Summary
A benign, clonable tag for the localization of proteins by electron microscopy of cells would be valuable, especially if it provided labelling with high signal-to-noise ratio and good spatial resolution. Here we explore the use of metallothionein as such a localization marker. We have achieved good success with desmin labelled in vitro and with a component of the yeast spindle pole body labelled in cells. Heavy metals added after fixation and embedding or during the process of freeze-substitution fixation provide readily visible signals with no concern that the heavy atoms are affecting the behaviour of the protein in its physiological environment. However, our methods did not work with protein components of the nuclear pore complex, suggesting that this approach is not yet universally applicable. We provide a full description of our optimal labelling conditions and other conditions tried, hoping that our work will allow others to label their own proteins of interest and/or improve on the methods we have defined.
Keywords: Clonable labels, electron microscopy, immuno-electron microscopy, metallothionein, nanoparticles, protein labels
Introduction
Widespread use of the green fluorescent protein (GFP) has demonstrated the tremendous scientific value of a clonable marker for protein localization in the light microscope (LM). Many believe that parallel value would devolve from an analogous label for electron microscopy (EM), particularly given the difficulties of obtaining successful localizations by immuno-EM (Ellisman et al., 2012). These difficulties include the frequent inactivation of epitopes by fixations that preserve cell fine structure, cellular damage wrought by the lysis required for antibody addition before sectioning, the limited labelling that occurs when only the section surface is available for antibody binding and the significant distance between antigen and label when indirect immune labelling is employed.
An ideal clonable tag for EM would have several readily identifiable properties: (1) a good signal-to-noise ratio (SNR), (2) a small size and low molecular weight, so as not to disrupt protein kinetics/function in vivo and (3) a tightly focused signal. If it could also be used to do correlative LM and EM, it would be additionally valuable. There have been several efforts to develop such labels for EM. One of the first was based on converting GFP into a signal visible in the EM by photobleaching in the presence of diaminobenzidine (Grabenbauer et al., 2005; Meiblitzer-Ruppitsch et al., 2008), which captures singlet oxygens generated by photobleaching and becomes an osmiophilic material that serves as an electron-dense stain suitable for EM. This approach provides an excellent way to correlate images from the LM and the EM, and GFP is a relatively compact protein tag that does not perturb protein function in many cases. Photobleaching turns this small protein into an electron scattering signal that is strong enough to visualize in cells, but it has emerged that this method has a rather low SNR and is comparatively insensitive, so only proteins of high abundance have been visualized successfully. Even then, it helps if the tag is situated in a membrane-bounded compartment, probably because this helps to concentrate reaction product (Gaietta et al., 2011). Finally, the reaction product generated by oxidation of diaminobenzidine is not as tightly localized to the source of singlet oxygen as one might like.
Two more efficient protein labels have been developed to precipitate diaminobenzidine by photo-oxidation:ReASH (Gaietta et al., 2002; Gaietta et al., 2006) and miniSOG (Shu et al., 2011). These have proven useful (Ludwig et al., 2013; Boassa et al., 2013), albeit they have not yet reached widespread use, partly because of practical difficulties and partly because of the diffuse localization of the electron-dense stain they produce. A cleverly engineered ascorbate peroxidase has recently been developed as a promising clonable tag (Martell et al., 2012), though it too uses the oxidation of diaminobenzidine to produce an electron scattering signal, so it lacks the tightly focused label one can obtain with a metallic nanoparticle. The value of a tag that produces a single particle of electron scattering atoms has been seen when such labels are practical, as in the localization of immunoglobulin while it is being transported across the gut epithelium of a neonatal mammal (He et al., 2008).
New and better labels for EM are so highly desired that there have been several efforts to obtain discrete, darkly staining labels by harnessing metallic particles as protein tags. Although this approach loses the value of a clonable tag, quantum dots are visible in the EM and can also be excellent fluorophores, making them attractive for correlative LM and EM (Deerinck et al., 2007). Bacterial ferritin has also been tried as an EM stain (Wang et al., 2011). However, with labels big enough to provide good SNR for cell biological EM there is a serious concern that so much additional mass will affect the kinetics of the labelled macromolecule, and therefore its function, yielding misleading results. These approaches lack the elegance of a small tag that is somehow developed for view in the EM at high SNR. A more promising label would be based on a metal binding protein or peptide of low molecular weight whose electron scattering properties could be developed after the protein’s functions had already been accomplished. One such protein is metallothionein (MTH), a name applied to a family of cysteine-rich, metal-binding proteins whose molecular weights range from 0.5 to 14 kDa. Some of these proteins will bind ~20 gold atoms, enough to make a particle that is visible in the electron microscope (Sano et al., 1992; Mercogliano & DeRosier, 2006), even in the context of a cell (Diestra et al., 2009; Risco et al., 2012). However, the tag has so far been used in cells only on abundant proteins, which do not test the ability of the method to yield sufficient SNR to make a practical method for labelling low-abundance proteins, as is necessary for widespread use. Moreover, in published work, the metal to be bound, e.g. zinc or gold, has been applied to living cells; with this approach one must be concerned about the physiological state of the cell in the presence of heavy metals at potentially toxic levels.
Here we build on previous work with MTH to develop a method that circumvents many of the above-mentioned problems and has proven to be impressively effective in some cases, though not all. Our work is grounded on Mercagliano’s demonstration that when MTH is conjugated to a protein, it can still bind enough gold to be seen in the EM, though two MTH’s in tandem make a bigger particle that is more readily visible by standard EM methods (Mercogliano & DeRosier, 2007). Others have shown that MTH used in this way can be a useful label for specific proteins assembled in vitro (Bouchet-Marquis et al., 2012). Our approach is designed to work on either macromolecular assemblies or cells but to avoid all issues of heavy metal toxicity by delaying the addition of metal until the samples that include a protein-MTH chimera have been frozen and are in preparation for EM. Here, we describe two successful procedures: (1) adding heavy metal to sections of samples that have already been rapidly frozen, fixed by freeze substitution, and embedded in a hydrophilic plastic and (2) adding metal during the process of freeze substitution.
As test systems for this approach, we have employed one protein polymer (desmin) and two intracellular sites where protein localization is well known, so we can assess the precision of labelling and the resulting SNR. For our cellular work we have employed yeast cells, whose genotypes can be precisely defined and where endogenous genes can be tagged by the insertion of DNA that encodes the label right into the chromosomal copy of the gene under study. Under these conditions, the tagged protein is expressed under the control of the wild-type promoter, so issues of overexpression are avoided and there is no unlabelled material to compete with the product of the labelled allele. In yeast, it is easy to assess the efficacy with which the labelled protein supports cellular function, and many cells can be processed comparatively quickly to obtain good statistics on both the distribution of label and the SNR. There is the potential difficulty of ‘noise’ introduced by the cell’s endogenous MTH, but this protein is largely confined to peroxisomes. Our results show that MTH can indeed be a useful, clonable label for the cellular localization of some proteins. We therefore provide protocols for use on your favourite protein and for assessing MTH labelling effectiveness in your own system.
Results
MTH can label protein assemblies with high SNR
The first specimen used to test MTH labelling was desmin, the subunit of intermediate filaments in muscle cells, as reviewed in (Herrmann et al., 2007). The intermediate filament proteins are readily purified, solubilized in 8 M urea and can be induced to polymerize in vitro by reducing the concentration of urea (Herrmann et al., 1996). Desmin in its wild-type form or with a single MTH added to either its N- or C-terminus was expressed in bacteria and purified for polymerization into filaments, as previous described (Kirmse et al., 2010); the proteins used here were generous gifts from R. Kirmse (Carl Zeiss Microscopy GmbH, Jena, Germany). Filaments of desmin stuck well enough to a sapphire disk to allow their preparation for EM by rapid freezing and freeze-substitution fixation, followed by embedding in Lowicryl K4M, a comparatively hydrophilic plastic. Sections of both wild-type and MTH-labelled desmin were treated with 1 mM aurothiomalate, followed by a gold enhancement protocol (Nanoprobes, Inc. Yaphank, NY 11980), then imaged as a series of tilted views to enable tomographic reconstruction. Computational slices of these tomograms show a rather dense distribution of small metal particles on the MTH sample but no such staining on wild-type desmin treated in an identical manner (Fig. 1, see also Movies S1 and S2, respectively).
Fig. 1.
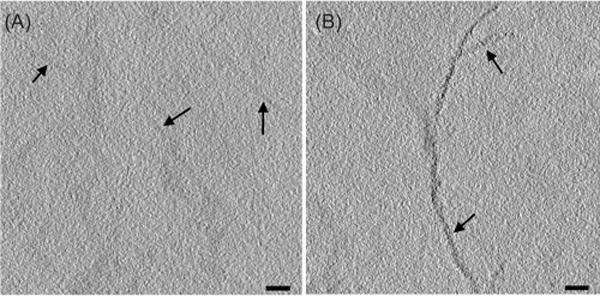
Section labelling of wild-type (A, arrows) and MTH-labelled (B, arrows) desmin. Samples prepared as described (Table S1, condition #10) were treated first with aurothiomalate, then a gold-enhancing solution Nanoprobes™ GoldEnhance EM. They were imaged without additional heavy metal staining. The labelling brought on by the attached MTH is evident in these 5-nm-thick tomographic slices. Bars = 50 nm. The distribution of label for this and other figures in the paper is more clearly seen in movies that show the 3D structures with serial computed sections. See Supplementary Materials.
The difference in gold deposition between labelled and unlabelled protein encouraged the belief that MTH was providing specific labelling at high SNR. We were not, however, able to detect a periodic distribution of label, as one might have hoped for a probe attached to a specific site on a polymerizing protein. This lack may be due to the MTH having been added at the protein’s C-terminus, a moiety that probably extends out from the core of the intermediate filament, providing a sufficiently flexible location that periodicity is not visible. It may also have to do with the polymeric chemical nature of aurothiomalate, which creates a more diffuse (less punctate) concentration of gold atoms (Bau, 1998). We concluded, however, that MTH could provide a labelling that is benign in the sense that protein function is not compromised, and that heavy metal can be added after the tagged protein has performed its function (e.g. assembly into an element of the cytoskeleton); the heavy metals needed for EM visualization can be added while the sample is being processed for EM, not to a living cell or active protein.
MTH can localize a tagged protein in the complex environment of a cell
To see if MTH could provide labelling with similar SNR in cells, we used budding yeast cells in which two copies of the DNA encoding MTH were cloned onto the 3′ end of the chromosomal copy of the gene for SPC42 whose protein product is a component of the yeast spindle pole body (SPB), the centrosome of this cell type (Jaspersen and Winey, 2004). The location of Spc42 has previously been determined by a combination of immuno-EM (Adams & Kilmartin, 1999), cryoEM (Bullitt et al., 1997) and fluorescence resonance energy transfer between tagged SPB components in the LM (Muller et al., 2005). This protein resides in a planar, hexagonal lattice at the centre of the SPB, about the level of the nuclear envelope (Fig. 2A). The protein is essential, since cells from which SPC42 has been deleted are not viable. On the other hand, cells that lack SPC42 but are expressing Spc42-2xMTH at wild-type levels grow at normal rates, showing that the tag does not compromise protein function.
Fig. 2.
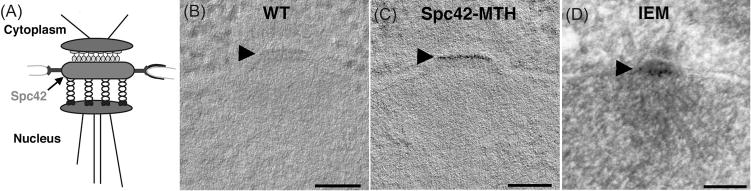
MTH labelling of Spc42 at the yeast spindle pole body. (A) A diagram of SPB organization. Spc42 resides in the central core of the SPB roughly at the plane of the nuclear envelope. The protein’s C-terminus, where our MTH tag has been put, is at the outer of two layers visible in the EM. (B) A computational slice from a tomogram of wild-type yeast treated with gold, as described (Table S1, condition #3). This and all other tomographic slices presented are 5-nm thick. (C) A similar slice from a yeast cell expressing Spc42-2xMTH. Both these samples contained uranyl acetate in the freeze substitution to provide a general contrast of the biological material (Table S1, condition #3). Arrowhead indicates the region containing Spc42. (D) EM image of a 70-nm section cut from a yeast SPB in which Spc42, tagged with GFP at its C-terminus, has been labelled by indirect immuno-EM with colloidal gold. Bar = 100 nm. Movies of tomographic volumes corresponding to (B) and (C) can be found in the supplementary materials.
Yeast cells expressing either the wild-type allele or Spc42-2xMTH were prepared for EM by high pressure freezing and freeze-substitution fixation in low levels of glutaraldehyde (0.1%) and/or uranyl acetate (0.02%), then embedded in K4M, sectioned, treated with aurothiomalate and gold enhancement, as described above for desmin, then imaged with serial tilts suitable for tomographic reconstruction. An example of the highly localized labelling achieved with this approach is shown in Figures 2(B) and (C). The precision of the localization is demonstrated by the sharp band of heavy metal associated with only the upper of the two visible bands in the SPB. This is the site of the C-terminus of Spc42 (Fig. 2A), the place where MTH is attached, indicating a spatial precision better than the size of a single protein. For comparison we also show a particularly favourable example of immunostaining of the same SPB component (Fig. 2D), achieved by using yeasts expressing an allele of Spc42 in which one GFP was placed at the protein’s C-terminus. These cells were prepared for EM just like the samples expressing Spc42-2xMTH then embedded in LowicrylHM20 and immunostained on sections with an affinity purified, rabbit polyclonal antibody against GFP, followed by an Fab fragment of goat antirabbit IgG, conjugated to 10 nm colloidal gold (BB International, Cardiff, UK; Zeng et al., 1999). Although this labelling is strong and convincing, the localization is less precise that that achieved with MTH.
It has proven difficult to measure the SNR of the MTH labelling because all label seen was where Spc42p is known to reside; we saw no ‘noise’. This statement is, however, only qualitative because there is obviously heavy metal binding elsewhere in the cell, e.g. the ribosomes, thanks in part to the uranyl acetate added during freeze substitution. Assessing whether gold or uranium was causing the electron scattering from ‘nonspecific’ regions of the cell did not seem worth pursuing in this case. Instead, we explored ways in which we might improve the staining and/or achieve it more quickly and easily. Using the same pair of yeast cell strains, we explored ranges of conditions for cell fixation, embedding and staining. Initial experiments followed the general protocol described above but varied fixation, embedding plastic, the gold salt used, duration and temperature of staining and the method for gold enhancement.
The results of 50 experiments on yeasts or desmin fibres, abstracted in Table S1, can be summarized as follows. Freeze-substitution fixation with glutaraldehyde and/or uranyl acetate gave good preservation of general cell structure. The uranyl ions provided clearer images of cell membranes but at the cost of giving a more general staining of cellular structure, which reduced the apparent SNR. However, when the concentration of uranyl ions was kept low, the identification of gold deposition at the SPB was easy. Other water-soluble gold salts, such as Au(I)Cl, gave a worrisome ‘noise’ in the form of heavy metal precipitate. This was usually easy to distinguish from the staining of the SPB, but it suggested that aurothiomalate was a preferable source of gold atoms. However, the use of aurothiomalate alone, without a secondary enhancement with the Nanoprobes kit, did not yield an electron dense label (Fig. 3A). Resins less hydrophilic than K4M, such as HM20 or Epon, would not permit labelling to occur, although we never tried very thin sections with these media. With the appropriate resin, the Nanoprobes gold enhancement kit could give reasonable SNR with no other source of gold to bind the MTH (Fig. 3B), though slightly better staining was usually obtained when aurothiomalate and gold enhancement were used sequentially (Fig. 3C). Similar results were obtained with desmin-MTH samples (data not shown).
Fig. 3.
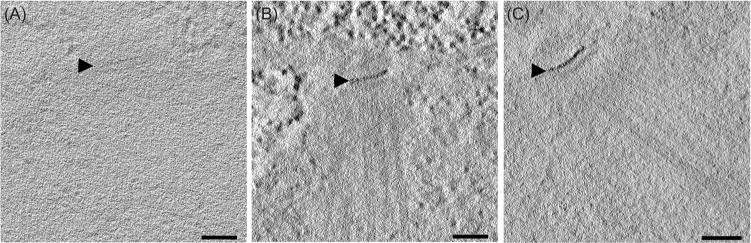
Staining of Spc42-2xMTH with a gold enhancement kit. Yeast cells expressing the tagged version of SPC42 were high-pressure frozen, freeze-substitution fixed in acetone with low concentrations of glutaraldehyde or uranyl acetate and embedded in K4M. (A) Sections incubated with aurothiomalate alone did not show appreciable staining (Table S1, condition #8). (B) Labelling was observed on sections stained only with the Nanoprobes gold enhancement solutions for 5 min at room temperature (Table S1, condition #7). Note that the heavy metal in this protocol has worked as a stain for ribosomes and spindle microtubules as well as the band of Spc42, emphasizing the problem of SNR in obtaining convincing localizations in the EM. (C) A better SNR was achieved by staining first with aurothiomalate for 1 h at 37°C, then enhancing that gold deposition with the Nanoprobes kit (Table S1, condition #3). Bars = 100 nm.
Heavy metal delivery during freeze substitution
Because of prior experience with the efficacy of enhancing heavy metal staining during freeze substitution (Morphew et al., 2008), we tried adding gold to samples at low temperatures while cellular water was being replaced by acetone. The addition of Au(III)Cl3 to the cold acetone used for freeze substitution resulted in excellent labelling of the SPB, again providing high SNR (Fig. 4). Indeed, both the mother SPB and the smaller, daughter SPB forming beside it exhibited strong staining in the Spc42-2xMTH preparations (Fig. 4B, arrowheads). Desmin-MTH samples also showed strong staining when gold salts were added during freeze substitution (Fig. 4C, D).
Fig. 4.
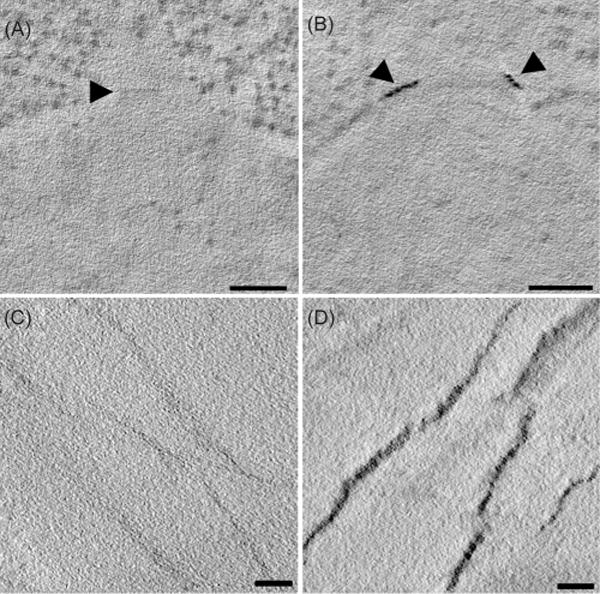
Staining of Spc42-2xMTH during freeze substitution. Samples were high-pressure frozen, then freeze substituted in 0.1% glutaraldehyde and 1 mM Au(III)Cl, then embedded in HM20 resin (Table S2, condition #6). Control cells (A) did not show staining of the SPB (arrowhead) whereas the SPBs of cells expressing Spc42-2xMTH (B) were nicely stained. This cell is from an α-factor arrested population, showing good staining of both the mother and forming daughter SPBs (arrowheads) (B). (C) and (D) show the results of similar treatments applied to wild-type desmin and desmin with one MTH at its N-terminus. The difference in labelling is clear. Bars = 100 nm for A and B; 50 nm for C and D. Movies of tomographic volumes corresponding to (A)—(D) can be found in Supplementary Materials.
Again, multiple conditions were tried to optimize this kind of stain localization by MTH. We explored several gold salts that are soluble in organic solvents, such as the phosphines (chloro[trimethylphosphine]Au[I] and chloro[triphenylphosphine]Au[I]). None that we tried other than Au(III)Cl3 gave good staining. Embedding these samples in different resins had a noticeable impact on subsequent image quality. EPON gave low general contrast, whereas K4M gave comparatively poor preservation of general cell structure. HM20 gave the nicest mixture of contrast, structure preservation and general image quality, but Durcupan ACM gave images with the most contrast. Although there was some general graininess in cell staining with this resin, we used it for many experiments, given its convenience of use (Table S2).
We also tried several ‘enhancements’ of labelling that work in nonaqueous conditions, such as a silver enhancement protocol (Morphew et al., 2008) and the one described in He et al. (2007). The He enhancement gave a slight contrast improvement but seemed to reduce the image SNR because it worked more like a stain than an enhancement of the label (Fig. 5B). However, samples freeze substituted in diglyme followed by silver enhancement resulted in the strongest staining of the SPB, and now distinct particles could be detected (Fig. 5C, inset). No staining was detected in control SPBs under the same conditions (Fig. 5D). The particles visible in the Spc42-2xMTH strain are in a roughly square array with a spacing of ~13 nm. Previous studies of periodic structures in the budding yeast SPB have found a hexagonal, 13.5 nm lattice that the authors attributed to Spc42 (Bullitt et al., 1997). The similarity in spacing between these two studies is a strong indication that the electron-dense particles in our images are a result of heavy metals deposited on Spc42-2xMTH; the approximately square lattice that we see, versus the hexagonal lattice in the frozen-hydrated SPBs viewed en face is probably a result of the collapse of plastic sections that occurs during the electron beam irradiation preceding tomographic imaging, as discussed in O’Toole et al. (1999).
Fig. 5.
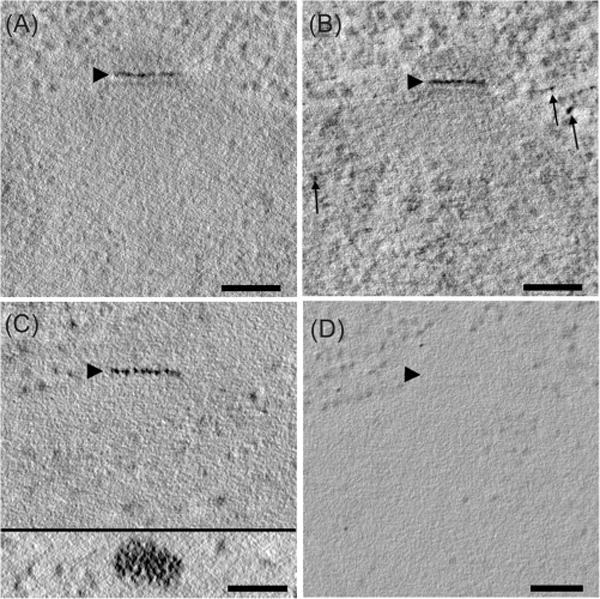
Optimization of Spc42-MTH staining during freeze substitution. (A) Au(III)Cl3 was added to acetone during freeze substitution, and it provided staining with good SNR (Table S2, condition #3, Lowicryl HM20 resin). This staining could be enhanced by the addition of more heavy metals, as described in He et al., 2008, but a few additional particles of heavy metal are now seen not associated with the SPB (B, arrows; Table S2, condition #12, Lowicryl HM20 resin). Samples that had been treated with diglyme followed by silver enhancement (as described in Morphew et al. 2008) gave the strongest label (C) and distinct particles could now be detected (C, inset; Table S2, condition #8, Durcupan resin). Control cells freeze treated with diglyme followed by silver enhancement showed no SPB staining (D; Table S2, condition #8, Durcupan resin). Bars = 100 nm. Movies of tomographic volumes corresponding to (A)—(D) can be found in the supplementary materials.
Exploration of MTH as a label for proteins of low abundance
To evaluate our now-optimized labelling methods as a tool for localization of low-abundance proteins, we studied protein components of the nuclear pore complex (NPC) in budding yeast. Each cell contains many nuclear pores, their locations are well known, NPCs are easy to spot and these complexes are exceptionally well characterized in Saccharomyces cerevisiae (Alber et al., 2007). The Jaspersen lab constructed several MTH-labelled alleles of genes encoding essential NPC components of known stoichiometry. Thus, we knew both how many copies of each protein were situated at each pore and how many copies of MTH were associated with each of these proteins. By tagging the genomic copy of each gene studied we could assess the viability of cells with this label in place and know that no unlabelled protein was present. Sadly, none of the labelled NPC proteins we tried in this way showed detectable levels of label at any of the NPCs (Fig. 6). During this work, we did learn that the addition of ammonium salts reduced the background staining that resulted from the metals used to develop gold particles in the MTH, but none of our methods gave convincing labelling of the NPCs above background. (See the two Supplementary Tables for a more complete account of what was tried).
Fig. 6.
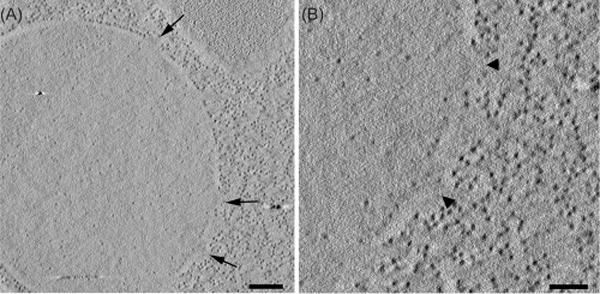
A yeast nuclear envelope showing pores but no labelling. A strain containing NUP100-8MTH was freeze substituted in glutaraldehyde and treated with diglyme followed by silver enhancement (Table S2, condition #8, Durcupan resin). (A) Tomographic slice showing an overview of a yeast nucleus with several nuclear pore complexes (arrows). Bar = 200 nm. (B) The arrowheads indicate two of the nuclear pores shown in (A), but no labelling above background is visible. Similar results were obtained both by labelling with other freeze substitution protocols and on sections, even when the two methods were combined. Similar results were obtained with multiple NPC proteins. Bar = 100 nm. A movie of the tomographic volume can be found in Supplementary Materials.
Discussion
The MTH labels studied here have produced some exciting results. The approach of employing a relatively small, clonable label to tag the protein of interest, then adding heavy metals only after the tagged protein has performed its function has proven effective in two cases. A priori, this strategy would seem to minimize the likelihood that the heavy metals needed as stains for EM would cause distortion of a physiological process. Our results also show that gold can be added either during freeze substitution or on sections of an appropriately hydrophilic plastic, making the tag visible with impressive SNR in two test situations: desmin polymerized in vitro and the centrosomal protein, Spc42 in budding yeasts. Moreover, by eliminating the need for a primary and secondary antibody, and relatively large gold particles commonly used for immuno-EM, our method significantly increases the precision of the localization. For example, the MTH-tagged Spc42 is present in only the outer layer of the central plaque of the mature yeast SPB, whereas colloidal gold particles bound to the SPB by traditional primary + secondary antibodies and gold immuno-EM are generally spread over a wider band, so specific localization is ambiguous. Of critical importance for electron tomography, the metal deposition occurs throughout the section and labelling of the tagged protein is not limited to the number of antigens that happen to be at the section’s surface.
Previous work on the use of MTH to create densities visible for EM was focused more on the practical realization of a useful tag than on understanding the mechanism underlying the localization of Au at this metal chelating protein. The work presented here enables some speculation about the chemical mechanisms underlying the formation of localized heavy metal spots with high contrast. These likely arise from either a high local concentration of Au(I) atoms nucleated by interaction with MTH, or from Au(0) nanoparticles that form on the MTH. The lower oxidation state Au(0) corresponds to the oxidation state of bulk gold; it represents the highest possible concentration of Au atoms, resulting, presumably, in the best possible SNR. For Au(0) based nanoparticles to form at MTH, the Au(I)-thiomalate or Au(III) must be exposed to a reducing agent. In this work, the reducing power may have been supplied by the Nanoprobes Gold Enhance kit, by quinone in the He protocol, and/or perhaps by the acetone used for freeze substitution.
To optimize SNR in the images, the binding of the Au(I) or Au(III)precursor to MTH must be more favourable than background reactivity. Biomacromolecules, especially proteins, can be excellent though relatively nonspecific coordinators, chelators or reductants of Au(III) and Au(I) ions (Xavier et al., 2012; Baksi et al., 2013a, b). We sought to avoid these potential ‘background’ chelation events, which would have reduced SNR in the resulting tomograms, by using more carefully chosen ligands for Au(I) or Au(III) precursors, testing the hypothesis that appropriate ligation may minimize background cellular reactions against the Au salt while maintaining specific binding to MTH.
Thiomalate Au(I) was successful for on section labelling in MTH based experiments, presumably because it satisfies the criteria suggested above. In the freeze-substitution experiments, however, the gold salt must be dissolved in acetone; the experiments with phosphine ligated Au(I) salts suggested that these salts are too inert for our purposes – reacting neither with background protein nor with MTH.
Our experiments showed that Au(III) in acetone during freeze substitution gave as good or better SNR than aurothiomalate applied to sections in aqueous solutions. Our best results were obtained using diglyme in acetone followed by silver enhancement (no gold) resulting in what appeared to be discrete nanoparticle densities. We suggest that under these conditions diglyme functions both as a heavy metal chelator and also as a solvent. In this instance, the diglyme–metal complexes are competent for ‘ligand exchange’ onto MTH, while still being stabilized against background reactivity with the general protein/biomolecule population in the cell. The appearance of nanoparticles thus may follow from an initial preconcentration of the metal on MTH, followed by reduction of the metal by the enhancement steps, followed by deposition of additional metal on the particles during the enhancement steps.
Overall this work suggests that aurothiomalate is a reasonably successful inorganic ion precursor for on section labelling, but an improvement on aurothiomalate may result from using diglyme as a chelating solvent to deliver heavy metal to MTH during freeze substitution.
Given the strength of labelling in these two cases, we were surprised at our failure with similar tags applied to proteins of the NPC. From the tables in Supplementary Material, it is clear that we tried many conditions, but none was effective. It is possible that the environment of the nuclear pore is in some unknown way particularly disadvantageous for this kind of labelling, but our explorations failed to identify an effective protocol. Other scientists with other questions may be able to build on our work, both to localize proteins of moderate abundance and perhaps to find ways in which MTH can provide the kind of labelling we all would like to see for informative cell biology with EM localization of low-abundance proteins.
Materials and methods
Generating and preparing protein assemblies
Wild-type and MTH-labelled desmins were prepared and polymerized into IFs as described in Bouchet-Marquis et al. (2012). Samples of the polymerized proteins were placed on 3-mm sapphire disks that had previously been coated with poly-L-lysine. After adsorption of protein, the disks were rapidly frozen by high pressure freezing at ~2050 bar in a Wohlwend HPF 020 high pressure freezing machine (Technotrade International, Manchester, NH, USA). Frozen discs were, transferred to acetone chilled to −80° C containing 0.1% glutaraldehyde or 0.02% uranyl acetate and fixed by freeze substitution (Giddings et al., 2001). NB, the source of each reagent used in this work is given in Table S3. After warming to room temperature, the samples were embedded in Lowicryl K4M and polymerized according to the manufacturer’s instructions. Sections ~200 nm thick were cut from both forms of desmin on a Lieca Ultracut UCT ultramicrotome (Leica Microsystems, Buffalo Grove, IL, USA) and collected onto formvar-coated, gold slot grids.
Tagging strains of yeast cells
MTH plasmids were generously provided by Drs. Chris Mercogliano and David DeRosier (Brandeis University). Standard techniques were used to generate plasmids and yeast strains. Briefly, two copies of MTH in tandem were PCR amplified and subcloned into the plasmid pFA6a-GFP-KMX (Longtine et al., 1998) replacing GFP to produce the plasmid pFA6a-2xMTH-KanMX6 (MW2756). DNA was generated by PCR and transformed into yeast (W303 background) at the C-terminus of SPC42. The Spc42-2xMTH strains were generated in a W303 background of the genotype leu2-3112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15.
Yeast strains generated in this study: JM281 – mat a, Spc42-2xMTH-KanMX6 MW5054 –a/α, Spc42-2xMTH-KanMX6/Spc42-2xMTH-KanMX6; Mlp2-ProA HIS3 URA3/Mlp2-ProA HIS3 URA3.
To generate tagged versions of proteins from the NPC, versions of MTH, codon optimized for S. cerevisiae, were synthesized by GeneWiz (South Plainfield, NJ, USA) and cloned into the PacI-AscI sites of pKT128-eGFP-HIS3MX (Sheff & Thorn, 2004) to create pSJ1584 (2xMTH-HIS3MX), pSJ1585 (8xMTH-HIS3MX) and pSJ1586 (2xMYC-8xMTH-HIS3MX). F5 and R3 primers were designed for each nucleoporin gene as follows: F5, 5′-(40 bp before stop)-GGTGACGGTGCTGGTTTA-3′ and R3, 5′-(40 bp after stop reverse complement)-TCGATGAATTCGAGCTCG-3′. Amplified templates were transformed into yeast using standard methods and correct integration was verified by PCR. All strains were derivatives of W303, with the exception of SLJ9109, which was made in the GFP collection strain background (Huh et al., 2003) by integrating the 8xMTH cassette before GFP. Sequence analysis of the resulting transformants showed that only three copies of the MTH were integrated in this strain. For a list of genotypes made and used, see Table S4.
Preparing yeasts for EM
Suspensions of yeast S. cerevisiae were prepared by rapid freezing and freeze substitution as previously described (Giddings et al., 2001). Briefly, cells grown to mid-log phase were harvested from liquid culture by vacuum filtration onto a 0.2 μm Millipore filter, transferred to brass freezing cups and frozen as above. Frozen samples were freeze substituted into acetone containing 0.1% glutaraldehyde and/or 0.02% uranyl acetate at −80°C for 2–3 days. Samples were gently warmed to −30°C over 24 h, embedded in methacrylate resin Lowicryl K4M and sectioned and collected as described above.
In some experiments, the pheromone α-factor was used to arrest cells as new spindle pole bodies (satellites) are forming. Yeast cells were grown at 30°C to log phase. α-factor (custom peptide synthesis by Macromolecular Resources, Colorado State University, Ft. Collins, Colorado, USA) was added to a final concentration of 11 μg mL−1. When 85–90% of the cells displayed the characteristic schmoo shape (~1.5–2 h), the cells were high-pressure frozen.
Staining sections of samples containing MTH
Gold grids with sections of desmin or yeast were incubated with their section-side down onto 50 μL drops of 1 mM sodium aurothiomalate dissolved in 25 mM Tris for 10–60 min at either room temperature or at 37°C, rinsed three times on drops of Tris buffer followed by an optional incubation in 25 μL Nanoprobes™ GoldEnhance EM for 5 min at room temperature. Grids were then rinsed three times on drops of distilled water and poststained (optional) in 2% uranyl acetate in water and Reynold’s lead citrate.
Freeze substitution-based labelling
Following high pressure freezing, samples in brass cups or on sapphire discs were transferred to frozen solutions of acetone containing either 0.1% anhydrous glutaraldehyde or 0.02% uranyl acetate (made from a 5% uranyl acetate stock solution in anhydrous methanol), substituted at −80°C for 2–3 days, warmed to −30°C over 24 h and rinsed with acetone. At this point, a variety of reagents and time schedules were applied, either to increase the labelling of MTH (i.e. Au(lll)Cl3 gold or silver enhancement, reductases (diglyme or NaBH4) or to reduce noise (i.e. borate salts). See Tables S2 and S3. Samples were rinsed in acetone at −30°C and embedded in Lowicryl HM20 or K4M or warmed to room temperature for epoxy embedding.
Electron tomography
Sections (250 nm) were cut using a Leica Ultracut UCT microtome and were collected onto formvar-coated, copper slot grids. Tilt series were acquired at room temperature with 1 nm pixels and tilt increment of 1°. The tilt range was ±60°, using either a Tecnai F20 or F30 electron microscope (FEI Corp, Hillsboro, OR, USA). On average, the samples were exposed to a total electron dose of 10000 e Å−2, which is typical for a single axis tilt series at this pixel size. Tomograms were generated using the IMOD software package (Kremer et al, 1996). The tomograms of desmin, both wild type and conjugated to MTH, were filtered by nonlinear, anisotropic diffusion.
Supplementary Material
Movie_Fig3C. Movie through a tomographic volume of a yeast spindle pole body containing Spc42-2MTH (Fig. 3C). Sample was high-pressure frozen, freeze substituted in 0.02% uranyl acetate in acetone and embedded in Lowicryl K4M resin. Sections were incubated with 1mM aurothiomalate for 1 h at 37°C followed by Nanoprobes gold enhancement for 5 min at room temperature. Movie was created using Quicktime Pro using H.264 compression. Bar = 100 nm.
Movie_Fig1A. Movie through a tomographic volume of a sample containing wild-type desmin (Fig. 1A). Sample was high-pressure frozen on a sapphire disc, freeze substituted in 0.2% glutaraldehyde in acetone and embedded in Lowicryl K4M resin. Sections were incubated with 1 mM aurothiomalate for 1 h at 37°C, followed by 5 min in Nanoprobes gold enhancement kit at room temperature. Movie was created using Quicktime Pro using H.264 compression. Bar = 50 nm.
Movie_Fig1B. Movie through a tomographic volume of a sample containing desmin-1MTH at its C-terminus (Fig. 1B). Sample was high-pressure frozen on a sapphire disc, freeze substituted in 0.2% glutaraldehyde in acetone and embedded in Lowicryl K4M resin. Sections were incubated with 1 mM aurothiomalate for 1 h at 37°C, followed by 5 min in Nanoprobes gold enhancement kit at room temperature. Movie was created using Quicktime Pro using H.264 compression. Bar = 50 nm.
Movie_Fig2B. Movie through a tomographic volume of a yeast spindle pole body containing wild-type Spc42 (Fig. 2B). Sample was high-pressure frozen, freeze substituted in 0.02% uranyl acetate in acetone and embedded in Lowicryl K4M resin. Sections were incubated with 1 mM aurothiomalate for 1 h at 37°C, followed by 5 min in Nanoprobes gold enhancement kit at room temperature. Movie was created using Quicktime Pro using H.264 compression. Bar = 100 nm.
Movie_Fig2C. Movie through a tomographic volume of a yeast spindle pole body containing Spc42-2MTH (Fig. 2C). Sample was high-pressure frozen, freeze substituted in 0.02% uranyl acetate and embedded in Lowicryl K4M resin. Sections were incubated with 1 mM aurothiomalate for 1 h at 37°C, followed by 5 min in Nanoprobes gold enhancement kit. Movie was created using Quicktime Pro using H.264 compression. Bar = 100 nm.
Movie_Fig3A. Movie though a tomographic volume of a yeast spindle pole body containing Spc42-2MTH (Fig. 3A). Sample was high-pressure frozen, freeze substituted in 0.1% glutaraldehyde in acetone and embedded in Lowicryl K4M resin. Sections were incubated with 1 mM aurothiomalate for 1 h at 37°C and imaged without further enhancement. Movie was created using Quicktime Pro using H.264 compression. Bar = 100 nm.
Movie_Fig3B. Movie through a tomographic volume of a yeast spindle pole body containing Spc42-2MTH (Fig. 3B). Sample was high-pressure frozen, freeze substituted in 0.1% glutaraldehyde in acetone and embedded in Lowicryl K4M resin. Sections were incubated with Nanoprobes gold enhancement kit for 5 min at room temperature. Movie was created using Quicktime Pro using H.264 compression. Bar = 100 nm.
Movie_Fig4A: Movie through a tomographic volume of a yeast spindle pole body containing wild-type Spc42 (Fig. 4A). Sample was high-pressure frozen, freeze substituted in 0.1% glutaraldehyde and 1 mM Au(III)Cl in acetone and embedded in Lowicryl HM20 resin. Movie was created using Quicktime Pro using H.264 compression. Bar = 100 nm.
Movie_Fig4B: Movie through a tomographic volume of a yeast spindle pole body containing Spc42-2MTH (Fig. 4B). Sample was high-pressure frozen, freeze substituted in 0.1% glutaraldehyde and 1 mM Au(III)Cl in acetone and embedded in Lowicryl HM20 resin. Movie was created using Quicktime Pro using H.264 compression. Bar = 100 nm.
Movie_Fig4C: Movie through a tomographic volume of a sample containing wild-type desmin (Figure 4C). Sample was high-pressure frozen, freeze substituted in 0.1% glutaraldehyde and 1 mM Au(III)Cl in acetone and embedded in Lowicryl HM20 resin. Movie was created using Quicktime Pro using H.264 compression. Bar = 50 nm.
Movie_Fig4D: Movie through a tomographic volume of a sample containing desmin-1MTH at its N-terminus (Fig. 4D). Sample was high-pressure frozen, freeze substituted in 0.1% glutaraldehyde and 1 mM Au(III)Cl in acetone and embedded in Lowicryl HM20 resin. Movie was created using Quicktime Pro using H.264 compression. Bar = 50 nm.
Movie_Fig6: Movie through a tomographic volume of a yeast cell containing nup100-8MTH shown (Fig. 6). Sample was high-pressure frozen, freeze substituted in 0.1% glutaraldehyde and diglyme in acetone, followed by silver enhancement and embedded in durcapan resin. Movie was created using Quicktime Pro using H.264 compression. Bar = 100 nm.
Movie_Fig5A: Movie through a tomographic volume of a yeast spindle pole body containing SPC42-2MTH (Fig. 5A). Sample was high-pressure frozen, freeze substituted in 0.02% uranyl acetate and 1 mM Au(III)Cl in acetone and embedded in Lowicryl HM20 resin. Movie was created using Quicktime Pro using H.264 compression. Bar = 100 nm.
Movie_Fig5B: Movie through a tomographic volume of a yeast spindle pole body containing SPC42-2MTH (Fig. 5B). Sample was high-pressure frozen, freeze substituted in 0.02% uranyl acetate and 1 mM Au(III)Cl in acetone, followed by He enhancement and embedded in Lowicryl HM20 resin. Movie was created using Quicktime Pro using H.264 compression. Bar = 100 nm.
Movie_Fig5C: Movie through a tomographic volume of a yeast spindle pole body containing SPC42-2MTH (Fig. 5C). Sample was high-pressure frozen, freeze substituted in 0.1% glutaraldehyde and diglyme in acetone, followed by silver enhancement and embedded in durcapan resin. Movie was created using Quicktime Pro using H.264 compression. Bar = 100 nm.
Movie_Fig5D: Movie through a tomographic volume of a yeast spindle pole body containing wild-type Spc42 (Fig. 5D). Sample was high-pressure frozen, freeze substituted in 0.1% glutaraldehyde and diglyme in acetone, followed by silver enhancement and embedded in durcapan resin. Movie was created using Quicktime Pro using H.264 compression. Bar = 100 nm.
Table S4. Yeast strains used for study of nuclear pore complexes.
Table S1. Section labelling of SPC42-MTH.
Table S2. Freeze substitution labelling of SPC42-MTH.
Table S3. Reagents and vendors.
Acknowledgments
This work was supported in part by NIH grant R01GM80993 to MW and AH and by P41GM103431 to AH. We thank Alex Stemm-Wolf for contributing his expertise in plasmid construction and for helpful discussions and Cindi Schwartz for imaging some of the yeast data sets.
Footnotes
Supporting Information
Additional Supporting information may be found in the online version of this article at the publisher’s website:
References
- Adams IR, Kilmartin JV. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cere-visiae. J Cell Biol. 1999;145:809–823. doi: 10.1083/jcb.145.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Baksi A, Xavier PL, Chaudhari K, Goswami N, Pal SK, Pradeep T. Protein-encapsulated gold cluster aggregates: the case of lysozyme. Nanoscale. 2013;5:2009–2016. doi: 10.1039/c2nr33180b. [DOI] [PubMed] [Google Scholar]
- Bau R. Crystal Structure of the Antiarthritic Drug Gold Thiomalate (Myochrysine): A Double-Helical Geometry in the Solid State. J Am Chem Soc. 1998;120:9380–9381. [Google Scholar]
- Boassa D, Berlanga ML, Yang MA, et al. Mapping the sub-cellular distribution of alpha-synuclein in neurons using genetically encoded probes for correlated light and electron microscopy: implications for Parkinson’s disease pathogenesis. J Neurosci. 2013;33:2605–2615. doi: 10.1523/JNEUROSCI.2898-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchet-Marquis C, Pagratis M, Kirmse R, Hoenger A. Metallothionein as a clonable high-density marker for cryo-electron microscopy. J Struct Biol. 2012;177:119–127. doi: 10.1016/j.jsb.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullitt E, Rout MP, Kilmartin JV, Akey CW. The yeast spindle pole body is assembled around a central crystal of Spc42p. Cell. 1997;89:1077–1086. doi: 10.1016/s0092-8674(00)80295-0. [DOI] [PubMed] [Google Scholar]
- Deerinck TJ, Giepmans BN, Smarr BL, Martone ME, Ellisman MH. Light and electron microscopic localization of multiple proteins using quantum dots. Methods Mol Biol. 2007;374:43–53. doi: 10.1385/1-59745-369-2:43. [DOI] [PubMed] [Google Scholar]
- Diestra E, Fontana J, Guichard P, Marco S, Risco C. Visualization of proteins in intact cells with a clonable tag for electron microscopy. J Struct Biol. 2009;165:157–168. doi: 10.1016/j.jsb.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Ellisman MH, Deerinck TJ, Shu X, Sosinsky GE. Picking faces out of a crowd: genetic labels for identification of proteins in correlated light and electron microscopy imaging. Methods Cell Biol. 2012;111:139–155. doi: 10.1016/B978-0-12-416026-2.00008-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaietta G, Deerinck TJ, Adams SR, et al. Multicolor and electron microscopic imaging of connexin trafficking. Science. 2002;296:503–507. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]
- Gaietta GM, Deerinck TJ, Ellisman MH. Correlated live cell light and electron microscopy using tetracysteine tags and biarsenicals. Cold Spring Harb Protoc. 2011;2011 doi: 10.1101/pdb.top94. pdb top94. [DOI] [PubMed] [Google Scholar]
- Gaietta GM, Giepmans BN, Deerinck TJ, et al. Golgi twins in late mitosis revealed by genetically encoded tags for live cell imaging and correlated electron microscopy. Proc Natl Acad Sci U S A. 2006;103:17777–17782. doi: 10.1073/pnas.0608509103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings TH, Jr, O’Toole ET, Morphew M, Mastronarde DN, McIntosh JR, Winey M. Using rapid freeze and freeze-substitution for the preparation of yeast cells for electron microscopy and three-dimensional analysis. Methods Cell Biol. 2001;67:27–42. doi: 10.1016/s0091-679x(01)67003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabenbauer M, Geerts WJ, Fernadez-Rodriguez J, Hoenger A, Koster AJ, Nilsson T. Correlative microscopy and electron tomography of GFP through photooxidation. Nat Methods. 2005;2:857–862. doi: 10.1038/nmeth806. [DOI] [PubMed] [Google Scholar]
- He W, Kivork C, Machinani S, et al. A freeze substitution fixation-based gold enlarging technique for EM studies of endocytosed Nanogold-labeled molecules. J Struct Biol. 2007;160:103–113. doi: 10.1016/j.jsb.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Ladinsky MS, Huey-Tubman KE, Jensen GJ, McIntosh JR, Bjorkman PJ. FcRn-mediated antibody transport across epithelial cells revealed by electron tomography. Nature. 2008;455:542–546. doi: 10.1038/nature07255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann H, Bar H, Kreplak L, Strelkov SV, Aebi U. Intermediate filaments: from cell architecture to nanomechanics. Nat Rev Mol Cell Biol. 2007;8:562–573. doi: 10.1038/nrm2197. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Haner M, Brettel M, et al. Structure and assembly properties of the intermediate filament protein vimentin: the role of its head, rod and tail domains. J Mol Biol. 1996;264:933–953. doi: 10.1006/jmbi.1996.0688. [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Winey M. The budding yeast spindle pole body: structure, duplication, and function. Annu Rev Cell Dev Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- Kirmse R, Bouchet-Marquis C, Page C, Hoenger A. Three-dimensional cryo-electron microscopy on intermediate filaments. Methods Cell Biol. 2010;96:565–589. doi: 10.1016/S0091-679X(10)96023-8. [DOI] [PubMed] [Google Scholar]
- Kremer J, Mastronarde DN, McIntosh DN. Computer visualization of three-dimensional image data using IMOD. J Structural Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification on Saccharmyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Ludwig A, Howard G, Mendoza-Topaz C, Deerinck T, Mackey M, Sandin S, Ellisman MH, Nichols BJ. Molecular composition and ultrastructure of the caveolar coat complex. PLoS Biol. 2013;11:e1001640. doi: 10.1371/journal.pbio.1001640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell JD, Deerinck TJ, Sancak Y, Poulos TL, Mootha VK, Sosinsky GE, Ellisman MH, Ting AY. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol. 2012;30:1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiblitzer-Ruppitsch C, Vetterlein M, Stangl H, Maier S, Neumuller J, Freissmuth M, Pavelka M, Ellinger A. Electron microscopic visualization of fluorescent signals in cellular compartments and organelles by means of DAB-photoconversion. Histochem Cell Biol. 2008;130:407–419. doi: 10.1007/s00418-008-0429-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercogliano CP, DeRosier DJ. Gold nanocluster formation using metallothionein: mass spectrometry and electron microscopy. J Mol Biol. 2006;355:211–223. doi: 10.1016/j.jmb.2005.10.026. [DOI] [PubMed] [Google Scholar]
- Mercogliano CP, DeRosier DJ. Concatenated metallothionein as a clonable gold label for electron microscopy. J Struct Biol. 2007;160:70–82. doi: 10.1016/j.jsb.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morphew M, He W, Bjorkman PJ, McIntosh JR. Silver enhancement of Nanogold particles during freeze substitution for electron microscopy. J Microsc. 2008;230:263–267. doi: 10.1111/j.1365-2818.2008.01983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller EG, Snydsman BE, Novik I, et al. The organization of the core proteins of the yeast spindle pole body. Mol Biol Cell. 2005;16:3341–3352. doi: 10.1091/mbc.E05-03-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole ET, Winey M, McIntosh JR. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:2017–2031. doi: 10.1091/mbc.10.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risco C, Sanmartin-Conesa E, Tzeng WP, Frey TK, Seybold V, de Groot RJ. Specific, sensitive, high-resolution detection of protein molecules in eukaryotic cells using metal-tagging transmission electron microscopy. Structure. 2012;20:759–766. doi: 10.1016/j.str.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano T, Glazer AN, Cantor CR. A streptavidin-metallothionein chimera that allows specific labeling of biological materials with many different heavy metal ions. Proc Natl Acad Sci U S A. 1992;89:1534–1538. doi: 10.1073/pnas.89.5.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21:661–670. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- Shu X, Lev-Ram V, Deerinck TJ, et al. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 2011;9:e1001041. doi: 10.1371/journal.pbio.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Mercogliano CP, Lowe J. A ferritin-based label for cellular electron cryotomography. Structure. 2011;19:147–154. doi: 10.1016/j.str.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Xavier PL, Chaudhari K, Baksi A, Pradeep T. Protein-protected luminescent noble metal quantum clusters: an emerging trend in atomic cluster nanoscience. Nano Rev. 2012 Feb 3;3 doi: 10.3402/nano.v3i0.14767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Kahana JA, Silver PA, Morphew MK, McIntosh JR, Fitch IT, Carbon J, Saunders WS. Slk19p is a centromere protein that functions to stabilize mitotic spindles. J Cell Biol. 1999;146:415–425. doi: 10.1083/jcb.146.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie_Fig3C. Movie through a tomographic volume of a yeast spindle pole body containing Spc42-2MTH (Fig. 3C). Sample was high-pressure frozen, freeze substituted in 0.02% uranyl acetate in acetone and embedded in Lowicryl K4M resin. Sections were incubated with 1mM aurothiomalate for 1 h at 37°C followed by Nanoprobes gold enhancement for 5 min at room temperature. Movie was created using Quicktime Pro using H.264 compression. Bar = 100 nm.
Movie_Fig1A. Movie through a tomographic volume of a sample containing wild-type desmin (Fig. 1A). Sample was high-pressure frozen on a sapphire disc, freeze substituted in 0.2% glutaraldehyde in acetone and embedded in Lowicryl K4M resin. Sections were incubated with 1 mM aurothiomalate for 1 h at 37°C, followed by 5 min in Nanoprobes gold enhancement kit at room temperature. Movie was created using Quicktime Pro using H.264 compression. Bar = 50 nm.
Movie_Fig1B. Movie through a tomographic volume of a sample containing desmin-1MTH at its C-terminus (Fig. 1B). Sample was high-pressure frozen on a sapphire disc, freeze substituted in 0.2% glutaraldehyde in acetone and embedded in Lowicryl K4M resin. Sections were incubated with 1 mM aurothiomalate for 1 h at 37°C, followed by 5 min in Nanoprobes gold enhancement kit at room temperature. Movie was created using Quicktime Pro using H.264 compression. Bar = 50 nm.
Movie_Fig2B. Movie through a tomographic volume of a yeast spindle pole body containing wild-type Spc42 (Fig. 2B). Sample was high-pressure frozen, freeze substituted in 0.02% uranyl acetate in acetone and embedded in Lowicryl K4M resin. Sections were incubated with 1 mM aurothiomalate for 1 h at 37°C, followed by 5 min in Nanoprobes gold enhancement kit at room temperature. Movie was created using Quicktime Pro using H.264 compression. Bar = 100 nm.
Movie_Fig2C. Movie through a tomographic volume of a yeast spindle pole body containing Spc42-2MTH (Fig. 2C). Sample was high-pressure frozen, freeze substituted in 0.02% uranyl acetate and embedded in Lowicryl K4M resin. Sections were incubated with 1 mM aurothiomalate for 1 h at 37°C, followed by 5 min in Nanoprobes gold enhancement kit. Movie was created using Quicktime Pro using H.264 compression. Bar = 100 nm.
Movie_Fig3A. Movie though a tomographic volume of a yeast spindle pole body containing Spc42-2MTH (Fig. 3A). Sample was high-pressure frozen, freeze substituted in 0.1% glutaraldehyde in acetone and embedded in Lowicryl K4M resin. Sections were incubated with 1 mM aurothiomalate for 1 h at 37°C and imaged without further enhancement. Movie was created using Quicktime Pro using H.264 compression. Bar = 100 nm.
Movie_Fig3B. Movie through a tomographic volume of a yeast spindle pole body containing Spc42-2MTH (Fig. 3B). Sample was high-pressure frozen, freeze substituted in 0.1% glutaraldehyde in acetone and embedded in Lowicryl K4M resin. Sections were incubated with Nanoprobes gold enhancement kit for 5 min at room temperature. Movie was created using Quicktime Pro using H.264 compression. Bar = 100 nm.
Movie_Fig4A: Movie through a tomographic volume of a yeast spindle pole body containing wild-type Spc42 (Fig. 4A). Sample was high-pressure frozen, freeze substituted in 0.1% glutaraldehyde and 1 mM Au(III)Cl in acetone and embedded in Lowicryl HM20 resin. Movie was created using Quicktime Pro using H.264 compression. Bar = 100 nm.
Movie_Fig4B: Movie through a tomographic volume of a yeast spindle pole body containing Spc42-2MTH (Fig. 4B). Sample was high-pressure frozen, freeze substituted in 0.1% glutaraldehyde and 1 mM Au(III)Cl in acetone and embedded in Lowicryl HM20 resin. Movie was created using Quicktime Pro using H.264 compression. Bar = 100 nm.
Movie_Fig4C: Movie through a tomographic volume of a sample containing wild-type desmin (Figure 4C). Sample was high-pressure frozen, freeze substituted in 0.1% glutaraldehyde and 1 mM Au(III)Cl in acetone and embedded in Lowicryl HM20 resin. Movie was created using Quicktime Pro using H.264 compression. Bar = 50 nm.
Movie_Fig4D: Movie through a tomographic volume of a sample containing desmin-1MTH at its N-terminus (Fig. 4D). Sample was high-pressure frozen, freeze substituted in 0.1% glutaraldehyde and 1 mM Au(III)Cl in acetone and embedded in Lowicryl HM20 resin. Movie was created using Quicktime Pro using H.264 compression. Bar = 50 nm.
Movie_Fig6: Movie through a tomographic volume of a yeast cell containing nup100-8MTH shown (Fig. 6). Sample was high-pressure frozen, freeze substituted in 0.1% glutaraldehyde and diglyme in acetone, followed by silver enhancement and embedded in durcapan resin. Movie was created using Quicktime Pro using H.264 compression. Bar = 100 nm.
Movie_Fig5A: Movie through a tomographic volume of a yeast spindle pole body containing SPC42-2MTH (Fig. 5A). Sample was high-pressure frozen, freeze substituted in 0.02% uranyl acetate and 1 mM Au(III)Cl in acetone and embedded in Lowicryl HM20 resin. Movie was created using Quicktime Pro using H.264 compression. Bar = 100 nm.
Movie_Fig5B: Movie through a tomographic volume of a yeast spindle pole body containing SPC42-2MTH (Fig. 5B). Sample was high-pressure frozen, freeze substituted in 0.02% uranyl acetate and 1 mM Au(III)Cl in acetone, followed by He enhancement and embedded in Lowicryl HM20 resin. Movie was created using Quicktime Pro using H.264 compression. Bar = 100 nm.
Movie_Fig5C: Movie through a tomographic volume of a yeast spindle pole body containing SPC42-2MTH (Fig. 5C). Sample was high-pressure frozen, freeze substituted in 0.1% glutaraldehyde and diglyme in acetone, followed by silver enhancement and embedded in durcapan resin. Movie was created using Quicktime Pro using H.264 compression. Bar = 100 nm.
Movie_Fig5D: Movie through a tomographic volume of a yeast spindle pole body containing wild-type Spc42 (Fig. 5D). Sample was high-pressure frozen, freeze substituted in 0.1% glutaraldehyde and diglyme in acetone, followed by silver enhancement and embedded in durcapan resin. Movie was created using Quicktime Pro using H.264 compression. Bar = 100 nm.
Table S4. Yeast strains used for study of nuclear pore complexes.
Table S1. Section labelling of SPC42-MTH.
Table S2. Freeze substitution labelling of SPC42-MTH.
Table S3. Reagents and vendors.


