Abstract
Cell identity signals influence the invasive capability of tumor cells, as demonstrated by the selection for programs of epithelial-to-mesenchymal transition (EMT) during malignant progression. Breast cancer cells retain canonical epithelial traits and invade collectively as cohesive groups of cells, but the signaling pathways critical to their invasive capabilities are still incompletely understood. Here we report that the transcription factor ΔNp63α drives the migration of basal-like breast cancer (BLBC) cells by inducing a hybrid mesenchymal/epithelial state. Through a combination of expression analysis and functional testing across multiple BLBC cell populations, we determined that ΔNp63α induces migration by elevating the expression of the EMT program components Slug and Axl. Interestingly, ΔNp63α also increased the expression of miR205, which can silence ZEB1/2 to prevent the loss of epithelial character caused by EMT induction. In clinical specimens, co-expression of various elements of the ΔNp63α pathway confirmed its implication in motility signaling in BLBC. We observed that activation of the ΔNp63α pathway occurred during the transition from noninvasive ductal carcinoma in situ to invasive breast cancer. Notably, in an orthotopic tumor model, Slug expression was sufficient to induce collective invasion of E-cadherin expressing BLBC cells. Together, our results illustrate how ΔNp63α can drive breast cancer cell invasion by selectively engaging pro-migratory components of the EMT program while, in parallel, still promoting the retention of epithelial character.
Introduction
The invasive phenotypes of tumor cells are dependent on signaling pathways which control cell identity (1). For example, extracellular stimuli can alter cell state by promoting an epithelial-to-mesenchymal transition (EMT) (2). The EMT program involves the silencing of epithelial traits, such as the expression of cell-cell adhesion proteins, and the induction of mesenchymal traits, including pro-migratory cytoskeletal proteins and proteases (3). Cells that complete the EMT process acquire a mesenchymal like phenotype and can invade as single cells. However, while a full EMT can promote aggressive single cell invasion, tumor cells may also invade while retaining epithelial traits (4). For instance, at least 50% of invasive breast tumors have epithelial characteristics, including the expression of E-cadherin (5,6), claudin-family tight junctions proteins (7), and EpCAM (8). Instead of invading as single cells, epithelial-like breast cancer cells frequently engage in a process called collective invasion (9,10), in which tumor cells invade as cohesive groups through paths in the ECM (11). There is heterogeneity with respect to the autonomous invasive traits of epithelial-like breast cancer populations derived from different patient tumors (9,10). Notably, basal-like breast cancer (BLBC) cells are intrinsically motile and can collectively invade into paths generated by fibroblasts while maintaining epithelial features, including the expression of E-cadherin (9). By comparison, luminal-type breast cancer (LBC) cells are an epithelial-like cell type with relative weak intrinsic migratory and invasive ability (9). Importantly, BLBC cells are distinguished from LBC cells by patterns of gene expression (7,12), indicating that these populations represent two distinct epithelial-like cell identities and that the BLBC identity can be an epithelial-like cell state that has an enhanced invasive behavior. Thus, we sought to determine the nature of the cell signaling networks that can confer BLBC cells with an invasive phenotype.
BLBC is diagnosed in approximately 15% of patients, with most BLBC tumors being classified as triple-negative breast cancer (TNBC; no detectable estrogen receptor (ER), progesterone receptor or HER2 expression) (13). The motility and invasion of BLBC cells can require EGFR and ERK1/2 kinase activity (9); however, the mechanistic basis of the requirement of these kinases is unknown. Interestingly, BLBC cells can be a hybrid cell type that maintains canonical epithelial traits, such as E-cadherin expression, while also expressing a subsets of mesenchymal genes, including transcription factors and cytoskeletal proteins that are increased in expression during the induction of EMTs (14–16). While tumor cells in a hybrid state can have enhanced invasive traits (17), whether elements of hybrid states functionally contribute to invasive phenotypes is not known. It is also unclear how a hybrid state is induced in BLBC cells. Thus, the cell signaling pathways that confer BLBC cells with invasive traits are poorly understood.
MicroRNAs (miRNAs) regulate cell identity by inducing the post-transcriptional silencing of genes through mRNA destabilization and antagonizing translation. Therefore, to define signaling pathways essential for BLBC migration, we determined the wound closure rate of a model BLBC cell line transfected with 879 miRNAs. Functional requirements for migration in wound healing assays can reflect necessary traits for invasion and metastasis in mouse models of breast cancer (18,19), thus this approach had the potential to reveal key elements of BLBC invasion in vivo. By combining the results of our wounding screen with miRNA expression profiling, we found that miR203a was highly expressed in LBC cells and can suppress BLBC motility by silencing the transcription factor ΔNp63α. Interestingly, ΔNp63α enhances the expression of miR205, which increases BLBC migration rate and can block cells from converting to a mesenchymal state by silencing ZEB1 and ZEB2. Further investigation revealed that ΔNp63α promoted motility by inducing the transcription factor Slug (SNAI2) and the tyrosine kinase Axl, both of which can contribute to the EMT programs. Thus, ΔNp63α confers BLBC cells with a migratory phenotype through inducing a hybrid state in which components of EMT programs promote migration while the parallel activation of a miRNA maintains key features of epithelial character.
Materials and Methods
See Supplementary Methods for additional details.
Cell Culture and reagents
MCFDCIS cells were purchased from Asterand. T47D and MCF7 cells were purchased from American Type Culture Collection. HCC1428, HCC1806 and HCC1954 cells were a gift from John Minna (UTSW). All cell lines were validated by Powerplex genotyping before use. Cells were cultured as described (9).
Wounding assay
Seventy-two hours after transfection, confluent cells in glass bottom 96-well plates (BD Biosciences) were wounded with 96 identical teflon pins mounted together to create a wounding tool (V&P Scientific). An automated image analysis protocol that used a threshold of pixel intensity to define cell-free space was used to quantify wound closure.
Transfection of siRNAs and miRNAs
Cells were transfected with 5–50 nM of siRNA or 10–50 nM of miRNA mimic using RNAiMax (Invitrogen). Control cells in siRNA-based experiments were transfected with a pool of siRNAs that does not target human genes. Control cells in miRNA-based experiments were transfected with miR545, which produced no phenotype compared to mock transfected cells (no siRNA) or control siRNA transfected cells (Supplementary Fig. 1A).
Quantitative real-time PCR
Performed as described (20).
Immunoblotting, immunofluorescence and live-imaging
All experiments were performed as described (9). Immunoblotting was performed on lysates from cells transfected with siRNAs or miRNAs for 72 h.
Xenografts
MCFDCIS or MCFDCIS-Slug cells were injected in the fourth mammary fat pad NOD/SCID female mice as described (9). Mice were sacrificed and the tumors were removed three weeks after injection.
Gene and miRNA expression profiling
The mRNA and miRNA expression data are available at the GEO (GSE58643, GSE62569).
Breast cancer patient survival analysis
The correlation between p63 expression and breast cancer patient survival time was performed using the KM-plotter meta-analysis database (21). ER−/HER2− (basal-type) and ER+/HER2− (Luminal A) patients were stratified into “p63-low” and “p63-high” groups based on the lower tertile of p63 expression (Gene ID 209863). Survival differences were compared by log-rank (Mantel-Cox) test.
Statistical Methods
Unpaired student's t-tests (2-tailed) were used unless otherwise indicated.
Results
Identification of microRNAs that regulate basal-type breast cancer cell migration
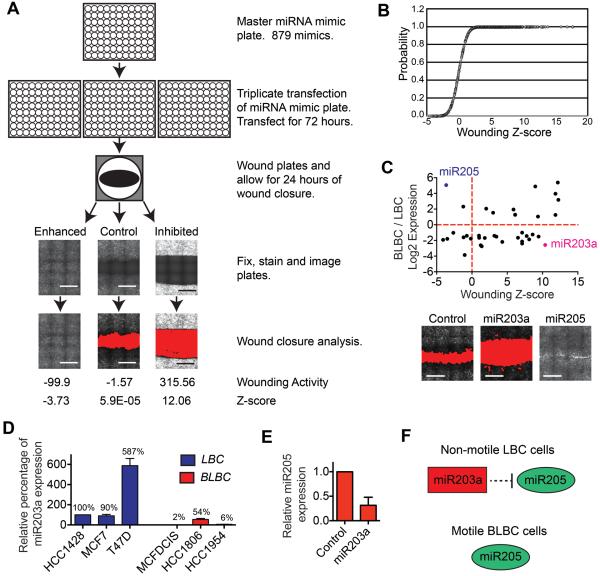
To define signaling pathways required for BLBC migration, we determined the wound closure rate of MCFDCIS cells transfected with 879 miRNA mimics in a one-condition/one-well format (Fig. 1A). MCFDCIS cells are a BLBC population (22) that completes wound closure within 24 h, which reduces the contribution of proliferation to observed phenotypes. Importantly, MCFDCIS cells invade in organotypic culture and in vivo while maintaining epithelial character (9,22,23). Cells were cultured for 72 h after reverse transfection, which allowed miRNAs to directly silence target genes and induce indirect changes in gene expression and signaling pathway characteristics. Equivalent wounds were introduced using a 96-pin wounding tool (Fig. 1A) and allowed to close for 24 h, after which cells were fixed, imaged and analyzed (Fig. 1A, Supplementary Fig. S1B and Supplementary Table S1). The total fluorescence of the wounded cells served as an indicator of the relative cell number for each condition (Supplementary Fig. S1C and Supplementary Table S1). Because significant reductions in cell number can reduce the extent of wound closure (18,24), we focused on the 574 miRNAs which induced a ≤50% reduction in fluorescent signaling intensity (Fig. 1B, Supplementary Fig. S1C and Supplementary Table S2). Re-testing of 132 miRNA mimics showed a correlation in wounding response (Supplementary Fig. S1D and Supplementary Table S3) and miRNA mimics with identical seed sequences (positions 2–7) had similar phenotypes (Supplementary Fig. S1E).
Figure 1. Results from the miRNA wounding screen.
(A) Model figure depicting the screening procedure. Representative images show cells stained with phalloidin 24 h after wounding. The area of the wound calculated by image analysis software is shown in red in the bottom images. Control= mock transfected cells. Raw activity values and z-scores are shown. Scale bars, 1 mm. (B) Cumulative distribution plot of the wound healing z-scores for miRNA mimics that nominally suppressed cell viability. (C) Graph of wound healing z-scores (x) and relative expression (y) for the 41 miRNAs with ≥2 fold difference in expression in motile BLBC cells (MCFDCIS) compared to non-motile LBC cells. (HCC1428). Representative wound healing images from the miRNA screen are shown. Scale bars, 1 mm. (D) Graph shows relative miR203a expression in nonmotile LBC (HCC1428, MCF7 and T47D, blue) and motile BLBC (MCFDCIS, HCC1806 and HC1954, red) cells as determined by q-PCR (mean+range, n=2). Values are relative to HCC1428 miR203a expression. (E) Graph showing relative miR205 expression as determined by q-PCR in MCFDCIS cells transfected as indicated (mean+range, n=2). (F) Graphic showing the regulation of cell state by miR203a and miR205.
To further prioritize analysis, we determined which miRNAs that inhibited wound closure may maintain LBC cells in a non-motile state. Of the 41 miRNAs expressed ≥ 2-fold higher in HCC1428 LBC cells (9) compared to MCFDCIS cells, miR203a most potently suppressed wound closure with a nominal inhibition of cell growth (Fig 1C and Supplementary Tables S4 and S5). MiR203a was also more highly expressed in nonmotile MCF7 and T47D LBC cells (9), compared to motile HCC1806 and HCC1954 BLBC cells (Fig. 1D and Supplementary Fig. S2A) (9). These results indicate that miR203a may maintain a nonmotile state in LBC cells by silencing signaling pathways that confer BLBC cells with a migratory ability.
Interestingly, our analysis also revealed that miR205 was the only miRNA expressed ≥2-fold higher in MCFDCIS cells that enhanced wound closure rate. MiR205 also increased the spontaneous motility of SUM149 BLBC cells (Supplementary Fig. S2B), further indicating that miR205 can endogenously function to promote BLBC motility. In addition, miR203a transfection reduced miR205 expression (Fig. 1E), suggesting that miR205 is a component of a signaling pathway that promoted the motile phenotype of BLBC cells and could be suppressed by miR203a (Fig. 1F).
ΔNp63α regulates cell migration
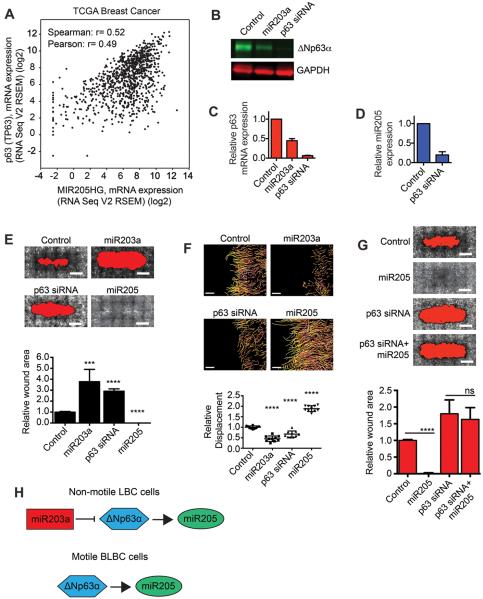
To define how miR203a controlled miR205 expression, we determined which predicted targets of miR203a were co-expressed with miR205 in breast cancer patient tumors (Supplementary Fig. S2C, D and E and Supplementary Table S6). The transcription factor p63 was one of 4 predicted miR203a target genes that were co-expressed with miR205 (Fig. 2A). P63 is necessary for miR205 expression in bladder cancer cells and miR203a can suppress p63 expression in normal mammary epithelial cells (25,26), suggesting that miR203a may regulate miR205 levels by silencing p63. Of the 6 p63 isoforms (27), ΔNp63α was the dominant isoform expressed in the MCFDCIS cells (Supplementary Fig. S2F). ΔNp63α was also silenced by miR203a (Fig. 2B and C) and ΔNp63α was necessary for miR205 expression (Fig. 2D).
Figure 2. ΔNp63α is required for cell motility.
(A) Graph showing expression of miR205 (x) and p63 (y) in breast tumors. (B) Immunoblot showing expression of ΔNp63α in MCFDCIS cells transfected as indicated. (C) Graph shows relative expression of ΔNp63α as determined by q-PCR in cells transfected as indicated (mean + range, n=2). (D) Graph shows relative miR205 expression as determined by q-PCR in MCFDCIS cells transfected as indicated (mean + SD, n=3). (E) Wound healing of MCFDCIS cells transfected as indicated. Scale bars, 1 mm. Graph shows relative wound area (n=6 wounds from 2 independent experiments). ***p < 0.001, ****p< 0.0001, unpaired Student's t-test. (F) MCFDCIS cells transfected as indicated were wounded and imaged for 7 h. Tracking of cell movement is shown. Color changes indicate increasing time. Scale bars, 100 μm. Graph shows quantification of cell displacement (mean ± SD, n=10 x,y positions over 2 independent experiments). ****p< 0.0001, unpaired Student's t-test. (G) Wound healing of MCFDCIS cells transfected as indicated. Scale bars, 1 mm. Graph shows relative wound area (mean + SD, n=6 wounds from 2 independent experiments). ****p< 0.0001, unpaired Student's t-test. (H) Graphic showing miR203a regulation of ΔNp63α and miR205.
The requirement of ΔNp63α for miR205 expression suggested that ΔNp63α may promote BLBC motility. Indeed, p63 siRNAs reduced MCFDCIS and HCC1806 motility (Fig. 2E and F and Supplementary Fig. S3A–C), with the targeting specificity of the individual p63 siRNAs indicating that migration specifically required ΔNp63α (Supplementary Fig. S3D). Exogenous miR205 was not sufficient to promote wound closure in ΔNp63α depleted MCFDCIS cells (Fig. 2G), indicating that additional ΔNp63α regulated signaling components were required for motility. Together, these results suggest that ΔNp63α promotes the motile phenotype of BLBC cells through the induction of miR205 (Fig. 2H). In addition, miR203a may sustain LBC identity by antagonizing the expression of a ΔNp63α dependent signaling network (Fig. 2H).
ΔNp63α regulates parallel signaling pathways that are required for collective migration
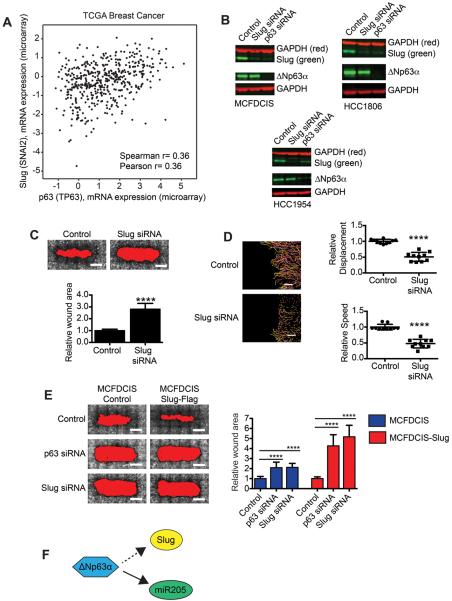
To define the additional ΔNp63α-regulated events required for migration, we determined that 181 genes were dependent on ΔNp63α for expression (≥2 fold reduction, p< 0.05) in both MCFDCIS and HCC1806 cells (Supplementary Fig. S4A and Supplementary Table S7). Sixty-one of these ΔNp63α regulated genes were expressed at a higher level in motile MCFDCIS and HCC1806 BLBC cells compared to nonmotile HCC1428 and MCF7 LBC cells (≥2 fold, p< 0.05), suggesting that they may confer BLBC cells with migratory ability (Supplementary Fig. S4A and Supplementary Tables S7–9). To further prioritize analysis, we determined that 11 of the ΔNp63α regulated “motility” genes were co-expressed with ΔNp63α in breast cancer patient tumors, indicating that they may contribute to ΔNp63α dependent cell behaviors in vivo (Supplementary Fig. S4A and B and Supplementary Table S10). One of these ΔNp63α regulated genes was the transcription factor Slug (SNAI2) (Fig. 3A), which can promote EMT and invasion (28) by binding to an E-Box site in the E-cadherin promoter to silence E-cadherin expression (29). However, because BLBC cells can express E-cadherin, it was not clear whether Slug expression was innocuous in BLBC cells, or if Slug contributed to migration through a different mechanism. Therefore, to determine if factors that contribute to EMT could promote the motile phenotype of hybrid mesenchymal/epithelial BLBC cells, we further investigated the regulation of Slug by ΔNp63α.
Figure 3. ΔNp63α promotes cell motility by regulating Slug expression.
(A) Graph showing expression of p63 (x) and Slug (y) in breast tumors. (B) Immunoblots showing expression of Slug and ΔNp63α in cell lines transfected as indicated (n=2). (C) Wound healing of MCFDCIS cells transfected as indicated. Scale bars, 1 mm. Graph shows relative wound area (mean + SD, n=6 wounds from 2 independent experiments). ****p< 0.0001, unpaired Student's t-test. (D) MCFDCIS cells transfected as indicated were wounded and imaged for 7 h. Scale bars, 100 μm. Graphs show cell speed and displacement (mean ± SD, n=10 x,y positions over 2 independent experiments). ****p< 0.0001, unpaired Student's t-test. (E) Wound healing of MCFDCIS and MCFDCIS-Slug cells transfected as indicated. Scale bars, 1 mm. Graph shows quantification of wound area (mean + SD, n=9 wounds from 3 independent experiments). ****p< 0.0001, unpaired Student's t-test. (F) Graphic showing ΔNp63α regulation of Slug and miR205.
Consistent with our gene expression analysis, ΔNp63α was necessary for Slug protein expression in MCFDCIS, HCC1806 and HCC1954 cells (Fig. 3B and Supplementary Fig. S5A). Importantly, Slug was required for MCFDCIS and HCC1806 motility (Fig. 3C and D and Supplementary Fig. S5B–D) and Slug overexpression increased the rate of MCFDCIS wound closure (Fig. 3E). However, MCFDCIS-Slug cells remained dependent on ΔNp63α for migration (Fig. 3E), demonstrating that Slug was one of multiple genes regulated by ΔNp63α that were necessary for BLBC motility (Fig. 3F).
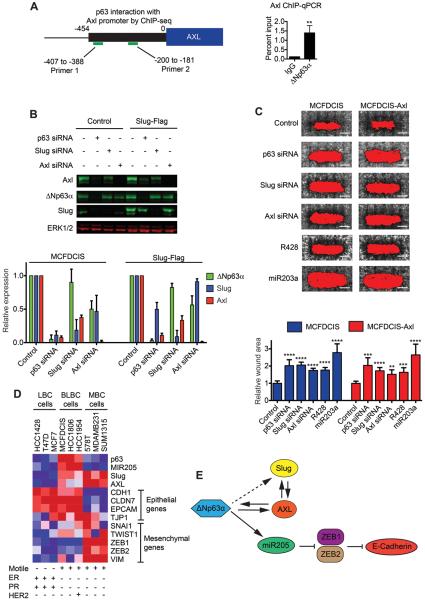
Our discovery that ΔNp63α could regulate Slug expression suggested that ΔNp63α may promote motility through the regulation of additional genes that can contribute to EMTs. The EMT-related tyrosine kinase Axl (30) was one of the 61 “motility” genes dependent on ΔNp63α for expression. In addition, a potential ΔNp63α interaction site was detected within the Axl promoter (Fig. 4A and Supplementary Fig. S6A) based on ChIP-seq experiments performed in keratinocytes (31,32). Indeed, ΔNp63α could bind to the Axl promoter in MCFDCIS cells (Fig. 4A) and ΔNp63α was necessary for Axl protein expression in MCFDCIS, HCC1806 and HCC1954 BLBC cells (Fig. 4B, Supplementary Fig. S6B). Slug depletion also partially reduced Axl levels and exogenous Slug partially sustained Axl expression in the absence of ΔNp63α (Fig. 4B), indicating that in addition to direct binding to the Axl promoter, ΔNp63α may control Axl expression through the induction of Slug. Interestingly, ΔNp63α and Slug were partially dependent on Axl for expression (Fig. 4B), suggesting that Axl functions within a positive feedback loop that contributes to ΔNp63α and Slug regulation. Axl siRNAs and a pharmacological inhibitor of Axl, R428 (33), reduced MCFDCIS wound closure, demonstrating that ΔNp63α induced Axl expression contributed to BLBC motility (Fig. 4C and Supplementary Fig. S6C). However, exogenous Axl did not rescue the migration defects of ΔNp63α depleted MCFDCIS cells (Fig. 4C), indicating that Slug and Axl each have distinct functions that promote BLBC motility.
Figure 4. ΔNp63α directly regulates Axl expression to promote cell motility.
(A) Schematic summarizes analysis of ΔNp63α binding to the Axl promoter. Graph shows quantification of ChIP qPCR of the Axl promoter (mean + SD, n=3). **p < 0.01, unpaired Student's t-test. (B) Immunoblots show Axl, ΔNp63α and Slug expression in MCFDCIS and MCFDCIS-Slug cells transfected as indicated. Graph shows relative expression (mean + SD, n=3). (C) Wound healing of MCFDCIS and MCFDCIS-Axl cells transfected as indicated. Scale bars, 1 mm. Graph shows relative wound area (mean + SD, n=6 wounds from 2 independent experiments). **p < 0.01, ***p < 0.001, ****p < 0.0001, unpaired Student's t-test. (D) Heatmap showing relative expression of the indicated genes. Red = high expression, blue = low expression. ER= estrogen receptor, PR= progesterone receptor, MBC= mesenchymal-type breast cancer. (E) Graphic showing ΔNp63α regulation of miR205, Slug and Axl.
Although Slug or Axl overexpression can induce a complete EMT and conversion to a mesenchymal state (34,35), their combined expression did not trigger the BLBC cells to shed their epithelial traits (Fig. 4D). ΔNp63α can promote the retention of epithelial character through inducing miR205, which can silence ZEB1 and ZEB2 to prevent a conversion to a mesenchymal-state (36,37). As has been previously observed (36,38), transfection of mesenchymal-like breast cancer cells with miR205 suppressed ZEB1 and ZEB2 expression (Supplementary Fig. S6D) and reduced cell motility (Supplementary Fig. S6E). Similar to observations in prostate and bladder cancer (25,37), ΔNp63α and miR205 were also expressed at a low level in mesenchymal-like breast cancer cells (Fig. 4D). These results suggest that high levels ΔNp63α and miR205 contribute to the retention of epithelial character in BLBC cells and that low levels of ΔNp63α and miR205 are necessary for breast cancer cells to adopt a mesenchymal state. Interestingly, Axl and Slug were expressed at a high level in the mesenchymal-like cells (Fig. 4D). Given the low ΔNp63α expression in these cells, Slug and Axl may be induced by p63-independent pathways in mesenchymal-type cells, which suggests the mechanism of Slug and Axl activation may influence cell identity. Together these results suggest that ΔNp63α can increase Slug and Axl expression to promote motility, while simultaneously inducing miR205, which can silence signaling pathways that suppress epithelial traits (Fig. 4E).
ΔNp63α is not sufficient to induce motility, Slug or Axl expression in LBC cells
We next determined if ΔNp63α was sufficient to confer intrinsically nonmotile HCC1428 LBC cells with a migratory phenotype. Exogenous ΔNp63α increased miR205 expression (Fig. 5A), indicating that ΔNp63α was capable of interacting with DNA and promoting transcription in LBC cells. However, ΔNp63α overexpression did not increase Slug and Axl protein levels (Fig. 5B), or accelerate the rate of HCC1428 cell wound closure (Fig. 5C). MiR203a can directly silence Slug expression (39), and based on the Targetscan algorithm (40), miR203a is predicted to target the 3'UTR of Axl (Supplementary Fig. S6F). This suggested that the high level of endogenous miR203a in HCC1428 cells may prevent ΔNp63α from inducing Slug and Axl expression, thereby impinging on the ability of ΔNp63α to induce a motile state. To test this possibility, we measured the expression of Slug and Axl in MCFDCIS cells expressing exogenous ΔNp63α (Fig. 5D) after transfection with miR203a. Whereas the exogenous ΔNp63α was resistant to depletion by miR203, both Slug and Axl expression were silenced (Fig. 5E). Therefore miR203a can silence Slug and Axl, even when there is a high level ΔNp63α expression. Together, these results indicate that the low expression of miR203a in BLBC cells permits ΔNp63α to induce Slug and Axl expression. Thus, the miRNA content of a cell may influence the expression of ΔNp63α target genes and the nature of ΔNp63α induced cell phenotypes (Fig. 5F).
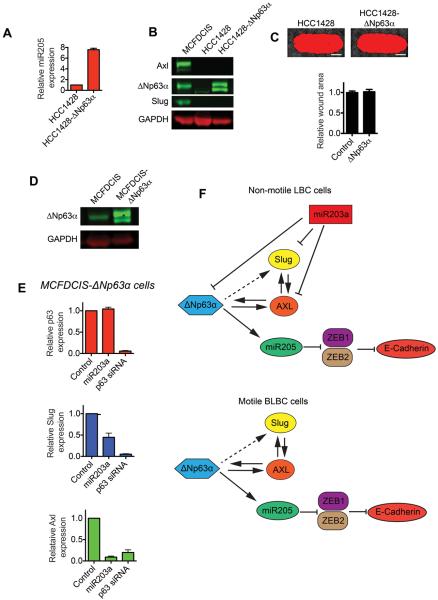
Figure 5. ΔNp63α is not sufficient to induce LBC motility.
(A) Graph shows relative miR205 expression in HCC1428 and HCC1428-ΔNp63α cells, as determined by q-PCR (mean + range, n=2). (B) Immunoblots show Axl, ΔNp63α and Slug expression in MCFDCIS, HCC1428 and HCC1428-ΔNp63α cells. (C) Wound healing of HCC1428 and HCC1428-ΔNp63α cells. Scale bars, 1 mm. Graph shows relative wound area (mean + SD, n=6 wounds from 2 independent experiments). (D) Immunoblot showing the expression of ΔNp63α in MCFDCIS and MCFDCIS-ΔNp63α cells. (E) Graphs show relative ΔNp63α, Slug and Axl expression as determined by q-PCR in MCFDCIS-ΔNp63α cells transfected as indicated (mean + SD, n=3). (F) Graphic summarizing the regulation of the ΔNp63α pathway.
ΔNp63α and Slug promote collective invasion in vivo
We next investigated how ΔNp63α expression correlated with breast cancer patient survival time. Interestingly, high ΔNp63α expression correlated with shorter overall survival time in ER-/HER2- patients (Fig. 6A); however, no correlation between ΔNp63α expression and ER+/HER2- patient survival was observed (Fig. 6A). These clinical observations are consistent with our results showing that ΔNp63α can contribute to the motile state of ER-/HER2- breast cancer cells, which frequently are classified as BLBC tumors (13).
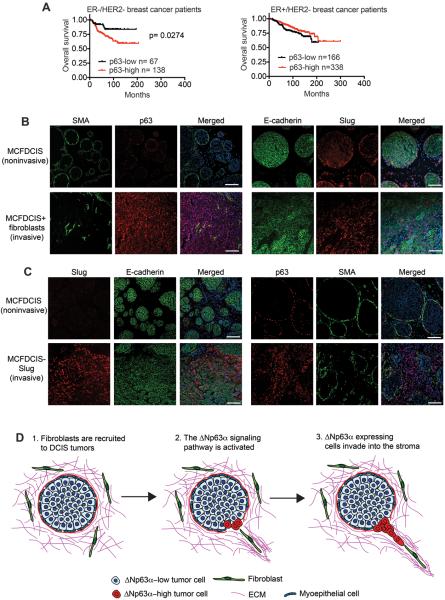
Figure 6. The ΔNp63α pathway promotes invasion in vivo.
(A) Kaplan-Meier curves showing overall survival of ER-/HER2- and ER+/HER- patients classified as “p63-high” and “p63-low” based on p63 mRNA expression. Survival differences were compared by log-rank (Mantel-Cox) test. Analysis of publicly available data sets was performed using KM-Plotter. (B) Immunostaining of noninvasive tumors formed by MCFDCIS cells or invasive tumors formed by MCFDCIS cells co-injected with mammary fibroblasts (n=10 mice, each condition). Scale bars, 100 μm. (C) Immunostaining of noninvasive tumors formed by MCFDCIS cells or invasive tumors formed by MCFDCIS-Slug cells (n=20 mice, each condition). Scale bars, 100 μm. (D) Model for ΔNp63α induced invasion.
To determine how ΔNp63α signaling contributes to cell phenotypes in primary tumors, we examined ΔNp63α and Slug expression in MCFDCIS orthotopic xenografts. MCFDCIS cells are a unique cell type that forms noninvasive DCIS lesions (22,23). ΔNp63α and Slug were expressed in the smooth muscle actin (SMA)-positive myoepithelial cell layer that forms around the xenograft DCIS lesions (Fig. 6B). However, ΔNp63α and Slug were rarely detected in the central luminal epithelial populations (Fig. 6B). ΔNp63α was expressed in all cells in monolayer culture (Supplementary Fig. S7A) and is positively regulated by cell attachment (22) and contact with ECM (10), which suggests that ΔNp63α levels are reduced in the luminal MCFDCIS cells due to a lack of cell-ECM contact (22). Consistent with this possibility, myoepithelial cells, basal mammary epithelial cells and mammary stem cells, all interact with ECM components and express ΔNp63α, Slug and miR205 (41–46). Interestingly, ΔNp63α and Slug expression were high in SMA-negative MCFDCIS cells induced to invade by fibroblasts, (Fig. 6B), suggesting that the induction of ΔNp63α and Slug was contributing to the transition from DCIS to invasive breast cancer. However, we were unable to stably reduce ΔNp63α and Slug expression with shRNAs to determine if either gene was required for MCFDCIS invasion. This may be because ΔNp63α and Slug were both necessary for long-term cell growth (Supplementary Fig. S7B). Nevertheless, our results showed that the ΔNp63α pathway was activated during the initiation of invasion.
We next determined if the ΔNp63α pathway was sufficient to induce invasion. While we were able to exogenously express ΔNp63α in MCFDCIS cells in monolayer culture (Supplementary Fig. S2E), the overexpressed ΔNp63α was not detected in the xenografts tumors, consistent with previous observations (22). This suggests that ΔNp63α levels are controlled by a post-transcriptional regulatory mechanism (22), or that a precise level of ΔNp63α expression is needed for tumor formation. Because ΔNp63α was potentially promoting invasion by increasing Slug expression, we determined if Slug could induce invasion. Indeed, exogenous Slug was sufficient to induce the collective invasion of MCFDCIS into the ECM (Fig. 6C and Supplementary Fig. S7C), demonstrating that the activation of a component of the ΔNp63α regulated signaling network was sufficient to promote invasion in vivo. Importantly, invasive MCFDCIS-Slug cells expressed E-cadherin, indicating cells retained their epithelial character (Fig. 6C). Like we observed during fibroblast induced invasion, ΔNp63α expression was increased in the invasive MCFDCIS-Slug cells (Fig. 6C), consistent with ΔNp63α being essential for Slug induced motility and invasion. It is possible that ΔNp63α expression is increased when the MCFDCIS cells come in contact with the ECM, or that Slug contributes to the induction of ΔNp63α expression in vivo. Together, these results demonstrate that increased Slug expression is sufficient to promote the collective invasion of DCIS cells.
Discussion
In defining how miR203a maintained a nonmotile luminal-type state, we uncovered a ΔNp63α regulated signaling network that conferred BLBC breast cancer cells with a motile phenotype. ΔNp63α promoted migration by inducing the expression of Slug and Axl, two genes that can facilitate EMT. Interestingly, ΔNp63α also directly increased the expression miR205, which can enhance the rate of BLBC motility and defend against the loss of epithelial traits. Thus, ΔNp63α promoted motility through the induction of a hybrid mesenchymal/epithelial state. Hybrid states can be induced by sub-threshold levels of TGFβ, suggesting that some hybrid states may be the result of a partial completion of an EMT program (47). By comparison, ΔNp63α, Slug and miR205 are all expressed in mammary stem cells and myoepithelial cells (41–46), which raises the possibility that this ΔNp63α induced hybrid state may be a pre-existing biological program that has evolved to allow EMT inducing genes, like Slug and Axl, to contribute to cell behavior within an epithelial lineage. Therefore, hybrid states may be conferred by signaling pathways that define normal cell identity and are not necessarily unstable transition states (3) on a path towards a conversion to a fully mesenchymal phenotype.
Our results suggest that fibroblasts can promote collective invasion of BLBC cells by inducing ΔNp63α expression in DCIS tumors (Fig. 6D). The tumor associated fibroblasts may trigger ΔNp63α expression through paracrine communication or by physically disrupting the myoepithelial cell layer, which could result in the DCIS tumor cells gaining direct contact with activating factors located within the stromal ECM. The ability of Slug to induce MCFDCIS invasion suggests that, once activated, ΔNp63α dependent signaling confers BLBC cells with a motile phenotype. Interestingly, because a mesenchymal phenotype can be incompatible with growth in distant tissues (48,49), the ability of ΔNp63α to confer migratory ability through the induction of a hybrid mesenchymal/epithelial state may promote the development of metastasis more potently than signaling mechanisms that induce a complete and sustained EMT. Consistent with this possibility, circulating tumor cells (50) and metastases (3) frequently display canonical epithelial traits. Together, these findings support the investigation of ΔNp63α regulatory programs that may be reawakened during neoplastic progression and contribute to tumor cell invasion.
Supplementary Material
Acknowledgements
We thank Xiaoyu Wang for providing assistance with gene expression analysis and the UTSW High-Throughput Screening Laboratory for assistance with the miRNA wounding screen.
Grant Support This work was supported by NIH grants 1RO1CA155241 (G.W.P) and 5T32CA124334 (M.E.), CPRIT training grant, RP10496 (T.T. Dang), the Mary Kay Foundation (G.W.P.) and UTSW institutional support funds (G.W.P.).
Footnotes
The authors declare no conflicts of interest.
References
- 1.Grunert S, Jechlinger M, Beug H. Diverse cellular and molecular mechanisms contribute to epithelial plasticity and metastasis. Nat Rev Mol Cell Biol. 2003;4:657–65. doi: 10.1038/nrm1175. [DOI] [PubMed] [Google Scholar]
- 2.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–96. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19:1438–49. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rorth P. Collective cell migration. Annu Rev Cell Dev Biol. 2009;25:407–29. doi: 10.1146/annurev.cellbio.042308.113231. [DOI] [PubMed] [Google Scholar]
- 5.Kowalski PJ, Rubin MA, Kleer CG. E-cadherin expression in primary carcinomas of the breast and its distant metastases. Breast Cancer Res. 2003;5:R217–22. doi: 10.1186/bcr651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qureshi HS, Linden MD, Divine G, Raju UB. E-cadherin status in breast cancer correlates with histologic type but does not correlate with established prognostic parameters. Am J Clin Pathol. 2006;125:377–85. [PubMed] [Google Scholar]
- 7.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soysal SD, Muenst S, Barbie T, Fleming T, Gao F, Spizzo G, et al. EpCAM expression varies significantly and is differentially associated with prognosis in the luminal B HER2(+), basal-like, and HER2 intrinsic subtypes of breast cancer. Br J Cancer. 2013;108:1480–7. doi: 10.1038/bjc.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dang TT, Prechtl AM, Pearson GW. Breast cancer subtype-specific interactions with the microenvironment dictate mechanisms of invasion. Cancer Res. 2011;71:6857–66. doi: 10.1158/0008-5472.CAN-11-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung KJ, Gabrielson E, Werb Z, Ewald AJ. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell. 2013;155:1639–51. doi: 10.1016/j.cell.2013.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedl P, Locker J, Sahai E, Segall JE. Classifying collective cancer cell invasion. Nat Cell Biol. 2012;14:777–83. doi: 10.1038/ncb2548. [DOI] [PubMed] [Google Scholar]
- 12.Charafe-Jauffret E, Ginestier C, Monville F, Finetti P, Adelaide J, Cervera N, et al. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene. 2006;25:2273–84. doi: 10.1038/sj.onc.1209254. [DOI] [PubMed] [Google Scholar]
- 13.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5:5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, et al. Core epithelial-tomesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci U S A. 2010;107:15449–54. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blick T, Widodo E, Hugo H, Waltham M, Lenburg ME, Neve RM, et al. Epithelial mesenchymal transition traits in human breast cancer cell lines. Clin Exp Metastasis. 2008;25:629–42. doi: 10.1007/s10585-008-9170-6. [DOI] [PubMed] [Google Scholar]
- 16.Lu M, Jolly MK, Levine H, Onuchic JN, Ben-Jacob E. MicroRNA-based regulation of epithelial-hybrid-mesenchymal fate determination. Proc Natl Acad Sci U S A. 2013;110:18144–9. doi: 10.1073/pnas.1318192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schliekelman MJ, Taguchi A, Zhu J, Dai X, Rodriguez J, Celiktas M, et al. Molecular portraits of epithelial, mesenchymal and hybrid states in lung adenocarcinoma and their relevance to survival. Cancer Res. 2015 doi: 10.1158/0008-5472.CAN-14-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simpson KJ, Selfors LM, Bui J, Reynolds A, Leake D, Khvorova A, et al. Identification of genes that regulate epithelial cell migration using an siRNA screening approach. Nat Cell Biol. 2008;10:1027–38. doi: 10.1038/ncb1762. [DOI] [PubMed] [Google Scholar]
- 19.Mouneimne G, Hansen SD, Selfors LM, Petrak L, Hickey MM, Gallegos LL, et al. Differential remodeling of actin cytoskeleton architecture by profilin isoforms leads to distinct effects on cell migration and invasion. Cancer Cell. 2012;22:615–30. doi: 10.1016/j.ccr.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bookout AL, Cummins CL, Mangelsdorf DJ, Pesola JM, Kramer MF. High-throughput real-time quantitative reverse transcription PCR. Curr Protoc Mol Biol. 2006;Chapter 15(Unit 15 8) doi: 10.1002/0471142727.mb1508s73. [DOI] [PubMed] [Google Scholar]
- 21.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast cancer research and treatment. 2010;123:725–31. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 22.Hu M, Yao J, Carroll DK, Weremowicz S, Chen H, Carrasco D, et al. Regulation of in situ to invasive breast carcinoma transition. Cancer Cell. 2008;13:394–406. doi: 10.1016/j.ccr.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller FR, Santner SJ, Tait L, Dawson PJ. MCF10DCIS.com xenograft model of human comedo ductal carcinoma in situ. J Natl Cancer Inst. 2000;92:1185–6. doi: 10.1093/jnci/92.14.1185a. [DOI] [PubMed] [Google Scholar]
- 24.Vitorino P, Meyer T. Modular control of endothelial sheet migration. Genes Dev. 2008;22:3268–81. doi: 10.1101/gad.1725808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tran MN, Choi W, Wszolek MF, Navai N, Lee IL, Nitti G, et al. The p63 protein isoform DeltaNp63alpha inhibits epithelial-mesenchymal transition in human bladder cancer cells: role of MIR-205. J Biol Chem. 2013;288:3275–88. doi: 10.1074/jbc.M112.408104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeCastro AJ, Dunphy KA, Hutchinson J, Balboni AL, Cherukuri P, Jerry DJ, et al. MiR203 mediates subversion of stem cell properties during mammary epithelial differentiation via repression of DeltaNP63alpha and promotes mesenchymal-to-epithelial transition. Cell death & disease. 2013;4:e514. doi: 10.1038/cddis.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su X, Chakravarti D, Flores ER. p63 steps into the limelight: crucial roles in the suppression of tumorigenesis and metastasis. Nature reviews Cancer. 2013;13:136–43. doi: 10.1038/nrc3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nieto MA. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu Rev Cell Dev Biol. 2011;27:347–76. doi: 10.1146/annurev-cellbio-092910-154036. [DOI] [PubMed] [Google Scholar]
- 29.Hajra KM, Chen DY, Fearon ER. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002;62:1613–8. [PubMed] [Google Scholar]
- 30.Gjerdrum C, Tiron C, Hoiby T, Stefansson I, Haugen H, Sandal T, et al. Axl is an essential epithelial-to-mesenchymal transition-induced regulator of breast cancer metastasis and patient survival. Proc Natl Acad Sci U S A. 2010;107:1124–9. doi: 10.1073/pnas.0909333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zarnegar BJ, Webster DE, Lopez-Pajares V, Vander Stoep Hunt B, Qu K, Yan KJ, et al. Genomic profiling of a human organotypic model of AEC syndrome reveals ZNF750 as an essential downstream target of mutant TP63. Am J Hum Genet. 2012;91:435–43. doi: 10.1016/j.ajhg.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kouwenhoven EN, van Heeringen SJ, Tena JJ, Oti M, Dutilh BE, Alonso ME, et al. Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genet. 2010;6:e1001065. doi: 10.1371/journal.pgen.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holland SJ, Pan A, Franci C, Hu Y, Chang B, Li W, et al. R428, a selective small molecule inhibitor of Axl kinase, blocks tumor spread and prolongs survival in models of metastatic breast cancer. Cancer Res. 2010;70:1544–54. doi: 10.1158/0008-5472.CAN-09-2997. [DOI] [PubMed] [Google Scholar]
- 34.Bolos V, Peinado H, Perez-Moreno MA, Fraga MF, Esteller M, Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J Cell Sci. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- 35.Asiedu MK, Beauchamp-Perez FD, Ingle JN, Behrens MD, Radisky DC, Knutson KL. AXL induces epithelial-to-mesenchymal transition and regulates the function of breast cancer stem cells. Oncogene. 2014;33:1316–24. doi: 10.1038/onc.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 37.Tucci P, Agostini M, Grespi F, Markert EK, Terrinoni A, Vousden KH, et al. Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer. Proc Natl Acad Sci U S A. 2012;109:15312–7. doi: 10.1073/pnas.1110977109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu H, Zhu S, Mo YY. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 2009;19:439–48. doi: 10.1038/cr.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding X, Park SI, McCauley LK, Wang CY. Signaling between transforming growth factor beta (TGF-beta) and transcription factor SNAI2 represses expression of microRNA miR-203 to promote epithelial-mesenchymal transition and tumor metastasis. J Biol Chem. 2013;288:10241–53. doi: 10.1074/jbc.M112.443655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 41.Chakrabarti R, Wei Y, Hwang J, Hang X, Andres Blanco M, Choudhury A, et al. DeltaNp63 promotes stem cell activity in mammary gland development and basal-like breast cancer by enhancing Fzd7 expression and Wnt signalling. Nat Cell Biol. 2014;16:1004–15. doi: 10.1038/ncb3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barbareschi M, Pecciarini L, Cangi MG, Macri E, Rizzo A, Viale G, et al. p63, a p53 homologue, is a selective nuclear marker of myoepithelial cells of the human breast. Am J Surg Pathol. 2001;25:1054–60. doi: 10.1097/00000478-200108000-00010. [DOI] [PubMed] [Google Scholar]
- 43.Greene SB, Gunaratne PH, Hammond SM, Rosen JM. A putative role for microRNA-205 in mammary epithelial cell progenitors. Journal of cell science. 2010;123:606–18. doi: 10.1242/jcs.056812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, et al. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–20. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 45.Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–28. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phillips S, Prat A, Sedic M, Proia T, Wronski A, Mazumdar S, et al. Cell-State Transitions Regulated by SLUG Are Critical for Tissue Regeneration and Tumor Initiation. Stem cell reports. 2014;2:633–47. doi: 10.1016/j.stemcr.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J, Tian XJ, Zhang H, Teng Y, Li R, Bai F, et al. TGF-beta-induced epithelial-to-mesenchymal transition proceeds through stepwise activation of multiple feedback loops. Science signaling. 2014;7:ra91. doi: 10.1126/scisignal.2005304. [DOI] [PubMed] [Google Scholar]
- 48.Ocana OH, Corcoles R, Fabra A, Moreno-Bueno G, Acloque H, Vega S, et al. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell. 2012;22:709–24. doi: 10.1016/j.ccr.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 49.Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 2012;22:725–36. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–4. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.