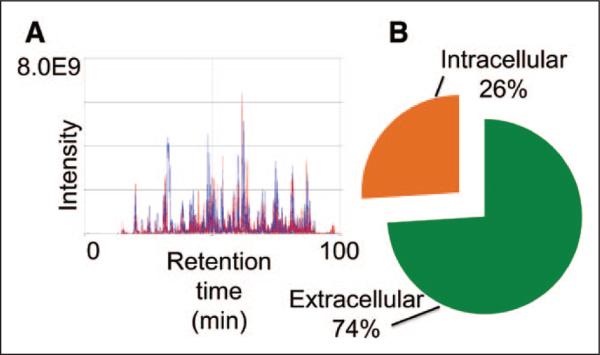
MS identified 501 glycopeptides in plasma from patients with sepsis, representative of 234 distinct proteins. Complete glycoproteomic results for the 501 peptides are shown in Supplemental Digital Content 2 (http://links.lww.com/CCM/B335). For each individual peptide identified, the annotated MS/MS spectra are provided in Supplemental Digital Content 3 (http://links.lww.com/CCM/B336). The top 20 glycopeptides uniquely identified in survivors and nonsurvivors are summarized in Table 2. The chromatography profiles showed tight alignment, indicating high consistency in the biological replicates within the groups (Fig. 1A). As expected, glycopeptide isolation enriched for extracellular proteins, as 74% of the peptides mapped to the extracellular compartment (Fig. 1B).
TABLE 2.
Top 20 Glycopeptides Identified in Plasma From Sepsis Nonsurvivors and Survivors and Associated Protein Names
| Peptide Rank | Peptide Sequence | Protein |
|---|---|---|
| Nonsurvivors | ||
| 1 | SPYEmFGDEEVmcLnGnWTEPPQcK | Complement factor H-related protein 1a,b,c,d (26) |
| 2 | QLDmLDLSnnSLASVPEGLWASLGQPNWDmR | Leucine-rich alpha-2-glycoproteinb,d |
| 3 | WVSnKTEGR | Antithrombin-IIIb,c,d (27) |
| 4 | MQLLKDVVGNDTYRInnK | Phospholipid-metabolizing enzyme A-C1b |
| 5 | YKVDYESQSTDTQnFSSESK | Insulin-like growth factor-binding protein 3b,c,d (28) |
| 6 | LQLDcRPLEGnSTVTGQPR | Inhibin beta E chainb |
| 7 | LSVATnVSATLTFnTSK | Lipopolysaccharide-binding proteinb,c (6) |
| 8 | eEQFnSTFR | FLJ00385 proteinb,d |
| 9 | HGIQYFNnNTQHSSLFTLNEVK | Kininogen-1b,c (21) |
| 10 | EEQYnSTYR | Human IgG1 Fcb |
| 11 | eEQFnSTYR | Immunoglobulin heavy chain, constant regionb |
| 12 | EEQYnSTFR | Gamma-3 immunoglobulin constant heavy chainb |
| 13 | YKnNSDISSTR | Ig mu chain C regionb,d |
| 14 | LSLHRPALEDLLLGSEAnLTcTLTGLR | Immunoglobulin heavy chainb,d |
| 15 | LAGKPTHVnVSVVmAEVDGTcY | Immunoglobulin heavy chainb,d |
| 16 | GLTFQQnASSmcVPDQDTAIR | Ig mu chain C regionb,d |
| 17 | TPLTAnITK | Immunoglobulin heavy chainb,d |
| 18 | TKPREEQFnSTFR | FLJ00385 proteinb,d |
| 19 | VHnGSEILFSYFQDLVITLPFELR | Apolipoprotein B-100b,c,d (19) |
| 20 | SIEDRISnLKGIISmK | Copper-transporting ATPase 2b |
| Survivors | ||
| 1 | KLINDYVKnGTR | Alpha-1-antichymotrypsina,b,d |
| 2 | GVTSVSQIFHSPDLAIRDTFVnASR | Plasma protease C1 inhibitora,b,c,d (23) |
| 3 | LINDYVKnGTR | Alpha-1-antichymotrypsina,b,d |
| 4 | LLETVEYnISGAER | Tenascin Ca,b,c (22) |
| 5 | EWLPLnR | Attractinb,d |
| 6 | RGPEcSQnYTTPSGVIK | Neuropilin-1b |
| 7 | YnFSFR | Complement C5b,c (29) |
| 8 | KVTVRPGESVmVnISAK | IgGFc-binding proteinb,c,d (30) |
| 9 | FQLLnFSSSELK | Cell adhesion molecule 1b,c,d (31) |
| 10 | SDFSQTmLFQAnTTR | Receptor-type tyrosine-protein phosphatase gammab |
| 11 | VQnVTEFDDSLLR | Adenosine deaminase CECR1b,d |
| 12 | GKEGHFYYnISEVK | Phospholipid transfer proteinb,c,d (32) |
| 13 | GLnVTLSSTGRnGFK | Complement C4-Bb,c,d (27) |
| 14 | TWnQSIALR | Coagulation factor Vb,c,d (33) |
| 15 | nYTLTGR | Complement component C7b,c,d (34) |
| 16 | KGcVLLSH LnETVTVSASLESGR | Pregnancy zone proteinb,d |
| 17 | TcGnKSSVnSTVLVKnTK | Junctional adhesion molecule-likeb |
| 18 | FLTEVEKnATALYHVEAFK | Multiple inositol polyphosphate phosphatase 1b,c (33) |
| 19 | YEFcPFHnVTQHEQTFR | N-acetylglucosamine-1-phosphotransferase subunit gammab |
| 20 | FDFQGTcEYLLSAPcHGPPLGAEnFTVTVANEHR | IgGFc-binding proteinb,c,d (30) |
p < 0.05 by Student t test.
p < 0.05 by Wilcoxon rank-sum test.
Previously associated with sepsis.
Multiple peptides identified.
Of the 501 glycopeptides identified, 66 were present only in survivors and 60 were present only in nonsurvivors. The top 20 from each group are shown here, ranked by p value. Of the 40 proteins listed, only 17 have previously been associated with sepsis. Nonsurvivor peptides 1–20 correspond to Supplemental Digital Content 2 peptides 1–20 (http://links.lww.com/CCM/B335), whereas survivor peptides 1–20 correspond to Supplemental Digital Content 2 peptides 435–454 (http://links.lww.com/CCM/B335).
Lower case amino acids indicate modifications.
Figure 1.
Glycopeptide isolation was reproducible and enriched for extracellular proteins. A, The solid-phase extraction of glycopeptides protocol was robust, highly reproducible, and enriched for low-abundance proteins. Glycopeptides in sepsis survivors and nonsurvivors were isolated. B, The majority of isolated glycopeptides (74%) identified by mass spectrometry were extracellular proteins.
Determination of Potential Effector Glycopeptides
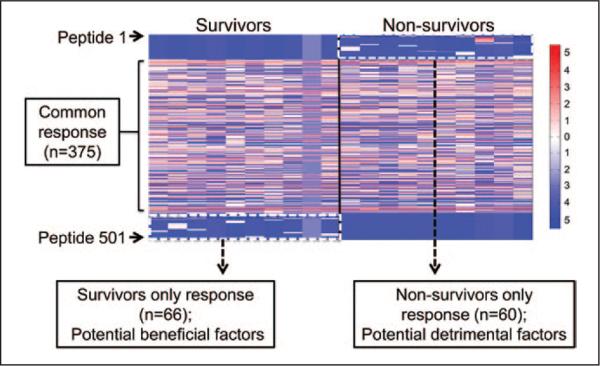
Expression levels of 501 glycopeptides identified by MS were visualized in a heat map, with red representing high expression and blue representing low expression. No change was seen for 375 of the peptides, demonstrating that many aspects of the sepsis response were in common between the two groups. One group of peptides (n = 66 peptides) was identified only in survivors, whereas another group (n = 60 peptides) was identified only in nonsurvivors (Fig. 2). Of note, one of the patients in the survivor group showed low levels for all visualized glycopeptides. This could be caused by overall low glycoprotein expression in this particular patient or inefficient capture of the glycopeptides.
Figure 2.
Heat mapping illustrates glycopeptides common to patients with sepsis and differential glycopeptide profiles in survivors and nonsurvivors. A total of 375 glycopeptides were in common between groups, whereas a total of 126 glycopeptides were present only in survivors or only in nonsurvivors. Glycopeptides in red represent high expression, green represent no difference in expression, and blue represent low expression. One group of 66 peptides was highly present in survivors, and another group of 60 peptides was highly present in nonsurvivors. One survivor showed low overall response.
Validating Glycoprotein Expression
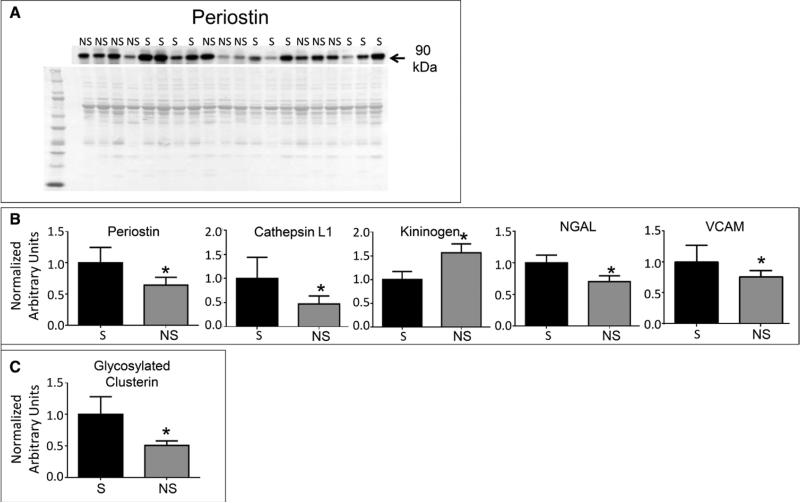
MS is not readily available in many clinical laboratories. To determine whether the proteins of interest could be quantified by immunoblotting, we evaluated 23 proteins (Table 3). We selected 16 proteins that were significantly different at the glycopeptide level to validate. We also selected eight proteins that were not different at the glycopeptide level as additional controls are known to be associated with sepsis. Of the 22 proteins analyzed, five proteins showed differences at the total level by immunoblotting (Fig. 3), whereas 17 proteins showed no differences. All immunoblot data are shown in Supplemental Digital Content 4 (http://links.lww.com/CCM/B337). Compared with survivors, cathepsin L-1, vascular cell adhesion molecule (VCAM), periostin, and neutrophil gelatinase–associated lipocalin (NGAL) were decreased at the protein level in nonsurvivors (all p < 0.05) (Fig. 3). Conversely, kininogen was increased in nonsurvivors compared with survivors (p < 0.05). Of these six proteins, only VCAM was decreased at both the glycopeptide and protein levels.
TABLE 3.
Comparison of Mass Spectrometry and Immunoblotting Analyses
| Protein Name | Mass Spectrometrya | IBb | Lectin IBb | Previously Associated With Sepsis |
|---|---|---|---|---|
| α-1-antichymotrypsin | ↓ ↓ | ↔ | ↔ | |
| Antithrombin III | ↑ | ↔ | NE | (27) |
| Attractin | ↑ ↓ | ↔ | ↔ | |
| Cathepsin L | ↑ | ↓ | NE | |
| CD44 | ↔ | ↔ | NE | (31) |
| CD276 | ↓ ↓ ↓ | ↔ | NE | (30) |
| Cell adhesion molecule 1 | ↓ | ↔ | NE | |
| Clusterin | ↔ | ↔ | ↓ | (27) |
| Complement factor H | ↑ | ↔ | NE | (29) |
| Fc fragment of IgG binding protein | ↓ ↓ | ↔ | NE | (35) |
| Fibronectin | ↓ | ↔ | NE | (36) |
| Fibrinopeptide A | ↔ | ↔ | NE | (37) |
| Galectin-3 | ↔ | ↔ | NE | (38) |
| Kininogen | ↔ | ↑ | NE | (21) |
| Leucine-rich glycoprotein 1 | ↑ | ↔ | NE | |
| Neutrophil gelatinase–associated lipocalin | ↔ | ↓ | NE | (21) |
| Periostin | ↑ | ↓ | NE | |
| Tenascin-C | ↓ | ↔ | ↔ | (22) |
| Tissue factor protease inhibitor | ↓ | ↔ | NE | (39) |
| Vascular cell adhesion molecule | ↓ ↓ ↓ | ↓ | NE | (40) |
| Vascular endothelial growth factor C | ↔ | ↔ | NE | (41) |
| Vitronectin | ↓ | ↔ | NE | (42) |
IB = immunoblotting, NE = not evaluated.
Arrows indicate the number of peptides and the direction of change in nonsurvivors.
Arrows indicate direction of change in nonsurvivors compared with survivors.
References for proteins previously associated with sepsis are included.
Figure 3.
Validation of glycopeptide and protein expression by immunoblotting. A, Representative immunoblot of periostin. B, Graphs representing densitometric units normalized to the total protein stain of the entire lane represent protein levels of periostin, cathepsin-L1, vascular cell adhesion molecule (VCAM), neutrophil gelatinase–associated lipocalin (NGAL), and kininogen, measured by immunoblotting. C, Immunoblotting for clusterin, isolated using lectin precipitation, revealed that densitometric units of glycosylated clusterin normalized to the total protein stain of the entire lane were decreased in nonsurvivors (NS) (n = 4) compared with survivors (S) (n = 4). Data are represented as mean ± sem.
Lectin precipitation coupled with immunoblotting was used to determine whether glycosylated protein levels matched glycopeptide levels for four proteins (α-1-antichymotrypsin, attractin, clusterin, and tenascin-C), with all immunoblot data shown in Supplemental Digital Content 5 (http://links.lww.com/CCM/B338). Only glycosylated clusterin was different between survivors and nonsurvivors (Fig. 3B). However, the clusterin glycopeptide identified by MS was decreased in non-survivors, whereas total clusterin glycoprotein levels increased in nonsurvivors.
Gene Ontology and Pathway Analysis
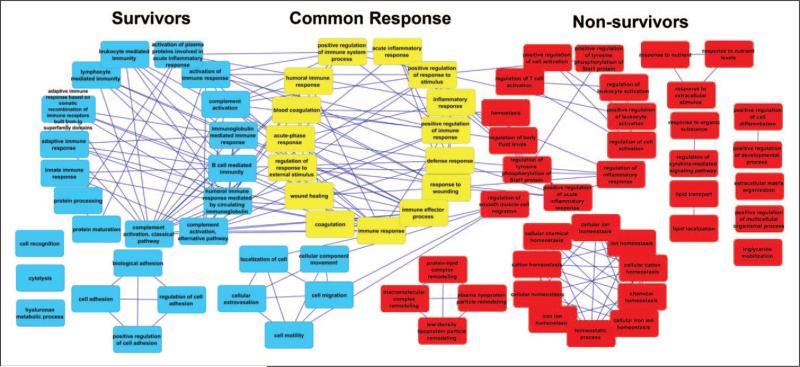
The UniProt database provides information on protein sequence and function and is divided into entries that have been reviewed (in which the sequence and functional information are manually updated) or unreviewed (in which the sequence and functional information are automatically annotated). UniProt identified 66 peptides in survivors that correlated with 54 reviewed proteins and 60 peptides in non-survivors that correlated with 43 reviewed proteins. Proteins overlapping between either survivors and common response or nonsurvivors and common response were used to identify enriched biological processes (Supplemental Digital Content 6, http://links.lww.com/CCM/B339; Supplemental Digital Content 7, http://links.lww.com/CCM/B340). A network of enriched GOBPs was constructed, with GO shared terms in yellow, distinct survivor terms in blue, and distinct nonsurvivor terms in red (Fig. 4). The survivor and common response GOBPs have overlapping pathways, including cell adhesion and complement activation using the classical pathway. Pathways that overlap between nonsurvivor and common response GOBPs include lipid transport, regulation of leukocyte activation, and chemical homeostasis.
Figure 4.
Pathways activated in sepsis survivors and nonsurvivors. The functional network shows interaction of pathways associated with survival, the common response, and nonsurvivors.
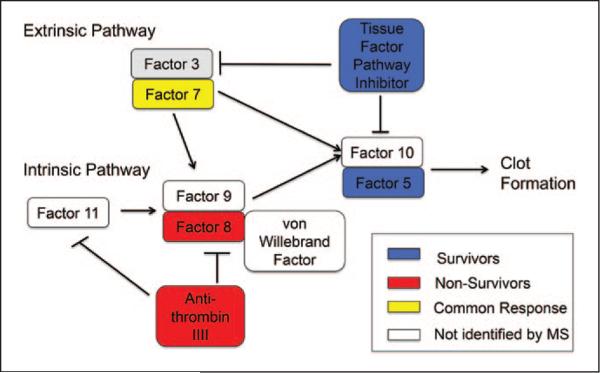
KEGG analysis was used to determine pathways associated with sepsis survival and revealed six pathways (complement cascades, coagulation cascade, cell adhesion molecules, ECM-receptor interaction, focal adhesion, and prion diseases) enriched in survivors and one pathway (systemic lupus erythematosus) enriched in nonsurvivors (Supplemental Digital Content 8, http://links.lww.com/CCM/B341). Interestingly, both survivors and nonsurvivors used complement and coagulation cascades, but the proteins identified within that pathway were different between groups. For example, anti-thrombin-III and complement factor VIII were both increased in nonsurvivors and are part of the intrinsic pathway. By contrast, tissue factor pathway inhibitor and complement factor V were both increased in survivors and are part of the extrinsic pathway (Fig. 5). This suggests that sepsis survivors and nonsurvivors use different proteins within common pathways to direct outcomes.
Figure 5.
Schematic of the complement and coagulation cascade, which is activated in the sepsis response. Proteins in the intrinsic pathway were present in the nonsurvivors, while the extrinsic pathway was increased in survivors. MS = mass spectrometry.
DISCUSSION
The goal of this study was to identify circulating glycopeptides present in the early stage of sepsis that linked to survival outcomes. In the current study, we identified 375 glycopeptides common to both groups that may play a role in survival in septic patients. Sepsis involves an extensive systemic inflammatory response that causes the release of a milieu of cytokines and chemokines into the bloodstream to combat the source of infection. Thus, many signaling cascades are activated to rid the body of infection, resulting in necessary increases in a shared group of glycopeptides that do not direct patient outcomes. This global inflammatory response is managed clinically through the use of antibiotics, fluid replacement, and respiratory support in the ICU.
Our study revealed a set of 66 glycopeptides that were unique to the survivor group and 60 unique to the nonsurvivor group. These data provide evidence that sepsis survivors and nonsurvivors have a distinct pattern of plasma glycopeptides that can be used as diagnostic tools. Analysis of biological processes using GOBPs revealed two overlapping biological processes between proteins unique to the survivor group and proteins that were identified in both survivor and nonsurvivor groups (common responders), including cell adhesion and complement activation, and three overlapping biological processes between nonsurvivor and common responders, including lipid transport, regulation of leukocyte activation, and chemical homeostasis. Survivor and common response GOBPs shared the classical complement activation pathway and coagulation cascade, although the two groups used different mechanisms within this signaling cascade. Activation of the complement and coagulation system is crucial for immune suppression of infections (34, 37, 39). At the same time, this is a double-edged sword as excessive activation may cause severe injury to the host (43). Our study illustrates specific pathways within the complement and coagulation cascade that regulate survival in patients with sepsis. Interestingly, pathways associated with cell adhesion and cell-matrix interaction were enriched in the survivor group. Our results reveal glycopeptides that distinguish survival in patients with sepsis, which provide candidate predictors of patient outcomes that could be targeted therapeutically.
NGAL and VCAM are increased in patients who have sepsis compared to healthy controls, and VCAM levels further increase with sepsis severity (44, 45). When evaluating VCAM levels within the sepsis group and comparing by survival, we observed decreases in both the glycopeptides identified, as well as total protein levels, in nonsurvivors compared with survivors. A similar decrease was also seen for NGAL protein levels in our study. Thus, even though VCAM and NGAL increase in sepsis, our results suggest that these increases may be part of a beneficial response that is necessary for survival.
There were several strengths to this study. Compared with whole proteome analysis, glycoproteomics greatly enriches for extracellular proteins (11). The enrichment of extracellular N-linked glycopeptides allows for identification of circulating factors associated with the sepsis response. A whole proteome analysis would have selected for the identification of the most abundant proteins but not necessarily the most important proteins (46). Using the glycopeptide selection strategy greatly reduced the complexity of plasma proteins, allowing for identification of low-abundance proteins. Because of the technical challenges of transporting therapeutic agents across cellular membranes (47), identification of extracellular therapeutic targets using glycoprotein enrichment also provides a promising avenue for drug development that may help to improve sepsis outcomes. Future use of data-independent analysis or targeted analysis with upfront enrichment of glycopeptides may further increase the sensitivity of detection and allow the reliable quantification of plasma proteins at the nanogram/milliliter level (48). The use of human samples was also a strength. Identification of therapeutic targets in rodent models of sepsis has been problematic, in part, because the inflammatory response in rodents models some, but not all, of the human responses (49, 50).
The LactATEs cohort enrolled patients and collected blood samples at the early stage of diagnosis of sepsis, which allowed us to query for potential predictors of patient outcomes and eliminated the possibility that patients whose condition improves rapidly were excluded, minimizing selection bias. The rapid onset and progression of severe sepsis highlights the necessity for the earliest possible identification of predictors of patient outcome, and for this reason, samples taken at admission were used. The LactATEs cohort was a large study, whereas the current study had a small sample size and lacked serial assessment results. Future studies that include a larger sample size and temporal evaluation of the sepsis response are warranted to determine how glycoprotein levels evolve. This may identify late targets for intervention. For example, the Mayr laboratory has used a tissue-based proteomics approach to identify long pentraxin 3 as a lead candidate for endotoxemia-induced myocar-dial inflammation (51). They further validated that conversion of octameric to monomeric pentraxin 3 was linked to greater day 28 survival in humans and was superior to NT-pro brain natriuretic peptide, troponin I, or troponin T as a predictor of outcomes (51). We did not observe differences in pentraxin 3, either because it has only one glycosylation site or because its increase does not occur until 2 days after admission.
Although sepsis remains a predominant cause of mortality in the ICU, identification of early changes in proteins that lead to detrimental inflammatory signaling cascades and result in mortality may advance therapeutics. Identifying proteins that predict adverse patient outcomes and trigger unfavorable downstream pathways provide candidates for inhibition. By contrast, proteins linked to improved survival or initiation of advantageous signaling cascades provides candidates for stimulation. Our study identified factor VIII as a possible target for early inhibition of poor sepsis outcome, whereas tissue factor pathway inhibitor or factor V stimulation might prove beneficial by preventing progression to septic shock. Both inhibition of proteins activating detrimental signaling cascades and up-regulation of advantageous proteins are promising avenues to pursue for improving sepsis outcomes (29–31).
CONCLUSIONS
Our glycoprotein data provide evidence that distinct patterns of plasma glycopeptides are seen in sepsis survivors and non-survivors. These plasma glycopeptides can be used as a diagnostic tool as well as predictors of patient outcomes that could be targeted therapeutically.
Supplementary Material
Acknowledgments
Supported, in part, by American Heart Association (13POST14350034), National Institutes of Health (T32HL105324, HL101430, HL075360, HL051971, GM104357, K23GM76652, P01HL051971, P20GM104357, and HHSN 268201000036C [N01-HV-00244]), San Antonio Cardiovascular Proteomics Center, and the Biomedical Laboratory Research and Development Service of the Veterans Affairs Office of Research and Development Award (5I01BX000505).
Dr. Puskarich's institution received grant support from NIGMS (platelet activation in septic shock [K23 GM113041]) and Emergency Medicine Foundation (Career Development Award–Mitochondrial dysfunction in early severe sepsis). Dr. Jones served as a board member for SAEM (Board of Directors) and received support for article research from the National Institutes of Health (NIH). His institution received grant support from the NIH.
Footnotes
This work was performed at University of Mississippi Medical Center and University of Texas San Antonio.
Drs. DeCoux, Tian, Nguyen, Flynn, Cannon, Griswold, Jin, and Lindsey contributed to data collection, analysis, and interpretation. Drs. Griswold, Jin, Puskarich, Jones, and Lindsey contributed to formulation of hypothesis and experimental design. All authors contributed to writing and revision of the article.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website (http://journals.lww.com/ccmjournal).
The remaining authors have disclosed that they do not have any potential conflicts of interest.
REFERENCES
- 1.Pavon A, Binquet C, Kara F, et al. EPIdemiology of Septic Shock (EPISS) Study Group: Profile of the risk of death after septic shock in the present era: An epidemiologic study. Crit Care Med. 2013;41:2600–2609. doi: 10.1097/CCM.0b013e31829a6e89. [DOI] [PubMed] [Google Scholar]
- 2.Gaieski DF, Edwards JM, Kallan MJ, et al. Benchmarking the incidence and mortality of severe sepsis in the United States. Crit Care Med. 2013;41:1167–1174. doi: 10.1097/CCM.0b013e31827c09f8. [DOI] [PubMed] [Google Scholar]
- 3.Dellinger RP, Levy MM, Rhodes A, et al. Surviving Sepsis Campaign Guidelines Committee including the Pediatric Subgroup: Surviving sepsis campaign: International guidelines for management of severe sepsis and septic shock: 2012. Crit Care Med. 2013;41:580–637. doi: 10.1097/CCM.0b013e31827e83af. [DOI] [PubMed] [Google Scholar]
- 4.Gygi SP, Corthals GL, Zhang Y, et al. Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proc Natl Acad Sci U S A. 2000;97:9390–9395. doi: 10.1073/pnas.160270797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu WW, Wang G, Baek SJ, et al. Comparative study of three proteomic quantitative methods, DIGE, cICAT, and iTRAQ, using 2D gelor LC-MALDI TOF/TOF. J Proteome Res. 2006;5:651–658. doi: 10.1021/pr050405o. [DOI] [PubMed] [Google Scholar]
- 6.Ratzinger F, Schuardt M, Eichbichler K, et al. Utility of sepsis bio-markers and the infection probability score to discriminate sepsis and systemic inflammatory response syndrome in standard care patients. PLoS One. 2013;8:e82946. doi: 10.1371/journal.pone.0082946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Echan LA, Tang HY, Ali-Khan N, et al. Depletion of multiple high-abundance proteins improves protein profiling capacities of human serum and plasma. Proteomics. 2005;5:3292–3303. doi: 10.1002/pmic.200401228. [DOI] [PubMed] [Google Scholar]
- 8.Gundry RL, White MY, Nogee J, et al. Assessment of albumin removal from an immunoaffinity spin column: Critical implications for proteomic examination of the albuminome and albumin-depleted samples. Proteomics. 2009;9:2021–2028. doi: 10.1002/pmic.200800686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alley WR, Jr, Mann BF, Novotny MV. High-sensitivity analytical approaches for the structural characterization of glycoproteins. Chem Rev. 2013;113:2668–2732. doi: 10.1021/cr3003714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brooks SA. Strategies for analysis of the glycosylation of proteins: Current status and future perspectives. Mol Biotechnol. 2009;43:76–88. doi: 10.1007/s12033-009-9184-6. [DOI] [PubMed] [Google Scholar]
- 11.Tian Y, Koganti T, Yao Z, et al. Cardiac extracellular proteome profiling and membrane topology analysis using glycoproteomics. Proteomics Clin Appl. 2014;8:595–602. doi: 10.1002/prca.201400009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnold RC, Shapiro NI, Jones AE, et al. Emergency Medicine Shock Research Network (EMShockNet) Investigators: Multicenter study of early lactate clearance as a determinant of survival in patients with presumed sepsis. Shock. 2009;32:35–39. doi: 10.1097/shk.0b013e3181971d47. [DOI] [PubMed] [Google Scholar]
- 13.Jones AE, Shapiro NI, Trzeciak S, et al. Emergency Medicine Shock Research Network (EMShockNet) Investigators: Lactate clearance vs central venous oxygen saturation as goals of early sepsis therapy: A randomized clinical trial. JAMA. 2010;303:739–746. doi: 10.1001/jama.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vincent JL, Moreno R, Takala J, et al. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive Care Med. 1996;22:707–710. doi: 10.1007/BF01709751. [DOI] [PubMed] [Google Scholar]
- 15.Tian Y, Zhou Y, Elliott S, et al. Solid-phase extraction of N-linked glycopeptides. Nat Protoc. 2007;2:334–339. doi: 10.1038/nprot.2007.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian Y, Kelly-Spratt KS, Kemp CJ, et al. Mapping tissue-specific expression of extracellular proteins using systematic glycoproteomic analysis of different mouse tissues. J Proteome Res. 2010;9:5837–5847. doi: 10.1021/pr1006075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, Li XJ, Martin DB, et al. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 18.Deleon-Pennell KY, de Castro Brás LE, Lindsey ML. Circulating Porphyromonas gingivalis lipopolysaccharide resets cardiac homeostasis in mice through a matrix metalloproteinase-9-dependent mechanism. Physiol Rep. 2013;1:e00079. doi: 10.1002/phy2.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanza-Jacoby S, Wong SH, Tabares A, et al. Disturbances in the composition of plasma lipoproteins during gram-negative sepsis in the rat. Biochim Biophys Acta. 1992;1124:233–240. doi: 10.1016/0005-2760(92)90134-h. [DOI] [PubMed] [Google Scholar]
- 20.Su CM, Cheng HH, Tsai TC, et al. Elevated serum vascular cell adhesion molecule-1 is associated with septic encephalopathy in adult community-onset severe sepsis patients. Biomed Res Int. 2014;2014:598762. doi: 10.1155/2014/598762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsieh YC, Hsu C, Yang RC, et al. Isolation of bona fide differentially expressed genes in the 18-hour sepsis liver by suppression subtractive hybridization. Shock. 2004;21:549–555. doi: 10.1097/01.shk.0000126148.83935.6a. [DOI] [PubMed] [Google Scholar]
- 22.Piccinini AM, Midwood KS. Endogenous control of immunity against infection: Tenascin-C regulates TLR4-mediated inflammation via microRNA-155. Cell Rep. 2012;2:914–926. doi: 10.1016/j.celrep.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Igonin AA, Protsenko DN, Galstyan GM, et al. C1-esterase inhibitor infusion increases survival rates for patients with sepsis. Crit Care Med. 2012;40:770–777. doi: 10.1097/CCM.0b013e318236edb8. [DOI] [PubMed] [Google Scholar]
- 24.Huber-Lang M, Barratt-Due A, Pischke SE, et al. Double blockade of CD14 and complement C5 abolishes the cytokine storm and improves morbidity and survival in polymicrobial sepsis in mice. J Immunol. 2014;192:5324–5331. doi: 10.4049/jimmunol.1400341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian Y, Esteva FJ, Song J, et al. Altered expression of sialylated glyco-proteins in breast cancer using hydrazide chemistry and mass spec-trometry. Mol Cell Proteomics. 2012;11:M111.011403. doi: 10.1074/mcp.M111.011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haapasalo K, Vuopio J, Syrjänen J, et al. Acquisition of complement factor H is important for pathogenesis of Streptococcus pyogenes infections: Evidence from bacterial in vitro survival and human genetic association. J Immunol. 2012;188:426–435. doi: 10.4049/jimmunol.1102545. [DOI] [PubMed] [Google Scholar]
- 27.Ocasio FM, Jiang Y, House SD, et al. Chronic morphine accelerates the progression of lipopolysaccharide-induced sepsis to septic shock. J Neuroimmunol. 2004;149:90–100. doi: 10.1016/j.jneuroim.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 28.Papastathi C, Mavrommatis A, Mentzelopoulos S, et al. Insulin-like Growth Factor I and its binding protein 3 in sepsis. Growth Horm IGF Res. 2013;23:98–104. doi: 10.1016/j.ghir.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 29.Yuan Y, Ren J, Wu X, et al. Exogenous C3 postpones complement exhaustion and confers organ protection in murine sepsis. J Surg Res. 2011;168:e87–e94. doi: 10.1016/j.jss.2011.01.062. [DOI] [PubMed] [Google Scholar]
- 30.Zhang G, Wang J, Kelly J, et al. B7-H3 augments the inflammatory response and is associated with human sepsis. J Immunol. 2010;185:3677–3684. doi: 10.4049/jimmunol.0904020. [DOI] [PubMed] [Google Scholar]
- 31.Hasan Z, Palani K, Rahman M, et al. Targeting CD44 expressed on neutrophils inhibits lung damage in abdominal sepsis. Shock. 2011;35:567–572. doi: 10.1097/SHK.0b013e3182144935. [DOI] [PubMed] [Google Scholar]
- 32.Levels JH, Pajkrt D, Schultz M, et al. Alterations in lipoprotein homeostasis during human experimental endotoxemia and clinical sepsis. Biochim Biophys Acta. 2007;1771:1429–1438. doi: 10.1016/j.bbalip.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 33.Yan SB, Nelson DR. Effect of factor V Leiden polymorphism in severe sepsis and on treatment with recombinant human activated protein C. Crit Care Med. 2004;32:S239–S246. doi: 10.1097/01.ccm.0000126122.34119.d1. [DOI] [PubMed] [Google Scholar]
- 34.Debard AL, Lamy B, Monneret G, et al. FcgammaRIIIb and complement component C7 codeficiency in a patient with recurrence of fulminant meningococcal septic shock. Clin Infect Dis. 2005;40:1679–1683. doi: 10.1086/430065. [DOI] [PubMed] [Google Scholar]
- 35.Sun D, Raisley B, Langer M, et al. Anti-peptidoglycan antibodies and Fcγ receptors are the key mediators of inflammation in Gram-positive sepsis. J Immunol. 2012;189:2423–2431. doi: 10.4049/jimmunol.1201302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruiz Martín G, Prieto Prieto J, Veiga de Cabo J, et al. Plasma fibronectin as a marker of sepsis. Int J Infect Dis. 2004;8:236–243. doi: 10.1016/j.ijid.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 37.Mavrommatis AC, Theodoridis T, Orfanidou A, et al. Coagulation system and platelets are fully activated in uncomplicated sepsis. Crit Care Med. 2000;28:451–457. doi: 10.1097/00003246-200002000-00027. [DOI] [PubMed] [Google Scholar]
- 38.ten Oever J, Giamarellos-Bourboulis EJ, van de Veerdonk FL, et al. Circulating galectin-3 in infections and non-infectious inflammatory diseases. Eur J Clin Microbiol Infect Dis. 2013;32:1605–1610. doi: 10.1007/s10096-013-1919-4. [DOI] [PubMed] [Google Scholar]
- 39.Tang H, Ivanciu L, Popescu N, et al. Sepsis-induced coagulation in the baboon lung is associated with decreased tissue factor pathway inhibitor. Am J Pathol. 2007;171:1066–1077. doi: 10.2353/ajpath.2007.070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaynar AM, Yende S, Zhu L, et al. Effects of intra-abdominal sepsis on atherosclerosis in mice. Crit Care. 2014;18:469. doi: 10.1186/s13054-014-0469-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Lu Y, Ma L, et al. Activation of vascular endothelial growth factor receptor-3 in macrophages restrains TLR4-NF-κB signaling and protects against endotoxin shock. Immunity. 2014;40:501–514. doi: 10.1016/j.immuni.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 42.Ren Y, Xie Y, Jiang G, et al. Apoptotic cells protect mice against lipopolysaccharide-induced shock. J Immunol. 2008;180:4978–4985. doi: 10.4049/jimmunol.180.7.4978. [DOI] [PubMed] [Google Scholar]
- 43.Charchaflieh J, Wei J, Labaze G, et al. The role of complement system in septic shock. Clin Dev Immunol. 2012;2012:407324. doi: 10.1155/2012/407324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zonneveld R, Martinelli R, Shapiro NI, et al. Soluble adhesion molecules as markers for sepsis and the potential pathophysiological discrepancy in neonates, children and adults. Crit Care. 2014;18:204. doi: 10.1186/cc13733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Macdonald SP, Stone SF, Neil CL, et al. Sustained elevation of resistin, NGAL and IL-8 are associated with severe sepsis/septic shock in the emergency department. PLoS One. 2014;9:e110678. doi: 10.1371/journal.pone.0110678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maslove DM, Wong HR. Gene expression profiling in sepsis: Timing, tissue, and translational considerations. Trends Mol Med. 2014;20:204–213. doi: 10.1016/j.molmed.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schiess R, Wollscheid B, Aebersold R. Targeted proteomic strategy for clinical biomarker discovery. Mol Oncol. 2009;3:33–44. doi: 10.1016/j.molonc.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, Hüttenhain R, Surinova S, et al. Quantitative measurements of N-linked glycoproteins in human plasma by SWATH-MS. Proteomics. 2013;13:1247–1256. doi: 10.1002/pmic.201200417. [DOI] [PubMed] [Google Scholar]
- 49.Takao K, Miyakawa T. Genomic responses in mouse models greatly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2015;112:1167–1172. doi: 10.1073/pnas.1401965111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seok J, Warren HS, Cuenca AG, et al. Inflammation and Host Response to Injury, Large Scale Collaborative Research Program: Genomic responses in mouse models poorly mimic human inflamma-tory diseases. Proc Natl Acad Sci U S A. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cuello F, Shankar-Hari M, Mayr U, et al. Redox state of pentraxin 3 as a novel biomarker for resolution of inflammation and survival in sepsis. Mol Cell Proteomics. 2014;13:2545–2557. doi: 10.1074/mcp.M114.039446. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.