In this study, Arenzana et al. perform a detailed analysis of the functional abnormalities at five sequential stages of thymocyte development in an Ikaros mutant mouse strain followed by RNA-seq to document the gene expression changes seen in the mutant cells. By combining the quantitative power of the RNA-seq method with the analysis of five sequential stages of development, the findings demonstrate a unique function for Ikaros in supporting dynamic changes in gene expression during developmental transitions.
Keywords: gene regulation, transcription, lymphocyte development, leukemogenesis, Ikaros
Abstract
The DNA-binding protein Ikaros is a potent tumor suppressor and hematopoietic regulator. However, the mechanisms by which Ikaros functions remain poorly understood, due in part to its atypical DNA-binding properties and partnership with the poorly understood Mi-2/NuRD complex. In this study, we analyzed five sequential stages of thymocyte development in a mouse strain containing a targeted deletion of Ikaros zinc finger 4, which exhibits a select subset of abnormalities observed in Ikaros-null mice. By examining thymopoiesis in vivo and in vitro, diverse abnormalities were observed at each developmental stage. RNA sequencing revealed that each stage is characterized by the misregulation of a limited number of genes, with a strong preference for stage-specific rather than lineage-specific genes. Strikingly, individual genes rarely exhibited Ikaros dependence at all stages. Instead, a consistent feature of the aberrantly expressed genes was a reduced magnitude of expression level change during developmental transitions. These results, combined with analyses of the interplay between Ikaros loss of function and Notch signaling, suggest that Ikaros may not be a conventional activator or repressor of defined sets of genes. Instead, a primary function may be to sharpen the dynamic range of gene expression changes during developmental transitions via atypical molecular mechanisms that remain undefined.
Lymphocyte development is dependent on the proper expression of large numbers of genes in a lineage- and stage-specific manner. The regulation of these genes depends on transcription factors that act via diverse molecular mechanisms. During B-cell development, for example, E2A, Pax5, and EBF appear to function primarily as transcriptional activators that bind the promoters and enhancers of target genes and presumably promote their activation by interacting with the general transcription machinery and chromatin regulators (Mandel and Grosschedl 2010). In contrast, PU.1 functions as a broadly acting pioneer factor that promotes the initial opening of chromatin at enhancers for hematopoietic-specific genes (Ghisletti et al. 2010; Heinz et al. 2010).
Ikaros, encoded by the Ikzf1 gene, is another DNA-binding protein that plays critical roles during lymphopoiesis (Georgopoulos et al. 1994; Wang et al. 1996; Kirstetter et al. 2002). Ikaros mutant mice also develop T-cell lymphoma with high penetrance as early as 3 mo of age (Winandy et al. 1995; Kirstetter et al. 2002). Notably, deletions of the human IKZF1 gene are frequently observed in patients with BCR-ABL1+ B-progenitor acute lymphoblastic leukemia (B-ALL) and pediatric patients with high-risk B-ALL, demonstrating that Ikaros is also a potent tumor suppressor in humans (Mullighan et al. 2008, 2009).
Although Ikaros plays broad roles in gene regulation in most cells in which it is expressed, its mechanisms of action remain poorly defined. A small number of genes, including Cd4 and Cd8, are frequently misregulated in Ikzf1 mutant cells and appear to be directly regulated by Ikaros (Harker et al. 2002; Naito et al. 2007). Evidence has also been presented that Ikaros directly regulates Notch target genes and other genes involved in development and cell cycle progression (Dumortier et al. 2006; Chari and Winandy 2008; Geimer Le Lay et al. 2014). However, the properties of Ikaros observed in vivo and in vitro have made it difficult to obtain a clear view of its full range of targets and mechanisms of action. For example, recent genome-wide chromatin immunoprecipitation (ChIP) combined with deep sequencing (ChIP-seq) experiments revealed the binding of Ikaros to 9878 genomic sites in progenitor B (pro-B) cells, including 60% of all active promoters and 30% of all active enhancers (Schwickert et al. 2014). In this same study, 61% of genes misregulated in Ikzf1 mutant cells were bound by Ikaros, demonstrating that Ikaros binding is distributed broadly and exhibits no enrichment at Ikaros-dependent genes. Moreover, earlier experiments demonstrated that a substantial fraction of Ikaros molecules is localized to foci of pericentromeric heterochromatin (Brown et al. 1997; Cobb et al. 2000); it was hypothesized that this localization may allow Ikaros to recruit silent target genes to a repressive chromatin environment, but the significance of its pericentromeric localization remains unknown.
The biochemical properties of Ikaros add further uncertainty regarding its mechanisms of action. In particular, Ikaros is associated most prominently with the Mi-2/NuRD complex (Kim et al. 1999; Sridharan and Smale 2007), which combines ATP-dependent nucleosome remodeling and histone deacetylase activities; unfortunately, the mechanisms of action of the Mi-2/NuRD complex remain as poorly understood as those of Ikaros. In addition, although Ikaros proteins are expressed as stable dimers (Trinh et al. 2001), it is not known how the two subunits recognize genomic DNA. In most dimeric transcription factors, the dimerization domain is adjacent to the DNA-binding domain, leading to strict spacing constraints between the DNA half-sites recognized by the two subunits. In contrast, the dimerization and DNA-binding domains of Ikaros are located at opposite ends of the protein, leading to considerable flexibility in DNA recognition (B Cobb and ST Smale, unpubl.). Indeed, Ikaros ChIP-seq peaks generally exhibit enrichment of only an Ikaros half-site (Zhang et al. 2011; Ferreiros-Vidal et al. 2013; Schjerven et al. 2013; Schwickert et al. 2014), raising the possibility that the two subunits associate with sequences separated by large distances or even on different chromosomes. Additional findings suggest that Ikaros dimers assemble into multimeric structures in vivo (Sun et al. 1996; Trinh et al. 2001).
Despite our limited knowledge of the mechanisms of action of Ikaros, the well-defined biological abnormalities observed in Ikzf1 mutant cells typically coincide with extensive misregulation of gene expression. To gain additional mechanistic insights, we recently generated mutant mouse strains in which Ikzf1 exons encoding the first and fourth zinc fingers of the four-finger DNA-binding domain were deleted (Schjerven et al. 2013). Each mutant strain, Ikzf1ΔF1/ΔF1 and Ikzf1ΔF4/ΔF4, exhibited a subset of the developmental abnormalities observed with Ikzf1null mice, consistent with earlier evidence that these two fingers are important for binding to only a subset of Ikaros recognition sequences. Notably, only Ikzf1ΔF4/ΔF4 mice exhibited extensive abnormalities during T-cell development and highly penetrant T-cell lymphoma. Gene expression profiling revealed limited misregulation of gene expression in the mutant strains, raising the possibility that further examination of this misregulation may help uncover the mechanisms of action of Ikaros.
In this study, we used Ikzf1ΔF4/ΔF4 mice for a detailed examination of five sequential stages of thymocyte development in vivo and in vitro. By RNA sequencing (RNA-seq) analysis, we found that almost all Ikaros-regulated genes exhibited Ikaros dependence at only a subset of developmental stages at which the genes were expressed or silent. Furthermore, at each developmental stage, Ikaros exhibited a strong bias toward the regulation of genes whose expression levels increased or decreased in wild-type cells in comparison with the preceding or subsequent stage. Most interestingly, the vast majority of genes whose expression levels changed during defined developmental transitions exhibited reduced dynamic ranges of expression in the Ikzf1ΔF4/ΔF4 cells, explaining most of the aberrant gene expression observed in these cells. An analysis of the impact of deregulated Notch1 expression on the functions of Ikaros demonstrated that, although Ikaros regulates genes in multiple developmental and signaling pathways, its functions cannot readily be assigned to defined pathways. Together, these results suggest that Ikaros may not be a conventional transcription factor that is consistently dedicated to the regulation of defined sets of genes. Instead, a primary function may be to reinforce or sharpen dynamic changes in gene expression levels during developmental transitions.
Results
Reduced double-negative (DN) thymocytes in Ikzf1ΔF4/ΔF4 mice
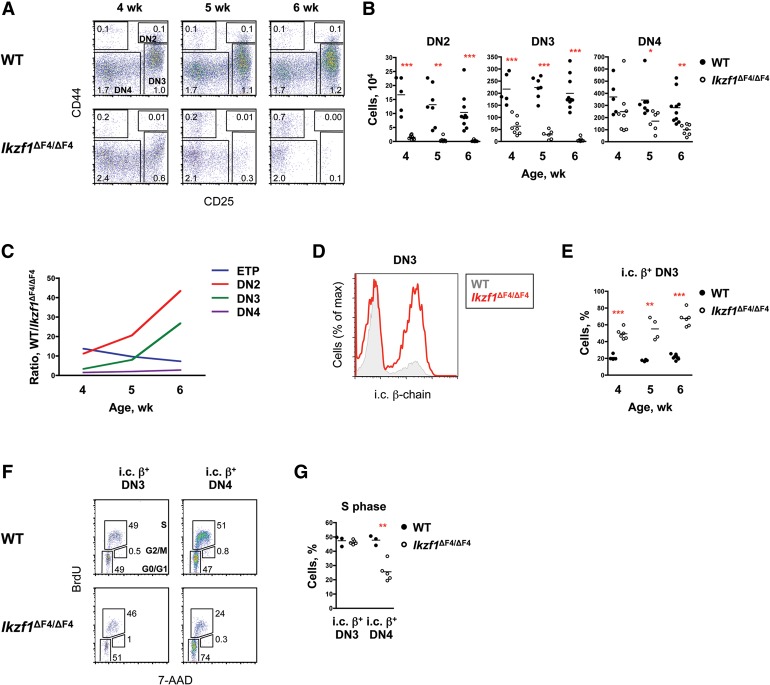
To extend our analysis of T-cell development in Ikzf1ΔF4/ΔF4 mice, we first examined mice at 4, 5, and 6 wk of age, prior to the onset of lymphoma. We previously reported reductions of early thymic progenitors (ETPs), subsequent DN populations, and CD4+CD8+ double-positive (DP) cells in 4-wk-old Ikzf1ΔF4/ΔF4 mice (Schjerven et al. 2013). Interestingly, the magnitudes of the reductions increased with age in the DN2 and DN3 populations (Fig. 1A–C). DN2 and DN3 cell numbers in Ikzf1ΔF4/ΔF4 mice in comparison with wild type were reduced by 11-fold and threefold, respectively, at 4 wk and worsened to 44-fold and 24-fold by 6 wk (Fig. 1B,C; Supplemental Fig. S1A). In contrast, the ETP deficit became slightly less severe with age (Fig. 1C; Supplemental Fig. S1B,C). The selective decline in DN2 and DN3 cells with age could be due to defective migration of progenitor cells to the thymus, an age-dependent decline in ETP maturation, an increase in cell death, or an increased rate of maturation.
Figure 1.
Altered development and response of Ikzf1ΔF4/ΔF4 DN thymocytes at the pre-TCR signaling checkpoint. (A) Shown are staining profiles of lineage− (lin−) gated thymocytes in wild-type (WT) and Ikzf1ΔF4/ΔF4 mice at 4, 5, and 6 wk of age. The absolute mean percentages are indicated (n = 5–10). (B) Shown are absolute numbers of the indicated thymocyte populations in wild-type and Ikzf1ΔF4/ΔF4 mice at 4 wk (n = 5–8), 5 wk (n = 6–7), and 6 wk (n = 7–10). Each symbol represents an individual mouse, and the bar shows the mean. (C) Shown are the ratios of the average cell numbers of the indicated populations in wild type over Ikzf1ΔF4/ΔF4 as they relate to age. (D,E) Shown are histograms of lin− CD44− CD25+ DN3 thymocytes in wild-type and Ikzf1ΔF4/ΔF4 mice at 5 wk of age (D) and the proportion of intracellular (i.c.) β-chain+ DN3 thymocytes in wild-type and Ikzf1ΔF4/ΔF4 mice at 4 wk (n = 4–6), 5 wk (n = 4), and 6 wk (n = 5–6) (E). Each symbol represents an individual mouse, and the bar shows the mean. (F,G) Shown are cell cycle analysis of lin− CD44− CD25+ DN3 cells and lin− CD44− CD25− DN4 cells (F) and proportions of the indicated populations in S phase in 4- to 5-wk-old wild-type and Ikzf1ΔF4/ΔF4 mice (n = 3–5) (G). Each symbol represents an individual mouse, and the bar shows the mean. For B, E, and G, (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
Abnormalities during the β-selection checkpoint
Although DN2 and DN3 cells exhibited strong age-dependent reductions in Ikzf1ΔF4/ΔF4 mice, DN4 cell numbers were reduced to a lesser extent, and the DN4 population did not exhibit an age-dependent decline (Fig. 1B,C; Supplemental Fig. S1A). During normal development, productive rearrangement of the TCRβ locus in DN2 and DN3 cells leads to the assembly of the pre-TCR complex. Signaling from the pre-TCR complex then allows passage through the β-selection checkpoint between the DN3 and DN4 stages of development, resulting in a transient phase of proliferation and down-regulation of the cell surface marker CD25 (Aifantis et al. 2006).
To investigate the DN3-to-DN4 transition in Ikzf1ΔF4/ΔF4 mice, DN3 cells were analyzed for intracellular expression of TCRβ. Surprisingly, a greater proportion of Ikzf1ΔF4/ΔF4 DN3 cells expressed intracellular β protein compared with wild-type DN3 cells (Fig. 1D,E), suggesting that a larger fraction of DN3 cells may have undergone successful β selection in Ikzf1ΔF4/ΔF4 mice than in wild-type mice. Indeed, TCRβ was expressed on half of the Ikzf1ΔF4/ΔF4 DN3 cells, similar to the prevalence of this protein on DN4 cells (Supplemental Fig. S1D). The pre-TCR on Ikzf1ΔF4/ΔF4 DN3 cells appeared to be functional, as BrdU incorporation revealed similar cell cycle distributions within the intracellular β-expressing DN3 populations from wild-type and Ikzf1ΔF4/ΔF4 mice (Fig. 1F,G). However, a smaller percentage of intracellular β-expressing Ikzf1ΔF4/ΔF4 DN4 cells was in S phase in comparison with intracellular β-expressing wild-type DN4 cells (Fig. 1F,G), suggesting that Ikaros finger 4 helps sustain multiple rounds of division at the DN4 stage of development. Thus, the combined results show a dramatic age-dependent decline in DN2 and DN3 numbers in Ikzf1ΔF4/ΔF4 mice and an equally dramatic increase in the percentage of proliferative intracellular β-expressing cells within the DN3 population.
The DN-to-DP transition proceeds through an intermediate CD4 immature single-positive (ISP) step
Ikaros is involved in Cd4 silencing and Cd8 activation during T-cell development, with increased percentages of mature CD4+ single-positive (SP) cells in the thymus of Ikaros mutant mice (Wang et al. 1996; Urban and Winandy 2004). In addition, Ikaros mutant strains contain a thymic CD4 SP population that exhibits immature characteristics, including low levels of TCR expression and the absence of maturation markers (Harker et al. 2002; Naito et al. 2007). The presence of this immature CD4 population was attributed to the failure of DP cells to activate the Cd8 gene or repress the Cd4 gene when Ikaros levels were reduced or absent.
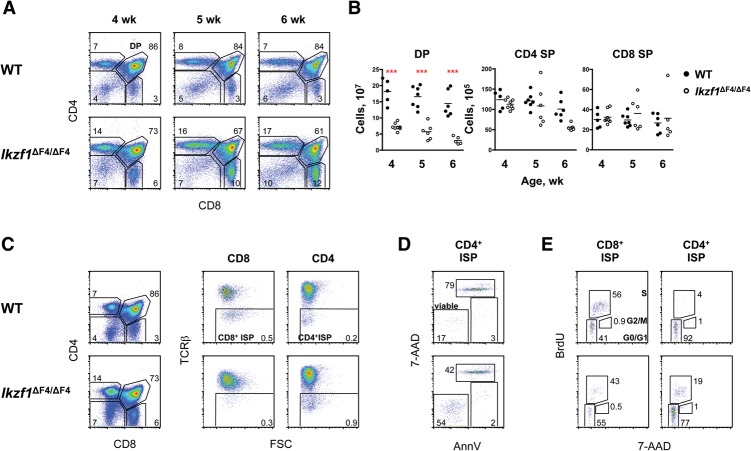
As previously described (Schjerven et al. 2013), Ikzf1ΔF4/ΔF4 mice contain moderately reduced numbers of DP cells in comparison with wild-type mice (Fig. 2A,B). The magnitude of this reduction became more pronounced with age but was not as severe as the age-dependent decline of DN2 and DN3 cells (Fig. 2A,B; Supplemental Fig. S1A,E). Despite the mild age-dependent decline of DP cells, mature TCRβhi CD4 SP and TCRβhi CD8 SP cell numbers were normal in Ikzf1ΔF4/ΔF4 mice (Fig. 2B; Supplemental Fig. S1A,E). This finding suggests that Ikaros finger 4 does not play a major role in the differentiation of DP cells to mature SP cells or in the regulation of Cd4 and Cd8 in mature cells.
Figure 2.
T-cell differentiation in Ikzf1ΔF4/ΔF4 mice progresses through a CD4+ ISP stage instead of a CD8+ ISP stage. (A) Shown are staining profiles of thymocytes in wild-type (WT) and Ikzf1ΔF4/ΔF4 mice at 4, 5, and 6 wk of age. The absolute mean percentages are indicated (n = 5–8). (B) Shown are absolute numbers of the indicated thymocyte populations in wild-type and Ikzf1ΔF4/ΔF4 mice at 4 wk (n = 5–8), 5 wk (n = 6–7), and 6 wk (n = 5–10). SP cells are gated on TCRβhi. (C) Shown are staining profiles of total (left), CD8+ gated (middle), and CD4+ gated (right) thymocytes of 4-wk-old wild-type and Ikzf1ΔF4/ΔF4 mice. The absolute percentages indicated are representative of five to eight mice. (D) Shown are staining profiles of Annexin V (AnnV) and 7-AAD (DNA content) for wild-type and Ikzf1ΔF4/ΔF4 TCRβlo CD4+ ISP thymocytes; the fractions of AnnV− 7-AAD− viable cells, AnnV+ 7-AAD− early apoptotic cells, and AnnV+ 7-AAD+ dead cells are indicated. Data are representative of three independent experiments. (E) Shown are cell cycle analyses of TCRβlo CD8+ gated (left) and TCRβlo CD4+ gated (right) cells using BrdU and 7-AAD (DNA content) staining, with cells in the G0/G1, S, and G2/M phases gated. Percentages are representative of three independent experiments.
Although the abnormality in the CD4 SP population observed in other Ikaros mutant strains was not observed in Ikzf1ΔF4/ΔF4 mice, these mice exhibited abnormalities in immature TCRβlo-expressing cells that were reminiscent of those in other mutant strains (Harker et al. 2002; Naito et al. 2007). During the DN-to-DP transition, DN4 cells in wild-type mice first differentiate into a proliferative TCRβlo CD8 ISP stage prior to becoming DP cells (MacDonald et al. 1988). Similar to observations in other mutant strains (Harker et al. 2002; Naito et al. 2007), this TCRβlo CD8 ISP population was difficult to detect in Ikzf1ΔF4/ΔF4 mice (Fig. 2C). Instead, a viable CD4 ISP population was observed that was larger than an equivalent population in wild-type mice (Fig. 2C,D). BrdU staining revealed that this CD4 ISP population was cycling in Ikzf1ΔF4/ΔF4 mice but not in wild-type mice, although to a lesser extent than the CD8 ISP population (Fig. 2E). Thus, Ikzf1ΔF4/ΔF4 DN cells appear to transit more frequently through an intermediate CD4 ISP stage than a CD8 ISP stage prior to becoming DP cells. These results suggest that Ikaros finger 4 contributes to Cd4 silencing and/or Cd8 activation but only during a specific stage of thymocyte development.
Delayed progression through T-cell differentiation in Ikzf1ΔF4/ΔF4 mice
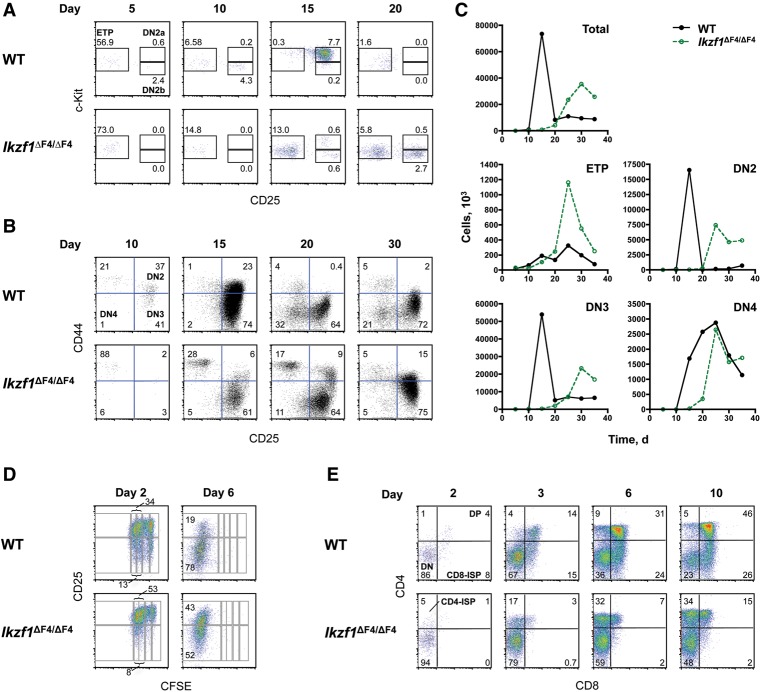
To study thymocyte development in greater depth, we used an in vitro T-cell differentiation system in which hematopoietic stem cell-enriched Lin−Sca-1+c-Kit+CD150+ progenitors from the bone marrow of wild-type and Ikzf1ΔF4/ΔF4 mice were cocultured with OP9-DL1 cells in the presence of Flt-3L and IL-7 (Schmitt and Zuniga-Pflucker 2002). As shown in Figure 3, A–C, wild-type progenitors generated ETPs by day 5 of differentiation, with peak abundance of DN2 and DN3 cells at day 15; at this time point, most DN2 cells appeared to be present at the DN2a stage of differentiation (Fig. 3A). DN4 cells were also readily detected at 15 d but reached their peak abundance at 25 d (Fig. 3C). Similar to the wild-type ETPs, Ikzf1ΔF4/ΔF4 ETPs were readily detected 5 d after the progenitors were cultured (Fig. 3A). However, the mutant ETPs continued to accumulate until the 25-d time point (Fig. 3A,C), suggesting a defect in their ability to mature. Consistent with this model and with studies performed with Ikzf1null cells (Yoshida et al. 2006), DN2, DN3, and DN4 cells accumulated with delayed kinetics in the Ikzf1ΔF4/ΔF4 cultures in comparison with wild-type cultures (Fig. 3A–C). DN2 and DN3 cell numbers were also reduced in the mutant cultures. The delayed kinetics of thymocyte development may be related to the age-dependent decline in DN2 and DN3 cells observed in vivo (Fig. 1), although the precise relationship cannot be determined from these results.
Figure 3.
Aberrant T-cell differentiation in vitro in the absence of finger 4 of Ikaros. (A–C) FACS-sorted lin− Sca-1+ c-Kit+ CD150+ LSK cells from 6-wk-old wild-type (WT) and Ikzf1ΔF4/ΔF4 bone marrow were cultured on OP9-DL1 stromal cells in the presence of 5 ng/mL Flt-3L and 5 ng/mL IL-7 for the indicated numbers of days. Shown are staining profiles of GFP− CD45+ lin− CD44+ c-Kit+ cells (A) and GFP− CD45+ lin− cells (B) at the indicated time points. Data are representative of three independent experiments. (C) Shown are absolute numbers of the indicated thymocyte populations in wild-type and Ikzf1ΔF4/ΔF4 cultures. Data are representative of two independent experiments. (D,E) Delayed T-cell differentiation in vitro when initiated from Ikzf1ΔF4/ΔF4 DN3 cells. (D) FACS-sorted carboxyfluorescein succinimidyl ester (CFSE)-labeled lin− CD44− CD25+ DN3 cells from 5-wk-old wild-type and Ikzf1ΔF4/ΔF4 mice were cultured on OP9-DL1 stromal cells in the presence of 5 ng/mL Flt-3L and 2.5 ng/mL IL-7 for the indicated numbers of days. Shown are staining profiles of GFP− CD45+ lin− cells representative of three independent experiments. (E) FACS-sorted lin− CD44− CD25+ DN3 cells from 4-wk-old wild-type and Ikzf1ΔF4/ΔF4 mice were cultured on OP9-DL1 stromal cells in the presence of 5 ng/mL Flt-3L and 1 ng/mL IL-7 for the indicated numbers of days. Shown are staining profiles of GFP− gated cells. The percentages indicated are representative of three mice.
To examine later stages of T-cell differentiation in Ikzf1ΔF4/ΔF4 cells, sorted DN3 cells from 4-wk-old wild-type and Ikzf1ΔF4/ΔF4 mice were seeded onto OP9-DL1 cells in the presence of IL-7. As with the cultures initiated with early hematopoietic progenitors, maturation differences between wild-type and mutant cells were observed. First, when sorted DN3 cells labeled with carboxyfluorescein succinimidyl ester (CFSE) were seeded onto an OP9-DL1 culture, a greater proportion of Ikzf1ΔF4/ΔF4 CD25+ DN3 cells in comparison with wild-type cells had progressed through two or three rounds of division by day 2 (Fig. 3D). This result is consistent with our finding that a larger fraction of DN3 cells expresses the pre-TCR in Ikzf1ΔF4/ΔF4 mice in comparison with wild-type mice (Fig. 1D); presumably, a larger number of mutant cells are competent for rapid proliferation in the in vitro cultures.
It is also noteworthy that a larger fraction of DN3 cells retained CD25 expression at day 6 in Ikzf1ΔF4/ΔF4 cultures in comparison with wild-type cultures (Fig. 3D). Because CD25 is down-regulated during the DN3-to-DN4 transition, this transition appeared to be delayed in the Ikzf1ΔF4/ΔF4 cultures. Delayed maturation of the mutant cells was also apparent when monitoring CD4 and CD8 expression (Fig. 3E). In wild-type cultures, DP cells were first detected as early as day 2 and reached ∼15% of the culture by day 3 (Fig. 3E; data not shown). In contrast, DP cells were not detected in mutant cultures until day 3 and did not reach 15% of the culture until day 10 (Fig. 3E).
Last, a kinetic analysis of cultures seeded with DN3 cells adds support to the hypothesis that the development of Ikzf1ΔF4/ΔF4 DP cells is often preceded by a CD4 ISP stage rather than a CD8 ISP stage. For example, at day 3, when only 3% of the Ikzf1ΔF4/ΔF4 cells had matured to the DP stage, a much larger fraction of cells expressed CD4 but not CD8 (Fig. 3E). This CD4 population remained more abundant than the DP cells at days 6 and 10. In contrast, a CD8+ population was observed during the differentiation of wild-type cells (Fig. 3E).
Thus far, we showed that deletion of a single zinc finger of Ikaros results in several T-cell abnormalities that have been observed in other Ikaros mutant strains, including reduced thymocyte cellularity across various developmental stages, deregulation of CD4 and CD8 expression in immature thymocytes, and abnormal T-cell differentiation from hematopoietic progenitors in vitro. We also uncovered previously undefined abnormalities, including an age-dependent decline of thymocyte populations, a greater proportion of dividing pre-TCR-expressing DN3 cells, and a reduction of proliferating DN4 cells. These results support the view that Ikaros plays an important role in establishing the identity and precise properties of T-lineage cells at every stage of T-cell development. Ikaros is not essential for any developmental transition, but its normal expression is clearly critical for proper development and the suppression of lymphomagenesis. With these results in mind, a major challenge is to elucidate the mechanisms by which Ikaros regulates such a diverse range of events.
Aberrant expression of small sets of genes in Ikzf1ΔF4/ΔF4 progenitors
Toward the goal of understanding how Ikaros mutations lead to a broad range of abnormalities, a central issue is the extent to which thymocytes at each developmental stage have retained or lost their identity in the Ikzf1ΔF4/ΔF4 mice. Of particular relevance, the analyses described above relied on the supposition that the cell surface markers used to define each developmental stage are not themselves aberrantly regulated in the mutant mice. A second and closely related supposition is that mutant cells have not altered their identity to such a great extent that the comparison with wild-type cells becomes uninformative.
To address these issues, we used RNA-seq to compare the gene expression profiles of wild-type and Ikzf1ΔF4/ΔF4 thymocytes at five developmental stages, including OP9-DL1-derived ETP and DN2 cells and thymic DN3, DN4, and DP cells. Previous RNA-seq studies have demonstrated that each of these stages is characterized by a highly unique gene expression signature (Zhang et al. 2012). The RNA-seq profiles were compared by calculating Pearson correlation values (R-values). Two independent experiments were performed with each developmental stage from both wild-type and mutant mice and the statistical comparison focused on RPKM (reads per kilobase per million mapped reads) values for all annotated RefSeq genes that reached an RPKM value of four or more in at least one of the 20 samples (n = 10,177 genes).
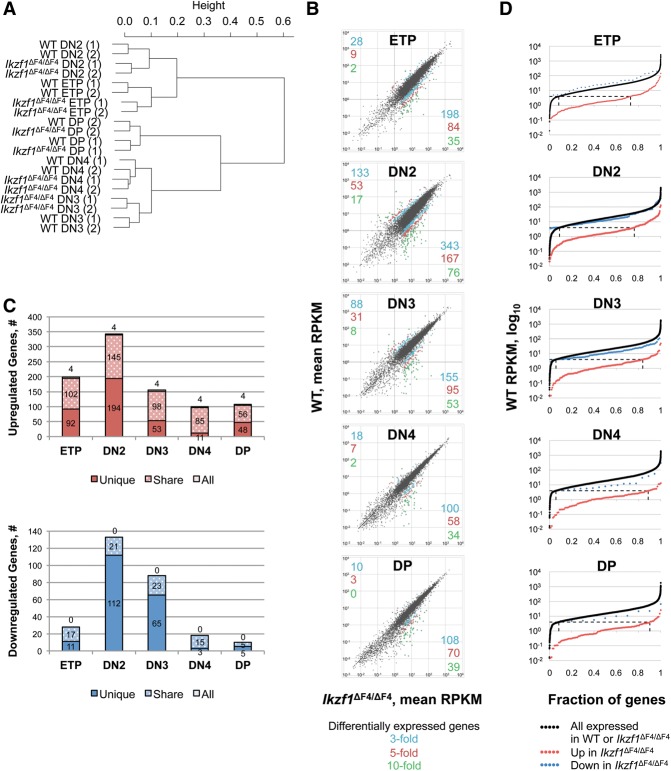
Importantly, hierarchical clustering based on the R-values showed that each Ikzf1ΔF4/ΔF4 thymocyte subset correlated more closely with its own wild-type counterpart than with any other thymocyte subset (Fig. 4A; Supplemental Fig. S2). These results suggest that the cell surface markers used to define each stage have not changed their expression patterns to an extent that diminishes their utility for the isolation of comparable cell populations from wild-type and mutant mice. The results further suggest that, despite many abnormalities, the gene expression profiles that define each developmental stage remain quite similar in wild-type and Ikzf1ΔF4/ΔF4 mice.
Figure 4.
Stage specificity of genes differentially expressed in Ikzf1ΔF4/ΔF4 thymocyte subsets. (A) Hierarchical clustering of expressed genes (>200 base pairs, four or more RPKM in at least one sample; n = 10,177 genes) is shown. (B) RPKM scatter plots of expressed genes of the indicated thymocyte subsets in wild-type (WT) and Ikzf1ΔF4/ΔF4 mice are shown. Colored dots and numbers indicate differentially expressed genes (P ≤ 0.001) up-regulated (bottom right quadrant) or down-regulated (top left quadrant) by threefold (blue), fivefold (red), and 10-fold (green) in Ikzf1ΔF4/ΔF4 T cells. (C) Genes differentially expressed (P ≤ 0.001) in any one Ikzf1ΔF4/ΔF4 thymocyte subset compared with wild type were divided into three categories based on whether the gene was also differentially expressed in any other thymocyte subset. (All) Genes that are differentially expressed in all five thymocyte subsets; (share) genes that are differentially expressed in two or more thymocyte subsets; (unique) genes that are differentially expressed only in the thymocyte subset indicated. (D) The majority of genes up-regulated in Ikzf1ΔF4/ΔF4 cells are not expressed in wild-type cells. RPKM values of wild-type cells were plotted for all expressed genes (four or more RPKM in wild-type or Ikzf1ΔF4/ΔF4 cells), for genes up-regulated by threefold (P ≤ 0.001) in Ikzf1ΔF4/ΔF4 cells compared with wild-type cells, and for genes down-regulated by threefold (P ≤ 0.001) in Ikzf1ΔF4/ΔF4 cells compared with wild-type cells. The dotted horizontal line shows where four RPKM intersects the graphs.
A closer examination of the RNA-seq profiles revealed three fundamentally important observations. First, only a moderate number of genes were aberrantly expressed in Ikzf1ΔF4/ΔF4 cells at each developmental stage, with most of these genes misregulated by <10-fold. However, as indicated in Figure 4B, the number of genes up-regulated or down-regulated by threefold, fivefold, or 10-fold (P ≤ 0.001) varied widely at the different stages.
Second, the aberrantly expressed genes usually exhibited aberrant expression at only a subset of the developmental stages. Only four genes (Dock1, Myoe1, Uaca, and Laptm4b) were up-regulated by threefold, with none down-regulated by threefold, in all five stages (Fig. 4C). A much larger number of genes were aberrantly expressed in two or more stages but not in all stages of development (Fig. 4C, “share” genes). Nevertheless, many genes were aberrantly expressed at only one stage (Fig. 4C, “unique”). One example is Klhl24; the transcript level for this gene is similar in wild-type DN2, DN3, and DN4 cells but is significantly altered only in the mutant DN2 cells (data not shown). Thus, finger 4 of Ikaros rarely alters the expression of a given gene in a consistent manner throughout thymocyte development but rather helps dictate the precise expression levels of distinct sets of genes at each developmental stage.
Last, most of the aberrantly expressed genes were up-regulated rather than down-regulated in the Ikzf1ΔF4/ΔF4 cells. Specifically, 64%–92% of aberrantly expressed genes at the various stages were up-regulated in the mutant cells (Fig. 4B). Furthermore, consistent with our previous observations with DP cells (Schjerven et al. 2013), the genes up-regulated at each developmental stage were usually up-regulated from a low transcript level. Figure 4D shows the RPKM distributions in wild-type cells (Y-axis) for all genes up-regulated (red) or down-regulated (blue) in the Ikzf1ΔF4/ΔF4 cells by threefold or more at each stage of development, in comparison with the RPKM distribution for all expressed genes (black). Seventy percent to 90% of the genes up-regulated in the Ikzf1ΔF4/ΔF4 cells at each developmental stage were expressed in wild-type cells at less than four RPKM, and 30%–46% were expressed at less than one RPKM. In contrast, the distributions of RPKMs in wild-type cells for genes that were down-regulated in the Ikzf1ΔF4/ΔF4 cells were comparable with the RPKM distributions for all expressed genes (Fig. 4D). These results are most consistent with models in which Ikaros primarily acts as a repressor of transcription. However, the Ikaros mutation does not generally lead to transcription of genes that could be characterized as “silent” in wild-type cells, as the majority of the genes up-regulated in the mutant cells are expressed at a level exceeding one RPKM in wild-type cells (Fig. 4D).
To summarize, these results show that finger 4 of Ikaros influences the expression of a limited and distinct subset of genes at each stage of thymocyte maturation. Furthermore, Ikaros appears to directly or indirectly repress a larger number of genes than it activates, but the Ikaros-mediated repression is usually moderate in its magnitude.
Finger 4 of Ikaros controls gene expression in a stage-specific manner
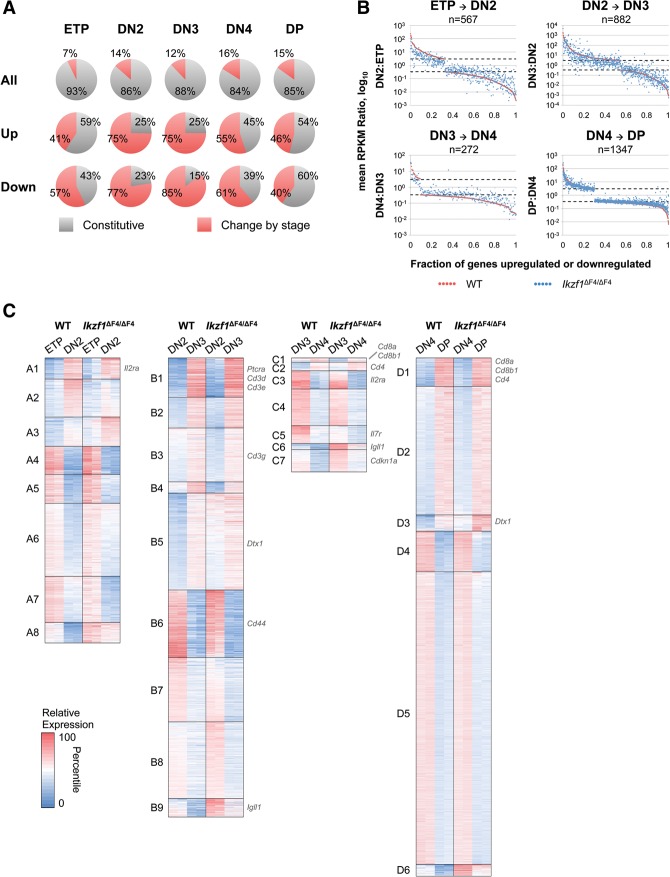
We next asked whether the aberrantly expressed genes were expressed in a sustained or stage-specific manner during the development of wild-type thymocytes. As a starting point, we found that, in wild-type mice, only a small percentage (7%–16%) of expressed RefSeq genes were up-regulated or down-regulated by at least threefold (P ≤ 0.001) in any given T-cell subset from the previous developmental stage or during maturation to the subsequent stage (Fig. 5A, top). These percentages are similar to those reported previously (Zhang et al. 2012). Surprisingly, a large percentage of the genes that were aberrantly expressed in Ikzf1ΔF4/ΔF4 cells were among this small group of stage-specific genes (Fig. 5A, middle and bottom). For example, only 14% of all expressed genes in wild-type DN2 cells were up-regulated or down-regulated by threefold or more in wild-type DN2 cells relative to their expression level in wild-type ETPs or wild-type DN3 cells; however, 75% of genes that were up-regulated in Ikzf1ΔF4/ΔF4 DN2 cells relative to wild-type DN2 cells and 77% of genes that were down-regulated in Ikzf1ΔF4/ΔF4 DN2 cells relative to wild-type DN2 cells were among the group of genes that exhibited variable expression in wild-type DN2 cells relative to wild-type ETPs and DN3 cells (Fig. 5A). This result suggests that Ikaros is strongly biased toward the regulation of genes that are dynamically modulated during thymocyte development. Notably, most of these dynamically modulated genes are not T-lineage-restricted genes (data not shown).
Figure 5.
Genes differentially expressed in Ikzf1ΔF4/ΔF4 thymocyte subsets are biased toward genes modulated by stage. (A) All expressed genes or genes differentially regulated in Ikzf1ΔF4/ΔF4 thymocyte subsets were divided into two categories: constitutive (expression of gene does not change during stage transition) and change by stage (expression of gene changes [greater than or equal to threefold; P ≤ 0.001] during stage transition). (B) Stage-dependent fold changes of all genes that were up-regulated or down-regulated by at least threefold (P ≤ 0.001) during the indicated stage transition in wild-type (WT) cells were plotted for wild-type (red) and Ikzf1ΔF4/ΔF4 (blue) stage transitions. (C) Expressed genes differentially regulated by threefold (P ≤ 0.001) at each stage transition of wild-type or Ikzf1ΔF4/ΔF4 cells were divided into a variable number of k-means clusters. Colors indicate the percentile of relative expression. ETP-to-DN2 transition, n = 796 genes; DN2-to-DN3 transition, n = 1389 genes; DN3-to-DN4 transition, n = 345 genes; DN4-to-DP transition, n = 1568 genes.
Strikingly, quantitative analyses revealed that, of the genes whose expression levels were dynamically modulated in wild-type cells by stage, the majority was up-regulated or down-regulated by a smaller magnitude in Ikzf1ΔF4/ΔF4 cells than in wild-type cells (Fig. 5B). This bias toward a reduced fold change in expression level in the mutant cells was readily apparent from a visual inspection of the ETP–DN2 transition in Figure 5B. The ETP–DN2 graph displays all genes that are up-regulated or down-regulated during the ETP-to-DN2 transition in wild-type cells (X-axis), with the fold change in transcript level for each gene (log10; Y-axis) represented by a red dot, and the fold change for the same gene in Ikzf1ΔF4/ΔF4 cells during the ETP-to-DN2 transition represented by a blue dot (Fig. 5B). Notably, the vast majority of genes exhibit a larger fold change in the wild-type cells than the mutant cells during this transition.
The strong bias toward a larger fold change in wild-type cells relative to mutant cells was also apparent during the DN3–DN4 transition as well as during the DN2–DN3 transition for genes that are expressed more highly in wild-type DN3 cells (Fig. 5B). Although this bias is not immediately apparent in Figure 5B during the DN4–DP transition, a quantitative analysis of the expression profiles revealed a small bias toward reduced fold changes in the mutant cells during this transition (Supplemental Fig. S3A). Thus, the only set of genes that failed to show any bias toward a reduced fold change in the mutant cells is the set of genes that are down-regulated during the DN2–DN3 transition (Supplemental Fig. S3A). It is relevant that these analyses include all genes that were dynamically modulated by stage by threefold or more in wild-type cells, not just those genes that were misregulated in Ikzf1ΔF4/ΔF4 cells; thus, the reduced fold change in expression level in Ikzf1ΔF4/ΔF4 cells during these developmental transitions is a widespread phenomenon that broadly impacts dynamically regulated genes.
It is important to note that, at some genes, the reduced fold change in expression level in the mutant cells is due to increased expression at the developmental stage at which the gene is expressed at a low level in wild-type cells; however, at many other genes, the reduced fold change is due to reduced expression at the developmental stage at which the gene is more highly expressed (data not shown). In other instances, increased expression was observed at both developmental stages but to a greater extent at one stage than the other, leading to a reduced fold change in the mutant cells (data not shown).
A closer examination of the bias toward a reduced fold change in the expression of stage-specific genes in Ikzf1ΔF4/ΔF4 cells revealed that the median magnitude with which genes are up-regulated during the ETP–DN2, DN2–DN3, and DN3–DN4 transitions was twice as high in wild-type cells as in Ikzf1ΔF4/ΔF4 cells (Supplemental Fig. S3A,B). In fact, a large percentage of genes that were up-regulated from one developmental stage to the next (by at least threefold) were up-regulated in wild-type cells at least twice as strongly as in Ikzf1ΔF4/ΔF4 cells (61% of up-regulated genes during the ETP–DN2 transition, 48% during the DN2–DN3 transition, and 52% during the DN3–DN4 transition) (Supplemental Fig. S3C,D). Together, these data suggest that not only is Ikaros biased toward the regulation of stage-specific genes, but a major role of Ikaros is to help establish the dynamic range of gene expression changes during developmental transitions.
To examine these stage-specific gene expression changes in greater depth, cluster analyses were performed with genes whose expression levels changed by threefold or more (P ≤ 0.001) during each developmental transition in either wild-type or Ikzf1ΔF4/ΔF4 cells (Fig. 5C). Although Figure 5B shows that the vast majority of stage-specific genes during some of the developmental transitions exhibit smaller dynamic changes in expression level in the mutant cells, the magnitudes of the differences between wild type and mutant were often too small to stand out in the cluster analysis. Notably, the genes encoding the cell surface markers used to define each stage of thymocyte development appeared in clusters in which the stage transitions were only minimally altered in Ikzf1ΔF4/ΔF4 cells (Il2ra in cluster A1, Cd44 in cluster B6, Il2ra in cluster C3, and Cd8a, Cd8b1, and Cd4 in cluster D1) (Fig. 5C).
In contrast to the majority of clusters that showed only small differences between wild-type and Ikzf1ΔF4/ΔF4 cells, larger differences were observed in other clusters. Clusters A3, B5, C2, and D3 contain genes that are up-regulated at the latter stage in both wild-type and Ikzf1ΔF4/ΔF4 cells, but expression levels were substantially higher in Ikzf1ΔF4/ΔF4 cells compared with wild-type cells (Fig. 5C; Supplemental Fig. S4). The Notch signaling target Deltex1 (Dtx1) falls into this category during the DN2-to-DN3 transition and the DN4-to-DP transition. It is important to note that Dtx1 was not constitutively up-regulated in Ikzf1ΔF4/ΔF4 cells; instead, it was up-regulated by threefold in DN2 cells, down-regulated by twofold in DN3 cells, and up-regulated by sixfold in DP cells (Supplemental Fig. S5). Interestingly, Dtx1 was up-regulated in Ikzf1ΔF4/ΔF4 cells when expression was low in wild-type cells and down-regulated when expression was high in wild-type cells, consistent with the evidence in Figure 5B that Ikaros helps maintain the dynamic range of gene expression changes during differentiation.
Interesting gene expression patterns were also observed in clusters A8, B9, C6, C7, and D6, where genes were down-regulated at the latter stage in both wild-type and Ikzf1ΔF4/ΔF4 cells; in these clusters, expression levels were generally higher in Ikzf1ΔF4/ΔF4 cells compared with wild-type cells (Fig. 5C; Supplemental Fig. S4). Igll1, a gene previously reported to be a direct target of Ikaros (Sabbattini et al. 2001; Thompson et al. 2007), was categorized into clusters B9 and C6, showing that a B-lineage gene was still expressed in Ikzf1ΔF4/ΔF4 DN3 cells. The DN3 stage consists of cells that are committed to the T-cell lineage (Yui et al. 2010), raising the possibility that genes involved in alternative lineages are not properly turned off in Ikzf1ΔF4/ΔF4 T cells. The cell cycle inhibitor Cdkn1a also appeared in these clusters. Categorized into cluster C7 during the DN3-to-DN4 transition, Cdkn1a was up-regulated by fivefold in Ikzf1ΔF4/ΔF4 DN4 cells, which is consistent with the reduced proliferation of Ikzf1ΔF4/ΔF4 DN4 cells.
Clusters B7 and C5 contain genes that are down-regulated from one stage to the next, but expression levels are lower at both stages in Ikzf1ΔF4/ΔF4 cells compared with wild-type cells (Fig. 5C; Supplemental Fig. S4). Il7r (cluster C5) expression was lower in both the DN3 and DN4 subsets of Ikzf1ΔF4/ΔF4 mice compared with wild-type mice (Supplemental Fig. S5). Similar to intracellular TCRβ protein expression on Ikzf1ΔF4/ΔF4 DN3 cells, Ikzf1ΔF4/ΔF4 DN3 cells expressed IL-7Rα protein at levels intermediate between wild-type DN3 and wild-type and Ikzf1ΔF4/ΔF4 DN4 cells (Fig. 1D; Supplemental Fig. S6). IL-7 signaling is thought to confer a survival advantage for proliferating cells that pass the β-selection checkpoint (Trigueros et al. 2003). Lower expression levels of Il7r in Ikzf1ΔF4/ΔF4 DN3 and DN4 cells would then suggest that Ikzf1ΔF4/ΔF4 DN3 and DN4 cells may not be receiving adequate survival signals needed to progress in differentiation beyond this stage or that these cells do not require IL-7 signaling and may be receiving survival signals elsewhere.
Finally, clusters B4 and C1 contain genes that are up-regulated in the latter stage, but expression levels are lower in Ikzf1ΔF4/ΔF4 cells compared with wild-type cells (Fig. 5C; Supplemental Fig. S4). Both Cd8a and Cd8b1 were classified into cluster C1, as both Cd8a and Cd8b1 were down-regulated at least twofold in DN3 and DN4 cells of Ikzf1ΔF4/ΔF4 mice compared with wild-type mice (Supplemental Fig. S5). Interestingly, Cd4 was classified into cluster C2, where its expression is eightfold higher in Ikzf1ΔF4/ΔF4 DN3 cells compared with wild-type DN3 cells (Supplemental Fig. S5); a similar finding was previously observed at the protein level in Ikzf1null DN3 cells (Naito et al. 2007). This early up-regulation of Cd4 expression and the corresponding down-regulation of Cd8a and Cd8b1 in Ikzf1ΔF4/ΔF4 DN3 cells likely account for the appearance of CD4 ISP cells seen in Ikzf1ΔF4/ΔF4 mice in vivo and in Ikzf1ΔF4/ΔF4 T-cell differentiation cultures in vitro (Fig. 2C, 3E). As is the case with the genes mentioned above, Cd4, Cd8a, and Cd8b1 were not constitutively deregulated in Ikzf1ΔF4/ΔF4 cells, as expression of all of these genes was normal in DP cells.
Altogether, global expression analysis of Ikzf1ΔF4/ΔF4 thymocyte subsets has provided clues regarding the role of Ikaros during thymocyte differentiation. Ikaros rarely regulates a gene in all developmental stages at which the gene is expressed or is not expressed. In addition, Ikaros is biased toward the regulation of genes that are modulated by stage and often helps maintain the dynamic range of gene expression changes during developmental transitions. The complex, stage-specific manner in which Ikaros controls gene expression in developing T cells provides insight into its observed contribution to such a broad range of developmental events and helps explain why its precise mechanisms of action have been difficult to elucidate.
Cooperation between the Ikzf1ΔF4/ΔF4 mutation and activating Notch mutations
In the above analysis, we were unable to find a set of deregulated genes that would be obvious drivers of the lymphomagenesis that characterizes older Ikzf1ΔF4/ΔF4 mice. Because several studies have indicated that T-lineage lymphomagenesis is driven by Ikaros loss of function in cooperation with activating Notch mutations, we examined how the introduction of a constitutively active Notch protein affects the differentiation and growth of nontransformed Ikzf1ΔF4/ΔF4 DN T cells. Constitutively active intracellular Notch (ICN) was introduced into DN T-cell progenitors from wild-type and Ikzf1ΔF4/ΔF4 mice by transduction with a retrovirus expressing ICN1 and GFP (MigR1-ICN1) (Aster et al. 2000). Upon coculture with OP9-DL1 stromal cells, ICN1-GFP+ wild-type and ICN1-GFP+ Ikzf1ΔF4/ΔF4 T cells accumulated in the culture to a greater extent than the empty vector transduced controls (Fig. 6A). Interestingly, ICN1-GFP+ Ikzf1ΔF4/ΔF4 T cells expanded more quickly and robustly than ICN1-GFP+ wild-type T cells. By the end of the time course, ICN1-GFP+ Ikzf1ΔF4/ΔF4 T cells accounted for >80% of the culture, while ICN1-GFP+ wild-type T cells only reached ∼30% of the culture (Fig. 6A). Analysis of cell surface markers showed that the ICN1-GFP+ Ikzf1ΔF4/ΔF4 T cells were predominantly blocked at the DN3 stage of T-cell development, while ICN1-GFP+ wild-type T cells were composed of both DN and DP T cells (Fig. 6B,C). These results suggest that loss of Ikaros finger 4 enhances Notch signaling-induced growth expansion and enforces a Notch signaling-induced block at the DN3 stage of T-cell development.
Figure 6.
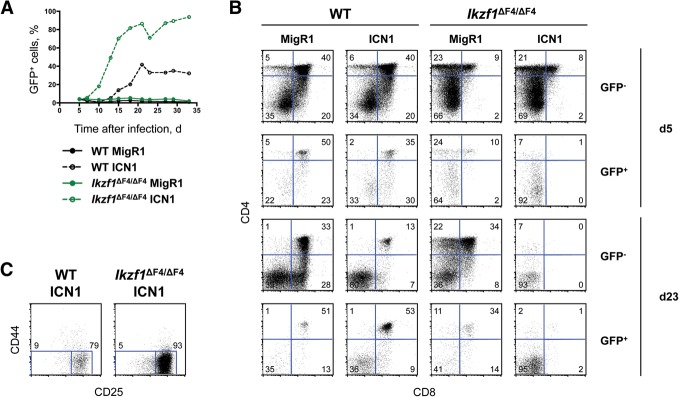
Enhanced Notch-induced growth expansion of Ikzf1ΔF4/ΔF4 DN T cells in vitro. (A) Lin-depleted thymocytes from 4- to 5-wk-old wild-type (WT) and Ikzf1ΔF4/ΔF4 mice were transduced with MigR1 (empty vector) or MigR1-ICN1 retroviruses and cultured on OP9-DL1 stromal cells in the presence of 5 ng/mL IL-7 for the indicated numbers of days. Shown is the proportion of GFP+ cells at the indicated time points. Data are representative of three independent experiments. (B) Shown are staining profiles of GFP− CD45+ and GFP+ CD45+ gated cells. The percentages indicated are representative of three independent experiments. (C) Shown are staining profiles of GFP+ CD45+ CD4− CD8− gated cells at day 13.
Enhanced Notch-induced gene expression program in Ikzf1ΔF4/ΔF4 T cells
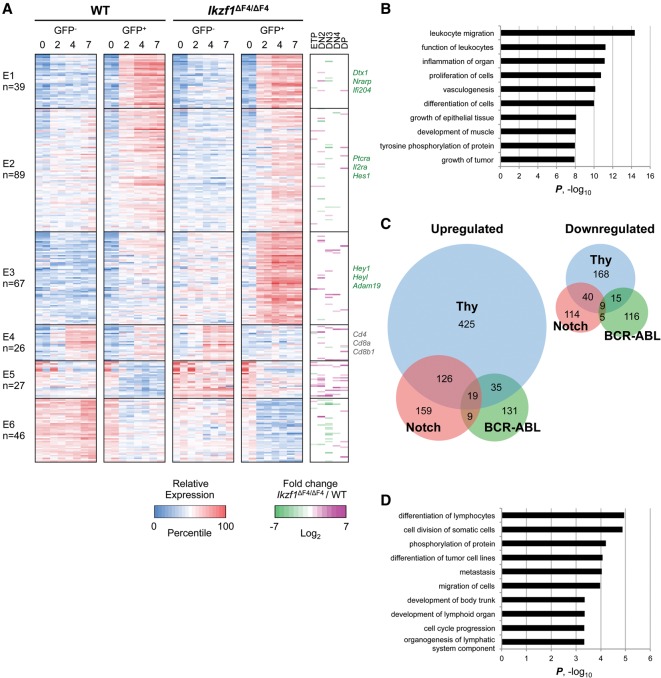
To investigate the gene expression changes that occur during the early events of Notch activation, we compared global expression profiles of ICN1-GFP− and ICN1-GFP+ fractions of wild-type and Ikzf1ΔF4/ΔF4 T-cell cultures at 0, 2, 4, and 7 d post-transduction in duplicate (see Supplemental Figs. S2B, S7A for statistical analysis). Two-hundred-ninety-four genes were identified that were differentially expressed by at least threefold (P ≤ 0.001) between ICN1-GFP+ and ICN1-GFP− cells and at least fivefold (P ≤ 0.001) at any time point relative to untransduced cells (i.e., day 0 cells). Separation of these 294 genes into six clusters revealed three induced (E1–E3) and three repressed (E4–E6) clusters (Fig. 7A; Supplemental Fig. S7B). More genes were activated by Notch than were repressed, as only a third of the 294 genes fell into clusters E4–E6. The largest two clusters, E1 and E2, contain those genes that were most potently activated by ICN1 in this assay system (Dtx1, Nrarp, and Ifi204 in cluster E1 and Ptcra, Il2ra, and Hes1 in cluster E2). Importantly, little difference in the ICN1-induced expression levels for these genes was observed in the wild-type and Ikzf1ΔF4/ΔF4 backgrounds (ICN1-GFP+ wild-type vs. ICN1-GFP+ Ikzf1ΔF4/ΔF4 samples), suggesting that the ability of ICN1 to induce the strongest Notch targets was unaffected by the Ikzf1ΔF4/ΔF4 mutation.
Figure 7.
Loss of finger 4 of Ikaros affects genes weakly induced or repressed by activated Notch. (A) Expressed genes differentially regulated by threefold (P ≤ 0.001) between ICN1-GFP+ and ICN1-GFP− cells and differentially regulated by fivefold (P ≤ 0.001) in any time point relative to day 0 were divided into six k-means clusters. The right panel shows the corresponding fold change of expression levels in Ikzf1ΔF4/ΔF4 over wild-type (WT) T-cell subsets. Colors indicate percentile of relative expression (blue to red) or log2 fold change (green to purple). (B) Gene ontology analysis is shown for genes up-regulated or down-regulated by fivefold (P ≤ 0.001) in ICN1-GFP+ Ikzf1ΔF4/ΔF4 cells compared with ICN1-GFP+ wild-type cells. (C) A Venn diagram is shown comparing the lists of genes differentially up-regulated (left panel) or down-regulated (right panel) by threefold in Ikzf1ΔF4/ΔF4 cells compared with wild-type cells in thymocytes (ETP, DN2, DN3, DN4, or DP), Notch-expressing cells (ICN1-GFP+ cells at day 2, 4, or 7), and BCR-ABL-expressing cells (day 14, 21, or 28). (D) A gene ontology analysis is shown for genes up-regulated or down-regulated by threefold in Ikzf1ΔF4/ΔF4 cells compared with wild-type cells in both Notch-expressing and BCR-ABL-expressing cells.
Cluster E4 is also of interest, as this cluster includes genes with reduced expression levels in ICN1-GFP+ Ikzf1ΔF4/ΔF4 T cells compared with all groups (Fig. 7A; Supplemental Fig. S7B). Some genes in this cluster, including Cd4, Cd8a, and Cd8b1, presumably were not expressed in the ICN1-GFP+ Ikzf1ΔF4/ΔF4 T cells because, as shown in Figure 6C, these cells were blocked at the DN3 stage of T-cell differentiation.
Cluster E3 contains genes that were up-regulated in the Ikaros mutant background in both the absence and presence of ICN1, with the highest expression observed when ICN1 was expressed in the Ikzf1ΔF4/ΔF4 cells (Fig. 7A). In other words, ICN1 appeared to cooperate with Ikaros loss of function to enhance the expression of cluster E3 genes, whereas cooperation was not observed under our assay conditions with genes in clusters E1 and E2. Interestingly, most genes in cluster E3 were induced by ICN1 only weakly in comparison with clusters E1 and E2 (Supplemental Fig. S7B), suggesting that Ikaros preferentially represses the weakest Notch target genes in this assay system.
As a mirror image of cluster E3, cluster E6 contains ICN1-repressed genes in which Notch signaling and Ikaros loss of function appear to cooperate to promote strong repression (Fig. 7A; Supplemental Fig. S7B). Therefore, both clusters E3 and E6 contain genes that appear to be regulated independently by Ikaros and Notch signaling, with Notch signaling and Ikaros loss of function cooperating with each other at these genes to promote the highest (cluster 3) or lowest (cluster 6) expression levels. From another perspective, wild-type Ikaros appears to contribute to the repression (cluster 3) or activation (cluster 6) of these genes, but ICN1 expression can only partially overcome this repression and activation in wild-type cells; to fully overcome the Ikaros-dependent repression or activation, an Ikaros loss-of-function mutation appears to be necessary.
Last, several Notch target genes (Hey1, Heyl, and Adam19) that are not normally expressed in T cells (two or less RPKM in all wild-type thymocyte subsets) (data not shown) were classified into cluster E3, where they were highly expressed in ICN1-GFP+ Ikzf1ΔF4/ΔF4 T cells, consistent with the previous finding that loss of Ikaros may lead to the misexpression of Notch targets in inappropriate cell types (Geimer Le Lay et al. 2014). Altogether, these results show a complex relationship between Notch activation and Ikaros loss of function. In particular, although the expression of the weakest Notch-regulated genes appeared to be altered by the Ikaros mutation, many of the strongest Notch-regulated genes remained unaffected.
Comparison of all expressed genes revealed 111 genes up-regulated and 80 genes down-regulated by fivefold or more (P ≤ 0.001) in ICN1-GFP+ Ikzf1ΔF4/ΔF4 T cells compared with ICN1-GFP+ wild-type T cells at any time point after day 0. Gene ontology analysis of these differentially expressed genes showed an enrichment of genes involved in leukocyte migration, leukocyte function, cell proliferation, and tyrosine phosphorylation of protein (Fig. 7B). Interestingly, among the genes involved in cell proliferation and tyrosine phosphorylation was Kit, which was previously shown to be aberrantly up-regulated in Ikzf1ΔF4/ΔF4 B cells compared with wild-type B cells in a mouse model of BCR-ABL+ B-ALL (Schjerven et al. 2013). BCR-ABL+ Ikzf1ΔF4/ΔF4 B cells not only expressed Kit but also expanded more robustly than their wild-type counterparts, similar to the enhanced Notch-induced expansion observed in ICN1-GFP+ Ikzf1ΔF4/ΔF4 T cells.
Comparing the lists of genes differentially expressed by at least threefold in Ikzf1ΔF4/ΔF4 untransformed thymocytes, Ikzf1ΔF4/ΔF4 Notch-expressing T cells, and Ikzf1ΔF4/ΔF4 BCR-ABL-expressing B cells (Schjerven et al. 2013) compared with their wild-type counterparts led to the identification of a small set of differentially expressed genes that were shared between all three lists (19 genes up-regulated and nine genes down-regulated in Ikzf1ΔF4/ΔF4 cells compared with wild-type cells) (Fig. 7C; Supplemental Figs. S8, S9). Gene ontology analysis of these 28 genes showed an enrichment for genes involved in various functions, making it unclear how they may contribute to the tumor suppressor function of Ikaros (Supplemental Fig. S10). However, Kit belonged to the small set of differentially expressed genes that are in common between the Notch-expressing T-cell and BCR-ABL-expressing B-cell lists (nine genes up-regulated and five genes down-regulated in Ikzf1ΔF4/ΔF4 cells compared with wild-type cells) (Fig. 7C; Supplemental Figs. S8, S9). Interestingly, BCR-ABL-expressing B cells also expressed high levels of the oncogenic Notch target gene Igf1r, which has been shown to be required for proliferation and leukemic-initiating activity in T-ALL (Medyouf et al. 2011; Witkowski et al. 2015). Altogether, these 14 genes common to the Notch and BCR-ABL data sets were found to be involved in lymphocyte differentiation, cell division, phosphorylation of protein, and cell cycle progression (Fig. 7D). These genes may contribute to the enhanced Notch-induced or BCR-ABL-induced expansion of Ikzf1ΔF4/ΔF4 cells compared with wild-type cells.
Discussion
For a lineage-restricted protein like Ikaros, which binds DNA in a sequence-specific manner and plays a critical role in development, the first potential mechanism of action that must be considered is that it binds the promoters and enhancers of a large number of critical lineage-restricted target genes and serves as an activator or repressor of those genes by modulating chromatin structure, enhancer–promoter communication, and/or the recruitment of the general transcription machinery. Some evidence supports the hypothesis that Ikaros can function as a typical transcriptional regulator, as a few lineage-restricted genes have been identified that exhibit a strong requirement for Ikaros for their activation or repression. However, as described above, several unusual properties of Ikaros raise the possibility that it may also regulate development and leukemogenesis via atypical mechanisms. The results described here support this possibility. In particular, Ikaros was found to be strongly biased toward the regulation of developmental stage-specific genes rather than lineage-specific genes, and the most consistent characteristic of the Ikzf1ΔF4/ΔF4 cells was a reduced fold change in expression for a large fraction of genes that are dynamically regulated during key developmental transitions. Thus, rather than serving as a dedicated activator or repressor, a primary function of Ikaros may be to support dynamic changes in gene expression during developmental transitions.
It is not yet known whether this function is directly or indirectly regulated by Ikaros. As an example of an indirect mechanism, Ikaros might function as a typical transcription factor to regulate one or more genes that control dynamic changes in gene expression. If Ikaros is instead a direct regulator of the dynamic changes in gene expression, the simplest hypothesis is that it carries out this function in cooperation with the Mi-2/NuRD complex, which is the primary interaction partner of Ikaros (Kim et al. 1999; Sridharan and Smale 2007). One possibility is that Ikaros and Mi-2/NuRD facilitate changes in the three-dimensional organization of the genome during developmental transitions, with these organizational changes needed for efficient gene expression changes. The fact that the dimerization and multimerization domains of Ikaros are at the opposite end of the protein from its DNA-binding domain and that Ikaros ChIP-seq peaks exhibit enrichment primarily for motifs recognized by an Ikaros monomer provide preliminary circumstantial support for the hypothesis that Ikaros participates in genome organization. An important goal for the future will be to compare the three-dimensional genome structure in wild-type and Ikzf1 mutant cells, although any differences observed would represent only a preliminary step toward establishing a direct role for Ikaros in genome organization. It also is tempting to speculate that the pericentromeric localization of Ikaros molecules might be relevant to the results described in this study.
It is noteworthy that the reduced fold changes in gene expression levels observed in the mutant cells could reflect uniform changes as cells progress from one developmental stage to the next. Alternatively, similar findings would be expected if the mutant cells exhibited developmental “blurring,” in which a fraction of the cells at each stage up-regulated or down-regulated stage-specific genes in a premature or delayed fashion. The elevated TCRβ expression levels observed in a fraction of the mutant DN3 cells is consistent with this hypothesis. However, the extent to which the reduced fold changes result from premature or delayed activation and repression remains to be determined and may benefit from single-cell RNA-seq experiments.
The relationship between our findings and previous ChIP-seq evidence that Ikaros associates with a large fraction of active genes is difficult to discern. Schwickert et al. (2014) showed that the genomic sites occupied by Ikaros differ at different stages of B-cell development. However, enrichment of Ikaros-binding events in the vicinity of genes that are misregulated in Ikzf1 mutant cells was not observed despite evidence that Ikaros regulates stage-specific genes during two stages of B-cell development, similar to our findings in thymocytes. Instead, the study by Schwickert et al. (2014), like our studies and those of others (Zhang et al. 2011; Schjerven et al. 2013), reported preferential binding in the vicinity of a large percentage of transcriptionally active genes or in the vicinity of control regions that exhibit histone modifications characteristic of transcriptionally active chromatin. These findings raise the possibility that the Ikaros-binding events observed by ChIP-seq may reflect a broader role in genome organization rather than a specific role in regulating genes in the vicinity of the binding sites. An alternative possibility is that many Ikaros-binding sites observed by ChIP-seq may represent nonfunctional, opportunistic binding events that obscure a smaller set of protein–DNA interactions that are critical for the functions of Ikaros.
The role of Ikaros as a tumor suppressor has been equally challenging to understand because it appears to contribute to both differentiation and cell growth (Merkenschlager 2010). Ikaros has previously been linked to repression of Notch target genes (Geimer Le Lay et al. 2014). Our results support the prior evidence of a relationship between Ikaros and Notch, although the relationship appears to be complex. In particular, introduction of active Notch into Ikzf1ΔF4/ΔF4 T cells led to a strong induction of the weakest Notch targets, while the most potently induced Notch targets were unaffected. These findings suggest that Ikaros may help establish activation thresholds in response to Notch signaling. Such a function would be reminiscent of the previously described role of Ikaros in establishing activation thresholds in response to antigen receptor signaling (Avitahl et al. 1999; Winandy et al. 1999). Last, we were able to identify a small subset of genes that were induced in Ikzf1ΔF4/ΔF4 lymphocytes in response to two different oncogenic factors: Notch and BCR-ABL. Although the Notch and BCR-ABL signaling pathways differ, Ikzf1ΔF4/ΔF4 Notch-expressing T cells and Ikzf1ΔF4/ΔF4 BCR-ABL-expressing B cells exhibit developmental blocks at similar stages, precisely before preantigen receptor checkpoints in both lineages. This observation raises the possibility that Ikaros uses similar mechanisms of action to enforce its tumor suppressor function in both the B- and T-cell lineages.
Materials and methods
Mice
The Ikzf1ΔF4/ΔF4 mouse strain, generated by deletion of Ikzf1 exon 6 (Schjerven et al. 2013), was backcrossed to the C57BL/6 background for at least 13 generations. Four-week-old to 6-wk-old mice of both sexes were used for the majority of experiments except transcriptome profiling experiments, for which only female mice were used. All mouse studies were approved by the University of California at Los Angeles Institutional Animal Care and Use Committee.
Antibodies and flow cytometry
Cells were stained for four- or five-color analysis with the following fluorochrome-conjugated antibodies directed against CD11b, B220, TCRγδ, NK1.1, TER119, Gr-1, CD11c, TCRβ, CD3ε, CD4, c-Kit, Sca-1, CD44, CD25, and IL-7Rα (eBioscience); CD8α (BD Biosciences); and CD150 (BioLegend). For intracellular TCRβ staining, thymocyte suspensions were surface-stained with the indicated fluorochrome-conjugated antibodies, fixed and permeabilized with 4% paraformaldehyde and 0.01% saponin, and then stained with an antibody against TCRβ. For steady-state cell cycle analysis, mice were injected intraperitoneally with 1 mg of BrdU (Sigma) and sacrificed after 50 min. To measure BrdU incorporation, single-cell suspensions were stained for surface markers, fixed, permeabilized, and then stained with an antibody against BrdU according to BD Pharmingen BrdU flow kit protocol. To measure apoptosis and cell viability, cells were stained with surface markers and then stained with FITC-conjugated Annexin-V in Annexin-V-binding buffer and 7-AAD (BD Biosciences). The samples were acquired using the FACSCalibur or LSR II flow cytometer (both BD Biosciences), sorted on the FACSAria II flow sorter (BD Biosciences), and analyzed using FlowJo software (Treestar, Inc.). The statistical significance (unpaired, two-tailed Student's t-test with 95% confidence intervals) of cell percentages and absolute numbers was estimated using GraphPad Prism software.
In vitro T-cell differentiation
OP9-DL1 stromal cells were cocultured with either CD150+ LSK cells (CD150+ Lin− Sca-1+ c-Kit+) from the bone marrow of 6-wk-old mice or DN3 cells (Lin− CD44− CD25+) from the thymuses of 4-wk-old mice in the presence of Flt-3L (5 ng/mL) and IL-7 (5 ng/mL for CD150+ LSK cocultures or 1 ng/mL for DN3 cocultures) (Schmitt and Zuniga-Pflucker 2002). For cell division tracking, sorted DN3 cells from the thymuses of 4-wk-old mice were loaded with CFDA-SE (Invitrogen) prior to coculturing with OP9-DL1 stromal cells. OP9-DL1 stromal cells were excluded from flow cytometry analysis by a combination of forward and side scatter characteristics, GFP expression, and/or lack of CD45 expression.
Retroviral transduction
Retroviral supernatant was produced by packaging MigR1 or MigR1-ICN1 (Aster et al. 2000) vectors into retroviruses by transient transfection of 293T cells; retroviral titers were determined with NIH3T3 cells. Lin-depleted (CD4− CD8− TCRβ− TCRγδ− B220− Gr-1− Ter119− DX5− CD11b− CD11c−) DN progenitors from the thymuses of 4-wk-old mice were transduced by a single spin infection with supernatant containing either MigR1 or MigR1-ICN1 retrovirus and 4 μg/mL polybrene at a low infection efficiency of 5% (for cell growth experiment) or a high infection efficiency of 50% (for transcriptome profiling). Infected cells were cultured on a pre-established layer of OP9-DL1 cells in the presence of 5 ng/mL IL-7.
Transcriptome analysis
The following T-cell populations were sorted by flow cytometry: ETP (CD45+ Lin− c-Kit+ CD44+ CD25−) and DN2 cells (CD45+ Lin− c-Kit+ CD44+ CD25+) derived from OP9-DL1 cocultures cocultured with CD150+ LSK cells from the bone marrow of 6-wk-old mice and DN3 (Lin− CD44− CD25+), DN4 (Lin− CD44− CD25−), and DP (CD4+ CD8+) cells from the thymuses of 4-wk-old mice. Cells (1 × 105 to 2.5 × 105) were harvested in TRI reagent LS (Molecular Research Center, Inc.), and total RNA was isolated using the RNeasy micro kit (Qiagen). Strand-specific cDNA libraries were constructed from a minimum of 200 ng of each RNA sample using the TruSeq RNA sample preparation kit (Illumina), quantified using the Qubit dsDNA BR assay kit (Life Technologies), and sequenced on an Illumina HiSeq 2000 instrument with a single-end sequencing length of 50 base pairs. Reads were mapped to the July 2007 mouse (Mus musculus) genome (mm9, National Center for Biotechnology Information Build 37 assembly) via TopHat (version 1.3.3) (Trapnell et al. 2010) through Galaxy (Goecks et al. 2010), with alignments restricted to uniquely mapped reads, allowing two possible mismatches. Expression levels (RPKM) for mm9 RefSeq genes were calculated (Mortazavi et al. 2008) using exonic reads obtained through SeqMonk (Babraham Bioinformatics) and mRNA lengths. Genes differentially expressed in two populations to a statistically significant degree were identified by analyzing exonic reads through the Bioconductor DESeq program (Anders and Huber 2010). Hierarchical clustering of the absolute Pearson correlation coefficients was generated in R Studio. Clustering analysis (k-means using Euclidean distance) was performed on log2 transformed, mean-centered RPKM values with Cluster 3.0 software (de Hoon et al. 2004). Gene ontology analysis was performed using Ingenuity Pathway Analysis (Qiagen) or Panther software (Thomas et al. 2006). All RNA-seq data are available on the Gene Expression Omnibus database under accession number GSE67572.
Supplementary Material
Acknowledgments
We thank Ellen Rothenberg for helpful discussions, Warren Pear for the MigR1-ICN1 plasmid, Jennifer Lam and Aleksandar Rivin for assistance with the mouse colony, the Broad Stem Cell Research Center High-Throughput Sequencing Core, and the University of California at Los Angeles Jonsson Comprehensive Cancer Center Flow Cytometry Shared Resource. This work was supported by National Institutes of Health grants R01 DK43726 (to S.T.S.) and T32 HL086345 (to T.L.A.).
Footnotes
Supplemental material is available for this article.
Article pubished online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.266999.115.
References
- Aifantis I, Mandal M, Sawai K, Ferrando A, Vilimas T. 2006. Regulation of T-cell progenitor survival and cell-cycle entry by the pre-T-cell receptor. Immunol Rev 209: 159–169. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aster JC, Xu L, Karnell FG, Patriub V, Pui JC, Pear WS. 2000. Essential roles for ankyrin repeat and transactivation domains in induction of T-cell leukemia by notch1. Mol Cell Biol 20: 7505–7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avitahl N, Winandy S, Friedrich C, Jones B, Ge Y, Georgopoulos K. 1999. Ikaros sets thresholds for T cell activation and regulates chromosome propagation. Immunity 10: 333–343. [DOI] [PubMed] [Google Scholar]
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. 1997. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell 91: 845–854. [DOI] [PubMed] [Google Scholar]
- Chari S, Winandy S. 2008. Ikaros regulates Notch target gene expression in developing thymocytes. J Immunol 181: 6265–6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb BS, Morales-Alcelay S, Kleiger G, Brown KE, Fisher AG, Smale ST. 2000. Targeting of Ikaros to pericentromeric heterochromatin by direct DNA binding. Genes Dev 14: 2146–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Hoon MJL, Imoto S, Nolan J, Miyano S. 2004. Open source clustering software. Bioinformatics 20: 1453–1454. [DOI] [PubMed] [Google Scholar]
- Dumortier A, Jeannet R, Kirstetter P, Kleinmann E, Sellars M, dos Santos NR, Thibault C, Barths J, Ghysdael J, Punt JA, et al. 2006. Notch activation is an early and critical event during T-cell leukemogenesis in Ikaros-deficient mice. Mol Cell Biol 26: 209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreiros-Vidal I, Carroll T, Taylor B, Terry A, Liang Z, Bruno L, Dharmalingam G, Khadayate S, Cobb BS, Smale ST, et al. 2013. Genome-wide identification of Ikaros targets elucidates its contribution to mouse B-cell lineage specification and pre-B-cell differentiation. Blood 121: 1769–1782. [DOI] [PubMed] [Google Scholar]
- Geimer Le Lay A-S, Oravecz A, Mastio J, Jung C, Marchal P, Ebel C, Dembélé D, Jost B, Le Gras S, Thibault C, et al. 2014. The tumor suppressor Ikaros shapes the repertoire of notch target genes in T cells. Sci Signal 7: ra28. [DOI] [PubMed] [Google Scholar]
- Georgopoulos K, Bigby M, Wang JH, Molnar A, Wu P, Winandy S, Sharpe A. 1994. The Ikaros gene is required for the development of all lymphoid lineages. Cell 79: 143–156. [DOI] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. 2010. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 32: 317–328. [DOI] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J. 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harker N, Naito T, Cortes M, Hostert A, Hirschberg S, Tolaini M, Roderick K, Georgopoulos K, Kioussis D. 2002. The CD8α gene locus is regulated by the Ikaros family of proteins. Mol Cell 10: 1403–1415. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, et al. 1999. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity 10: 345–355. [DOI] [PubMed] [Google Scholar]
- Kirstetter P, Thomas M, Dierich A, Kastner P, Chan S. 2002. Ikaros is critical for B cell differentiation and function. Eur J Immunol 32: 720–730. [DOI] [PubMed] [Google Scholar]
- MacDonald HR, Budd RC, Howe RC. 1988. A CD3− subset of CD4−8+ thymocytes: a rapidly cycling intermediate in the generation of CD4+8+ cells. Eur J Immunol 18: 519–523. [DOI] [PubMed] [Google Scholar]
- Mandel EM, Grosschedl R. 2010. Transcription control of early B cell differentiation. Curr Opin Immunol 22: 161–167. [DOI] [PubMed] [Google Scholar]
- Medyouf H, Gusscott S, Wang H, Tseng J-C, Wai C, Nemirovsky O, Trumpp A, Pflumio F, Carboni J, Gottardis M, et al. 2011. High-level IGF1R expression is required for leukemia-initiating cell activity in T-ALL and is supported by Notch signaling. J Exp Med 208: 1809–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M. 2010. Ikaros in immune receptor signaling, lymphocyte differentiation, and function. FEBS Lett 584: 4910–4914. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, White D, Hughes TP, Le Beau MM, Pui CH, et al. 2008. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature 453: 110–114. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, Ma J, Liu W, Cheng C, Schulman BA, et al. 2009. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med 360: 470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito T, Gomez-del Arco P, Williams CJ, Georgopoulos K. 2007. Antagonistic interactions between Ikaros and the chromatin remodeler Mi-2β determine silencer activity and Cd4 gene expression. Immunity 27: 723–734. [DOI] [PubMed] [Google Scholar]
- Sabbattini P, Lundgren M, Georgiou A, Chow C, Warnes G, Dillon N. 2001. Binding of Ikaros to the λ5 promoter silences transcription through a mechanism that does not require heterochromatin formation. EMBO J 20: 2812–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjerven H, McLaughlin J, Arenzana TL, Frietze S, Cheng D, Wadsworth SE, Lawson GW, Bensinger SJ, Farnham PJ, Witte ON, et al. 2013. Selective regulation of lymphopoiesis and leukemogenesis by individual zinc fingers of Ikaros. Nat Immunol 14: 1073–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt TM, Zuniga-Pflucker JC. 2002. Induction of T cell development from hematopoietic progenitor cells by Delta-like-1 in vitro. Immunity 17: 749–756. [DOI] [PubMed] [Google Scholar]
- Schwickert TA, Tagoh H, Gültekin S, Dakic A, Axelsson E, Minnich M, Ebert A, Werner B, Roth M, Cimmino L, et al. 2014. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat Immunol 15: 283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R, Smale ST. 2007. Predominant interaction of both Ikaros and Helios with the NuRD complex in immature thymocytes. J Biol Chem 282: 30227–30238. [DOI] [PubMed] [Google Scholar]
- Sun L, Liu A, Georgopoulos K. 1996. Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. EMBO J 15: 5358–5369. [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, Lazareva-Ulitsky B. 2006. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res 34: W645–W650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson EC, Cobb BS, Sabbattini P, Meixlsperger S, Parelho V, Liberg D, Taylor B, Dillon N, Georgopoulos K, Jumaa H, et al. 2007. Ikaros DNA-binding proteins as integral components of B cell developmental-stage-specific regulatory circuits. Immunity 26: 335–344. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigueros C, Hozumi K, Silva-Santos B, Bruno L, Hayday AC, Owen M, Pennington DJ. 2003. Pre-TCR signaling regulates IL-7 receptor α expression promoting thymocyte survival at the transition from the double-negative to double-positive stage. Eur J Immunol 33: 1968–1977. [DOI] [PubMed] [Google Scholar]
- Trinh LA, Ferrini R, Cobb BS, Weinmann AS, Hahm K, Ernst P, Garraway IP, Merkenschlager M, Smale ST. 2001. Down-regulation of TDT transcription in CD4+CD8+ thymocytes by Ikaros proteins in direct competition with an Ets activator. Genes Dev 15: 1817–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JA, Winandy S. 2004. Ikaros null mice display defects in T cell selection and CD4 versus CD8 lineage decisions. J Immunol 173: 4470–4478. [DOI] [PubMed] [Google Scholar]
- Wang JH, Nichogiannopoulou A, Wu L, Sun L, Sharpe AH, Bigby M, Georgopoulos K. 1996. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity 5: 537–549. [DOI] [PubMed] [Google Scholar]
- Winandy S, Wu P, Georgopoulos K. 1995. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell 83: 289–299. [DOI] [PubMed] [Google Scholar]
- Winandy S, Wu L, Wang JH, Georgopoulos K. 1999. Pre-T cell receptor (TCR) and TCR-controlled checkpoints in T cell differentiation are set by Ikaros. J Exp Med 190: 1039–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkowski MT, Cimmino L, Hu Y, Trimarchi T, Tagoh H, McKenzie MD, Best SA, Tuohey L, Willson TA, Nutt SL, et al. 2015. Activated Notch counteracts Ikaros tumor suppression in mouse and human T-cell acute lymphoblastic leukemia. Leukemia 29: 1301–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Ng SY, Zuniga-Pflucker JC, Georgopoulos K. 2006. Early hematopoietic lineage restrictions directed by Ikaros. Nat Immunol 7: 382–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui MA, Feng N, Rothenberg EV. 2010. Fine-scale staging of T cell lineage commitment in adult mouse thymus. J Immunol 185: 284–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Jackson AF, Naito T, Dose M, Seavitt J, Liu F, Heller EJ, Kashiwagi M, Yoshida T, Gounari F, et al. 2011. Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nat Immunol 13: 86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. 2012. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell 149: 467–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.