Summary
Neural crest cells are induced at the neural plate border by the combined action of transcription factors and signaling molecules. Here, we show that Axud1, a downstream effector of Wnt signaling, represents a critical missing link that integrates signaling and transcriptional cues to mediate neural crest formation. Axud1 is a transcription factor expressed in neural crest progenitors in a Wnt1/β-catenin dependent manner. Axud1 loss leads to downregulation of multiple genes involved in neural crest specification, similar to the effects of Wnt1 knockdown. Importantly, Axud1 is sufficient to rescue neural crest formation after disruption of Wnt signaling. Furthermore, it physically interacts with neural plate border genes Pax7 and Msx1 in vivo to directly activate transcription of stem cell factor FoxD3, initiating the neural crest program. Thus, Axud1 integrates Wnt signaling with transcriptional inputs to endow the neural crest with its unique molecular signature.
Keywords: neural crest, Wnt, gene regulatory network
Introduction
The neural crest is a progenitor cell population that gives rise to many defining features of vertebrates, including much of the peripheral nervous system, structural elements of the face and pigmentation of the skin(Green et al., 2015; Le Douarin and Kalcheim, 1999; Simoes-Costa and Bronner, 2013). The process of neural crest induction begins during gastrulation, and requires the interplay of transcriptional regulators, receptors, and signaling molecules at the neural plate border(Sauka-Spengler and Bronner-Fraser, 2008; Simoes-Costa et al., 2014). Neural crest formation is coordinated by a multistep neural crest gene regulatory network (NC GRN), which comprises multiple signals and transcriptional inputs that are active in the neural crest lineage(Meulemans and Bronner-Fraser, 2002; Simoes-Costa and Bronner, 2015).
Initiation of the NC GRN is thought to be driven by secreted growth factors such as Wnts, BMPs and FGFs, which induce expression of a set of transcription factors at the border between neural and non-neural ectoderm(Prasad et al., 2012). These neural plate border specifier genes, including Pax7(Basch et al., 2006) and Msx1(Monsoro-Burq et al., 2005), are critical for neural crest formation and directly activate transcription of bona fide neural crest markers like FoxD3(Simoes-Costa et al., 2012). It has been proposed that the border specifier genes cooperate with signaling events, and in particular the Wnt pathway(Garcia-Castro et al., 2002; Ikeya et al., 1997; LaBonne and Bronner-Fraser, 1998), to mediate neural crest specification(Sauka-Spengler and Bronner-Fraser, 2008; Simoes-Costa and Bronner, 2015). However, the molecular mechanism through which these different tiers of regulation are integrated to activate the neural crest molecular program has remained elusive.
Here, we probe how signaling and transcriptional inputs are coordinated during neural crest formation, focusing on the Wnt pathway. Our results show that Axud1 (also known as Csrnp1), proposed to be a Wnt effector, is critical for linking Wnt signaling with transcriptional regulators at the neural plate border to mediate neural crest specification. Axud1 was first uncovered in a microarray screen designed to identify Wnt targets of Axin1-induced LoVo colon cancer cells(Ishiguro et al., 2001). We show that Axud1 expression in neural crest precursors is controlled by Wnt signaling and that its loss in turn abrogates expression of multiple neural crest genes. Furthermore, Axud1 physically interacts with Msx1 and Pax7 at the neural plate border to directly activate transcription of FoxD3, one of the earliest markers of premigratory neural crest cells. These results clarify how signaling pathways are integrated with the transcriptional machinery to allow for emergence of nascent neural crest stem cells, uncovering a mechanism through which Wnt signaling functions in the induction of the neural crest.
Results
Expression pattern of Axud1 in the early chick embryo
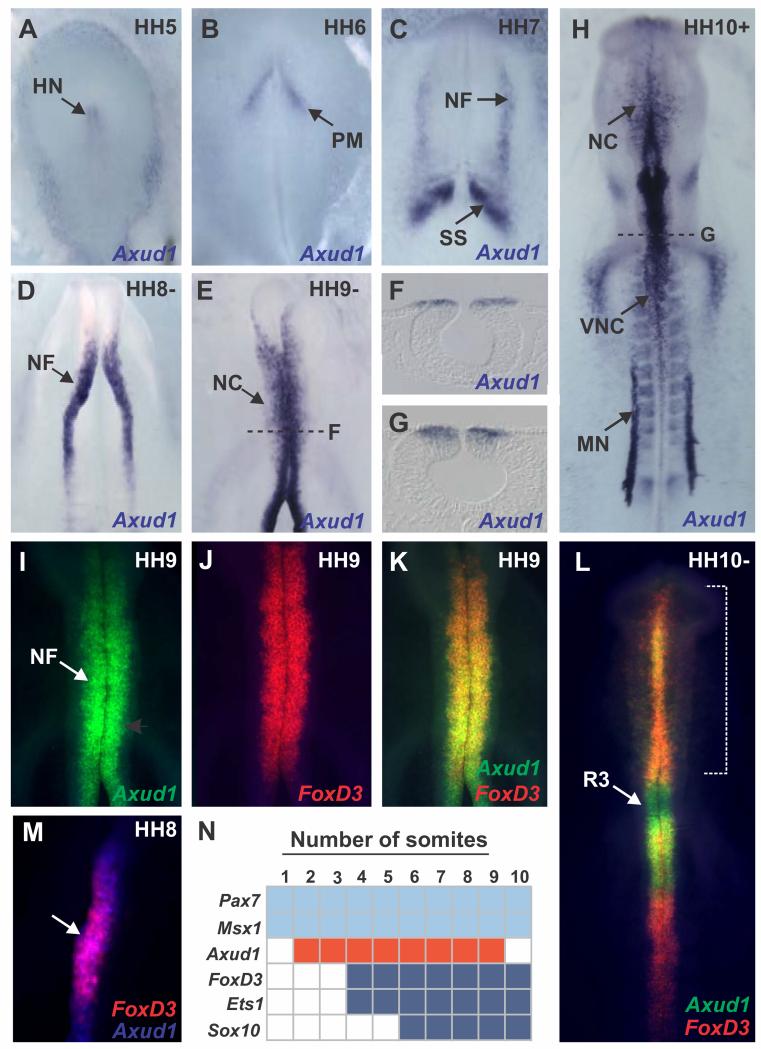
We identified transcriptional regulator Axud1 as a neural crest marker that is expressed in the neural plate border as well as in the dorsal neural folds (Fig. 1). In the chick embryo, Axud1 expression was first detectable during gastrulation in Hensen’s node and the paraxial mesoderm (Fig. 1A-B). At Hamburger and Hamilton stage (HH) 7, Axud1 was robustly expressed in the neural folds as they began to elevate (Fig. 1C). Expression was enhanced in the dorsal neural tube at stages HH8-9 (Fig. 1D), and persisted in early migratory neural crest cells (Fig. 1E-G). At stage HH10, Axud1 transcripts were detected in cranial and vagal neural crest cells (Fig. 1H). In addition to the neural folds and neural crest cells, Axud1 expression was observed in the first presumptive somite (Fig. 1C), mesonephros (Fig. 1H), and cardiac cushion, consistent with previous observations suggesting that Wnt signaling is ongoing in these tissues(Kühl, 2003).
Figure 1. Axud1 is a marker of the premigratory neural crest in chick embryos.
All embryos are shown from a dorsal aspect. (A-B) In situ hybridization showed Axud1 expression in the Hensen’s node (A) and the paraxial mesoderm (B) in gastrulating chick embryos. (C-E) At neurula stages, Axud1 transcripts were detected in the dorsal neural folds (NFs) as they elevate and converge to form the neural tube. (F-G) Cross sections of neurula stage embryos showed specific expression of Axud1 in the dorsal neural tube. (H) At later stages, Axud1 was expressed in the early migrating neural crest (NC) and other structures targeted by Wnt signaling such as the mesonephros (MN) and the first presumptive somite. (I-F) Double in situ hybridization for Axud1 (I) and FoxD3 (J) revealed co-expression of both genes in the premigratory (K) and early migrating cranial neural crest (L). (M) Onset of FoxD3 expression was observed at HH8 within the Axud1 expression domain. (N) Expression of Axud1 in neural crest progenitors initiates after the establishment of the neural plate border but before neural crest gene expression, making it a likely candidate regulator in neural crest formation. HH: Hamburger and Hamilton developmental stages. HH: Hensen’s node, MN: Mesonephros, NF: Neural folds, NC: Neural crest, PM: Paraxial mesoderm, R: Rhombomere, SS: Somites, VNC: Vagal neural crest.
To determine its position in the NC GRN relative to other transcription factors involved in neural plate border and neural crest specification, we performed double fluorescent in situ hybridization of Axud1 and FoxD3, one of the earliest expressed neural crest specifier genes and critical for initiating the neural crest program(Simoes-Costa et al., 2012). The results showed that Axud1 is co-expressed with FoxD3 in the cranial region in both the premigratory (Fig. 1I-K) and early-migrating (Fig. 1L) neural crest. At HH8, when FoxD3 appears, Axud1 preceded but then overlapped with FoxD3 (Fig. 1M), although it was expressed in regions of the neural axis where FoxD3 is later missing, like rhombomere (R) 3 and R5 (Fig. 1L). Thus, Axud1 co-localizes with both neural plate border and neural crest specifier genes. Importantly, onset of FoxD3 expression takes place within the Axud1-positive territory in the neural folds (Fig. 1M). Taken together with previous data on the expression of chick neural crest genes(Betancur et al., 2010; Khudyakov and Bronner-Fraser, 2009; Simoes-Costa et al., 2012), the results show that Axud1 expression initiates after neural plate border genes Pax7 and Msx1, but prior to neural crest specifier genes like FoxD3 and Sox10 (Fig. 1N). In conclusion, Axud1 is expressed at the correct time and place to be involved in neural crest specification.
Loss of function of Axud1 blocks expression of early neural crest specifier gene, FoxD3
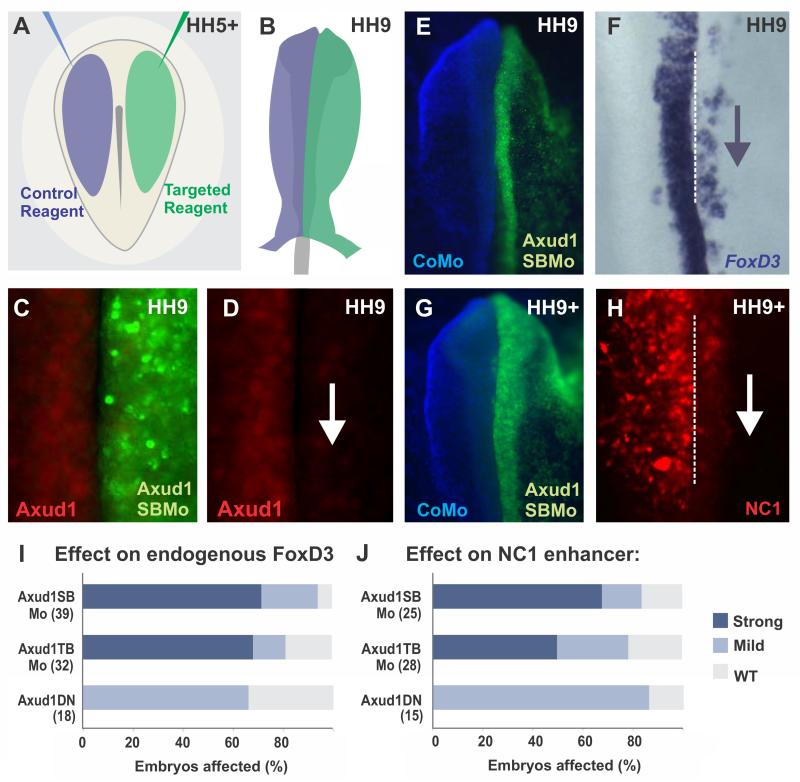
To test the role of Axud1 in neural crest specification, we performed loss-of-function assays using three different strategies: a translation-blocking morpholino, a splice-blocking morpholino, and a dominant negative construct, all of which yielded similar results. These were individually electroporated into the right side of HH5+ chick embryos, after establishment of the neural plate border but before onset of Axud1 expression, while the left side was electroporated with control constructs/morpholinos (Fig. 2A). Following electroporation, embryos were grown until pre-migratory neural crest stages (Fig. 2B), and gene expression on the experimental side (right) was compared to the control side (left). Electroporation of both targeted morpholinos resulted in strong knockdown of the Axud1 protein in the dorsal neural folds, when compared to the control side (Fig. 2C-D). To further demonstrate specificity, we performed rescue experiments, in which morpholinos were co-electroporated with an Axud1 expression vector. Whereas electroporation of the Axud1 expression vector was not sufficient to induce ectopic neural crest formation in other regions of the neural tube, it attenuated the loss of FoxD3 expression observed in Axud1 morpholino-treated embryos (Fig. S1A-G). We also verified that Axud1 knockdown did not alter the amount of cell death in electroporated embryos by conducting immunohistochemistry with the anti-active Caspase-3 antibody (Fig. S1H-L).
Figure 2. Axud1 is necessary for FoxD3 expression.
(A-B) Diagram of electroporation assays for loss-of-function experiments. (A) Control (blue) and targeted (green) reagents were injected in different sides of gastrulating embryos. (B) Following injection, embryos were electroporated and cultured until the desired stages. Phenotypes were scored by comparing the control and experimental sides of the same embryo. (C-D) Immunohistochemistry with an Axud1 antibody shows loss of the Axud1 protein in dorsal neural folds electroporated with Axud1 morpholino. (E-H) Morpholino-mediated knockdown of Axud1 resulted in a marked reduction of endogenous FoxD3 expression (E-F), as well as loss in activity of the NC1 enhancer, which controls cranial expression of FoxD3(Simoes-Costa et al., 2012) (G-H). (I-J) Quantitation of embryos following loss-of-function assays. The total number of embryos analyzed for each experiment is represented in parenthesis. See Fig. S1 for additional controls of loss of function experiments. CoMo: Control morpholino, Axud1DN: Axud1 dominant negative, Axud1SBMo: Axud1 splice-blocking morpholino, Axud1TBMo: Axud1 translation-blocking morpholino, HH: Hamburger and Hamilton developmental stages.
We analyzed the effects of Axud1 loss of function on neural crest specification by examining its requirement for expression of FoxD3, the first neural crest specifier gene to be transcribed in chick embryos(Simoes-Costa et al., 2012). Electroporation of either antisense morpholinos or dominant negative Axud1 led to elimination of endogenous FoxD3 in the cranial neural folds (Fig. 2E-F). Each of the two different morpholinos had a stronger effect than the dominant negative construct, likely because the latter must compete with endogenous protein. Next, we assayed the effects of Axud1 knockdown on reporter expression driven by the NC1 enhancer, which mediates FoxD3 expression in the cranial neural crest(Simoes-Costa et al., 2012). Similar to what was observed for endogenous FoxD3 expression, enhancer activity was lost following Axud1 knockdown (Fig. 2G-H). All three approaches for loss of function resulted in downregulation of FoxD3 expression and loss of enhancer activity (Fig. 2I-J). These results indicate that expression of Axud1 in the neural plate border is required for the onset of neural crest specification.
Axud1 is a direct input into the FoxD3 enhancer, NC1
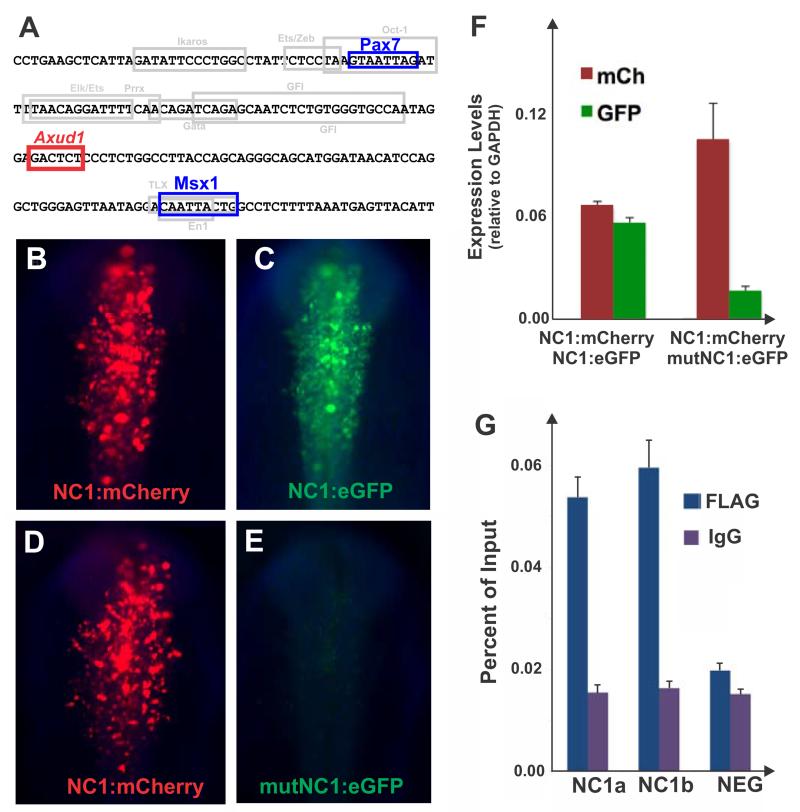
Since loss of Axud1 abrogates expression driven by the FoxD3 NC1 enhancer element, we asked whether Axud1 is a direct transcriptional regulator of FoxD3, as previously shown for neural plate border genes Pax7 and Msx1(Simoes-Costa et al., 2012). Axud1 is a member of the Cysteine-Serine Rich Nuclear Protein family, and interacts with DNA through the consensus binding sequence AGAGTS(Gingras et al., 2007). Intriguingly, analysis of the NC1 sequence revealed an Axud1 binding site located between the Hox (Pax7) and the Msx1 binding sites necessary for enhancer activity(Simoes-Costa et al., 2012) (Fig. 3A).
Figure 3. Axud1 is a direct regulator of neural crest specifier gene FoxD3.
(A) The NC1 enhancer core contains an Axud1 binding site between Pax7 and Msx1 sites. (B,C) Co-electroporation of a NC1 enhancer driving mCherry (NC1:mCherry) and an NC1 enhancer encoding eGFP (NC1:eGFP) result in similar levels of reporter gene expression in the same embryo. (D, E) Alternatively, co-electroporation of a NC1:mCherry with a mutated Axud-binding site encoding GFP (mut:NC1:eGFP) indicate the construct harboring the mutation (E) has negligible activity when compared to the wild type mCherry vector (D). (F) Quantitation of reporter gene expression levels through qPCR confirmed mutation of the Axud1 binding site results in loss of NC1 enhancer activity. Reporter gene levels are represented relative to GAPDH expression. (G) Chromatin immunoprecipitation assay performed in dissected dorsal neural folds of stage HH9 embryos electroporated with a FLAG-tagged Axud1 expression construct. Immunoprecipitation of the Axud1 protein with an anti-FLAG antibody, followed by site specific qPCR with primer pairs designed to amplify two fragments within the NC1 region (NC1a and NC1b) revealed significant enrichment of the NC1 region in the Axud1 pull-down when compared to IGG controls. No enrichment was observed for a negative control region upstream of the FoxD3 locus in the chicken chromosome 8 (NEG). Enrichment of amplicons is expressed as a percent of the total input chromatin used in the immunoprecipitation assay. Error bars on F and G represent standard deviation.
Consistent with the possibility that Axud1 is a direct input into FoxD3, mutation of the Axud1 site resulted in strong loss of enhancer activity in electroporated embryos (Fig. 3B-F). While co-electroporation of two constructs with the NC1 enhancer driving either mCherry or GFP in the same embryo result in equivalent levels of expression (Fig. 3B-C), the vector carrying a mutation in the Axud1 binding side (mut:NC1:eGFP) exhibited striking loss of activity when compared to the wild type version (Fig. 3D-E). Indeed, quantification of reporter genes by qPCR showed a five-fold reduction in expression driven by the mutated construct relative to the wild type vector (Fig. 3F).
Finally, we performed chromatin immunoprecipitation in dissected dorsal neural folds to explore whether Axud1 binds to the NC1 enhancer. The assay revealed that Axud1 directly interacts with this element (Fig. 3G) in vivo. In summary, enhancer analysis places Axud1 as a direct regulator of neural crest specification that is expressed after establishment of the neural plate border, and thus occupies an intermediate position in the neural crest gene regulatory network.
Axud1 is a regulator of multiple neural crest specifier genes
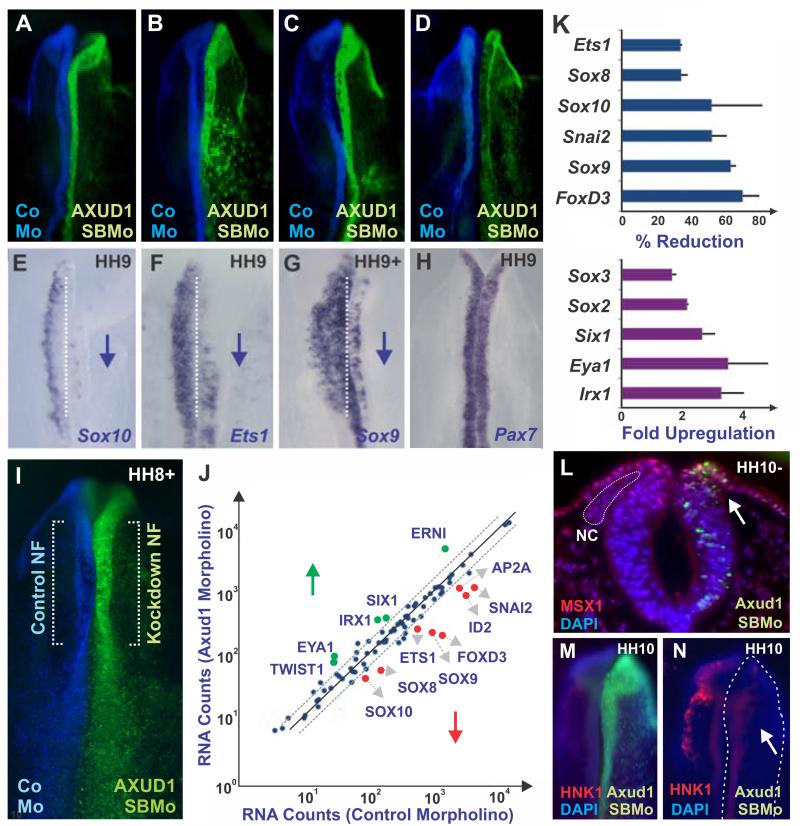
To assay whether Axud1 is a specific input for FoxD3 or more broadly regulates the neural crest specification program, we investigated how its loss impacts other members of the neural crest specifier module. In addition to FoxD3, Axud1 knockdown caused loss of expression of numerous neural crest specifier genes, including Sox9, Sox10, and Ets1, but not neural plate border gene Pax7 (Fig. 4A-H). Multiplex Nanostring analysis (Fig. 4I-J), which enabled quantitative analysis of transcript levels of hundreds of genes simultaneously, confirmed that Axud1 knockdown caused loss of neural crest specifier genes in the neural folds (Fig. 4K). Interestingly, this analysis revealed upregulation of genes important for placodal and central nervous system development, concomitant with loss of neural crest markers (Fig. 4K). This suggests that Axud1 drives neural plate border cells to a neural crest identity at the expense of neural and placodal fates. Consistent with this, immunostaining with the Sox2 antibody revealed a striking expansion of the neural marker into the neural crest forming region of the dorsal neural tube after Axud1 knockdown (Fig. S2A-B). In addition, we observed a medial expansion of the placodal marker Six1 into the dorsal neural tube (Fig. S2A, C). Indeed, without Axud1, cranial neural crest cells fail to delaminate from the neural tube (Fig. 4L) to become migratory (Fig. 4M-N). The results indicate that Axud1 acts as a critical regulator of genes involved in neural crest specification, such that its loss of function causes a global loss of neural crest specifier gene expression.
Figure 4. Axud1 is required for neural crest formation.
(A-D) Whole mount views of embryos after Axud1 morpholino (AXUD1SBMo) was introduced on the right side (green) and control morpholino (CoMo) on the left side (blue) of each embryo. (E-H) The same embryos as above after in situ hybridization for the indicated gene, with downward blue arrows indicating downregulation. Knockdown of Axud1 caused loss of neural crest specifier genes, including Sox10 (E), Ets1 (F) and Sox9 (G) while neural plate border specifiers such as Pax7 were not affected (H). (I-J) Multiplex Nanostring analyses highlighted loss of multiple genes involved in neural crest specification. (I) Regions of the embryo dissected for Nanostring analysis. (J) Graph comparing transcript levels between control MO and Axud1 MO electroporated sides of the same embryo assayed by Nanostring analysis. Transcripts above diagonal lines (upward arrow, in green) were upregulated; transcripts below diagonal lines (downward arrow, in red) were downregulated by loss of Axud1; e.g. depletion of Axud1 drastically reduced the expression levels of FoxD3, Snai2 and SoxE genes. Genes within diagonal lines were not significantly affected above background. (K) Quantitation of transcript levels averaged between different Nanostring experiments revealed downregulation of neural crest specifier genes, while neural genes and placodal markers were upregulated in neural folds after loss of Axud1 (see also Fig. S2). Error bars represent standard deviation. (L) Knockdown of Axud1 (right side; green) results in loss of the neural crest population, which failed to delaminate from the neural tube as seen in a midbrain-level transverse section. (M,N) Neural crest cells were barely detected at later stages as seen in whole mount views of embryos stained with HNK-1 antibody. Axud1SBMo: Axud1 splice-blocking morpholino, CoMo: Control morpholino, NF: neural fold, NC: neural crest.
Axud1 rescues neural crest formation caused by loss of Wnt1 or β-catenin
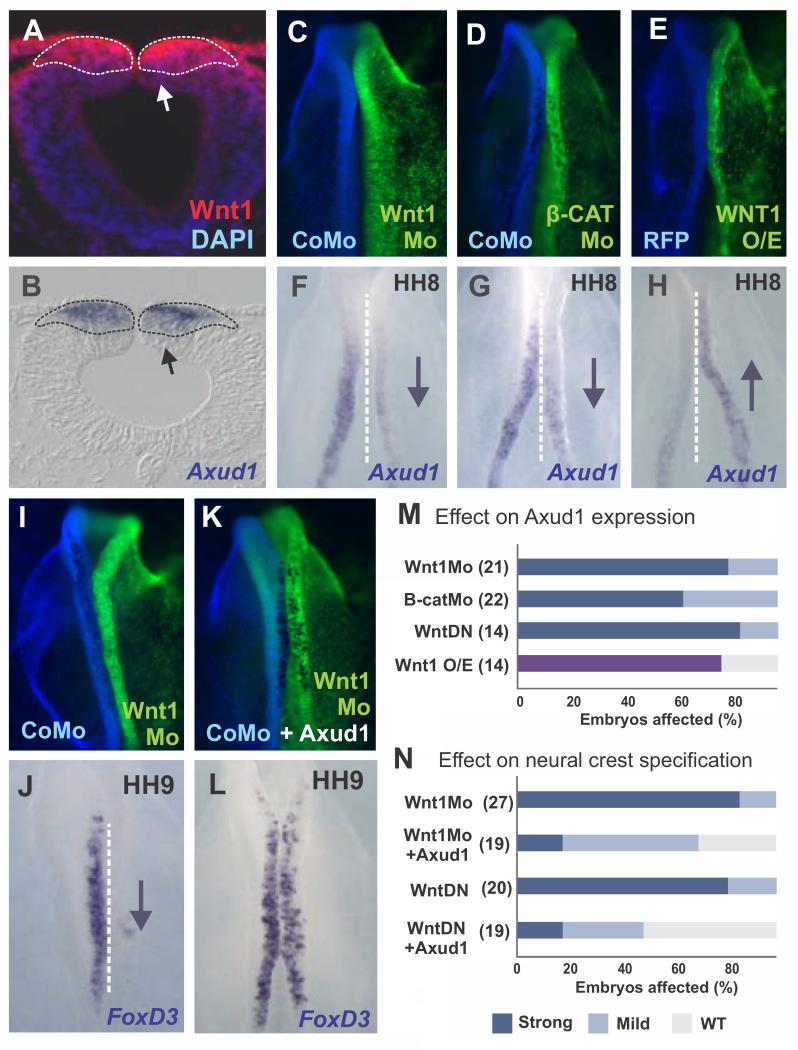
Axud1 has been proposed to be a Wnt target gene based on studies in cell lines and chick embryos(Ishiguro et al., 2001; Quinlan et al., 2009). Therefore, we tested the hypothesis that Axud1 is downstream of Wnt signaling in the dorsal neural folds. Immunohistochemistry with a Wnt1 antibody revealed that Axud1 is expressed within the Wnt1 domain in the dorsal neural tube (Fig. 5A-B). To test whether Wnt1 is an activator of Axud1, we disrupted Wnt signaling using morpholinos targeted to Wnt1 (Fig. 5C, F) and β-catenin (Fig. 5D, G), or a dominant negative (DN) Wnt1 construct. Knockdown of Wnt1 by any of these methods resulted in loss of Axud1 expression (Fig. 5M). Conversely, over-expression of Wnt1 increased Axud1 transcription (Fig 5E-H). Although confirmation of the direct control of Axud1 enhancers by TCF/Lef will require in-depth cis-regulatory analysis of this gene, these data indicate that Axud1 is downstream of Wnt signaling.
Figure 5. Axud1 acts as a Wnt effector during neural crest specification.
(A) Immunostaining of transverse sections of HH9 embryos revealed Wnt1 expression in the dorsal neural tube. DAPI staining was used to visualize cell nuclei. (B) In situ hybridization showed that Axud1 expression lies within the Wnt1 positive domain (dashed outline). (C-D) Whole mount dorsal views of embryo in which control morpholino was electroporated on the left (blue) and Wnt1 morpholino or β-Catenin morpholino was electroporated onto the right side of the same embryo. (F,G) The same embryos as above, showing effects on expression of Axud1. Inactivation of Wnt signaling results in downregulation (downward arrow) of Axud1 expression. (E,H) Conversely, over-expression of Wnt1 (Wnt1 O/E) results in elevation of Axud1 levels in the neural folds (upward arrow). (I, J) Whole mount views of an embryo with control morpholino on the left and Wnt1 morpholino on the right. Knockdown of Wnt1 resulted in downregulation of FoxD3 expression. (K, L) Whole mount views of an embryo with control morpholino on the left and Wnt1 morpholino plus Axud1 expression vector on the right, which was able to rescue FoxD3 expression to near-normal levels (compare J with L; see also Fig. S3). (M-N) Quantitation of embryos following loss of function and rescue assays. The purple bar in (M) denotes enhancement of Axud1 expression following Wtn1 overexpression, while the blue and light blue bars represent strong or mild downregulation following loss-of-function assays. Error bars in E and G represent standard deviation. HH: Hamburger and Hamilton developmental stages. β-CatMo: β-Catenin morpholino, Wnt1DN: Wnt1 dominant negative construct, Wnt1Mo: Wnt1 morpholino, Wnt1 O/E: Wnt1 overexpression.
Consistent with previous results suggesting a role for canonical Wnt signaling in neural crest induction(Garcia-Castro et al., 2002), we found that loss of either Wnt (Fig. 5I) or β-catenin (Fig. S3A-I) resulted in downregulation of expression of FoxD3 as well as other neural crest specifier genes. To firmly place Axud1 as a downstream effector of the Wnt pathway, we tested if it could rescue this loss of neural crest specification. The results show that co-electroporation of Axud1 expression vector with Wnt1 morpholino or Wnt1DN construct restored FoxD3 expression (Fig. 5K-L; quantification summarized in Fig. 5N). Similarly, the Axud1 expression vector was sufficient to rescue expression of neural crest specifier genes Sox9 and Sox10 following Wnt1 loss-of-function (Fig. S3J-R). These results suggest that Axud1 is a key player in Wnt1 signal transduction during formation of the neural crest, establishing its role as a critical effector of Wnt signaling during neural crest formation.
Axud1 interacts with neural plate border genes Pax7 and Msx1 in a transcriptional complex to activate neural crest gene expression
Our analysis showed that Axud1 is a downstream effector of Wnt signaling that is co-expressed with neural plate border genes Pax7 and Msx1 and directly regulates FoxD3 expression through the NC1 enhancer. Taken together with our previous data showing that Pax7 and Msx1 are direct inputs into the FoxD3 enhancers(Simoes-Costa et al., 2012), we hypothesized that Axud1 may interact directly with these neural plate border specifier genes to activate neural crest gene expression.
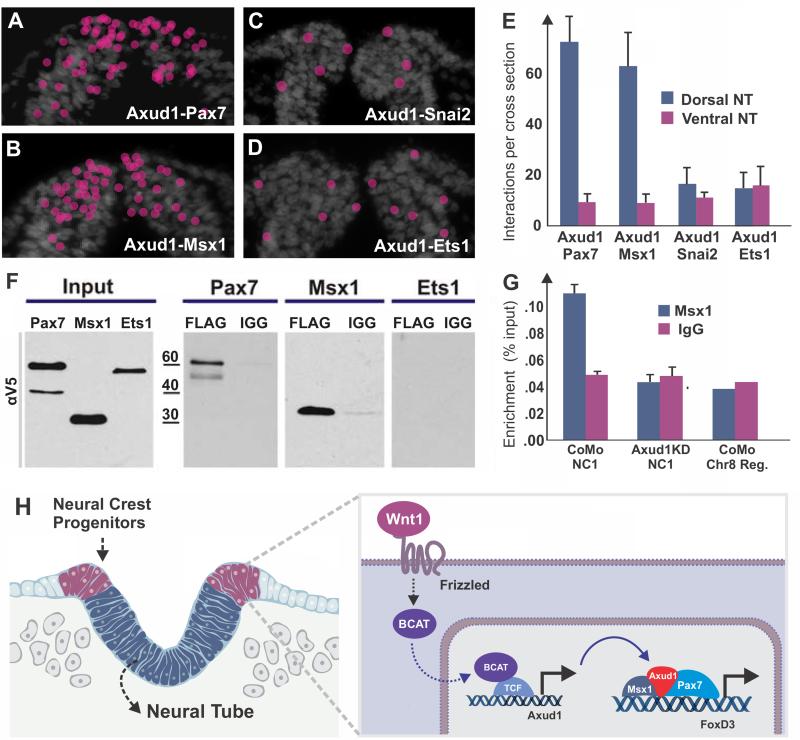
To test whether Axud1 might integrate Wnt signaling with transcriptional inputs to control cell identity and behavior, we tested if it directly interacts with neural plate border specifiers in vivo. To this end, we used proximity ligation assays (PLA)(Soderberg et al., 2006), which detect direct protein interactions in vivo with high specificity and sensitivity, to examine interactions between Axud1 with neural plate border and neural crest specifier genes. This approach uses two primary antibodies raised in different species that recognize the target proteins of interest and species-specific PLA secondary antibodies, each with an attached unique short DNA strand. Only when the PLA probes are in close proximity (less than 14 nm), do the DNA strands interact resulting in the production of a red punctum visible in tissue sections (Fig. S4). To quantitate the results, we counted puncta in the ventral and dorsal neural tubes (dividing the neural tube into four regions of the same size in the dorsal-ventral axis, and counting puncta in the two distal regions). The ventral region of the neural tube, where none of the genes are expressed, was used to define the inherent background levels of the assay. Using this approach, we examined pair-wise interactions between Axud1 and Pax7, Axud1 and Msx1, Axud1 and Snai2, and Axud1 and Ets1, shortly after cranial neural tube closure. The results showed that Axud1 interacted with both Pax7 and Msx1 proteins (Fig. 6A-B) within the newly closed neural folds. In contrast, Axud1 did not interact with Snai2 or Ets1, which are also expressed in this domain (Fig. 6C-E). Quantitation of the results is shown in Fig. 6E.
Figure 6. Axud1 physically interacts with neural plate border specifiers to regulate neural crest specification.
(A) Proximity ligation assays (PLA) visualized in transverse sections through the cephalic neural tube of HH9 embryos demonstrated that Axud1 interacts with Pax7 (positive interactions indicated by red dots; see also Fig. S4). (B) Similarly, Axud1 interacts with Msx1 (red dots). (C,D) In contrast, no interactions above background levels were detected between Axud1 and Snai2 or Ets1. (E) Quantification of PLA puncta revealed a large number of interactions in the dorsal neural tube between Axud1/Pax7 and Axud1/Msx1, whereas Axud1 interactions with Snai2 and Ets1 were within background levels. (F) Tagged constructs were introduced into HH5 embryos and co-immunoprecipitation pulldowns were performed at HH9. The results showed that Axud1 co-immunoprecipitates Pax7 and Msx1, but not Ets1. (G) Chromatin immunoprecipitation performed in embryos electroporated with control or Axud1 morpholinos. Under control conditions (CoMo), Msx1 directly binds to the NC1 enhancer, but was unable to do so in the absence of Axud1 (Axud1KD), indicating its requirement for recruitment of the transcriptional complex to the enhancer. (H) Model summarizing the results. Axud1 acts downstream of Wnt/β–catenin to integrate Wnt signaling with the activity of neural plate border specifiers by physically interacting with Msx1 and Pax7 to drive neural crest gene expression in the dorsal neural folds.
As a secondary test of the interaction between these transcriptional regulators, we used co-immunoprecipitation with protein isolated from dorsal neural folds of neurulating embryos. Embryos were electroporated with expression constructs encoding V5-tagged Axud1 and FLAG-tagged Pax7, Msx1, and Ets1. In the co-immunoprecipitation assays, Axud1 interacted with both Pax7 as well as Msx1 (Fig. 6F), further suggesting that they are members of a transcriptional complex responsible for neural crest gene transcription. In contrast, Ets1 was not able to bring down Axud1, consistent with the results obtained in the proximity ligation assay.
Finally, we used chromatin immunoprecipitation analysis to assess the binding of the neural plate border specifier Msx1 to the FoxD3 NC1 enhancer in the presence and absence of Axud1. This assay revealed that, after loss of Axud1, Msx1 failed to occupy the NC1 enhancer element to which it normally binds under control conditions (Fig. 6G). Thus, not only does Axud1 directly interact with neural plate border genes, but it also is necessary for their recruitment to neural crest enhancers. Taken together, these results firmly place Axud1 as a downstream effector of canonical Wnt signaling and critical component of the regulatory complex that, together with neural plate border transcription factors, initiates formation of the nascent neural crest (Fig 6H).
Discussion
Wnt signaling is used reiteratively throughout neural crest development, as it is involved in the activation of multiple transcriptional players that are part of different modules of the neural crest gene regulatory network(Raible and Ragland, 2005; Sauka-Spengler and Bronner-Fraser, 2008; Simoes-Costa and Bronner, 2015). First, the interplay of Wnt signaling with BMPs and FGFs elicits expression of a group of transcription factors that specify the neural plate border territory. Subsequently, these neural plate border specifiers cooperate with Wnt signaling to activate the neural crest specification program. During neural crest migration, non-canonical Wnt pathway controls formation of lamellipodia and filopodia required for cell motility(De Calisto et al., 2005). At later stages, Wnts also participate in the process of neural crest diversification, influencing the fate that neural crest cells will adopt as they differentiate into distinct derivatives. These multiple roles of Wnt signaling indicate that its function is context-dependent, and dictated by the regulatory state and competence of the target cells.
Our analyses of the expression, function, and regulation of Wnt effector, Axud1, reveal a critical role for this transcription factor in the process of neural crest specification. Axud1 has been shown to play important roles in several developmental systems, acting as a tumor suppressor in imaginal disc cells in Drosophila(Glavic et al., 2009) and in cell cycle control in the brain of zebrafish embryos(Feijoo et al., 2009). The finding that Axud knockout mice have craniofacial defects suggested a possible role for this transcription factor in neural crest development(Schmahl et al., 2007). Consistent with this possibility, our results show that integration of Wnt signaling and the neural plate border module is Axud1-dependent, such that a transcriptional complex comprised of Axud1, Pax7 and Msx1 directly activates FoxD3 gene expression. Thus, coordination of signaling and transcriptional inputs during neural crest formation takes place at the trans regulatory level, and depends upon protein-protein interactions between transcription factors. In particular, Axud1 was shown to be crucial for the recruitment of this transcriptional complex to the cis-regulatory element that mediates expression of cranial FoxD3 (Fig. 6H).
Furthermore, our model clarifies how the timing of neural crest specification is controlled. We previously showed that neural crest induction in the chick is ongoing during gastrulation(Basch et al., 2006). In fact, isolation of the presumptive neural crest domain from the gastrula subsequently led to production of neural crest cells in the absence of additional signals, suggesting that the domain was already induced by this time. Loss of Pax7 blocked the specification process, demonstrating a critical role for this transcription factor at the neural plate border. However, it was unclear why signaling and induction occurred at the gastrula stage whereas neural crest genes like FoxD3, Sox10 and others did not initiate expression until much later in the neurula. The present findings help to explain this conundrum regarding the timing of neural crest induction versus the manifestation of neural crest gene expression. Because onset of neural crest gene expression requires a transcriptional complex comprised of Pax7, Msx1 and Axud1, absence of any individual factor results in failure of neural crest formation. While Pax7 and Msx1 are already present in the neural plate border at gastrula stages, it is only after onset of Axud1 expression that the neural crest becomes specified. Thus, our uncovering of Axud1 as a critical neural crest input helps to resolve the gap in timing between neural crest induction and onset of specifier gene expression.
In summary, our results clarify the mechanism by which Wnts function in neural crest specification, and highlight how signaling pathways are integrated with the transcriptional machinery to allow for emergence of nascent neural crest cells. The findings also suggest that the Wnt1-Axud1 cascade is important for segregation of neural crest precursors from neural and placodal precursors within the neural plate border territory. In addition to playing critical roles in neural crest development, both the Wnt pathway and FoxD3 have been implicated in the maintenance of pluripotency of embryonic stem cells(Liu and Labosky, 2008; Marson et al., 2008; Sato et al., 2004) and important events such as the epithelial-to-mesenchymal transition (EMT)(Huber et al., 2005; Sauka-Spengler and Bronner-Fraser, 2008) and metastasis(Yook et al., 2006). Thus, our results showing that Axud1 acts as a molecular link between Wnt signaling and stem cell factor FoxD3 may reflect a general mechanism that functions in multiple cellular processes.
Experimental Procedures
Embryos
Fertile chicken eggs were purchased from commercial sources, and incubated at 37°C until embryos reached the desired stages. Chic ken embryos were collected according to Chapman and colleagues(Chapman et al., 2001) and staged according to the criteria of Hamburger and Hamilton(Hamburger and Hamilton, 1951). For in situ hybridization, embryos were fixed overnight at 4°C in phosphate buffered s aline (PBS) pH 7.4 containing 4% paraformaldehyde, dehydrated and stored in methanol. For immunohistochemistry, embryos were fixed for 15 minutes in 4% PFA in PBS and processed immediately.
Whole mount in situ hybridization
Whole mount in situ hybridization was performed as described (Wilkinson, 1992), with modifications including more extensive washes and a CHAPS-based hybridization buffer(Sauka-Spengler et al., 2007). Double fluorescent in situ hybridization was performed using the Tyramide TSA system from Perking Elmer (TSA Plus Cyanine 5 & Fluorescein, NEL754001KT), as described(Denkers et al., 2004). The templates for mRNA probe synthesis were the following: FoxD3(Kos et al., 2001), Pax7(Basch et al., 2006), Msx1 (ChEST900p21 (Boardman et al., 2002)), Snail2(Nieto et al., 1994), Sox9 and Sox10(Betancur et al., 2010), Ets1 (ChEST218o8 (Boardman et al., 2002)), Wnt1(Hollyday et al., 1995). Linearized DNA was used to synthesize digoxigenin- and fluorescein-labeled antisense probes with Promega RNA polymerases. RNA probes were purified with Illustra ProbeQuant G-50 Micro Columns (GE Healthcare, 28-9034-08).
Cryosectioning
For histological analysis, embryos were incubated in 5% sucrose (in PBS) for 2 hours at room temperature and subsequently transferred to 15% sucrose and incubated overnight at 4°C. Embryos were incubated in 17% gel atin for 3 hour at 37°C and frozen at −80°C. 10-15 μm transverse sections were obtained by cryosectioning. For imaging, slides were washed twice for 10 minutes in PBS with 0.1% Tween, rinsed in double-distilled water, and mounted with PermaFluor Mounting Medium (Thermo Electron Corporation, 434990).
Immunohistochemistry
For whole mount immunostaining we used the protocol described by Ezin and colleagues(Ezin et al., 2009). Briefly, after fixation embryos were dissected and washed in PBS with 0.3% Triton supplemented with 0.1% DMSO. Embryos were blocked for 2 hours in the same solution supplemented with 10% donkey serum. The following antibodies were used: Anti-CSRNP1 (Axud1) antibody produced in rabbit (Sigma, HPA045207, 1:300), anti-WNT1 rabbit polyclonal (Abcam, b15251, 1:200) and anti-HNK1 (DSHB, 1:10), anti-cleaved Caspase-3 antibody (Cell Signaling, 9661, 1:200), anti-Sox2 (R&D Systems, AF2018, 1:200) and anti-Six1 (Sigma, HPA001893, 1:200). Secondary antibodies used included donkey anti-rabbit IGG, or goat anti-mouse IGM, conjugated with Alexa Fluor 488/555/594/647 (1:2000, Molecular probes). For anti-WNT1(Serralbo and Marcelle, 2014), signal was amplified with the Tyramide TSA system from Perkin Elmer according to the manufacturer’s instruction.
Cloning of Axud1
Axud1 cDNA was PCR-amplified from a Gallus gallus cDNA library made from stage HH8 dorsal neural folds. Following amplification, the PCR product was cloned with the Invitrogen TA Cloning Kit Dual Promoter (Invitrogen, K2050), sequenced from both ends and used as template for RNA probe synthesis. To obtain the full-length sequence of the Axud1 UTR for morpholino design, we performed 5′ RACE using the GeneRacer Kit (Invitrogen, L150201). The gene specific primer used was AxudRaceInt (5′-TCTGCTAACGTGAACTGCCGGGAGGA-3′) and the RACE library was synthesized with RNA obtained from HH8 G. gallus embryonic heads.
Chick embryo electroporation
Chicken embryos were electroporated ex ovo at HH5+ using previously described techniques(Sauka-Spengler and Barembaum, 2008). Briefly, embryos were dissected from the eggs using a filter paper ring and submersed in Ringer’s buffer. Morpholinos or expression constructs were subsequently micro-injected in the space between the epiblast and vitelline membrane, and the embryos were electroporated with platinum electrodes (five 50ms pulses of 5.1V, with an interval of 100ms between pulses). Following electroporation, embryos were cultured at 37°C in sterile petri dishes containing fresh albumen until the desired stages. Embryo survival was >90%, with the vast majority of the embryos looking healthy prior to fixation. In all experiments, the two sides of the embryos were electroporated with different reagents. The left side (control) was injected with biotin-labeled control morpholinos or control expression vectors. The right side (experimental) was injected with FITC-labeled targeted morpholinos and expression vectors. Both morpholinos are injected at the same time, in the space between the epiblast and the vitelline membrane of gastrula stage embryos. At the time of injection, the left and right domains are sufficiently far apart that the morpholinos do not meet in the middle. Moreover, the midline often appears to act as barrier separating the injected morpholinos. As the embryos undergo neurulation, the two electroporated domains converge toward the midline. This approach allows for direct comparison between the two sides of the same embryo. All embryos were screened prior to further analysis, and embryos with weak or patchy electroporation, or with morphological abnormalities were discarded. Visualization of biotin-labeled control morpholino was performed by incubating morphants with 1ng/ul Streptavidin, Alexa Fluor 647 (Molecular Probes, S32357) in PBS Triton (0.3%) for 2 hours, followed by 3 washes of 5 minutes in PBST.
Axud1 loss-of-function
Axud1 knockdown experiments were performed by electroporation of targeted morpholinos (Gene Tools) in the experimental side of the embryos. We used a FITC-labeled translation-blocking morpholino (AxudTBmo 5′- TTTTCAACACCCCACTCATGGTGAC-3′) and a FITC-labeled splice-blocking morpholino (AxudSBmo 5′-CCCCCAATCCAGCCACTCACTTTCC-3′), which was targeted to the Exon2/Intron2 boundary. Morpholinos were electroporated at a concentration of 1.5uM and supplemented with carrier DNA (empty pTK-EGFP(Uchikawa et al., 2003) vector) at a concentration of 1ug/ul, and 10mM Tris pH8. To assess effects on the NC1 enhancer, control and targeted morpholinos were co-electroporated with the ptk-NC1:mCherry constructs(Simoes-Costa and Bronner, 2013).
Expression vectors
Axud1 expression vector was built by inserting full-length coding sequence of Axud1 (1.7kb) in a pCAGGS-EGFP backbone. The coding sequence of Axud1 was fused with a 2A linker and mCherry with fusion PCR. This Axud1-2A-mCherry cassette was then cloned into pCAGGS-EGFP with XhoI and BsrGI. To further support our knockdown experiments, we also developed an Axud1 dominant negative construct (Axud1DN). Previous studies indicate that Axud1 acts as an activator, containing a transactivation domain in the C-terminus of the protein(Gingras et al., 2007). To generate a dominant negative construct, we inserted fused the Engrailed Repressor (EnR)(Lee et al., 2004) domain to the C-terminus of the protein, such that it would override its activating properties. The Axud1-EnR cassette was subsequently subcloned in pCI:H2B-RFP with XhoI and NheI.
Enhancer analysis
Mutation of Axud1 binding site in the NC1 enhancer was performed as described (Simoes-Costa et al., 2012) with substitution of 20bp containing the site by 20bp of the coding region of GFP (tggagtacaactacaacagc). Amplified fusion fragments were cloned into ptkeGFP and sequenced to ensure no additional mutations were present. The mutated enhancer construct driving GFP was electroporated as described above into stage HH4 embryos together with wild type enhancer driving mCherry, and analyzed for expression of eGFP and mCherry at stages HH9–10. Co-electroporation with the wild type enhancer assures that differences in intensity are not a result of variation in electroporation efficiency. For qPCR analysis, we extracted RNA from embryos co-electroporated with wtNC1:eGFP and wtNC1:mCherry or mutNC1:eGFP and wtNC1:mCherry (all at a concentration of 1ug/ul), according to the manufacturer’s protocol. cDNA was synthesized using SuperScript III First Strand Synthesis kit (Invitrogen) with random primers for RT priming. qPCR was performed using SYBR Green (Bio-Rad) according to the manufacturer’s instructions. For reference genes, we used qPCR primers designed for the HPRT1 and GAPDH genes. Reporter gene levels are represented relative to GAPDH expression.
Chromatin Immunoprecipitation (ChIP)
Embryos were electroporated with a pCI:FLAG-Axud1 construct, and incubated until stage HH8. For each experiment, 20 embryos were dissected in Ringer’s solution, chromatin prepared from dorsal neural folds was immunoprecipitated as described(Karafiat et al., 2005; Simoes-Costa et al., 2012) using the Sigma M2 antibody. For endogenous chromatin immunoprecipitation in Axud1 morphants, embryos were electroporated with either control morpholino or the AxudSBmo at a concentration of 1.5mM. Chromatin was isolated from dorsal neural tube regions of HH8 chicken and immunoprecipitated with an Msx1 rabbit polyclonal antibody (Sigma M0944) and normal rabbit IgGs (ab27478, Abcam) as controls, as previously described(Simoes-Costa et al., 2012).
Nanostring analysis
To identify targets of Axud1, we compared the expression profiles of control and morpholino-treated neural folds obtained from the same embryo. Stage HH5+ chick embryos were electroporated with control morpholino and Axud1SBmo on the left and right side, respectively (Fig. 4J). Both morpholinos were injected in a concentration of 1.5uM. Embryos were incubated at 37°C for 12 hours until they reached stage HH8+. The dorsal cranial neural folds from these embryos were microdissected with iridectomy scissors and lysed in Ambion lysis buffer (RNAqueous®-Micro Kit, AM1931). Control and morpholino-treated neural folds were processed separately. RNA lysates were hybridized for 12 hours at 65°C to a Nanostring probe set containing ~100 probes for neural crest, placodal, neural and cell cycle markers(Hu et al., 2012). Samples were subsequently processed according to nCounter Prep Station Manual and the nCounter Digital Analyzer. Sample analysis and normalization were performed according to the guidelines presented by Barsi and colleagues(Barsi et al., 2014).
Wnt loss- and gain-of-function
For disruption of Wnt signaling, we used three different strategies: a dominant negative Wnt1 vector(Garcia-Castro et al., 2002), a splice-blocking Wnt1 morpholino (5′- GATGATGCCCCTACGGAGCGGGAAT-3′) and a translation-blocking β-catenin morpholino (5′-ATGTTCCTTGCTGTGCTCCCAGAAC-3′). Activation of the Wnt pathway was performed with a Wnt1 overexpression vector (Garcia-Castro et al., 2002). The morpholinos were electroporated at 1-1.5 mM, while the dominant negative construct and overexpression vectors were injected at 1ug/ul at HH5+. Both the dominant negative construct and the Wnt1 expression vector were co-electroporated with pCI-H2BeGFP at 1ug/ul vector as an electroporation control. After electroporation, embryos were cultured until HH7-9 and processed for in situ hybridization as described as above.
Proximity Ligation Assay
To identify interactions between Axud1 and neural plate border specifiers, we used the DuoLink Fluorescence approach (Sigma, DUO92101). Embryos were fixed for 15 minutes in 4% PFA, sectioned and processed for immunohistochemistry as described above. The primary antibodies used were the Anti-CSRNP1 (Axud1) antibody produced in rabbit (Sigma, HPA045207, 1:300), and the following monoclonal mouse antibodies: anti-Msx1/2 (DSHB 4G1, 1:50), anti-Pax7 produced in mouse (DSHB, 1:50), anti-Snail2 (DSHB 62.1E6, 1:50) anti-Ets1 (Santa Cruz Biotechnology, sc-56674, 1:300). After washing off excess of primary antibody, the assay was carried out according to the manufacturer’s instructions. PLA-positive puncta were quantified with a fluorescent microscope. We counted puncta in the ventral and dorsal neural tubes (dividing the neural tube in four regions of the same size in the dorsal-ventral axis, and counting puncta in the two distal regions). The ventral region of the neural tube was used to define the background levels of the assay. To facilitate visualization of the interactions in Fig. 6, a fuchsia circle was placed on each punctum.
Protein Immunoprecipitation (IP) and Co-IP
For Co-IP experiments, the coding sequences of Axud1, Ets1, Msx1 and Pax7 were fused to V5 and FLAG tags and cloned into pCI:H2B-RFP. Ets1(Betancur et al., 2010) and Pax7(Basch et al., 2000) were PCR amplified from the referenced templates. Axud1 and Msx1 were PCR amplified from cDNA. pCI:FLAG-Axud1 was co-electroporated into stage HH4 embryos with PCI:V5-Axud1, PCI:V5-Ets1, PCI:V5-Msx1 and PCI:V5-Pax7 constructs. Embryos were incubated until stage HH8, when their heads were dissected and used for preparation for IP. Electroporation efficiency was confirmed by RFP expression prior to dissection. Nuclear extracts were obtained by incubating and lysing embryonic heads (n=~30 per sample) in cell lysis buffer (10mM Hepes pH7.9, 1.5mM MgCl2, 10mM KCl solution). After centrifugation for 5 minutes at 2,500 g and 4°C, the supernatant was stored as cytosolic fraction. The pellet was then resuspended in 20mM HEPES, 1.5mM MgCl2, 0.2mM EDTA, 20% glycerol and 420mM KCL, supplemented with DTT, PMSF and protease inhibitor cocktail (Roche Applied Science, 11697498001). Samples were incubated for 20 minutes on ice and centrifuged for 3 minutes at 16,000 g and 4°C. The supernatant was stored as the nuclear fraction at −70°C. For co-immunoprec ipitation, nuclear protein extract was incubated in modified RIPA buffer with Dynabeads conjugated with the M2 anti-Flag antibody (Sigma-Aldrich, A8592) in rotation for 3 hours. Magnetic beads washed five times using modified RIPA, and proteins were eluted in Laemmli buffer. For Western blotting, proteins were separated by electroporation on SDS-PAGE gels for standard protein samples. Following immunoblotting on nitrocellulose membranes, bands were visualized by chemiluminescence (GE Healthcare, RPN2232). In co-IP experiments, V5 Mouse Monoclonal Antibody conjugated with HRP (Promega, R961-25) was used for target protein visualization.
Supplementary Material
Research Highlights.
Transcription factor Axud1 is expressed prior to neural crest markers.
Axud1 is required for the expression of multiple neural crest genes.
Axud1 acts as an effector of Wnt signaling during neural crest specification.
Axud1 physically interacts with neural plate border genes Pax7 and Msx1 in vivo.
Acknowledgements
We thank Joanna Tan-Cabugao, Brian Jun and Daniel S. E. Koo for technical assistance. This work was supported by NIH grants DE024157 and HD037105 to MEB. MS-C was funded by a fellowship from the Pew Fellows Program in Biomedical Sciences and by NIH grant K99DE024232. MS was funded by a Caltech SURF fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
MS-C and MEB conceived and designed the experimental approach. MS-C and MS performed the experiments and analyzed the data. MS-C and MEB wrote the manuscript.
References
- Barsi JC, Tu Q, Davidson EH. General approach for in vivo recovery of cell type-specific effector gene sets. Genome research. 2014;24:860–868. doi: 10.1101/gr.167668.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basch ML, Bronner-Fraser M, Garcia-Castro MI. Specification of the neural crest occurs during gastrulation and requires Pax7. Nature. 2006;441:218–222. doi: 10.1038/nature04684. [DOI] [PubMed] [Google Scholar]
- Basch ML, Selleck MA, Bronner-Fraser M. Timing and competence of neural crest formation. Dev Neurosci. 2000;22:217–227. doi: 10.1159/000017444. [DOI] [PubMed] [Google Scholar]
- Betancur P, Bronner-Fraser M, Sauka-Spengler T. Genomic code for Sox10 activation reveals a key regulatory enhancer for cranial neural crest. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3570–3575. doi: 10.1073/pnas.0906596107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boardman PE, Sanz-Ezquerro J, Overton IM, Burt DW, Bosch E, Fong WT, Tickle C, Brown WR, Wilson SA, Hubbard SJ. A comprehensive collection of chicken cDNAs. Curr Biol. 2002;12:1965–1969. doi: 10.1016/s0960-9822(02)01296-4. [DOI] [PubMed] [Google Scholar]
- Chapman SC, Collignon J, Schoenwolf GC, Lumsden A. Improved method for chick whole-embryo culture using a filter paper carrier. Developmental dynamics: an official publication of the American Association of Anatomists. 2001;220:284–289. doi: 10.1002/1097-0177(20010301)220:3<284::AID-DVDY1102>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- De Calisto J, Araya C, Marchant L, Riaz CF, Mayor R. Essential role of non-canonical Wnt signalling in neural crest migration. Development. 2005;132:2587–2597. doi: 10.1242/dev.01857. [DOI] [PubMed] [Google Scholar]
- Denkers N, Garcia-Villalba P, Rodesch CK, Nielson KR, Mauch TJ. FISHing for chick genes: Triple-label whole-mount fluorescence in situ hybridization detects simultaneous and overlapping gene expression in avian embryos. Developmental dynamics: an official publication of the American Association of Anatomists. 2004;229:651–657. doi: 10.1002/dvdy.20005. [DOI] [PubMed] [Google Scholar]
- Ezin AM, Fraser SE, Bronner-Fraser M. Fate map and morphogenesis of presumptive neural crest and dorsal neural tube. Developmental biology. 2009;330:221–236. doi: 10.1016/j.ydbio.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feijoo CG, Sarrazin AF, Allende ML, Glavic A. Cystein-serine-rich nuclear protein 1, Axud1/Csrnp1, is essential for cephalic neural progenitor proliferation and survival in zebrafish. Developmental dynamics: an official publication of the American Association of Anatomists. 2009;238:2034–2043. doi: 10.1002/dvdy.22006. [DOI] [PubMed] [Google Scholar]
- Garcia-Castro MI, Marcelle C, Bronner-Fraser M. Ectodermal Wnt Function As a Neural Crest Inducer. Science. 2002;13:13. doi: 10.1126/science.1070824. [DOI] [PubMed] [Google Scholar]
- Gingras S, Pelletier S, Boyd K, Ihle JN. Characterization of a family of novel cysteine- serine-rich nuclear proteins (CSRNP) PloS one. 2007;2:e808. doi: 10.1371/journal.pone.0000808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glavic A, Molnar C, Cotoras D, de Celis JF. Drosophila Axud1 is involved in the control of proliferation and displays pro-apoptotic activity. Mech Dev. 2009;126:184–197. doi: 10.1016/j.mod.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Green SA, Simoes-Costa M, Bronner ME. Evolution of vertebrates as viewed from the crest. Nature. 2015;520:474–482. doi: 10.1038/nature14436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. Journal of morphology. 1951;88:49–92. [PubMed] [Google Scholar]
- Hollyday M, McMahon JA, McMahon AP. Wnt expression patterns in chick embryo nervous system. Mech Dev. 1995;52:9–25. doi: 10.1016/0925-4773(95)00385-e. [DOI] [PubMed] [Google Scholar]
- Hu N, Strobl-Mazzulla P, Sauka-Spengler T, Bronner ME. DNA methyltransferase3A as a molecular switch mediating the neural tube-to-neural crest fate transition. Genes & development. 2012;26:2380–2385. doi: 10.1101/gad.198747.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Current opinion in cell biology. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Ikeya M, Lee SM, Johnson JE, McMahon AP, Takada S. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature. 1997;389:966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- Ishiguro H, Tsunoda T, Tanaka T, Fujii Y, Nakamura Y, Furukawa Y. Identification of AXUD1, a novel human gene induced by AXIN1 and its reduced expression in human carcinomas of the lung, liver, colon and kidney. Oncogene. 2001;20:5062–5066. doi: 10.1038/sj.onc.1204603. [DOI] [PubMed] [Google Scholar]
- Karafiat V, Dvorakova M, Krejci E, Kralova J, Pajer P, Snajdr P, Mandikova S, Bartunek P, Grim M, Dvorak M. Transcription factor c-Myb is involved in the regulation of the epithelial-mesenchymal transition in the avian neural crest. Cellular and molecular life sciences: CMLS. 2005;62:2516–2525. doi: 10.1007/s00018-005-5297-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khudyakov J, Bronner-Fraser M. Comprehensive spatiotemporal analysis of early chick neural crest network genes. Developmental dynamics: an official publication of the American Association of Anatomists. 2009;238:716–723. doi: 10.1002/dvdy.21881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos R, Reedy MV, Johnson RL, Erickson CA. The winged-helix transcription factor FoxD3 is important for establishing the neural crest lineage and repressing melanogenesis in avian embryos. Development. 2001;128:1467–1479. doi: 10.1242/dev.128.8.1467. [DOI] [PubMed] [Google Scholar]
- Kühl M. Wnt signaling in development. Landes Bioscience; Georgetown, Tex. Austin, Tex.: 2003. [Google Scholar]
- LaBonne C, Bronner-Fraser M. Neural crest induction in Xenopus: evidence for a two-signal model. Development. 1998;125:2403–2414. doi: 10.1242/dev.125.13.2403. [DOI] [PubMed] [Google Scholar]
- Le Douarin N, Kalcheim C. The neural crest. 2nd edn Cambridge University Press; Cambridge: 1999. [Google Scholar]
- Lee YH, Aoki Y, Hong CS, Saint-Germain N, Credidio C, Saint-Jeannet JP. Early requirement of the transcriptional activator Sox9 for neural crest specification in Xenopus. Developmental biology. 2004;275:93–103. doi: 10.1016/j.ydbio.2004.07.036. [DOI] [PubMed] [Google Scholar]
- Liu Y, Labosky PA. Regulation of embryonic stem cell self-renewal and pluripotency by Foxd3. Stem Cells. 2008;26:2475–2484. doi: 10.1634/stemcells.2008-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Foreman R, Chevalier B, Bilodeau S, Kahn M, Young RA, Jaenisch R. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell stem cell. 2008;3:132–135. doi: 10.1016/j.stem.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M. Amphioxus and lamprey AP-2 genes: implications for neural crest evolution and migration patterns. Development. 2002;129:4953–4962. doi: 10.1242/dev.129.21.4953. [DOI] [PubMed] [Google Scholar]
- Monsoro-Burq AH, Wang E, Harland R. Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during Xenopus neural crest induction. Developmental cell. 2005;8:167–178. doi: 10.1016/j.devcel.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Nieto MA, Sargent MG, Wilkinson DG, Cooke J. Control of cell behavior during vertebrate development by slug, a zinc finger gene. Science. 1994;264:835–839. doi: 10.1126/science.7513443. [DOI] [PubMed] [Google Scholar]
- Prasad MS, Sauka-Spengler T, LaBonne C. Induction of the neural crest state: control of stem cell attributes by gene regulatory, post-transcriptional and epigenetic interactions. Developmental biology. 2012;366:10–21. doi: 10.1016/j.ydbio.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan R, Graf M, Mason I, Lumsden A, Kiecker C. Complex and dynamic patterns of Wnt pathway gene expression in the developing chick forebrain. Neural development. 2009;4:35. doi: 10.1186/1749-8104-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raible DW, Ragland JW. Reiterated Wnt and BMP signals in neural crest development. Seminars in cell & developmental biology. 2005;16:673–682. doi: 10.1016/j.semcdb.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nature medicine. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Barembaum M. Gain- and loss-of-function approaches in the chick embryo. Methods in cell biology. 2008;87:237–256. doi: 10.1016/S0091-679X(08)00212-4. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nature reviews Molecular cell biology. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Meulemans D, Jones M, Bronner-Fraser M. Ancient evolutionary origin of the neural crest gene regulatory network. Developmental cell. 2007;13:405–420. doi: 10.1016/j.devcel.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Schmahl J, Raymond CS, Soriano P. PDGF signaling specificity is mediated through multiple immediate early genes. Nature genetics. 2007;39:52–60. doi: 10.1038/ng1922. [DOI] [PubMed] [Google Scholar]
- Serralbo O, Marcelle C. Migrating cells mediate long-range WNT signaling. Development. 2014;141:2057–2063. doi: 10.1242/dev.107656. [DOI] [PubMed] [Google Scholar]
- Simoes-Costa M, Bronner ME. Insights into neural crest development and evolution from genomic analysis. Genome research. 2013;23:1069–1080. doi: 10.1101/gr.157586.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes-Costa M, Bronner ME. Establishing neural crest identity: a gene regulatory recipe. Development. 2015;142:242–257. doi: 10.1242/dev.105445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes-Costa M, Tan-Cabugao J, Antoshechkin I, Sauka-Spengler T, Bronner ME. Transcriptome analysis reveals novel players in the cranial neural crest gene regulatory network. Genome research. 2014;24:281–290. doi: 10.1101/gr.161182.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes-Costa MS, McKeown SJ, Tan-Cabugao J, Sauka-Spengler T, Bronner ME. Dynamic and differential regulation of stem cell factor FoxD3 in the neural crest is Encrypted in the genome. PLoS genetics. 2012;8:e1003142. doi: 10.1371/journal.pgen.1003142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nature methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- Uchikawa M, Ishida Y, Takemoto T, Kamachi Y, Kondoh H. Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Developmental cell. 2003;4:509–519. doi: 10.1016/s1534-5807(03)00088-1. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG. In situ hybridization: a practical approach. IRL Press at Oxford University Press; Oxford; New York: 1992. [Google Scholar]
- Yook JI, Li XY, Ota I, Hu C, Kim HS, Kim NH, Cha SY, Ryu JK, Choi YJ, Kim J, et al. A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nat Cell Biol. 2006;8:1398–1406. doi: 10.1038/ncb1508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.