Váraljai et al. show that loss of the histone demethylase Kdm5a restores differentiation through increasing mitochondrial respiration. The regulatory functions of pRB in the cell cycle and differentiation are distinct because although restoring differentiation requires intact mitochondrial function, it does not necessitate cell cycle exit.
Keywords: pRB, differentiation, mitochondria, KDM5A, RBP2, histone methylation
Abstract
The retinoblastoma tumor suppressor protein pRb restricts cell growth through inhibition of cell cycle progression. Increasing evidence suggests that pRb also promotes differentiation, but the mechanisms are poorly understood, and the key question remains as to how differentiation in tumor cells can be enhanced in order to diminish their aggressive potential. Previously, we identified the histone demethylase KDM5A (lysine [K]-specific demethylase 5A), which demethylates histone H3 on Lys4 (H3K4), as a pRB-interacting protein counteracting pRB's role in promoting differentiation. Here we show that loss of Kdm5a restores differentiation through increasing mitochondrial respiration. This metabolic effect is both necessary and sufficient to induce the expression of a network of cell type-specific signaling and structural genes. Importantly, the regulatory functions of pRB in the cell cycle and differentiation are distinct because although restoring differentiation requires intact mitochondrial function, it does not necessitate cell cycle exit. Cells lacking Rb1 exhibit defective mitochondria and decreased oxygen consumption. Kdm5a is a direct repressor of metabolic regulatory genes, thus explaining the compensatory role of Kdm5a deletion in restoring mitochondrial function and differentiation. Significantly, activation of mitochondrial function by the mitochondrial biogenesis regulator Pgc-1α (peroxisome proliferator-activated receptor γ-coactivator 1α; also called PPARGC1A) a coactivator of the Kdm5a target genes, is sufficient to override the differentiation block. Overexpression of Pgc-1α, like KDM5A deletion, inhibits cell growth in RB-negative human cancer cell lines. The rescue of differentiation by loss of KDM5A or by activation of mitochondrial biogenesis reveals the switch to oxidative phosphorylation as an essential step in restoring differentiation and a less aggressive cancer phenotype.
The functional inactivation of the tumor suppressor pRB results in an inability of cells to respond to growth inhibitory signals and forms the basis of multiple cancers. Rb1 deficiency in mice (Rb1−/−) was found to be embryonic-lethal, and each tissue expressing a high level of pRb showed evidence of not only continuous cell proliferation but also defective terminal differentiation (Macleod 1999). However, there is a lack of consensus on whether the differentiation failure is simply a consequence of the cell cycle defect or even a cell-nonautonomous effect. Moreover, the idea that the main function of pRB is regulation of E2F-dependent genes and cell cycle exit (Frolov and Dyson 2004) was appealing, as it might explain why pRb is so important for tumor suppression. However, analysis of Rb1−/− embryos that were rescued from embryonic lethality with either wild-type placentas or low levels of the Rb1 gene showed marked skeletal muscle defects at birth (Zacksenhaus et al. 1996; de Bruin et al. 2003; MacPherson et al. 2003; Wu et al. 2003), thus underscoring the functional importance of pRB as a regulator of muscle differentiation and development. Consistent with in vivo findings, Rb1−/− mouse embryonic fibroblasts (MEFs) did not respond to the forced expression of the myogenic differentiation antigen (MyoD) (Novitch et al. 1996). Lysine (K)-specific demethylase 5A (KDM5A; also known as RBP2 and JARID1A) was isolated in a genetic screen with pRB derivatives deficient in interaction with E2F but retaining the ability to induce differentiation, suggesting that induction of differentiation is an E2F-independent pRB function (Benevolenskaya et al. 2005). Importantly, pRB mutants that failed to form complexes with KDM5A were also unable to induce differentiation. Surprisingly, the loss of this nonmyogenic epigenetic factor was able to rescue the differentiation defect. Specifically, loss of Kdm5a by knockdown or knockout in cells defective in pRb was sufficient to resume expression of markers associated with senescence and myogenic or adipogenic differentiation, as it increased transcription factor (TF) activity and restored gene expression, thus phenocopying the reintroduction of wild-type pRb (Benevolenskaya et al. 2005; Lin et al. 2011). These studies highlight the importance of the RB/KDM5A branch of the pathway in regulating differentiation. However, associating specific genes and biological processes regulated by pRB and KDM5A with differentiation rescue would be critical for understanding the link between differentiation and tumor suppression. This would establish whether differentiation induction is independent of cell cycle regulation and might lead the way to the design of new strategies counteracting malignant transformation.
Results
Kdm5a loss in Rb-negative cells rescues differentiation but not permanent cell cycle withdrawal
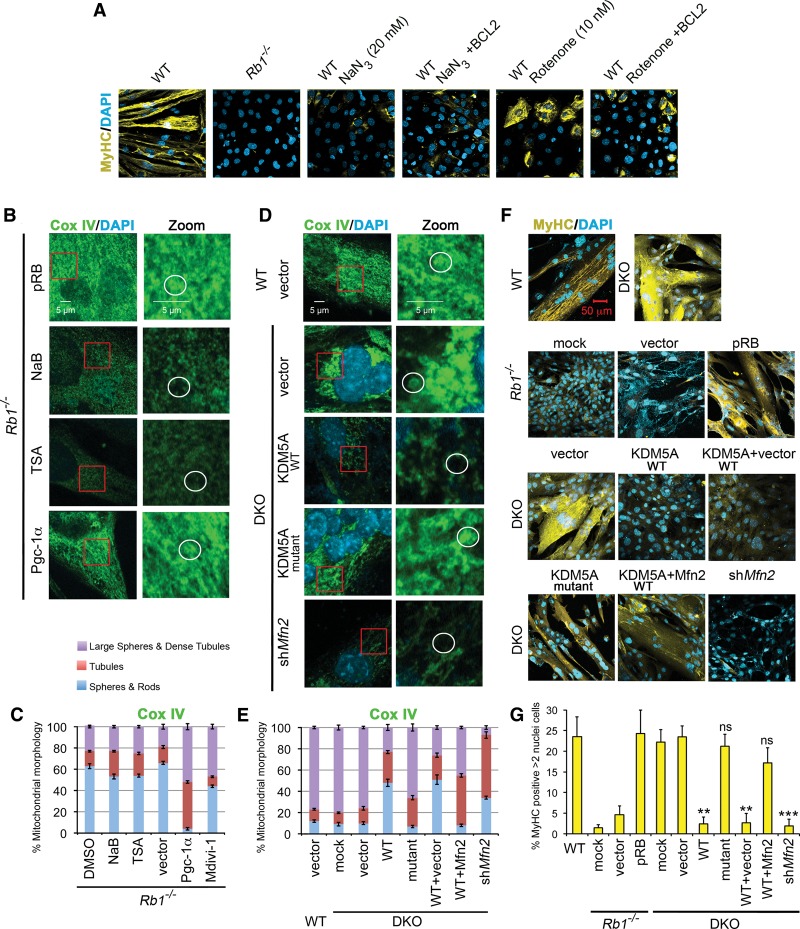
To study how pRB-mediated differentiation is rescued by Kdm5a loss, we adopted differentiation assays in MEFs isolated from Rb1 and Kdm5a knockout animals. MEFs were induced for myogenic differentiation using transduction with adenoviral (Adeno) or lentiviral (Lenti) MyoD and incubation in differentiation medium (DM) (Fig. 1A). A decrease in Kdm5a phenocopies reintroduction of pRB in myotube formation, as described previously by staining induced MEFs with DAPI and the late marker of myogenic differentiation myosin heavy chain (MyHC) (Benevolenskaya et al. 2005; Lin et al. 2011). However, pRb is essential for cell cycle exit in myoblasts, activating at least three distinct chromatin-based regulatory mechanisms (Blais et al. 2007; van Oevelen et al. 2008), and KDM5A cooperates with rather than opposes RB family protein complexes in repressing cell cycle genes during differentiation (Beshiri et al. 2012). Induced MEFs exhibit three hallmarks of differentiation: up-regulation of muscle gene expression, cell cycle arrest, and myoblast fusion with formation of multinucleated myotubes. To test whether Kdm5a loss fully phenocopies pRb during differentiation, cells were stained for MyHC and DAPI for detection of multinucleated myotubes and with EdU as an indicator of S-phase entry. Consistent with the dependence of myogenic differentiation on the Rb1 status (Novitch et al. 1996), the three hallmarks of myogenic differentiation were impaired in Rb1−/− MEFs (Fig. 1B). In particular, the MyHC protein level was negligible in Rb1−/− but highly elevated in the wild-type genotype (Fig. 1C). Transduction of Rb1−/− MEFs with a lentiviral pRB construct restored differentiation back to normal (Supplemental Fig. 1A–D). MEFs are mesenchymal stem cell-like multipotent progenitor cells capable of differentiation into several lineages, including bone, cartilage, adipose, and muscle. To test whether pRb function requires inhibition of Kdm5a during differentiation progression in cells committed to myogenic lineage, we used C2C12 cells, representing myogenic precursor cells. Transduction of C2C12 cells with Rb1 shRNAs blocked differentiation progression (Supplemental Fig. 1E–G). The block was released by simultaneous transduction with Kdm5a shRNA. Taken together, these experiments demonstrated that Kdm5a loss rescues Rb-dependent differentiation in cells with the properties of multipotent progenitors or committed progenitors.
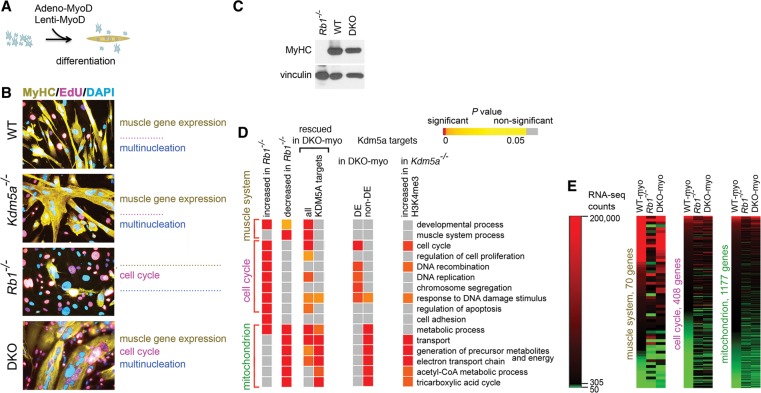
Figure 1.
Kdm5a loss in Rb-negative cells phenocopies specific functions of pRB. (A) Conversion of MEFs to myotubes using adenoviral or lentiviral expression of MyoD. Hereafter, this treatment is referred to as “induced” cells. (B) Induced MEFs show differences in muscle gene expression, cell cycle arrest, and multinucleation depending on Rb1 status. MEFs were isolated from wild-type, Kdm5a−/−, and double-knockout littermates and Rb1−/− embryos at the same genetic background as described previously (Lin et al. 2011). In order to convert them to myotubes, cells were first transduced with Adeno-MyoD and induced in DM for 96 h. To identify the differentiated cells, immunocytochemical (ICC) staining was performed for the late marker of differentiation MyHC. To assess permanent cell cycle withdrawal, cells were stimulated in high-serum medium and stained with nucleotide analog EdU and nuclear stain DAPI. Cells were visualized by confocal microscopy. (C) The expression level of MyHC is fully rescued in the double knockout. Immunoblot analysis of cell lysates prepared from MEFs of the indicated genotypes transduced with Lenti-MyoD and induced for 120 h. (D) Kdm5a loss results in rescued expression in gene groups associated with specific functions. Enrichment analysis for gene relations to gene ontology (GO) biological process terms is shown for the gene sets derived from an RNA sequencing (RNA-seq) experiment in induced MEFs with different Rb1 and Kdm5a statuses and from a chromatin immunoprecipitation (ChIP) combined with deep sequencing (ChIP-seq) assay in Kdm5af/f embryonic stem cells (for gene lists, see Supplemental Table 2) (this study; Beshiri et al. 2012). P-values are delineated in a colored heat map, where color-coding indicates the degree of significance: highest significance (red) to least significance (orange) and nonsignificant (values are in gray). Data are shown for GOs representing large gene sets (>20 genes) and with P-values of ≤10−5 in Rb1−/− MEFs and include all GOs that are also highly significantly enriched in rescued Kdm5a targets. (E) RNA-seq count values for genes in GO biological process terms “muscle system” and “cell cycle” and GO cellular location term “mitochondrion.” RNA-seq count values (see the Supplemental Material) are shown (for genes with ≥50 counts) as a heat map relative to the mean count value in wild type (WT-myo; 305 counts).
In order to identify molecular features of rescued myotubes, we performed an RNA sequencing (RNA-seq) experiment. To exclude the possibility that cells failing differentiation induction would obscure the results of analysis in total cell population, a pure population of myotubes was prepared using a size fractionation procedure (Supplemental Fig. 1H). RNA species for a majority of genes annotated to the gene ontology (GO) “muscle system process” were induced at a high level in both wild-type (WT-myo) and double knockout (Rb1−/− and Kdm5a−/−; DKO-myo) MEFs (Supplemental Table 1). In WT-myo, 34,000 RNA-seq counts were detected for the myogenin (Myog) gene, which is expressed early during myogenesis (Berkes and Tapscott 2005), and 169,000 counts were detected for the Myh3 gene expressed in embryonic muscle, which was the most abundant MyHC expressed in induced MEFs. In contrast, genes encoding transcriptional activators E2F1–3 or cyclins A and E reached only up to 700 counts, which was consistent with cell withdrawal from the cell cycle during differentiation. A comparison between induced Rb1−/− MEFs and WT-myo showed that 2766 genes were decreased at log2 0.76 in Rb1−/− MEFs. Strikingly, 2426 of these genes experienced more than half (log2 0.38) restoration of expression level in DKO-myo. To further define biological processes that were deregulated in the Rb1−/− genotype but rescued in DKO-myo, we performed GO enrichment analysis. Analysis of the 2766 genes that were decreased in Rb1−/− cells showed significant overrepresentation in the GO “muscle system process” (Fig. 1D; Supplemental Table 2). The expression levels for many muscle genes were similar in DKO-myo when compared with the WT-myo while different from Rb1−/− cells (Fig. 1E; Supplemental Table 1), suggesting that the myogenic transcription program was significantly rescued upon concomitant deletion of Kdm5a. Moreover, there was no delay in induction of differentiation markers in double-knockout MEFs. In fact, double-knockout cells showed more robust expression of myogenin than wild-type cells as early as 12 h after induction, and it persisted longer (Supplemental Fig. 2A–C). Chromatin immunoprecipitation (ChIP) experiments showed that MyoD was very highly enriched at the Myog promoter when compared with the control unbound intergenic region Igr2 at 24 h, which was similar to the time it is recruited to the Myog in mouse C2C12 myoblasts (Supplemental Fig. 2D,E).
While the muscle genes were significantly rescued in DKO-myo, cell cycle-related GOs were overrepresented among differentially expressed (DE) genes between DKO-myo and WT-myo (Fig. 1D). E2f1-3, cyclins, and other GO “cell cycle” genes were as highly expressed in double-knockout myotubes as in induced Rb1−/− fibroblasts (Fig. 1E; Supplemental Table 1). These data suggested that the deletion of Kdm5a in Rb1−/− MEFs rescued two hallmarks of myogenic differentiation; however, it did not rescue cell cycle withdrawal. Indeed, staining for EdU incorporation and the mitotic marker H3pS10 indicated that both induced Rb1−/− and double-knockout MEFs had active DNA synthesis and progressed through mitosis (Fig. 1B; Supplemental Fig. 3). In fact, there was a small increase in cell cycle entry in induced double-knockout compared with Rb1−/− MEFs, consistent with cooperative effects between KDM5A and pRB proteins (Beshiri et al. 2012), where pRB is likely playing a dominant role, as the loss of Kdm5a alone did not result in a cell cycle defect. This establishes KDM5A as a downstream target of pRB, regulating differentiation independently from cell cycle exit.
pRB is required for activation of genes encoding mitochondrial proteins
In order to distinguish direct and indirect consequences of Kdm5a loss, expression differences were studied in the genes with the locations closest to the Kdm5a peaks. As muscle gene and cell cycle gene expression coordinately changes during myogenic differentiation, we tested a set of developmental genes in which Kdm5a binds either proximal to the transcription start site (TSS) or in a distal regulatory region in proliferating cells and a set of cell cycle genes in which Kdm5a binds specifically during differentiation (Beshiri et al. 2012). RNA-seq and RT-qPCR analysis showed that none of the tested genes was rescued in double knockout and instead remained at the levels seen in Rb1−/− cells (see RNA-seq data GSE53528 for full gene list) (Fig. 2A). Some of these genes are related to myogenic differentiation, as MyoD bound to the same regions as Kdm5a in Ass1, Mstn, Gdf15, and Glis2 genes (when ENCODE/California Institute of Technology MyoD data from C2C12 myoblasts are compared with our ChIP-seq [ChIP combined with deep sequencing] data [GSE28343]). Conversely, the muscle genes that were decreased in Rb1−/− cells and significantly rescued in DKO-myo (Fig. 1E) are not known to be Kdm5a targets. Notably, the muscle genes were activated in the induced wild-type or double-knockout MEFs at least at the level of their activation in differentiating C2C12 myoblasts (Supplemental Figs. 4, 5A). In contrast to the more than log2 10-fold increase in expression of muscle genes, Kdm5a-bound developmental genes either intermittently or progressively decreased in expression during differentiation, falling far behind the median expression value, along with the Kdm5a/Rb/E2f targets Nusap1 and Kif2c (Supplemental Fig. 4B,C). Therefore, pRb-mediated differentiation does not require the activity of developmental genes occupied by Kdm5a.
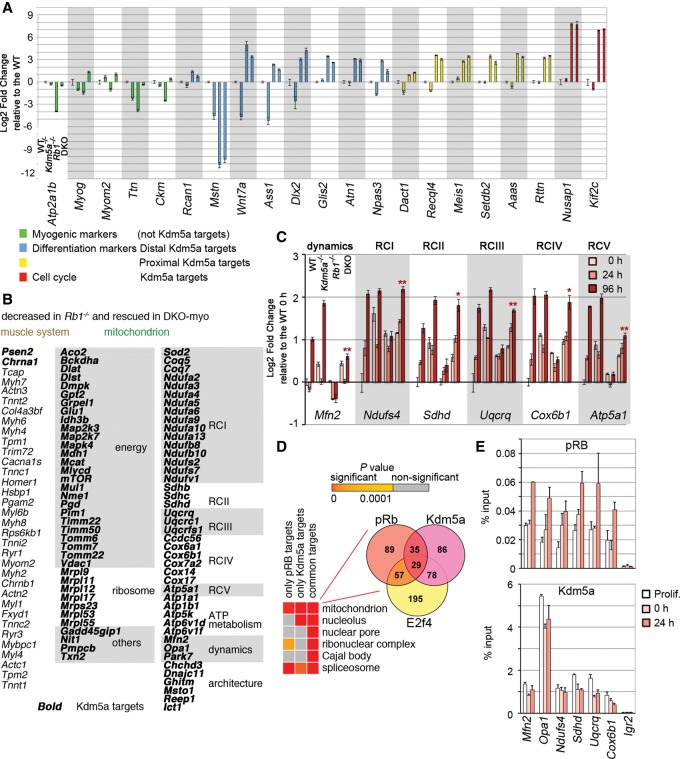
Figure 2.
Kdm5a loss in Rb-negative cells restores expression of muscle genes and Kdm5a targets with mitochondrial functions. (A) Myogenic markers but not differentiation and cell cycle genes occupied by Kdm5a are rescued in double knockout. MEFs of four different genotypes were induced for 72 h. The expression levels of the myogenic markers and Kdm5a targets (with Kdm5a location in a distal or proximal promoter region) with function in differentiation or cell cycle were determined by RT-qPCR analysis. The data were normalized to the Gapdh level and are presented relative to the wild type. Mean ± standard error of the mean (SEM) for n = 2 RT-qPCR replicates. (B) The list of rescued genes contains many Kdm5a targets in the GO “mitochondrion” but not in the “muscle system.” Genes in the mitochondrion are grouped based on their function: generation of energy, ribosome, respiratory chain complex I–V (RCI–V), ATP metabolism, mitochondrial dynamics, or architecture. (C) Activation of genes encoding mitochondrial components failed in Rb1−/− MEFs but were restored in double knockout back toward wild-type levels. MEFs were induced to differentiate for 0, 24, and 96 h. The expression of genes randomly chosen from the lists of Kdm5a targets with functions in the mitochondrion was analyzed at each time point in each genotype. RT-qPCR data was normalized to spike controls and is shown as a fold change relative to the wild type at 0 h for each gene. Mean ± SEM for n = 3 RT-qPCR replicates. The statistical significance for the condition double knockout compared with Rb1−/− at 96 h was determined by the Student's t-test: (*) P-value ≤ 0.005; (**) P-value ≤ 0.001. (D) Enrichment analysis reveals mitochondrion as the top GO term for common targets of pRb and Kdm5a. Top GOs are presented for overlapping genes, with P-values shown for both overlapping and nonoverlapping genes in a heat map. The overlap between pRb targets and Kdm5a targets is enriched in the GO term “mitochondrion.” A Venn diagram shows the number of genes in the GO “mitochondrion” between pRb and Kdm5a and with E2f4. (E) Genes with functions in the mitochondrion are direct targets of both pRB and Kdm5a. ChIP experiments in Rb1−/− MEFs that were transduced with Lenti-Flag-pRB and grown as proliferating, growth-arrested (0 h), or induced for 24 h. The ChIP antibodies used were Flag and anti-KDM5A. Igr2 is an intergenic control region. Mean ± SEM for n = 2 ChIPs.
We were surprised to find that the GOs that were most highly significantly overrepresented among rescued genes along with the muscle system related to functions in the mitochondrion (Fig. 1D). These included 80 genes related to the generation of precursor metabolites and energy and respiratory chain complex (RC) subunits and involved in ATP metabolism, mitochondrial dynamics, and architecture (Fig. 2B; Supplemental Table 1). Monitoring gene expression dynamics showed that genes encoding mitochondrial proteins that are bona fide Kdm5a targets increased in expression early during differentiation compared with the muscle signaling genes (e.g., Myog, Mef2c [myocyte enhancer factor 2c], and Ckm) and structural genes (e.g., Myh3, My6, Des, Tnni1, and Tnnc2) (Fig. 2C; Supplemental Fig. 5A,B; Supplemental Table 1). These genes represented not only myotube-specific isoforms such as the RC subunit Cox6a2, which displayed an almost log2 sevenfold rescue in DKO-myo, but also the ubiquitously expressed subunits. For example, the gene involved in mitochondrial fusion (Mfn2) and genes that we tested for each of the RCs exhibited increased expression levels in all induced MEFs except for Rb1−/− (Fig. 2C). Notably, the increase was also seen at the protein level, as exemplified by the Mfn2 increase in induced C2C12 cells and all MEFs except for Rb1−/− (Supplemental Fig. 5C,D). The decrease in the Mfn2 protein level was relevant to the pRb/Kdm5a axis, as Kdm5a shRNA mitigated the effect of acute loss of pRb by increasing Mfn2 expression (Supplemental Fig. 1G). To confirm that activation of genes encoding mitochondrial proteins accompanies differentiation into other cell types besides myotubes, MEFs were also induced to undergo adipogenic differentiation. Double-knockout MEFs displayed rescued Mfn2 and RC gene expression at 24 h of differentiation, paralleling expression changes of adipogenesis markers Agpat2, Fabp4, and Cebpα and followed by lipid accumulation (Supplemental Fig. 6). These data highlighted that mitochondrial function was central to the cell type-specific gene expression and differentiation rescue induced by the Kdm5a loss.
In order to identify direct effectors of pRb-mediated differentiation, we analyzed the whole-genome distribution of pRB (Chicas et al. 2010) with respect to Kdm5a distribution (Beshiri et al. 2012). Kdm5a occupied 651 pRB target genes (χ2 = 294.27; P < 10−16). A statistical test for binding of both proteins at the promoter and 5′ untranslated regions (UTRs) showed a P-value of <10−322, suggesting that pRb and Kdm5a are frequently colocalized at promoters. When enrichment analysis was run on the overlap, we observed that the top enriched (P < 10−15) GO was “mitochondrion” (Fig. 2D; Supplemental Table 3). Consistent with the observation that almost one-third of Kdm5a targets are shared with E2f4 (Beshiri et al. 2012), some mitochondrial genes were occupied by E2f4. ChIP experiments in induced MEFs showed that pRb becomes enriched at TSSs of the genes involved in mitochondrial dynamics (Mfn2 and Opa1) as well as genes encoding RC subunits, while Kdm5a binding to some of these genes decreased concomitantly with pRb recruitment (Fig. 2E). These results demonstrate that Kdm5a directly regulates genes encoding mitochondrial proteins and suggests that, during differentiation, a decrease in Kdm5a binding activates these genes.
Kdm5a catalytic activity is involved in inhibition of myogenic differentiation
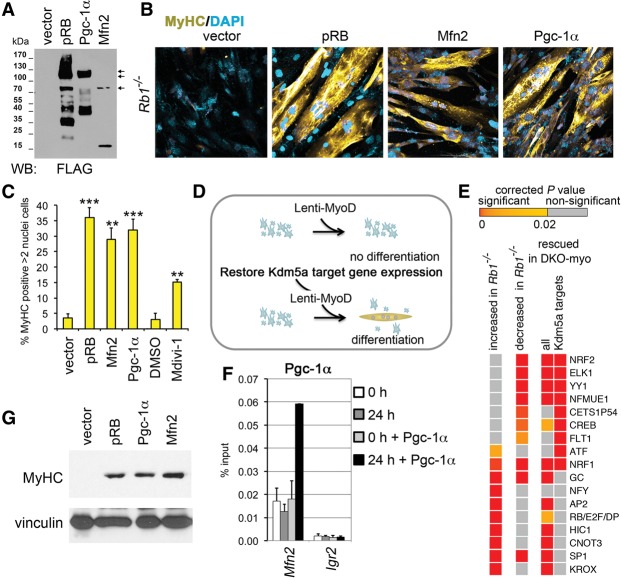
To test whether KDM5A inhibition of differentiation is dependent on KDM5A demethylase activity, we examined the double-knockout cells transduced with a lentiviral construct expressing wild-type or catalytic domain mutant KDM5A. The mutant was similar to the wild type in expression level and was properly localized to the nucleus (Fig. 3A; Supplemental Fig. 7). Only the cells with reintroduced wild-type KDM5A evaded their ability to differentiate and behaved similar to Rb1−/− MEFs (Fig. 3B–D; Supplemental Fig. 7B,C). Transduction with wild-type KDM5A, but not with the catalytic domain or frameshift KDM5A mutants, resulted in highly significantly decreased numbers of MyHC- and EdU-positive cells and loss of expression of myogenic genes Ckm, Tnnc2, and Tnni2. As the double-knockout MEFs have an increased global level of H3K4 methylation compared with the MEFs with functional Kdm5a (Beshiri et al. 2012), one possibility is that inducing more active chromatin generally would be sufficient to promote differentiation. However, histone deacetylase (HDAC) inhibitors sodium butyrate and TSA were unable to rescue differentiation defects in Rb1−/− MEFs (Fig. 3E). These experiments directly linked the inhibitory KDM5A function in differentiation to its demethylase activity.
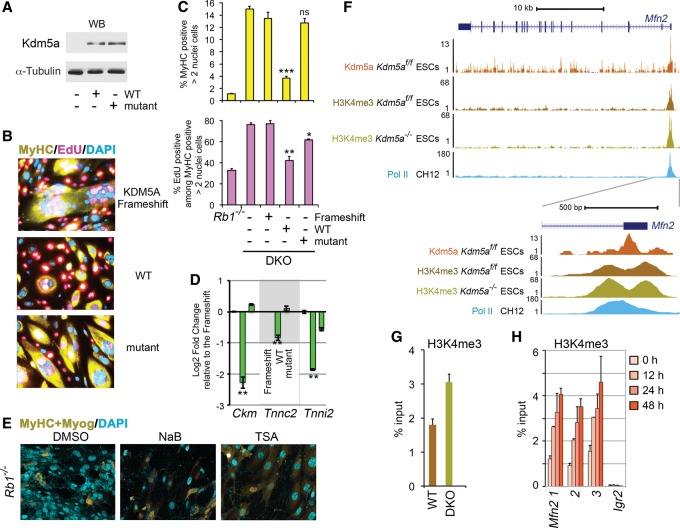
Figure 3.
Kdm5a catalytic activity is required for the inhibition of myogenic differentiation. (A) Expression level of KDM5A derivatives. The immunoblots were probed with the anti-KDM5A antibody and with α-tubulin antibody as a loading control. (B) KDM5A inhibition of differentiation is dependent on its demethylase activity. ICC was performed as in Figure 1B in induced double-knockout cells that were transduced with different KDM5A derivatives, including the frameshift control. (C) Analysis of MyHC expression and cell cycle arrest in double-knockout cells prepared as in B. Quantification of MyHC-positive cells and cells that re-entered the cell cycle, marked by EdU, was performed relative to the total number of cells. In order to avoid artifacts of chromosome segregation (Manning et al. 2010) and nuclear duplication, which is common to Rb-deficient myocytes (Ciavarra et al. 2011), only MyHC-positive cells containing more than two nuclei and lacking obvious aneuploidy were counted. Mean ± SEM for n = 4 replicate wells from three representative experiments is shown. (ns) P > 0.1; (*) P < 0.02; (**) P < 0.001; (***) P < 0.0001, calculated relative to the frameshift control. (D) Muscle gene expression is dependent on KDM5A demethylase activity. Double-knockout cells expressing different KDM5A derivatives were induced for 72 h. RNA was isolated, and transcript levels were determined relative to the frameshift control by RT-qPCR. The data are presented as the mean ± standard deviation (SD) of RT-qPCR replicates (n = 2) from a representative experiment. (**) P value ≤ 0.005 is shown relative to the frameshift control. Gene expression was normalized relative to the geometrical mean of two spike controls. (E) Inducing more active chromatin with HDAC inhibitors does not rescue differentiation. Rb1−/− cells were induced in the presence of 5 mM sodium butyrate or 50 nM TSA for the first 24 h and in DM for the total of 72 h and were compared with the control DMSO-treated cells. (F) Kdm5a loss increases H3K4me3 at the Mfn2 TSS region. Genome browser view of the Mfn2 gene region at 10-kb and 500-base-pair (bp) scale. The TSS region is depicted by RNA polymerase II (Pol II) and Kdm5a occupancy and H3K4me3 enrichment. (G) H3K4me3 enrichment is increased at the Mfn2 TSS Kdm5a peak in double-knockout compared with wild-type MEFs before induction of differentiation. ChIP was performed in 0-h MEFs. (H) The Mfn2 TSS region displays increasing H3K4me3 enrichment after the induction of myogenic differentiation in MEFs. ChIP was performed in MEFs transduced as in Figure 2E, and data are shown for three amplicons (1, 2, and 3) covering the Mfn2 TSS region, as presented at 500-bp scale in F. In G and H, mean ± SEM for n = 2 ChIP assays.
Kdm5a-binding peaks lie within TSS regions overlapping with H3K4me3 peaks (Beshiri et al. 2012). As investigating differences in H3K4me3 around Kdm5a peaks is informative for identification of the genes involved, we performed a H3K4me3 ChIP-seq assay in Kdm5af/f embryonic stem cells with deleted flox/flox alleles and the cells without a deletion. Enrichment analysis of genes in regions demonstrating increased H3K4me3 levels in cells with Kdm5a deletion showed that, again, the mitochondrial functions were highly enriched (Fig. 1D). The Mfn2 TSS region showed a peak of Kdm5a correlating with enriched H3K4me3, which further increased (1.4-fold change, P < 10−4) in Kdm5a−/− cells (Fig. 3F). Quantitation of H3K4me3 enrichment at the Kdm5a peak using ChIP-qPCR experiments showed an increase in MEFs lacking Kdm5a even in uninduced condition (Fig. 3G), correlating with an increase in the Mfn2 transcript level (Fig. 2C). This result is consistent with the data that MFN2 promoter activity is regulated by KDM5A through H3K4me3 demethylation in human osteosarcoma SAOS-2 cells (Lopez-Bigas et al. 2008). ChIP-qPCR experiments using three different primer pairs in the Mfn2 TSS regions showed that the H3K4me3 enrichment increased in a broad genomic region as early as 12 h in induced MEFs (Fig. 3H). Therefore, pRb and Kdm5a binding to genes encoding mitochondrial proteins likely plays a regulatory role during differentiation.
Kdm5a loss rescues the mitochondrial defect
Consistent with the gene expression changes, MEFs of the four genotypes undergoing differentiation displayed profound differences in mitochondrial mass and morphology. Analysis of the immunocytochemical (ICC) confocal microscopy images for the cytochrome c oxidase (RCIV) component Cox IV as well as the electron microscopy (EM) data revealed an increased number but decreased size of mitochondria in Rb1−/− cells (Fig. 4A,B). The defect was especially prominent in the induced condition, when mitochondrial mass and morphology (spheres/rods and tubules versus dense tubules and large spheres) were visualized by ICC for Cox IV or staining with MitoTracker, which covalently binds to mitochondrial proteins (Supplemental Fig. 8). This suggested that mitochondrial networking is affected by Rb1 loss, resulting in mitochondrial fragmentation. On the other hand, concomitant loss of Kdm5a rescued the observed changes in mitochondrial mass and morphology (Fig. 4A,B; Supplemental Figs. 7D, 8). To further characterize the difference in mitochondria between the Rb1−/− and other genotypes, the mitochondrial protein levels were studied by immunoblot analysis. The levels of Cox IV and Mfn2 were very low in induced Rb1−/− cells compared with the wild type but increased back to the normal level in double knockout (Fig. 4C; Supplemental Fig. 5D). The difference between Rb1−/− and double knockout was observed in the level of proteins encoding Kdm5a target genes—i.e., Cox IV and Mfn2—but not in proteins encoding genes not bound by Kdm5a, such as the outer mitochondrial membrane protein Vdac1. It should also be pointed out that only specific mitochondrial processes were rescued in double knockout (Fig. 2B). For example, genes associated with apoptosis or glutamine metabolism—such as manganese superoxidase dismutase sod2, known to protect cells against oxidative stress, or the transglutaminase tgm2—were not rescued (see RNA-seq data GSE53528 for a full gene list).
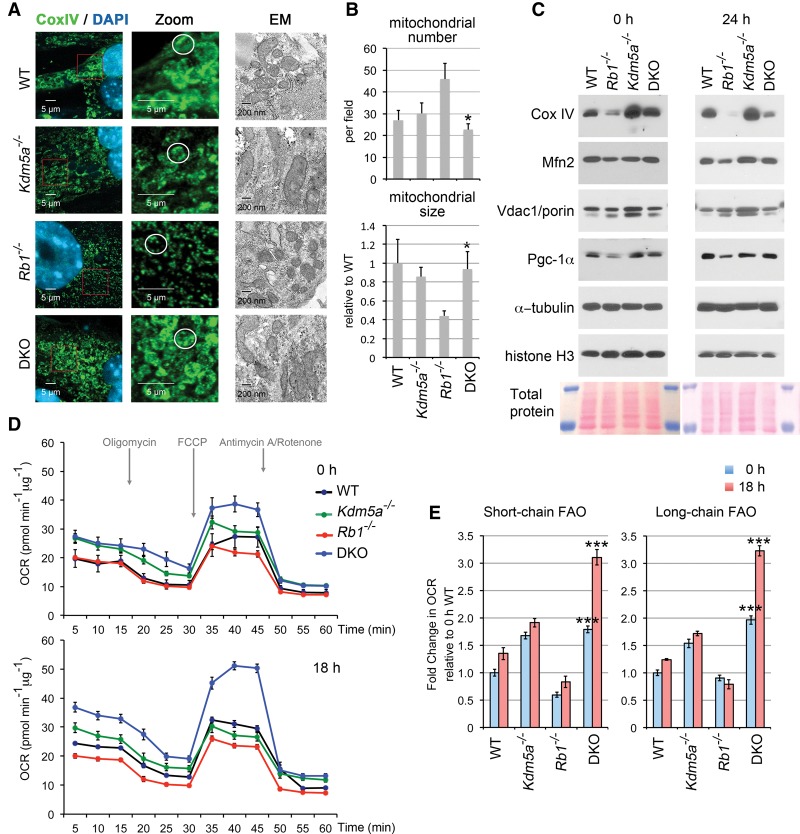
Figure 4.
Rescue of mitochondrial morphology and mitochondrial functions with Kdm5a knockout. (A) Comparison of induced MEFs of different genotypes reveals pRb and Kdm5a function in the mitochondrion. ICC for the RC component Cox IV and EM images are shown for four different genotypes. A representative mitochondrial structure for each class is circled in the zoomed panels. MEFs containing Lenti-CMV-MyoDER[T] (MyoDER[T]) lentiviruses were induced with 4-hydroxytamoxifen (OHT) for 72 h. (B) Quantitation of mitochondrial number and area (size) based on EM data. Mean ± SEM for n ≥ 5 EM images (>100 mitochondria per genotype). (*) P < 0.05 relative to Rb1−/−. (C) Selective mitochondrial components are rescued in induced double-knockout MEFs to the wild-type level. Immunoblot analysis was performed with the indicated antibodies from total cell extracts prepared from cells of four genotypes induced for 24 h and compared with the uninduced condition (0 h). (D) Kinetics of the OCR response to oligomycin to determine ATP-coupled respiration, FCCP to establish maximal respiratory capacity, and a mixture of antimycin and rotenone to determine mitochondrial respiration. MEFs of four genotypes were transduced with Lenti-empty vector (0 h) or Lenti-MyoD and induced for 18 h. Assays were performed using the XFe96 extracellular flux analyzer. Upon the completion of an assay, cells were lysed, and protein content was determined. Data were normalized to the protein content. (E) Substrate utilization in cells treated for 0 h and 18 h. The difference in OCR was measured acutely after injection of the short chain fatty acid butyrate or the long chain fatty acid palmitate by comparing the fourth and third measurements. Assays were performed as in D, and data are presented as a fold change relative to the 0-h wild-type MEFs. (***) P < 0.0001 is shown for double knockout relative to the Rb1−/−. In D and E, mean ± SEM for n ≥ 4 replicate wells.
To assess whether double-knockout MEFs have appropriately assembled RCs capable of oxidative phosphorylation, oxygen consumption was measured using a Seahorse Bioscience extracellular flux analyzer. A comparison of induced MEFs of four different genotypes showed that both the basal oxygen consumption rate and the maximal respiratory capacity as determined by addition of the uncoupler FCCP increased in all differentiating MEFs in contrast to Rb1−/− MEFs (Fig. 4D). The basal respiration was higher in all cells lacking Kdm5a, consistent with the increased levels of genes encoding mitochondrial proteins (Figs. 2C, 4C). The DKO-myo cells had a markedly high maximal respiratory capacity upon induction, thus demonstrating that loss of Kdm5a not only reversed the depletion of the mitochondrial reserve by Rb1 loss but even enhanced the respiratory reserve capacity. In a glycolysis stress test, loss of Kdm5a resulted in a higher basal extracellular acidification rate (ECAR), and glucose addition triggered a further increase (Supplemental Fig. 9A). However, glycolytic capacity decreased in induced wild-type and Kdm5a−/− cells to the Rb1−/− level, suggesting that it is unlikely to be a major contributor to the differentiation rescue. The ECAR increase indicated that added glucose metabolized to lactate. We inhibited oxidative phosphorylation with oligomycin to determine the contribution of TCA cycle-derived CO2 in nonglycolytic acidification. Oligomycin further increased glycolytic capacity, and it was abolished by the addition of the inhibitor of glycolysis 2-DG. Interestingly, the glycolytic capacity of double knockout was much higher than those of other genotypes, with basal levels of both glycolysis and glycolytic reserve after 2-DG injection persisting at the increased (P-value < 0.0001) level. In the glucose oxidation test, the difference between the four genotypes (Supplemental Fig. 9B) reflected the difference observed in the basal respiration rates (Fig. 4D), as addition of glucose failed to increase the oxygen consumption rate (OCR) (data not shown). These results suggest that glycolysis accounts for the majority of the ECAR observed in MEFs and profoundly decreases in differentiating cells to the level observed in cells incapable of differentiation. Next, we asked whether the Kdm5a-dependent increase in the OCR was due to differences in utilization of fatty acids of various chain lenghs, the major nutrient substrates in muscle tissue. We did not see OCR induction after glucose addition (data not shown), suggesting that none of our MEFs actively oxidize glucose. In contrast, MEFs exhibited active oxidation of fatty acids (FAO), as addition of butyrate or palmitate caused an increase in the OCR (Fig. 4E). When we compared the fold change in the OCR, we found that these fatty acids exhibited lower levels of oxidation in Rb1−/− MEFs. Metformin favors the mitochondrial FAO, and palmitate oxidation in differentiating C2C12 cells was increased after metformin treatment (Supplemental Fig. 9C). An increase in the OCR in response to acute palmitate addition was also specifically enhanced by metformin in Kdm5a−/− cells (Supplemental Fig. 9C). Metformin treatment resulted in the anticipated activation of AMP-activated protein kinase (AMPK), as evidenced by marked induction in one of its downstream targets, acetyl-CoA carboxylase (ACC) (Supplemental Fig. 9D). These findings indicate that one of the key metabolic consequences of Kdm5a loss during myogenesis is the propensity to increase FAO. Therefore, myogenic differentiation correlates with the increase in specific mitochondrial components and a shift to oxidative phosphorylation and oxidation of specific nutrients, a process that is impaired in Rb1−/− cells but restored in double-knockout cells.
Intact mitochondrial function is intimately linked to differentiation potential
The top GO terms identified during myogenesis were muscle system and mitochondrion (Fig. 1D), distinguishing double-knockout MEFs, which are proficient in differentiation, oxidative phosphorylation, and activity of the RCs, from Rb1−/− MEFs (Figs. 1B, 4D). Was the rescue of mitochondrial function a consequence or the cause of restoring myogenic differentiation? The intact mitochondrial functions are essential for myogenic differentiation in MEFs, as inhibitors of RCI rotenone, metformin, and phenformin as well as the inhibitor of RCIV sodium azide blocked myogenesis (Fig. 5A). The effect was not due to apoptosis, as inhibition of apoptosis with BCL2 still failed induction of differentiation in MEFs treated with the inhibitors (Fig. 5A). Moreover, treatments with phenformin and metformin, nontoxic RCI inhibitors, did not induce apoptosis, as assessed by staining for cleaved Caspase 3 (Supplemental Fig. 9E). Although the effect of phenformin and metformin was weaker than the effect of rotenone, it still resulted in significant differentiation inhibition (Supplemental Fig. 9E,F). These results showed that functional mitochondria are necessary for myogenic differentiation.
Figure 5.
Intact mitochondrial function is necessary for differentiation. (A) Mitochondrial “poisons” inhibit differentiation. Rotenone blocks the ubiquinone-binding pocket of RCI, disrupting the first step in the RC reaction. Sodium azide inhibits RCIV by binding to the heme cofactor of cytochrome c. All cells were transduced with Adeno-MyoD and induced for 96 h. To rescue apoptosis, cells were transduced with Adeno-BCL2 where indicated. (B) Pgc-1α (peroxisome proliferator-activated receptor γ-coactivator 1α; also called PPARGC1A) rescues the mitochondrial defect in Rb1−/− cells. Cox IV staining was performed in Rb1−/− cells that were transduced with lentiviral pRB or Pgc-1α constructs or treated with HDAC inhibitors. A representative mitochondrial structure for each class is circled in the zoomed panels. (C) Quantitation of mitochondrial morphology in Rb1−/− MEFs. Mean ± SD for n = 5 microscopic fields (∼100 cells per experiment) in two representative experiments. Mitochondrial morphology is changed toward tubular morphology and large spheres in Rb1−/− MEFs transduced with Pgc-1α lentiviruses. Inhibition of Drp1 by treatment with the inhibitor Mdivi-1 results in changes toward tubular morphology and large spheres. (D) Loss of Mfn2 by RNAi decreases the representation of large sphere/dense tubule mitochondria in induced double-knockout MEFs, similar to the overexpression of KDM5A. Wild-type cells and double-knockout cells with overexpression of the KDM5A demethylase mutant served as controls. (E) Quantitation of mitochondrial morphology in double-knockout MEFs. Double-knockout MEFs were transduced with lentiviruses expressing KDM5A derivatives, Mfn2, or shRNA to Mfn2. Mean ± SD for n = 5 microscopic fields (∼100 cells per experiment) in two representative experiments. (F) Knockdown of Mfn2 abrogates differentiation rescue in double knockout, while Mfn2 overexpression rescues differentiation from KDM5A inhibition. In B–F, MEFs were transduced with MyoDER[T] and analyzed after a 72-h OHT treatment. All images were generated by confocal microscopy. (G) Comparison of MyHC expression levels. Rb1−/− cells prepared as in F were analyzed after a 72-h treatment with OHT, and wild-type and double-knockout cells were analyzed after a 48-h treatment with OHT. (ns) P > 0.1; (**) P < 0.001; (***) P < 0.0001, relative to the double-knockout vector control. Cells were analyzed for MyHC or Cox IV expression by ICC and confocal microscopy.
Reintroduction of pRB in Rb1−/− cells rescued not only MyHC expression but also the mitochondrial phenotype (Fig. 5B,C). In contrast, introduction of KDM5A with intact demethylase function in double-knockout cells decreased the number of not only differentiated cells but also cells with normal mitochondria (Fig. 5D–G). Treatment of Rb1−/− cells with HDAC inhibitors had no effect (Fig. 5B,C). These experiments demonstrated that it was possible to correct the mitochondrial morphology along with promoting differentiation in Rb1−/− cells.
Mitochondrial morphology is determined by the balance between mitochondrial fusion and fission. The dense tubule networks and large spheres, which were observed in induced double-knockout compared with Rb1−/− MEFs (Fig. 4A), recognize a preference of fusion events (Chan 2012). Mfn2, a GTPase of the outer mitochondrial membrane, is commonly known for its regulatory role in mitochondrial fusion. Mfn2 knockdown affected the prevalent mitochondrial phenotypes correlating with differentiation rescue, as transduction of double-knockout MEFs with shMfn2 lentiviruses reduced Cox IV staining, which was similar to the KDM5A reintroduction (Fig. 5D–G). In contrast, cotransduction of KDM5A and Mfn2 lentiviruses restored the number of dense tubules as well as differentiation, further suggesting that KDM5A inhibits Mfn2 function to repress differentiation. Excessive mitochondrial fission is common in human lung cancer cells, where it was shown to be due to imbalance in the expression of Mfn2 and the main regulator of mitochondrial fission Drp1 (Rehman et al. 2012). Therefore, as Mfn2 is down-regulated in Rb1−/− cells (Fig. 4C), inhibition of Drp1 was attempted. Treatment of induced Rb1−/− cells with the Drp1 inhibitor Mdivi-1 increased large sphere/dense tubule formation (Fig. 5C). These findings demonstrate that Rb1−/− cells are responsive to modulation of mitochondrial fusion, and it is possible to at least partially restore the balance between mitochondrial fusion and fission toward the wild type.
Increased mitochondrial function rescues differentiation
We hypothesized that there is an intimate link between the failure of mitochondrial function and the failure of differentiation in Rb1−/− cells. Rb1−/− MEFs transduced with Mfn2 lentiviruses were tested for MyHC gene expression. Mfn2 significantly rescued MyHC when compared with the empty vector and reintroduced pRB (Figs. 5F,G, 6A–C). Treatment with Mdivi-1 also increased MyHC expression, while DMSO treatment had no effect (Fig. 6C). In contrast, shMfn2 resulted in impaired differentiation in double-knockout MEFs (Fig. 5F,G). As overexpression of Mfn2, a regulator of many other mitochondrial functions besides fusion, was sufficient for differentiation rescue, it is possible that redirecting energy metabolism away from glycolysis and toward the Krebs cycle and mitochondrial oxidative phosphorylation would have the same effect. However, treatment with inhibitors of glycolysis or with selective peroxisome proliferator-activated receptor agonists (see the Supplemental Material) failed to induce MyHC expression. Thus, rescue of pRb-mediated differentiation specifically requires rescue of mitochondrial functions.
Figure 6.
Increased mitochondrial function rescues differentiation. (A) Immunoblot analysis of lysates prepared from cells expressing Flag-tagged proteins pRB, Mfn2, and Pgc-1α. (B) Differentiation rescue with mitochondrial regulators. Images of ICC for MyHC were generated using confocal microscopy. Rb1−/− MEFs containing MyoDER[T] lentiviruses were transduced with lentiviral vectors expressing pRB, Mfn2, and Pgc-1α proteins as in A or with the empty vector and induced with OHT. (C) Cells with increased mitochondrial function show high MyHC expression. MEFs containing MyoDER[T] were transduced with lentiviruses expressing the mitochondrial regulators or treated with 30 µM Drp1 inhibitor Mdivi-1. Empty vector or DMSO treatment was used as a control. Cells were analyzed after a 72-h treatment with OHT. Mean ± SD for the number of MyHC-positive cells with more than two nuclei in n = 5 microscopic fields (∼100 nuclei per field) in three representative experiments; (**)P < 0.001; (***) P < 0.0001, relative to the vector. (D) Schematics of myogenic differentiation rescue. Rb1−/− cells carrying Lenti-MyoD (or MyoDER[T]) were transduced with lentiviruses that express the mitochondrial regulators, restoring the expression level of Kdm5a targets. (E) Occurrences of TF-binding sites (TFBSs) in the promoter regions of Kdm5a targets. Enriched TFBSs with corrected P-values of <10−16 are shown in the heat map, arranged in the order of significance for genes increased in Rb1−/− and for the Kdm5a target genes rescued in DKO-myo. The whole list of enriched TFBSs with P < 0.02 is shown in Supplemental Table 4. (F) Pgc-1α specifically binds to the Mfn2 promoter during differentiation. ChIP experiments in Rb1−/− cells that were transduced with Pgc-1α lentiviruses and analyzed at 0 h and 24 h after induction. Igr2 is an intergenic control region. Mean ± SEM for n = 2 ChIP assays. (G) Expression level of MyHC is fully rescued by mitochondrial regulators. Immunoblot analysis of cell lysates prepared from Rb1−/− MEFs transduced with Lenti-MyoD and lentiviral vectors expressing pRB, Mfn2, or Pgc1α and induced for 120 h.
As loss of Kdm5a, which directly inhibits multiple genes encoding mitochondrial proteins, rescued differentiation, we attempted to identify an alternative approach to restore this subset of Kdm5a targets through their ultimate regulatory transcriptional activator (Fig. 6D). In order to identify such a factor, we analyzed the TSSs of Kdm5a targets for possible occurrences of TF-binding motifs. Rb/E2f/Dp sites were enriched at cell cycle genes increased in Rb1−/− cells compared with the wild type, while muscle initiator sequence MINI, a motif commonly found in muscle genes, was characteristic of the genes decreased in Rb1−/− cells but not of the Kdm5a targets (Fig. 6E; Supplemental Table 4) as expected. Binding sites for eight different TFs—including Nrf1 and Nrf2 (major regulators of genes involved in mitochondrial biogenesis and oxidative phosphorylation), cAMP response element-binding protein (CREB), and Yin Yang 1 (Yy1)—were highly significantly enriched at the genes decreased in Rb1−/− cells as well as at the promoters of Kdm5a targets (P < 10−16) that were rescued in DKO-myo.
The activity of Nrf1, Nrf2, CREB, and Yy1 can be dramatically increased by direct interaction with peroxisome proliferator-activated receptor γ-coactivator 1α (PPARGC1A/PGC-1α) (Liang and Ward 2006; Blättler et al. 2012). Pgc-1α activity determines mitochondrial gene expression signature in differentiating mouse myoblast or intact muscle cells (Rangwala et al. 2010). In particular, the Mfn2 and Cox IV genes are regulated by Pgc-1, and mice with knockout of Pgc-1β display reduced mitochondrial size, which was associated with reduced Mfn2 expression (Puigserver et al. 1998; Liesa et al. 2008). Pgc-1α and Pgc-1β stimulate mitochondrial biogenesis and respiration in muscle cells (Liesa et al. 2008; Wenz et al. 2008). Both Pgc-1 proteins were expressed in induced MEFs, with about log2 1.6-fold difference in the Pgc-1α transcript in Rb1−/− compared with the double knockout and wild type (RNA-seq data GSE53528). Therefore, we reasoned that restoration of KDM5A target gene expression may be achieved through overexpression of the common coactivator of TFs identified in our TF-binding site (TFBS) study (Fig. 6E). Transduction of Rb1−/− MEFs with Pgc-1α lentiviruses increased the number of cells with tubular mitochondria (Fig. 5C), suggesting that Pgc-1α overexpression is sufficient for mitochondrial morphology rescue. In fact, Pgc-1α binding to the Mfn2 promoter highly enriched after induction of differentiation (Fig. 6F). Remarkably, when the multinucleation and expression of MyHC was examined in cells with Pgc-1α overexpression, it was very distinctive from the cells transduced with the control vector (Fig. 6B,C). Both Pgc-1α and Mfn2 phenocopied reintroduction of pRB in the level of MyHC expression (Fig. 6G). Therefore, overexpression of two different regulators that control a broad range of mitochondrial functions—mitofusin Mfn2 and the transcriptional coactivator Pgc-1α—restored the Rb-dependent differentiation.
Growth of human RB-negative cell lines is responsive to KDM5A and PGC-1α
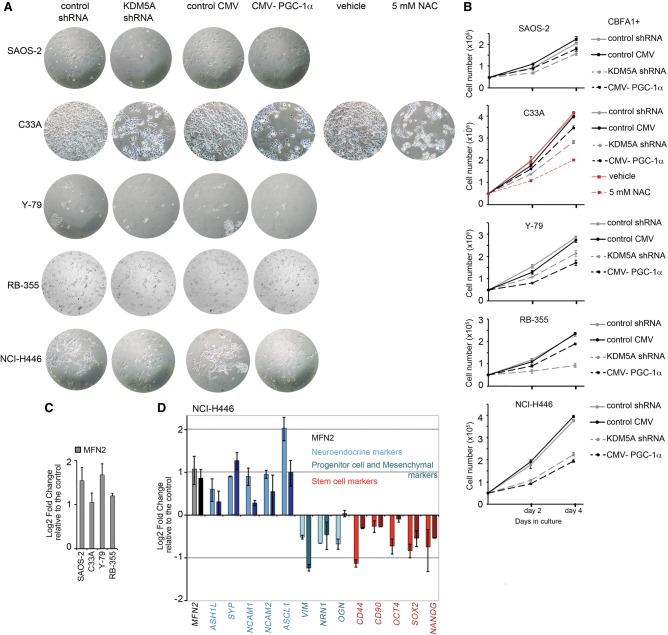
The obtained results predict that induction of mitochondrial functions through epigenetic or metabolic modulation will drive differentiation in RB1−/− cancer cells. We tested this idea by performing experiments on KDM5A knockdown by shRNA and Pgc-1α overexpression in cell lines that were established from five different human tumors with inactivated pRB function (Supplemental Fig. 10). The analysis included the Y79 cell line derived from a 2-yr-old girl with familial retinoblastoma and the RB-355 cell line derived from a nonfamilial unilaterally affected child. Remarkably, reduced proliferation was evident in all cancer cell lines that were transduced with KDM5A or Pgc-1α lentiviruses but not with the control lentiviruses (Fig. 7A,B). Osteosarcoma cell line SAOS-2, cervical carcinoma cell line C33A, and small-cell lung cancer (SCLC) cell line NCI-H446 displayed significant morphological changes with both KDM5A knockdown and Pgc-1α overexpression (Fig. 6A). This was consistent with the previous data, as SAOS-2 cells can be differentiated by a master regulator of osteoblast differentiation (RUNX2/CBFA1) along with reintroduction of pRB (Thomas et al. 2001) or KDM5A knockdown (Benevolenskaya et al. 2005). As a control for the C33A cell line, we used treatment with the antioxidant N-acetyl L-cysteine (NAC), which is known to induce keratinocyte differentiation in colon and ovarian cancer cell lines (Parasassi et al. 2005).
Figure 7.
Growth of human RB-negative cell lines is responsive to KDM5A RNA inhibition and Pgc-1α overexpression. (A) Growth inhibition in RB-negative cell lines. SAOS-2 osteosarcoma, C33A cervical carcinoma, Y79 and RB-355 retinoblastoma cell lines, and the SCLC cell line H446 were transduced with lentiviruses encoding KDM5A shRNA and Pgc-1α protein or with control lentiviruses. The NAC treatment, which was claimed to promote differentiation in several cancer cell models (Parasassi et al. 2005), was performed as another control along with the DMSO (vehicle) treatment. SAOS-2 were transfected with the master regulator of osteoblast differentiation RUNX2/CBFA1 in order to induce the osteoblast differentiation (Thomas et al. 2001; Benevolenskaya et al. 2005). Representative images were acquired using a Zeiss microscope. (B) Proliferation assay for cells transduced with KDM5A shRNA or Pgc-1α lentiviruses. Cell counting was performed at 2 and 4 d after plating of 5 × 104 transduced cells. Mean ± SD from two replicate wells. (C) MFN2 mRNA level is up-regulated in KDM5A shRNA-treated cells. (D) Gene expression changes in the levels of specific mRNAs reflecting self-renewal and differentiation status and of MFN2 mRNA in NCI-H446. Data for KDM5A shRNA-treated cells are in light colors, and data for cells transduced with Pgc-1α lentiviruses are in dark colors. In C and D, gene expression was normalized relative to the POLRIIA level and is shown compared with the control lentiviruses as mean ± SEM for n = 2 biological replicates.
To evaluate cell differentiation potential and mitochondrial status, we performed RT-qPCR analysis from transduced cells. As Mfn2 expression correlates with the promotion of Rb-mediated myogenic differentiation in MEFs, we used it as a readout of mitochondrial dysfunction in RB1−/− cancer cells. KDM5A shRNA significantly increased the MFN2 level in all examined cell lines (Fig. 7C,D). The NCI-H446 cell line contains subpopulations of neuroendocrine cells and cells with stem cell properties (Calbo et al. 2011). The levels of specific mRNAs encoding neuronal, progenitor cell, and mesenchymal markers as well as self-renewal genes was significantly changed in cells with KDM5A shRNA and Pgc-1α overexpression (Fig. 7D). Thus, KDM5A and Pgc-1α affect cell growth of RB-deficient human cancer cells, and decreasing heterogeneity of SCLC cell population indicates a shift to a more differentiated phenotype.
Discussion
The inability of cells to execute their full differentiation potential is a hallmark of cancer. Despite the existence of multiple cancer subtypes, the pRB tumor suppressor function is lost in the majority of human cancers, suggesting that pRB carries a number of essential functions that are shared by most tumors. Here we describe a pRB function in promoting differentiation that is independent of its well-established role in the cell cycle regulation. Our data show that the differentiation block observed in Rb-deficient cells may be explained by the repression of genes encoding mitochondrial proteins by the histone demethylase Kdm5a. pRB directly binds to and activates the Kdm5a targets with mitochondrial functions, resulting in the rescue of mitochondrial biological processes in cells lacking both pRb and Kdm5a. Kdm5a removal from genes encoding mitochondrial proteins occurs early during differentiation and is required for the activation of differentiation markers. Significantly, the differentiation rescue can be achieved through a bona fide transcriptional regulator of mitochondrial genes, showing that an increase in mitochondrial function is sufficient for induction of differentiation in cells lacking pRB.
Previous studies have attributed the reluctance of Rb-negative cells to undergo differentiation to the role of pRb as an activator of positive differentiation regulators. pRb directly activates the basic helix–loop–helix (bHLH) protein NeuroD1 (Batsche et al. 2005) and serves a coactivator role to CBFA1 (Thomas et al. 2001). pRb may promote activation of differentiation markers through degradation of the E1A-like inhibitor of differentiation 1 (EID-1) (Miyake et al. 2000) or inhibition of the bHLH protein Id2 (Iavarone et al. 2004) that restricts activity of other bHLH as well as Ets and Pax families of TFs. Although these experiments provided important insights into cell type-specific roles of pRB and the fundamental mechanisms of differentiation, they were typically limited to the study of biological processes that were necessary but not sufficient to rescue differentiation. Kdm5a bound to certain genes associated with differentiation and was displaced from their promoters by pRB (Benevolenskaya et al. 2005). This would suggest that pRB is playing a role in sequestering KDM5A from developmental genes, thus explaining the rescue of differentiation in cells lacking both Rb and Kdm5a. However, at odds with this simplistic model were data from ChIP-seq and RNA-seq studies revealing that expression of Kdm5a-bound developmental genes was generally not restored by the Kdm5a loss. Instead, the rescued GOs represented genes with functions in the mitochondrion. Enhancing mitochondrial biogenesis restored differentiation in a manner similar to Kdm5a loss, thus providing further evidence for the central role of mitochondrial function as a mediator of differentiation.
pRb function in mitochondrial gene expression has been previously described during erythropoiesis, where the up-regulation of mitochondrial biogenesis was linked to a cellular exit from the cell cycle (Sankaran et al. 2008). Because terminal differentiation in erythroblasts occurs concomitantly with the cell cycle exit, the requirement of mitochondrial burst specifically for differentiation could not be addressed. Our in vitro study using primary embryonic fibroblasts allowed us to distinguish cell cycle arrest from the expression of myogenic genes and acquisition of skeletal muscle structures. The muscle gene expression and multinucleation, but not the cell cycle exit, were rescued in Rb1−/− cells even with the deletion of only a single Kdm5a allele. In fact, a characteristic of differentiation induced upon combined Rb1 and Kdm5a loss in MEFs was the exacerbated cell cycle re-entry. A large number of E2F direct targets were deregulated in DKO-myo, consistent with the role of KDM5A, pRB, and E2F family proteins in repression of cell cycle genes in terminally differentiated cells (Beshiri et al. 2012). However, a common feature in differentiating erythroblasts, skeletal muscle cells, and adipocytes was the mitochondrial burst, which was not restricted to erythroblast maturation to provide high-level heme biosynthesis and increased ATP production for globin synthesis. These findings are not in contradiction to the previously reported role of pRB in highly specialized tissue functions, such as in adaptive thermogenesis in brown adipose tissue (Hansen et al. 2004). It is conceivable that the differentiation block of mitochondrial function released by pRB, while occurring in a cell type-specific manner, is as common in living cells as the pRB-mediated cell cycle control.
The dependence of differentiation rescue in Rb1−/− cells on the mitochondrial GO and its independence of E2F-dependent GOs changes the way we prioritize pRB functions in gene regulation and tumorigenesis. In accordance with our observations, Dyson's group (Nicolay et al. 2015) discovered that the extensive proteomic changes commonly occurring in Rb1−/− tissues affect proteins of the mitochondrion but not of the E2F target genes. The decrease in mitochondrial components observed in mouse tissues had an effect on both TCA cycle and oxidative phosphorylation that was independent of cell cycle withdrawal, similar to our in vitro model. Differentiation promotion thus represents a functional aspect of pRB regulation of the mitochondrion. This may help explain a recently established role of pRb in lineage commitment and cell fate determination (Calo et al. 2010). As mitochondrial function depends on pRB in different tissues, Rb loss may affect stem cell and progenitor cell populations in multiple tissues.
Our data imply that the release of differentiation block at the mitochondrial level can be achieved through changes in the activity of TFs directly regulating genes of the mitochondrion. Upon induction of myogenic differentiation in wild-type cells, pRb was recruited to genes encoding mitochondrial proteins such as Mfn2. This occurred along with the dissociation of Kdm5a, which correlated with the increased TSS enrichment for H3K4me3 and was required for increased Mfn2 expression. This mechanism explains why genes fail to activate in Rb1−/− cells (i.e., pRb playing a positive role) but become rescued by Kdm5a loss in double-knockout cells (i.e., Kdm5a playing a negative role). It is likely that Kdm5a is one of many transcriptional regulators of Mfn2, as the expression pattern of Mfn2 during differentiation is complex, with gene repression followed by gene activation (Lopez-Bigas et al. 2008). Consistent with this idea, the TFBS analysis predicted other TFs involved in the regulation of pRB-dependent genes like Mfn2. Importantly, the identified TFs are playing a critical role in mitochondrial function in Rb1−/− cells, as mitochondrial morphology was significantly improved in Rb1−/− MEFs that were induced for differentiation after overexpression of their common coactivator, Pgc-1α. While we identified Pgc-1α as a molecular mechanism for inducing differentiation through normalizing mitochondrial activity, Pgc-1α has a specific role in type I skeletal muscle (Liang and Ward 2006), indicating that other mitochondrial regulators may be more effective in other cell types. Recent studies on cardiomyocyte differentiation established the link between Mfn2 and Notch signaling (Kasahara et al. 2013). Consistent with this result, our RNA-seq data (GSE53528 with full gene list) show that Notch targets, genes involved in calcium-regulated signaling, and the gene encoding Mef2 are rescued in double-knockout MEFs. Whether the functional connection between Mfn2 and transcriptional nodes downstream from Notch signaling exists in all muscle cells and whether a calcium excitation transcription coupling generally operates during differentiation are highly speculative ideas at this point.
The respiratory capacity of the mitochondria during myogenic differentiation seems to be positively impacted by an increased mitochondrial fusion. Induced Rb1−/− MEFs had small mitochondria with distinctive Cox IV and Mfn2 expression. In contrast, induced double-knockout MEFs had mitochondrial morphology that was similar to the wild-type MEFs, and the mitochondria were rescued functionally; i.e., for RC activity and ATP production. Although Mfn2 is mostly known for its role in mitochondrial fusion, it also activates mitochondrial metabolism and increases the RC expression (Pich et al. 2005). Specifically, mfn2 loss in skeletal muscle was characterized by reduced RC expression level and activity and impaired respiratory function (Segalés et al. 2013). A loss of Kdm5a in Rb1−/− MEFs resulted in a significant shift from glycolytic to oxidative metabolism. MEFs lacking Kdm5a still had the highest glycolytic capacity, which seemingly did not interfere with successful differentiation. In response to acute glucose addition, the OCR not only failed to increase but in fact decreased, probably due to the Crabtree effect. Other substrate utilization experiments showed that cells lacking Kdm5a have a high rate of FAO, which can be further increased by pretreatment with metformin. Currently proposed mechanisms for metformin are consistent with not only the direct inhibition of RCI but also the stimulation of glucose uptake and FAO, which include activating AMPK. KDM5A loss results in increased phosphorylation of AMPK at T172 (Tzatsos et al. 2013). This suggests that Kdm5a−/− and double-knockout MEFs may have downstream consequences of the Kdm5a target gene up-regulation for sensing cellular energy status.
While molecular and chemical inhibition of fission promoted differentiation, further analysis will show to what extent increased mitochondrial fusion phenocopies the effect of a Kdm5a loss. Although restoration of the pRb-dependent mitochondrial function is both necessary and sufficient for the differentiation rescue in vitro, it is conceivable that Kdm5a is playing an additional pRb-dependent role as a repressor or activator of developmental genes. In particular, KDM5A has a direct effect on TF activity at some pRB target promoters; this may contribute to the characteristic morphological changes reminiscent of the effects seen by reintroducing pRB (Benevolenskaya et al. 2005). Kdm5a deletion failed to rescue the embryonic lethality of Rb1−/− mice and resulted in a twofold reduction in the number of Rb1+/− pups born (Lin et al. 2011), indicating that Kdm5a loss can accelerate embryonic defects in animals lacking Rb1.
As an indication that a Kdm5a-directed mechanism prevails in tumor suppression by pRB, data in knockout mice showed that the pituitary and thyroid tumors that normally spontaneously developed in Rb1+/− mice were unable to progress in Kdm5a−/−;Rb1+/− mice (Lin et al. 2011). Thus, genetic loss of Kdm5a results in the inhibition of tumor progression and the extension of cancer-free survival. Whether this was due to the rescued expression of mitochondrial proteins and/or differentiation remains to be tested. In particular, enhancing the mitochondrial network formation was shown to have a tumor-suppressive role, as injection of either MFN2-encoding adenovirus or Mdivi-1 in a lung adenocarcinoma xenograft model significantly reduced tumor volume (Rehman et al. 2012). However, there is no reason to believe that metabolic function of the RB pathway is restricted by regulation of KDM5A target genes. pRB is required for increased glycolysis and mitochondrial oxidative phosphorylation in cells that have undergone oncogene-induced senescence (Takebayashi et al. 2015). Intriguingly, the responsible mechanism may include changes in catalytic activity of histone-modifying enzymes, like the KDM5 family. In addition to pRB, other RB family members, upstream regulators of pRB, and E2F family proteins may directly regulate metabolism in order to maintain cellular homeostasis or drive cell proliferation, differentiation, and survival (Macleod 2008; Nicolay and Dyson 2013; Benevolenskaya and Frolov 2015). In particular, E2f1 orchestrates not only a proliferative response but also a metabolic response in conditions requiring adaptations to new energy demands (e.g., fasting and cold) (Blanchet et al. 2012).
Proliferation control by pRB has been considered central in its tumor suppression, although additional roles of pRB in apoptosis, maintenance of genome stability, and cellular senescence have also been described (Narita et al. 2003; Dick and Rubin 2013; Hilgendorf et al. 2013). Therefore, cytotoxic drugs used in chemotherapy were designed to arrest dividing cells by inducing DNA damage or targeting products of E2F-dependent genes (Whitfield et al. 2006). Mutations in mitochondrial proteins are rare in cancer cells, arguing the importance of mitochondrial functions in tumorigenesis. However, the direct link of mutated pRB to transcriptional regulation of genes encoding mitochondrial proteins that we described suggests that the impaired oxidative metabolism results from an inactivation of pRB, which is common in human tumors. As we found in Rb1−/− MEFs, moderate gene expression changes are likely reflective of severe mitochondrial and metabolic defects. The impairment of mitochondrial functions in Rb-negative cancer cells is not detrimental to cancer cell growth, suggesting that targeting dysfunctional mitochondria is not an easy task. Cells continue to engage in glycolysis and lactate production even under conditions of high oxygen, known as the Warburg effect; in addition, they adapt glutamine-dependent reductive carboxylation (Nicolay et al. 2013). However, the link between mitochondrial function and differentiation and its separation from cell cycle regulation may have implications in developing restorative, differentiation-based therapies.
In exploring a therapeutic strategy for the activation of mitochondrial function in order to differentiate cells derived from Rb-deficient tumors, we manipulated KDM5A and PGC-1α level in four main human cancer types with pRB inactivation. Both KDM5A shRNA and PGC-1α overexpression resulted in reduced cell growth in osteosarcoma, cervical carcinoma, retinoblastoma, and SCLC cell lines. Differentiation of retinoblastoma cell lines is manifested in increased cell adhesion and requires special techniques that were not applied in our analysis by light microscope (Laurie et al. 2009). However, in cases with the KDM5A knockdown, we recorded increased MFN2 expression, indicating that retinoblastoma cells respond to KDM5A loss by increasing mitochondrial fusion protein similar to other cell types. Importantly, KDM5A loss reduced the heterogeneity in SCLC, which remains the main challenge for cancer therapy, resulting in a high mortality rate in SCLC patients. Almost 95% of SCLC cell lines have alterations in the RB1 gene (Blanco et al. 2009), underscoring the importance of identification of downstream RB1 effectors for the development of successful SCLC drug therapies. Our work establishes the KDM5A metabolic axis as a critical mediator of the tumor-promoting effects secondary to pRB loss. This allows for the development of novel therapeutic approaches that diminish the aggressiveness of RB-deficient tumors by restoring their differentiation potential and could thus be used alongside existing cancer therapies targeting the cell cycle to augment their efficacy. The involvement of KDM5A and/or mitochondrial functions in determining the differentiation status can be further tested in vivo using recently generated mouse models of highly aggressive SCLC (Gazdar et al. 2015). Moreover, peculiarities of the mitochondrial response in RB-deficient tumors compared with normal tissue and RB-positive tumors are poorly understood. Exploiting anti-cancer agents for their effect on mitochondrial functions in tissues with different pRB statuses will thus be beneficial. Furthermore, KDM5A may represent an epigenetic regulator controlling the metabolic program that synchronizes energy homeostasis with a differentiation signal. In this regard, inhibition of KDM5A also provides a potential node for therapeutic intervention for diseases associated with reduced oxidative metabolism and a lower type I fiber content in skeletal muscle, such as cachexia and obesity.
Materials and methods
Cell culture and differentiation
MEFs were isolated from wild-type, Kdm5a−/−, and double-knockout littermates and from Rb1−/− embryos at the same genetic background as described (Lin et al. 2011). Adenoviral transductions were performed in a DM containing DMEM (CellGro), 2% horse serum (Gibco), and 10 µg mL−1 insulin from bovine pancreas (Sigma). After 18 h of incubation, the DM containing viruses was replaced with fresh DM. Lentiviral transductions were performed in DMEM growth medium (GM) containing 7.5 µg mL−1 polybrene (Sigma). GM was replaced with DM to induce differentiation 24–48 h afterward. For the differentiation of cells stably transduced with the inducible MyoD MyoDER[T], cells were first treated with DM containing 100 nM 4-hydroxytamoxifen (OHT), which, after 24 h, was replaced on DM without OHT. For the rescue experiments, Rb1−/− MEFs were first transduced with Lenti-CMV-MyoD or MyoDER[T] overnight as above. After that, cells were washed twice with PBS, medium was replaced with fresh GM, and, 8 h later, the second transduction with corresponding pLenti-CMV-Hygro-3xFlag lentiviruses was performed overnight. The medium was changed on GM for 2 h to allow cells recover, and then induction was started in DM supplemented with OHT if necessary. C2C12 myoblast cells were maintained in DMEM containing 20% FBS and induced in DM containing 2% horse serum and 10 µg mL−1 insulin. Induction of adipogenic differentiation was performed as described previously (Benevolenskaya et al. 2005). Cell lines were purchased from the American Type Culture Collection, and the RB-355 cell line was kindly provided by Dr. M.A. Dyer (Stewart et al. 2015).
RNA isolation and RNA-seq
Cells were induced for differentiation for 96 h as above with the following modifications. Cells of each genotype were plated at 2.5 × 105 cells per well in eight wells on 12-well dishes for RNA isolation from total cell population and on six-well dishes when the protocol involved purification of myotubes. The following day, the cells were transduced with Adeno-MyoD (2.2 × 108 plaque-forming units [PFU] per well). Each cDNA library was prepared from the pool of four wells. RNA was extracted with Trizol (Invitrogen) and purified by loading the aqueous phase containing the RNA premixed 1:1 with 70% ethanol on Qiagen RNeasy Micro (Qiagen, catalog no. 217084) column according to the Qiagen protocol. For mRNA (used for wild-type and double-knockout genotypes), 1.5 µg of RNA from each sample was subjected to two rounds of Oligo-dT purification with Dynal Oligo-dT beads (Invitrogen). For RiboMinus RNA (used for wild-type, Kdm5a−/−, Rb1−/−, and double-knockout genotypes), 1.7 µg of total RNA from each sample was processed using the RiboMinus eukaryote kit for RNA-seq (Ambion, catalog no. A10837-08) according to the Ambion protocol. RNA-seq counts represent baseMean values in Supplemental Table 1. The full data are deposited under GSE53528.
Functional enrichment analysis
Functional annotation of target genes was based on GO biological process or cellular location (http://www.geneontology.org). Analysis for differentially expressed genes was based on the GOseq package (Young et al. 2010). Resulting P-values were adjusted for multiple testing using Benjamin and Hochberg's method of false discovery rate (FDR) (Benjamini and Hochberg 1995). The heat map of corrected P-values was generated using GiTools (Perez-Llamas and Lopez-Bigas 2011).
Enrichment of TF binding
The occurrence of TF motifs in the promoter regions (600 base pairs [bp] upstream with 200 bp downstream with respect to the TSS) were predicted using the STORM algorithm (Schones et al. 2007) with a P-value cutoff of 0.0000125 as well as position frequency matrices (PFMs) from the TRANSFAC database (professional version release 2009.4) (Matys et al. 2003). Analysis of overrepresentation of the identified putative TF motifs on DE gene promoters against all promoters as background (i.e., enrichment of TF binding) was carried out using Gitools (Perez-Llamas and Lopez-Bigas 2011). FDR-corrected P-values (Benjamini and Hochberg 1995) were used for the heat map representation of enriched TFs.
ChIP-seq data
H3K4me3 ChIP-seq was performed in Kdm5af/f and Kdm5a−/− (with deleted Kdm5a alleles) embryonic stem (C57BL/6) cells. ChIP was performed with rabbit anti-H3K4me3 antibody (Millipore, 07-473) following our established protocol (Beshiri et al. 2010). The data have been deposited under GSE28348. Genomics targets for Kdm5a came from our previous study (Beshiri et al. 2012), and targets for E2F4 came from GSM516408 (Kim et al. 2010). ChIP-seq data for pRB (GSM497489) (Chicas et al. 2010) were processed from raw source data. Peak caller algorithm MACS (version 1.3.7.1) (Zhang et al. 2008) was used to determine the enriched peak regions. Enriched peaks were annotated to the nearest EnsEMBL gene using the Bioconductor package ChIPpeakAnno (Zhu et al. 2010). Human genes were converted to mouse orthologs using EnsEMBL Biomart. Data were annotated with the University of California at Santa Cruz mouse reference genome (mm9) (Mouse Genome Sequencing Consortium 2002).
Supplementary Material
Acknowledgments
We thank W. Kaelin Jr., N. Dyson, and B. Nicolay for the discussion and comments on the manuscript. We are grateful to K. Macleod for providing additional Rb1−/− MEFs, M.A. Dyer for the retinoblastoma RB-355 cells, and A. Vilkova for the generation of lentiviral constructs in destination vectors. This work was supported by R01CA138631 (to E.V.B.) and R01GM094220 (to J.R.) from the National Institutes of Health, educational grant SAF2009-06954 (to N.L.-B.) from the Spanish Ministry of Science, and a fellowship from the Agencia de Gestió d'Ajuts Universitaris i de Recerca of the Catalonian Government, Spain (to A.B.M.M.K.I.).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.264036.115.
References
- Batsche E, Moschopoulos P, Desroches J, Bilodeau S, Drouin J. 2005. Retinoblastoma and the related pocket protein p107 act as coactivators of NeuroD1 to enhance gene transcription. J Biol Chem 280: 16088–16095. [DOI] [PubMed] [Google Scholar]
- Benevolenskaya EV, Frolov MV. 2015. Emerging links between E2F control and mitochondrial function. Cancer Res 75: 619–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benevolenskaya EV, Murray HL, Branton P, Young RA, Kaelin WG. 2005. Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol Cell 18: 623–635. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57: 289–300. [Google Scholar]
- Berkes CA, Tapscott SJ. 2005. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol 16: 585–595. [DOI] [PubMed] [Google Scholar]
- Beshiri ML, Islam A, Dewaal DC, Richter WF, Love J, Lopez-Bigas N, Benevolenskaya EV. 2010. Genome-wide analysis using ChIP to identify isoform-specific gene targets. JoVE 41: 2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beshiri ML, Holmes KB, Richter WF, Hess S, Islam ABMMK, Yan Q, Plante L, Litovchick L, Gévry N, Lopez-Bigas N, et al. 2012. Coordinated repression of cell cycle genes by KDM5A and E2F4 during differentiation. Proc Natl Acad Sci 109: 18499–18504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais A, van Oevelen CJC, Margueron R, Acosta-Alvear D, Dynlacht BD. 2007. Retinoblastoma tumor suppressor protein-dependent methylation of histone H3 lysine 27 is associated with irreversible cell cycle exit. J Cell Biol 179: 1399–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchet E, Annicotte JS, Pradelli LA, Hugon G, Matecki S, Mornet D, Rivier F, Fajas L. 2012. E2F transcription factor-1 deficiency reduces pathophysiology in the mouse model of Duchenne muscular dystrophy through increased muscle oxidative metabolism. Hum Mol Genet 21: 3910–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco R, Iwakawa R, Tang M, Kohno T, Angulo B, Pio R, Montuenga LM, Minna JD, Yokota J, Sanchez-Cespedes M. 2009. A gene-alteration profile of human lung cancer cell lines. Hum Mutat 30: 1199–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blättler SM, Verdeguer F, Liesa M, Cunningham JT, Vogel RO, Chim H, Liu H, Romanino K, Shirihai OS, Vazquez F, et al. 2012. Defective mitochondrial morphology and bioenergetic function in mice lacking the transcription factor YY1 in skeletal muscle. Mol Cell Biol 32: 3333–3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calbo J, van Montfort E, Proost N, van Drunen E, Beverloo HB, Meuwissen R, Berns A. 2011. A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell 19: 244–256. [DOI] [PubMed] [Google Scholar]
- Calo E, Quintero-Estades JA, Danielian PS, Nedelcu S, Berman SD, Lees JA. 2010. Rb regulates fate choice and lineage commitment in vivo. Nature 466: 1110–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC. 2012. Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet 46: 265–287. [DOI] [PubMed] [Google Scholar]
- Chicas A, Wang X, Zhang C, McCurrach M, Zhao Z, Mert O, Dickins RA, Narita M, Zhang M, Lowe SW. 2010. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. 17: 376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciavarra G, Ho AT, Cobrinik D, Zacksenhaus E. 2011. Critical role of the Rb family in myoblast survival and fusion. PLoS One 6: e17682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin A, Wu L, Saavedra HI, Wilson P, Yang Y, Rosol TJ, Weinstein M, Robinson ML, Leone G. 2003. Rb function in extraembryonic lineages suppresses apoptosis in the CNS of Rb-deficient mice. Proc Natl Acad Sci 100: 6546–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick FA, Rubin SM. 2013. Molecular mechanisms underlying RB protein function. Nat Rev Mol Cell Biol 14: 297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov MV, Dyson NJ. 2004. Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J Cell Sci 117: 2173–2181. [DOI] [PubMed] [Google Scholar]
- Gazdar AF, Savage TK, Johnson JE, Berns A, Sage J, Linnoila RI, MacPherson D, McFadden DG, Farago A, Jacks T, et al. 2015. The comparative pathology of genetically engineered mouse models for neuroendocrine carcinomas of the lung. J Thorac Oncol 10: 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JB, Jørgensen C, Petersen RK, Hallenborg P, De Matteis R, Bøye HA, Petrovic N, Enerbäck S, Nedergaard J, Cinti S, et al. 2004. Retinoblastoma protein functions as a molecular switch determining white versus brown adipocyte differentiation. Proc Natl Acad Sci 101: 4112–4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgendorf KI, Leshchiner ES, Nedelcu S, Maynard MA, Calo E, Ianari A, Walensky LD, Lees JA. 2013. The retinoblastoma protein induces apoptosis directly at the mitochondria. Genes Dev 27: 1003–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iavarone A, King ER, Dai X-M, Leone G, Stanley ER, Lasorella A. 2004. Retinoblastoma promotes definitive erythropoiesis by repressing Id2 in fetal liver macrophages. Nature 432: 1040–1045. [DOI] [PubMed] [Google Scholar]
- Kasahara A, Cipolat S, Chen Y, Dorn GW, Scorrano L. 2013. Mitochondrial fusion directs cardiomyocyte differentiation via calcineurin and Notch signaling. Science 342: 734–737. [DOI] [PubMed] [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH. 2010. A myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell 143: 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie N, Mohan A, McEvoy J, Reed D, Schweers B, Ajioka I, Valentine V, Dyer MA. 2009. Changes in retinoblastoma cell adhesion associated with optic nerve invasion. Mol Cell Biol 29: 6268–6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Ward WF. 2006. PGC-1: a key regulator of energy metabolism. Adv Physiol Educ 30: 145–151. [DOI] [PubMed] [Google Scholar]
- Liesa M, Borda-d’Água B, Medina-Gómez G, Lelliott CJ, Paz JC, Rojo M, Palacín M, Vidal-Puig A, Zorzano A. 2008. Mitochondrial fusion is increased by the nuclear coactivator PGC-1β. PLoS One 3: e3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Cao J, Liu J, Beshiri ML, Fujiwara Y, Francis J, Cherniack AD, Geisen C, Blair LP, Zou MR, et al. 2011. Loss of the retinoblastoma binding protein 2 (RBP2) histone demethylase suppresses tumorigenesis in mice lacking Rb1 or Men1. Proc Natl Acad Sci 108: 13379–13386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bigas N, Kisiel TA, Dewaal DC, Holmes KB, Volkert TL, Gupta S, Love J, Murray HL, Young RA, Benevolenskaya EV. 2008. Genome-wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation. Mol Cell 31: 520–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod K. 1999. pRb and E2f-1 in mouse development and tumorigenesis. Curr Opin Genet Dev 9: 31–39. [DOI] [PubMed] [Google Scholar]
- Macleod KF. 2008. The role of the RB tumour suppressor pathway in oxidative stress responses in the haematopoietic system. Nat Rev Cancer 8: 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson D, Sage J, Crowley D, Trumpp A, Bronson RT, Jacks T. 2003. Conditional mutation of Rb causes cell cycle defects without apoptosis in the central nervous system. Mol Cell Biol 23: 1044–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AL, Longworth MS, Dyson NJ. 2010. Loss of pRB causes centromere dysfunction and chromosomal instability. Genes Dev 24: 1364–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gössling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. 2003. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res 31: 374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake S, Sellers WR, Safran M, Li X, Zhao W, Grossman SR, Gan J, DeCaprio JA, Adams PD, Kaelin WG. 2000. Cells degrade a novel inhibitor of differentiation with E1A-like properties upon exiting the cell cycle. Mol Cell Biol 20: 8889–8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. [DOI] [PubMed] [Google Scholar]
- Narita M, Nũnez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113: 703–716. [DOI] [PubMed] [Google Scholar]
- Nicolay BN, Dyson NJ. 2013. The multiple connections between pRB and cell metabolism. Curr Opin Cell Biol 25: 735–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolay BN, Gameiro PA, Tschöp K, Korenjak M, Heilmann AM, Asara JM, Stephanopoulos G, Iliopoulos O, Dyson NJ. 2013. Loss of RBF1 changes glutamine catabolism. Genes Dev 27: 182–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolay BN, Danielian PS, Kottakis F, Lapek JD Jr, Sanidas I, Miles WO, Dehnad M, Tschöp K, Gierut JJ, Manning AL, et al. 2015. Proteomic analysis of pRb loss highlights a signature of decreased mitochondrial oxidative phosphorylation. Genes Dev (this issue). 10.1101/gad.264127.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitch BG, Mulligan GJ, Jacks T, Lassar AB. 1996. Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle. J Cell Biol 135: 441–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parasassi T, Brunelli R, Bracci-Laudiero L, Greco G, Gustafsson AC, Krasnowska EK, Lundeberg J, Lundeberg T, Pittaluga E, Romano MC, et al. 2005. Differentiation of normal and cancer cells induced by sulfhydryl reduction: biochemical and molecular mechanisms. Cell Death Differ 12: 1285–1296. [DOI] [PubMed] [Google Scholar]
- Perez-Llamas C, Lopez-Bigas N. 2011. Gitools: analysis and visualisation of genomic data using interactive heat-maps. PLoS One 6: e19541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pich S, Bach D, Briones P, Liesa M, Camps M, Testar X, Palacín M, Zorzano A. 2005. The Charcot-Marie-Tooth type 2A gene product, Mfn2, up-regulates fuel oxidation through expression of OXPHOS system. Hum Mol Genet 14: 1405–1415. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92: 829–839. [DOI] [PubMed] [Google Scholar]
- Rangwala SM, Wang X, Calvo JA, Lindsley L, Zhang Y, Deyneko G, Beaulieu V, Gao J, Turner G, Markovits J. 2010. Estrogen-related receptor is a key regulator of muscle mitochondrial activity and oxidative capacity. J Biol Chem 285: 22619–22629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman J, Zhang HJ, Toth PT, Zhang Y, Marsboom G, Hong Z, Salgia R, Husain AN, Wietholt C, Archer SL. 2012. Inhibition of mitochondrial fission prevents cell cycle progression in lung cancer. FASEB J 26: 2175–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran VG, Orkin SH, Walkley CR. 2008. Rb intrinsically promotes erythropoiesis by coupling cell cycle exit with mitochondrial biogenesis. Genes Dev 22: 463–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Smith AD, Zhang MQ. 2007. Statistical significance of cis-regulatory modules. BMC Bioinformatics 8: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segalés J, Paz JC, Hernández-Alvarez MI, Sala D, Muñoz JP, Noguera E, Pich S, Palacín M, Enríquez JA, Zorzano A. 2013. A form of mitofusin 2 (Mfn2) lacking the transmembrane domains and the COOH-terminal end stimulates metabolism in muscle and liver cells. Am J Physiol Endocrinol Metab 305: E1208–E1221. [DOI] [PubMed] [Google Scholar]
- Stewart E, Federico S, Karlstrom A, Shelat A, Sablauer A, Pappo A, Dyer MA. 2015. The Childhood Solid Tumor Network: a new resource for the developmental biology and oncology research communities. Dev Biol 10.1016/j.ydbio.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebayashi S-I, Tanaka H, Hino S, Nakatsu Y, Igata T, Sakamoto A, Narita M, Nakao M. 2015. Retinoblastoma protein promotes oxidative phosphorylation through upregulation of glycolytic genes in oncogene-induced senescent cells. Aging Cell 14: 689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DM, Carty SA, Piscopo DM, Lee JS, Wang WF, Forrester WC, Hinds PW. 2001. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol Cell 8: 303–316. [DOI] [PubMed] [Google Scholar]
- Tzatsos A, Paskaleva P, Ferrari F, Deshpande V, Stoykova S, Contino G, Wong K-K, Lan F, Trojer P, Park PJ, et al. 2013. KDM2B promotes pancreatic cancer via Polycomb-dependent and -independent transcriptional programs. J Clin Invest 123: 727–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oevelen C, Wang J, Asp P, Yan Q, Kaelin WG Jr, Kluger Y, Dynlacht BD. 2008. A role for mammalian Sin3 in permanent gene silencing. Mol Cell 32: 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz T, Diaz F, Spiegelman BM, Moraes CT. 2008. Activation of the PPAR/PGC-1α pathway prevents a bioenergetic deficit and effectively improves a mitochondrial myopathy phenotype. Cell Metab 8: 249–256. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Whitfield ML, George LK, Grant GD, Perou CM. 2006. Common markers of proliferation. Nat Rev Cancer 6: 99–106. [DOI] [PubMed] [Google Scholar]
- Wu L, de Bruin A, Saavedra HI, Starovic M, Trimboli A, Yang Y, Opavska J, Wilson P, Thompson JC, Ostrowski MC, et al. 2003. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature 421: 942–947. [DOI] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacksenhaus E, Jiang Z, Chung D, Marth JD, Phillips RA, Gallie BL. 1996. pRb controls proliferation, differentiation, and death of skeletal muscle cells and other lineages during embryogenesis. Genes Dev 10: 3051–3064. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LJ, Gazin C, Lawson ND, Pagès H, Lin SM, Lapointe DS, Green MR. 2010. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11: 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.