Abstract
The tumor suppressor p53 prevents cancer development via initiating cell cycle arrest, cell death, repair, or anti-angiogenesis processes. Over 50% of human cancers harbor cancer-causing mutant p53. p53 mutations not only abrogate its tumor suppressor function, but also endow mutant p53 with a gain-of-function (GOF), creating a proto-oncogene that contributes to tumorigenesis, tumor progression and chemo- or radiotherapy resistance. Thus, targeting mutant p53 to restore a wild-type p53 signaling pathway provides an attractive strategy for cancer therapy. We demonstrate that small molecule NSC59984 not only restores wild-type p53 signaling, but also depletes mutant p53 GOF. NSC59984 induces mutant p53 protein degradation via MDM2 and the ubiquitin-proteasome pathway. NSC59984 restores wild-type p53 signaling via p73 activation specifically in mutant p53-expressing colorectal cancer cells. At therapeutic doses, NSC59984 induces p73-dependent cell death in cancer cells with minimal genotoxicity and without evident toxicity towards normal cells. NSC59984 synergizes with CPT11 to induce cell death in mutant p53-expressing colorectal cancer cells and inhibits mutant p53-associated colon tumor xenograft growth in a p73-dependent manner in vivo. We hypothesize that specific targeting of mutant p53 may be essential for anti-cancer strategies that involve the stimulation of p73 in order to efficiently restore tumor suppression. Taken together, our data identify NSC59984 as a promising lead compound for anti-cancer therapy that acts by targeting GOF mutant p53 and stimulates p73 to restore the p53 pathway signaling.
Keywords: Colorectal cancer, p53 rescue, cancer therapy, anti-tumor effect
Introduction
Tumor suppressor p53 protects cells from oncogenesis and promotes sensitivity to anti-cancer therapy. Over 50% of human cancers harbor mutant p53, which inactivates p53 pathway signaling and its tumor suppressor function (1). p53 DNA mutations not only abrogate the p53 tumor suppressor function, but can also endow mutant p53 with a gain-of-function (GOF), rendering it a proto-oncogene (2, 3). One property of mutant p53 GOF is to form aberrant protein complexes with numerous interacting protein factors, including a subset of transcription factors such as SP1, NF-Y, p53, and p63/p73, to perturb their activities (4). The GOF of mutant p53 contributes to tumorigenesis, tumor progression and resistance to therapy (3). Therefore, targeting mutant p53 is an attractive strategy to overcome drug resistance and to sensitize tumors to cancer therapy. This concept is particularly further developed and mechanistically explored in the present study with NSC59984.
Some small molecule compounds targeting mutant p53 have been selected based on putative conformational changes within mutant p53 to restore wild-type p53. For example, CP31398 (5), PRIMA-1 (6) and NSC319726 (7) have been proposed to cause a conformational shift from mutant to wild-type p53, reactivating p53 function in tumor suppression. Although several small molecules can restore the p53 pathway, the GOF of mutant p53 can remain in the tumor cell and can represent an obstacle to tumor suppression as well as therapeutic efficacy. Eliminating mutant p53 is an approach that we decided to pursue in an attempt to abolish the GOF properties of mutant p53 in tumor cells, and with the idea that mutant p53 may represent a challenge for the general approach to stimulate p73, given the ability of mutant p53 to quench the tumor suppressive activity of p73. Few compounds have been reported to destabilize mutant p53, including 17AAG, Saha, gambogic acid and Arsenic (8–11). However, those compounds are incapable of restoring the p53 pathway of mutant p53 in tumor cells, and they have many other targets and mechanisms, making them non-specific. Thus, small molecules with the dual capability to restore the p53 pathway and deplete mutant p53 GOF proteins represent a novel strategy for cancer therapy and appear desirable to pursue for further therapeutic development.
p73, a member of the p53 family, is a transcription factor with high structural and sequence homology with p53. p73 has been found to have similar functions as wild-type p53 (12). p73 can transactivate the vast majority of p53 transcriptional target genes by binding to their regulatory regions in the same manner as p53, thereby impacting on cell growth and cell death pathways (13). Unlike p53, p73 is rarely deleted or mutated in human cancer. p73 is activated by complex signaling pathways in mammalian cells under stress (14). Activated p73 induces apoptosis and enhances chemosensitivity. A large variety of chemotherapeutic agents, such as camptothecin, etoposide and cisplatin, can up-regulate p73 expression (15). However, mutant p53 inhibits p73 activation by binding with p73 to form an inhibited complex with respect to the transactivation of target genes (4). Therefore, p73 provides a legitimate and bona fide attractive target to restore the p53 pathway in cancer therapy. A peptide, 37AA, has been found to cause p73 dependent cancer cell apoptosis (16). A small molecule, RETRA, was shown to release p73 by disturbing interaction of p73 and mutant p53 (17). These studies support the strategy of bypassing dysfunctional p53 signaling in cancer therapy through stimulation of p73-dependent signaling, while at the same time attempting to eliminate mutant p53, as is reported here.
In this paper, we use a functional cell-based high throughput screening strategy to identify small molecule compounds that both destabilize mutant p53 and restore wild-type p53 pathway via the activation of p73 in cancer cells. One such compound, NSC59984, appears to have a favorable therapeutic index, is non-genotoxic at effective doses that preferentially kill tumor cells, stimulates p73 activity, targets mutant p53 for degradation and displays anti-tumor effects in vivo in a p73-dependent manner.
Materials and Methods
High-Throughput Screening
Functional cell-based screening for small molecules that can increase p53-transcriptional activity was performed using noninvasive bioluminescence imaging in human colorectal cancer cells that stably express a p53-regulated reporter(Supplementary Materials and Methods), as previously described (18).
Cell lines
SW480, DLD-1, HCT116 and p53-null HCT116 cells which stably express a p53-regulated luciferase reporter were generated in our laboratory in 2003 (18, 19). MRC5, Wi38, Hop92 and RXF393 were obtained from ATCC and cultured as recommended. Cells were regularly authenticated by bioluminescence, growth and morphological observation.
CellTiter-Glo luminescent Cell viability assay
Cells were seeded at 5000 cells/well on 96-well plates. Cells were mixed with an equal volume of CellTiter-Glo reagents (Promega, Madison, WI), following the manufacturer’s protocol, and bioluminescence imaging was measured using the IVIS imager.
Knocking down p73 expression by retroviral shRNA
Cells were infected with retrovirus containing the pSIREN-REtrpcQ retroviral vector recombinant with TAp73 RNAi. Cells were cultured with blasticidin for several weeks, and blasticidin-resistant clones were selected. Knock-down of p73 was detected by measuring p73 protein levels by Western blot (Supplementary Materials and Methods) with anti-p73 antibody (Bethyl laboratories Inc. USA).
Over-expression of p73 by adenovirus infection
Cells were infected with an adenovirus that expresses p73-beta (Ad-p73) or wild-type p53 (Ad-p53) and cultured for 24 hr, as previously described (20). Then, the infected cells were cultured in fresh medium and subjected to different treatments.
RNA isolation and semi-quantitative RT-PCR (qRT-PCR)
Total RNA was isolated from cells using RNeasy mini kit (Qiagen, USA). Reverse transcription used SuperScript II first-strand synthesis system (Invitrogen, USA) with random primers. qRT-PCR reactions used SYBR green master mix with the Real-Time PCR Detection systems (Bio-Rad, USA). Primers for qRT-PCR are in Supplementary Materials and Methods.
Colony formation assays (21)
500 cells/well on 6-well plate were treated with NSC59984 for 3 days, then, cells were cultured with drug-free complete medium for two weeks with fresh medium changed every 3 days. Cells were fixed with 10% formalin and stained with 0.05% crystal violet at the end of two weeks period of cell culture.
In vivo Anti-tumor assays
All animal experiments were approved by the Institutional Animal Care and Use Committee at Penn State University. 5 million DLD-1 and p73 knock-down DLD-1 cells were implanted subcutaneously in the opposite flanks in each CRL nude mouse (female, 4–6 weeks old). Treatment with NSC59984 (i.p. injection) was initiated when the tumor masses reached a size of 3–5 mm. NSC59984 (45mg/kg) was injected by i.p. route every 5 days. Fifteen days after treatment, the mice were euthanized.
Statistical analysis
All results were obtained from triplicate experiments, unless other indicated. Statistical analyses were performed using PRISM4 Software (GraphPad Software, Inc., San Diego, CA, USA), ANOVA and Student’s t-test. Statistical significances were determined by p<0.05. Combination indices were calculated using the Chou-Talalay method with CalcuSyn software (Biosoft).
Results
NSC59984 specifically restores p53 pathway signaling in mutant p53-expressing human colorectal cancer cells
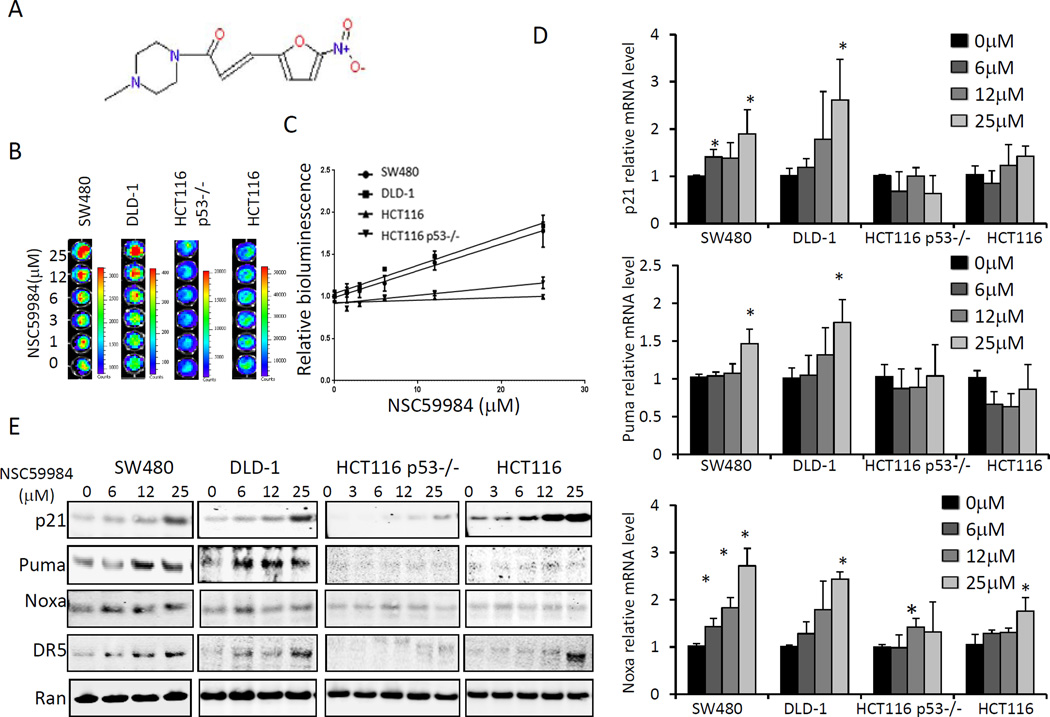
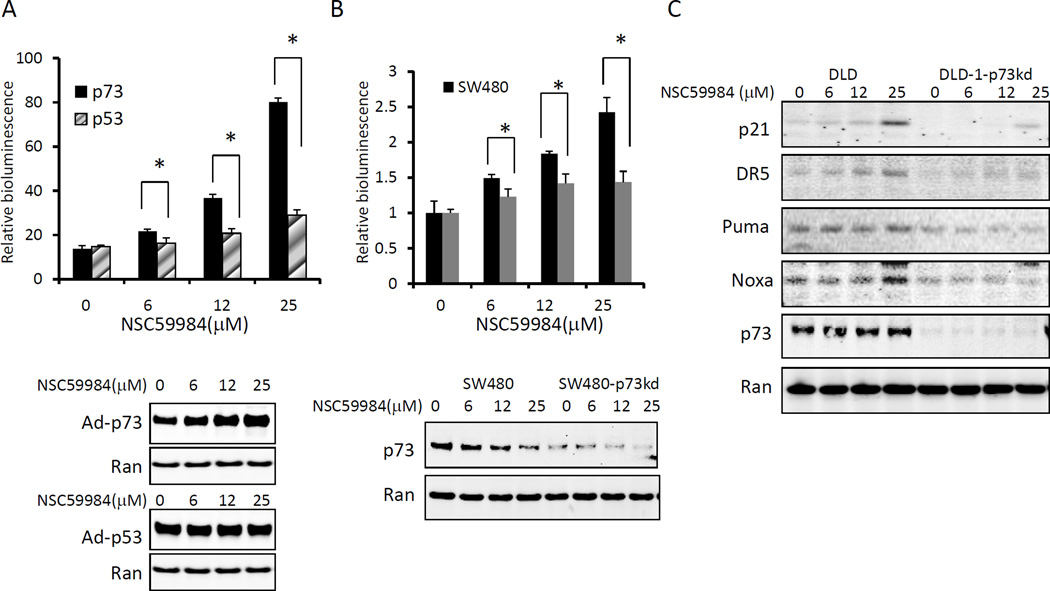
To identify small molecules that could restore p53 pathway signaling, we screened approximately 1990 small molecules from the National Cancer Institute (NCI) chemical diversity library II using a functional cell-based assay. A small molecular weight compound, NSC59984 (IPUA name is (E)-1-(4-methylpiperazin-1-yl)-3-(5-nitrofuran-2yl)prop-2-en-1-one, figure 1A) was found to increase p53-responsive reporter activity in both SW480 (mutant p53 R273H, P309S) and DLD-1(mutant p53 S241F) cells in a dose-dependent manner (1/slope = 31.37 in SW480, and 29.75 in DLD-1, P<0.01 compared to cells lacking mutant p53) (figure 1B and C). Consistent with p53 activation, endogenous protein levels of p21, Puma, Noxa and DR5, target genes of p53, were significantly up-regulated in SW480 cells and DLD-1cells in response to increasing doses of NSC59984 (figure 1E). Furthermore, mRNA levels of p21, Noxa and Puma were significantly increased in a dose-dependent manner in SW480 and DLD-1 cells at 3 hours after NSC59984 treatment (figure 1D). These results suggest that NSC59984 restores p53 pathway signaling in mutant p53-expressing SW480 and DLD-1 human colorectal cancer cells. To test whether the effect of NSC59984 on restoration of the p53 pathway was mutant p53-dependent, we treated HCT116 cells and p53-null HCT116 cells (figure 1) with NSC59984. Increasing doses of NSC59984 slightly induced p53-responsive bioluminescence in p53-null HCT116 cells (1/ slope=102.9), and no significant increase of p53-responsive bioluminescence was observed in wild-type p53-expressing HCT116 cells (1/slope=328.4) (figure 1B and C). Puma and p21 were not up-regulated at the mRNA level in these two cell lines, which lack mutant p53, in response to NSC59984 treatment. Noxa mRNA was slightly increased in response to 25 µM of NSC59984 in HCT116 and 12 µM of NSC59984 in p53-null HCT116 cells. However, Noxa mRNA was increased much less in these two cells than in mutant p53-expressing cancer cells DLD-1 and SW480 (figure 1D). Consistent with results showing lack of increase in mRNA levels of p53 target genes, protein levels of Puma, DR5 and Noxa were not up-regulated in HCT116 and p53-null HCT116 cells treated with NSC59984. Although p21 protein was up-regulated in HCT116 cells and p53-null HCT116 cells (figure 1E), the mRNA level of p21 was not significantly increased in response to NSC59984 treatment (figure 1D), suggesting that NSC59984-mediated up-regulation of p21 protein occurs at a post-translational level in HCT116 and p53-null HCT116 cells. Taken together, these results indicate that NSC59984 restores p53 pathway signaling specifically in mutant p53-expressing human cancer cells.
Figure 1.
NSC59984 restores p53-responsive transcriptional activity in mutant p53- expressing tumor cells. A. Structure of NSC59984. B. Imaging bioluminescence assay of p53-responsive transcriptional activity in cells at 24 hr after NSC59984 treatment. Data are representative of triplicate wells. C. The fold-increase of p53 responsive bioluminescence (B). D. mRNA levels of p21, Puma and Noxa in cells at 3 hr after NSC59984 treatment. mRNA levels were quantified by qRT-PCR. Data were normalized to GAPDH expression and plotted relative to cells treated with DMSO as control. Data are expressed as mean ± SD, *p<0.05 vs. control. E. Protein levels of p53 target genes in cells at 8 hr after NSC59984 treatment.
NSC59984 induces cell death in tumor cells but not normal cells with little or no genotoxicity
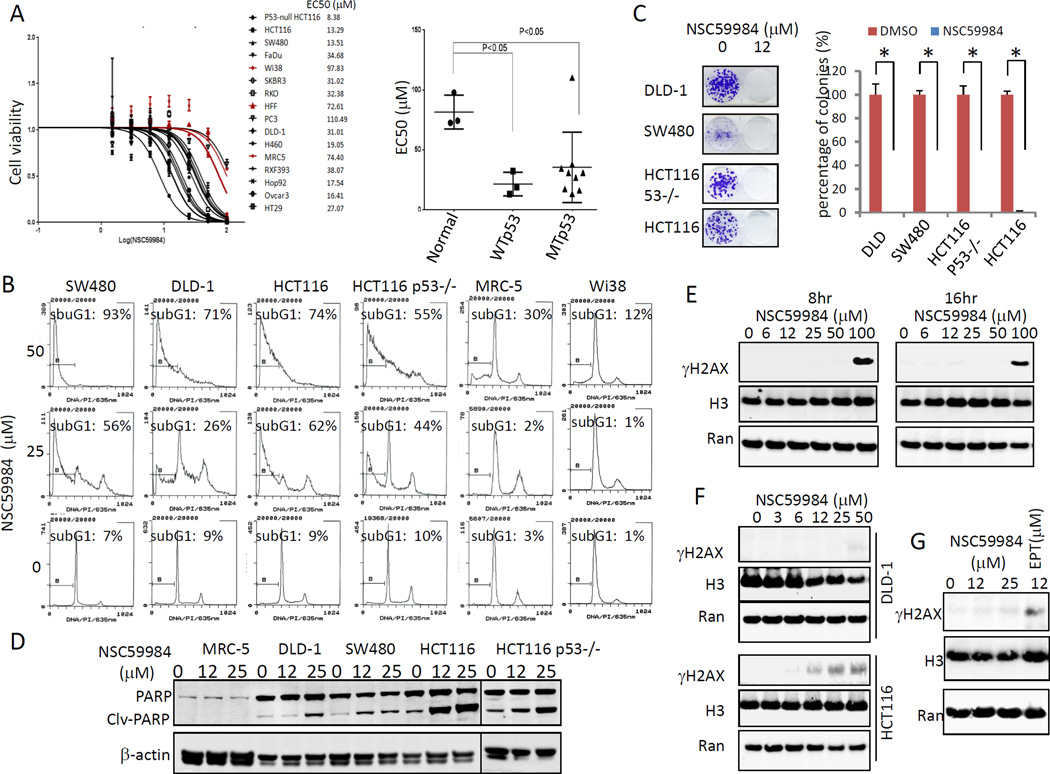
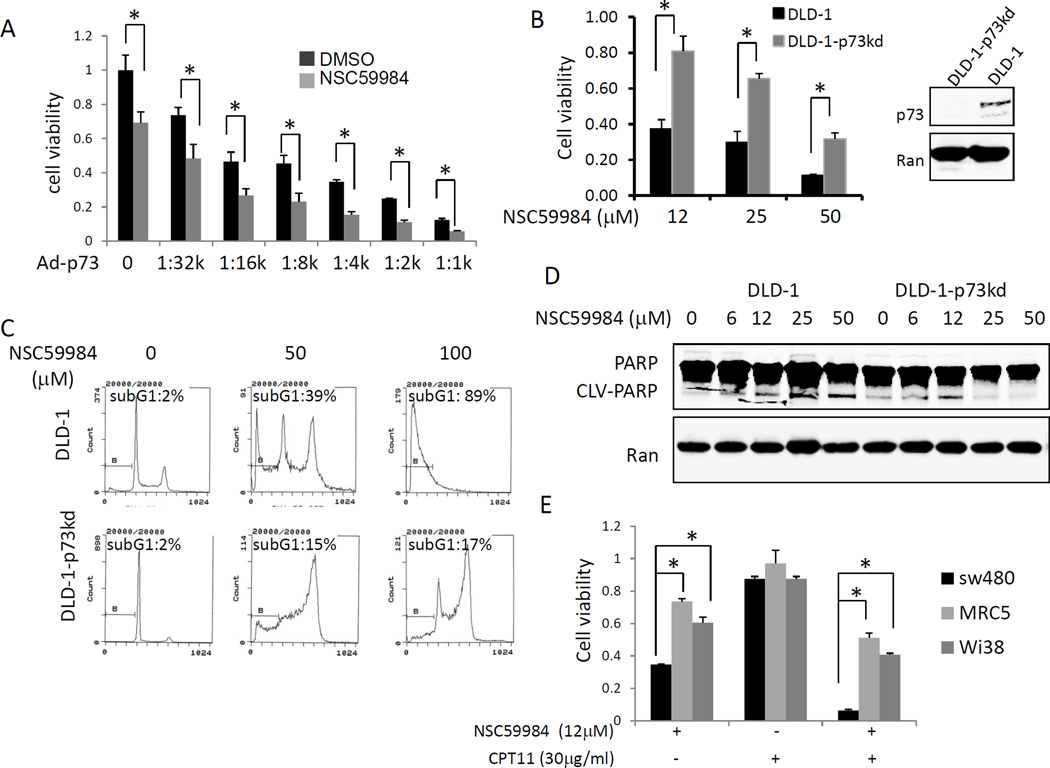
We investigated the effect of NSC59984 on cell death in tumor cells because NSC59984 restores the p53 pathway in mutant p53-expressing cancer cells. We first determined EC50 values for NSC59984 using a panel of cancer cell lines bearing different p53 mutations. The EC50 of NSC59984 varied among different cancer cell lines tested, which harbor different p53 mutations. The EC50 of NSC59984 in most cancer cells was found to be significantly lower than those of normal cells (figure 2A). FACS analysis showed that 25 µM of NSC59984 increased the sub-G1 DNA content (26–56%) in cancer cells, but not in normal cells at 72 hr after treatment (figure 2B). The high dose of NSC59984 (50µM) led to a 55–93% cancer cells to have sub-G1 content but only 12% and 30% of DNA content was found to be in sub-G1 in Wi38 and MRC5 normal human fibroblast cells (figure 2B). Taken together, these data suggest a favorable therapeutic index between normal and cancer cells. Colony formation assays further confirmed that NSC59984 was toxic toward cancer cells. Thus, NSC59984 significantly reduced colony numbers in cancer cells (figure 2C). We also examined cleaved PARP as a hallmark of caspase 3-dependent cell death in cells treated with NSC59984 for 30 hours. As shown in figure 2D, NSC59984 increased PARP cleavage in cancer cells in a dose-dependent manner, but PARP cleavage was not observed in normal cells at the same doses. On the basis of these findings, we conclude that NSC59984 induces cell death in cancer cells but displays little or no cytotoxicity toward normal cells at the doses tested.
Figure 2.
NSC59984 induces cell death in cancer cells with no genotoxicity. A. Cell viability of cells treated with NSC59984 for 72 hr. Cell viability data were normalized to those of DMSO treatment as control in each cell line and data analyses were performed using PRISM4 Software (left panel). EC50 data are expressed as mean ± SD in normal fibroblast cells (Normal, n=3), p53 mutant cancer cells (MTp53, n= 9) and cancer cells with wild-type p53 (WTp53, n=3) (right panel). B. Cell cycle profiles of cells at 72 hr after NSC59984 treatment. C. Colony formation of cancer cells. A percentage of colonies was obtained for each respective cell line treated with DMSO. Data represent mean ± SD, *p<0.05. D. Cleaved PARP protein level in cells treated with NSC59984 for 30 hr. E. Genotoxicty of NSC59984 in SW480 cells at 8 and 16 hr after NSC59984 treatment. F. Genotoxicty of NSC59984 in DLD-1 and HCT116 cells treated with NSC59984 for 8 hr. G. SW480 cells were treated with NSC59984 and DNA damage agent etoposide (EPT) for 8 hr. Genotoxicity was measured by γH2AX.
To determine whether NSC59984 has chemical genotoxicity that may be involved in cell death and p53 pathway restoration, we examined the level of γH2AX, a marker of genotoxicity due to DNA double-strand breaks (22). No γH2AX was found in SW480 and DLD-1 cancer cells treated with NSC59984 within 24 hr, even at high concentrations of 50µM (figure 2E–G). These data suggest that NSC59984 has little or no genotoxicity at the doses that effectively kill mutant p53-expressing cancer cells. We observed an increase of γH2AX in HCT116 cells at 8 hours after 12 µM of NSC59984 treatment.
NSC59984 induces mutant p53 protein degradation through MDM2-mediated ubiquitination in cancer cells
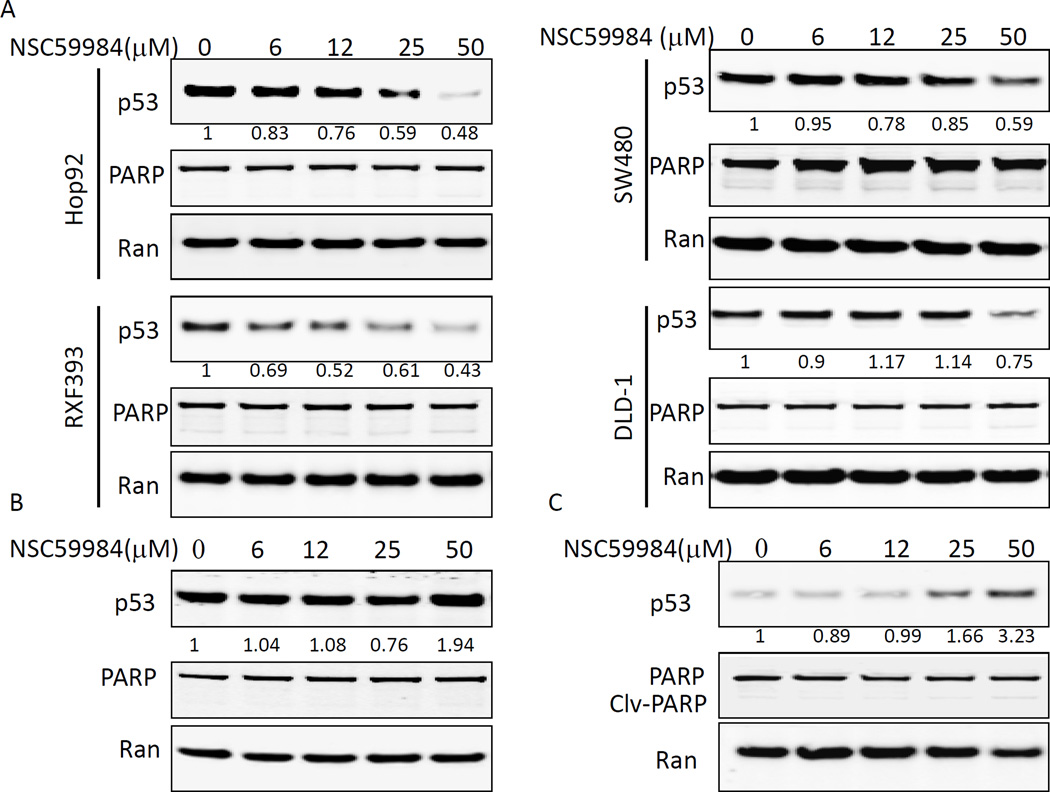
Given that NSC59984 restores p53 pathway signaling specifically in mutant p53-expressing cancer cells, we hypothesized that mutant p53 was the molecular target in NSC59984-treated cancer cells. NSC59984 treatment down-regulated various mutant p53 proteins in a dose-dependent manner in SW480, DLD-1, Hop92 (mutant p53 R175L) and RXF393 cells (mutant p53 R175H) (figure 3A). By contrast, wild-type p53 protein was up-regulated in MRC-5 normal cells treated with 50µM of NSC59984 (figure 3B) as well as in HCT116 cancer cells treated with 25 and 50µM of NSC59984 (figure 3C). The up-regulation of wild-type p53 protein was correlated with the increase of γH2AX (figure 2F) in HCT116 cells treated with NSC59984. These results suggest that NSC59984 specifically down-regulates the mutant p53 protein. Neither cleaved PARP (figure 3), nor subG1 DNA-content was increased in Hop92 and SW480 cells treated with 25 µM and 50 µM of NSC59984 (Supplementary figure S1) in the context of reduced mutant p53 protein expression. Increasing dose of NSC59984 (50 µM) slightly increased sub-G1 DNA content to 8.7 and 8.2% from 4.5% and 5.8% of DMSO treatment in RXF393 and DLD-1cells, respectively (Supplementary figure S1). At this dose, 25–50% of total mutant p53 protein was degraded in DLD-1 and RXF393 cells treated with NSC59984 (figure 3A). Taken together, these results suggest that cell death is not a significant mechanism by which NSC59984 reduces mutant p53. Thus, these results exclude the possibility of cell death as a major mechanism for the NSC59984-mediated depletion of mutant p53 protein.
Figure 3.
Effect of NSC59984 treatment on mutant p53 and wild-type p53 protein levels. Cells were treated with NSC59984 for 8 hours. A. Mutant p53 protein levels in cancer cells. B. Wild-type p53 protein level in MRC5 normal fibroblast cells. C. Wild-type p53 protein level in HCT116 cells. Data represent the fold-induction of p53 protein. The fold indicated is the ratio of p53: Ran as normalized to the DMSO control.
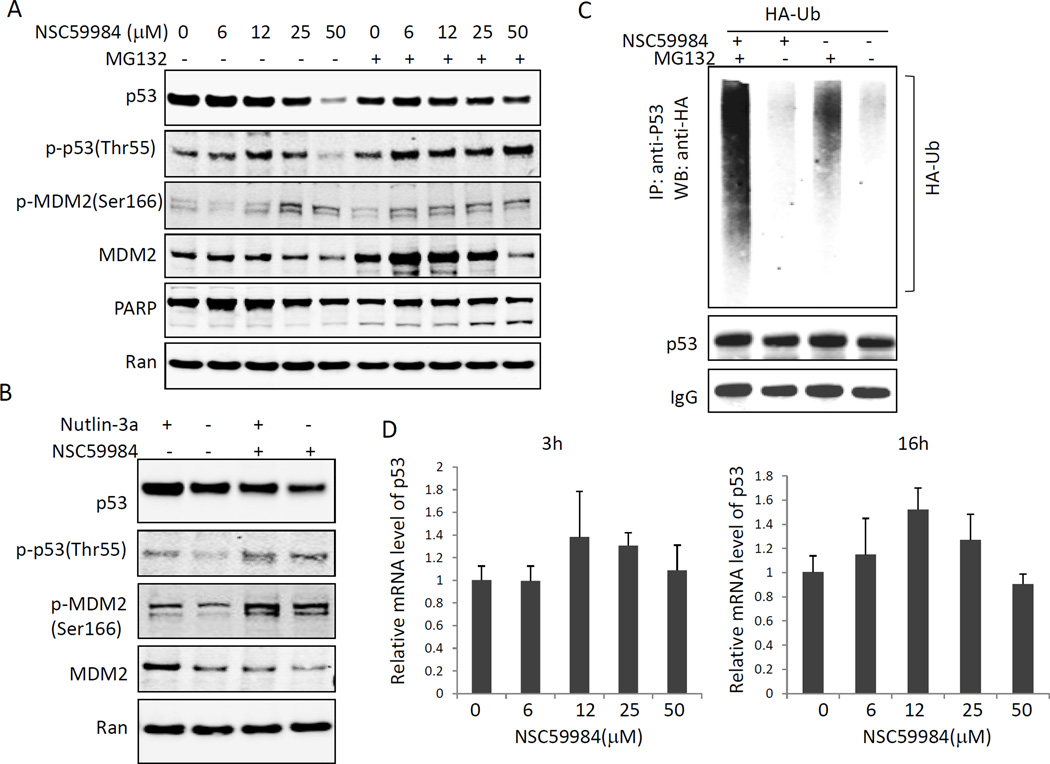
Ubiquitination is a major mechanism by which p53 protein stability is regulated (23). We treated mutant p53-expressing cancer cells with MG132, a proteasome inhibitor. MG132 treatment rescued the NSC59984-mediated down-regulation of mutant p53 (figure 4A). Moreover, we detected increased ubiquitination of mutant p53 in cancer cells treated with NSC59984 and MG132 (figure 4C). Taken together, our results suggest that NSC59984 causes mutant p53 protein ubiquitination and proteasomal degradation. To further determine whether p53 transcription contributes to the NSC59984-mediated decrease of mutant p53, we examined the mRNA level of mutant p53 in SW480 cancer cells. We found that p53 mRNA was not decreased in SW480 cells at 3 hr or at 16 hr of continuous NSC59984 treatment as compared to the DMSO control (figure 4D). These results taken together suggest that the effect of NSC59984 on decrease of mutant p53 protein occurs mostly at the post-translational level.
Figure 4.
NSC59984 induces mutant p53 degradation via MDM2-mediated ubiquitination. A. Mutant p53 protein levels in SW480 cells treated with 10µM of MG132 and NSC59984 for 16 hr. B. Mutant p53 protein levels in SW480 cells treated with 5µM of nutlin-3 and 25µM of NSC59984 for 16 hr. C. Ubiquitination (Ub) of mutant p53 in cells treated with NSC59984 and MG132. Cells were transfected with HA-Ub for 48 hr, followed by treatment with 25µM of NSC59984 and MG132 for 16 hr. Cell lysates were subjected to immunoprecipitation. D. Mutant p53 mRNA level in SW480 cells treated with NSC59984 for 3hr and 16 hr, mRNA was quantified by qRT-PCR. Data were normalized to GAPDH and plotted relative to cells treated with the DMSO control. Data are expressed as mean ± SD.
To investigate the role of MDM2 in NSC59984-mediated mutant p53 protein degradation, we treated mutant p53-expressing cancer cells with nutlin-3, an MDM2 inhibitor. Nutlin-3 treatment partially rescued the NSC59984-induced decrease in mutant p53 (figure 4B). We further found that mutant p53 was phosphorylated at Thr55 and MDM2 was phosphorylated at Ser166 in response to NSC59984 treatment in SW480 cells (figure 4A and B). Both phosphorylation of p53 at Thr55 and phosphorylation of MDM2 at Ser166 are important protein modifications that allow p53 degradation via MDM2 (24, 25). Taken together, these results suggest that NSC59984 induces mutant p53 protein degradation in part through an MDM2-mediated proteasomal mechanism.
To determine whether there is a wild-type conformational shift in p53 after NSC59984 treatment of mutant p53-expressing cancer cells, we examined the wild-type p53 protein in RXF393(p53 R175L) cells using the Pab1620 antibody. p53 R175L is a mutant that has been previously examined after exposure of p53 mutant conformation modifying agents (7). However, immunohistochemistry revealed no staining with Pab1620 in RXF393 cells before or after NSC59984 treatment. Immunoprecipitation assays further confirmed that no p53 protein bound with the Pab1620 antibody in RXF393 cell treated with NSC59984 (Supplementary figure S2). These results do not support a conformational shift towards wild-type of mutant p53 in cells after NSC59984 treatment, and they also exclude the possibility that mutant p53 degradation is due to a wild-type p53 conformational change.
Our results suggest that NSC59984 specifically induces mutant p53 protein degradation, at least in part, via the MDM2-mediated degradation through the ubiquitin-proteasome pathway.
NSC59984 restores p53 pathway signaling through activation of p73
It is possible that mutant p53 degradation leads to the release of p73 from their mutual complex. Therefore, we hypothesized that p73 function may be stimulated in NSC59984-treated cancer cells as a mechanism to restore p53 pathway signaling. To address this issue, we examined the effect of NSC59984 on the p53 pathway in mutant p53-expressing tumor cells in which p73 was either overexpressed by adenovirus infection or constitutively knocked-down by shRNA. NSC59984 treatment significantly increased p53-responsive bioluminescence interestingly to much higher levels in the p73-overexpressing SW480 cells than in the wild-type p53-overexpressing SW480 cells (figure 5A). The NSC59984-induced p53-responsive bioluminescence was abrogated by knock-down of p73 (figure 5B). We consistently observed that NSC59984-mediated up-regulation of endogenous of p21, Puma, DR5 and Noxa proteins was reduced by p73 knock-down (figure 5C). However, a small amount of p21 protein induction was still observed in p73 knock-down DLD-1 cells treated with 25 µM of NSC59984 (figure 5C). NSC59984 was found to up-regulate p21 expression at the protein level in HCT116 cells (figure 1). It is possible that p21 expression is regulated at transcriptional and protein levels in DLD-1 cells in which mRNA and protein levels of p21 were increased in response to NSC59984, although we believe only the transcriptional effects are p53-dependent (figure 1). Thus, knock-down of p73 only partially inhibited the NSC59984-mediated increase of p21 protein in DLD-1 cells (figure 5C). Taken together, these results indicate that p73 appears to be required for NSC59984 to restore the p53 pathway in mutant p53-expressing cancer cells.
Figure 5.
p73 is required for NSC59984 to restore the p53 pathway in mutant p53-expressing tumor cells. Cells were treated with NSC59984 for 8 hours. A. NSC59984-mediated p53-responsive reporter bioluminescence in p73-overexpressing SW480 cells and in wild-type p53-overexpressing SW480 cells. Relative bioluminescence was normalized to those of SW480 cells treated with DMSO as control. B. NSC59984-mediated p53-responsive reporter bioluminescence in p73 knock-down SW480 cells. Relative bioluminescence was normalized to those of DMSO treatment. Data (A and B) are expressed as mean ± SD. *p<0.05. C. Protein levels of p53 target gene expression in cells by Western blot.
NSC59984 induces p73-dependent cell apoptosis in cancer
Given the role of p73 in the NSC59984 restoration of the p53 pathway, we next investigated the impact of p73 on NSC59984-induced cell death. NSC59984 treatment synergized with exogenous p73 to reduce cell viability in DLD-1 cancer cells (figure 6A, supplementary table 1). By contrast, cell viability in p73 knock-down DLD-1 cells was found to be higher than those in DLD-1 cells after NSC59984 treatment (figure 6B). FACS assays revealed that the percentage of cells with sub-G1 content was increased by NSC59984; however, the effect of NSC59984 was significantly reduced by knock-down of p73 (figure 6C), suggesting that deficiency of p73 rescues cells from NSC59984-induced apoptosis. Consistent with these observations, NSC59984-induced PARP cleavage was partially abrogated by knock-down of p73 in DLD-1 cells at 24 hours of NSC59984 treatment (figure 6D). Taken together, these results suggest that NSC59984 induces p73-dependent cell death in mutant p53-expressing cancer cells.
Figure 6.
NSC59984 induces p73-dependent cell death in cancer cells. A. Cell viability of p73-overexpressing DLD-1 cells treated with NSC59984. DLD-1 cells were transiently infected with Ad-p73 (stock titer was 1:1000,1:1k) with double dilutions and followed with 12µM of NSC59984 treatment for 24 hr. B. Cell viability in p73 knock-down DLD-1 and DLD-1 cells treated with NSC59984 for 72 hr. Cell viability (A and B) was normalized to DLD-1 cells treated with DMSO as control. Data are expressed as mean ± SD. *p<0.05. C. Cell cycle profiles of DLD-1 and p73 knock-down DLD-1 cells treated with NSC59984 for 72 hr. D. Cleaved PARP protein level in DLD-1 and p73 knock-down DLD-1 cells treated with NSC59984 for 24 hr. E. Cell viability in SW480, MRC-5 and Wis38 cells treated with NSC59984 and CPT11 for 72 hr. Cell viability in cells treated with NSC59984 and CPT11 were normalized to those treated with DMSO as control. Data are expressed as mean ± SD. *p<0.05.
CPT11 is a DNA damaging agent used as cancer therapy in the clinic to treat colorectal cancer. CPT11 treatment has been reported to increase p73 protein levels in cancer cells (15). To test whether NSC59984 mediates the cellular sensitivity to conventional chemotherapy, the combination of NSC59984 and CPT11 was applied to cancer cells and normal human fibroblast cells. There was synergic activity of combinational treatment with NSC59984 and CPT11 in SW480 colon cancer cells as well as in normal fibroblast MRC5 and Wi38cells (supplementary table 2). Cell viability assay showed that combinational treatment with NSC59984 and CPT11 significantly reduced cell viability in SW480 cancer cells as compared to those in MRC5 and Wi38 normal cells (figure 6E).
NSC59984 represses xenograft tumor growth in vivo
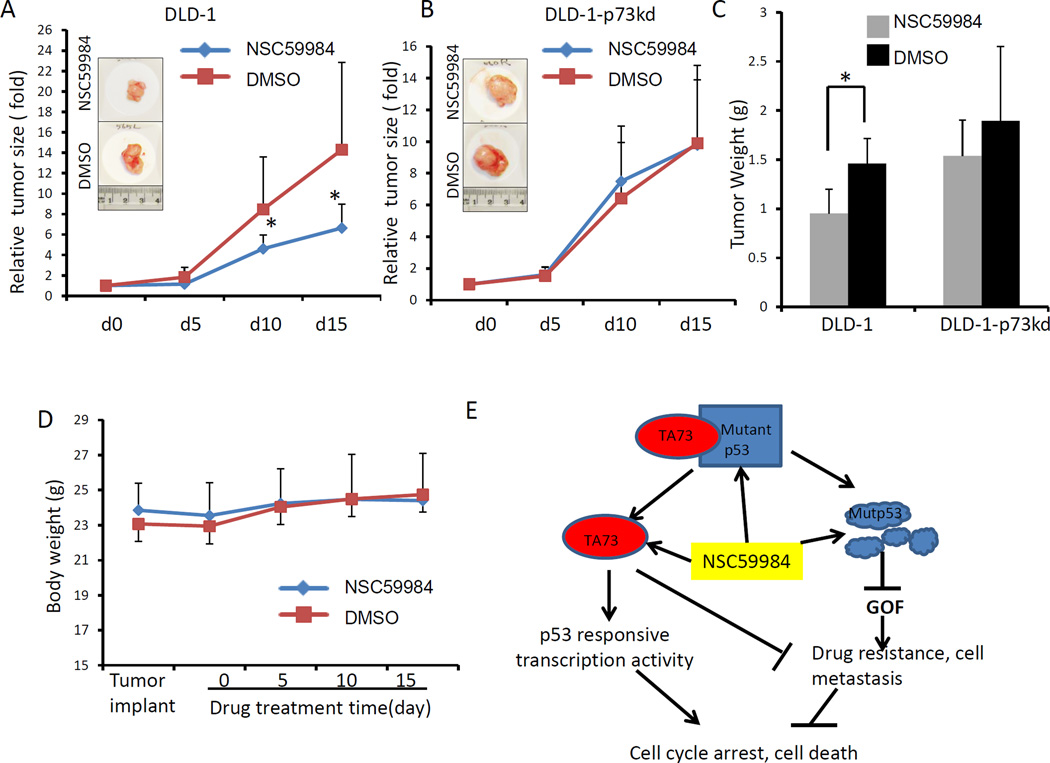
We further tested the potential therapeutic effects of NSC59984 in nude mice bearing colon-tumor xenografts. NSC59984 did not cause an obvious change in mouse body weights (figure 7D), and no overt toxic effect of NSC59984 was observed in mice with the tested dose. NSC59984 treatment significantly repressed DLD-1 xenograft tumor growth as compared to the DMSO control (figure 7A). We further measured tumor weight at day 15. Tumor weight was reduced by 34% with NSC59984 treatment in DLD-1 xenograft tumors, suggesting the NSC59984 suppresses tumor growth (p<0.05, figure 7C). By contrast, we did not observe tumor growth suppression in p73 knock-down DLD-1 xenograft tumors in response to NSC59984 at the same dose (figure 7B). NSC59984 treatment reduced tumor weight by 18% in p73 knock-down DLD-1 xenograft tumors (figure 7C). These results further confirm our observation in vitro that p73 is required for NSC59984 to induce tumor cell death in DLD-1 cells.
Figure 7.
Effects of NSC59984 injection on tumor growth and cell death in xenograft tumors in vivo. A. Relative tumor volumes of the DLD-1 xenograft tumors (n=7). B. Relative tumor volumes of the p73-knock-down DLD-1 xenograft tumors (n=7). C. Xenograft tumor weights at day 15 after treatment (n=7). D. Mouse body weights during the course of NSC59984 treatment (n=7). Data are expressed as the percent tumor growth in A and B and were normalized to tumor volume at the initiation of treatment. Tumor volumes and body weight were determined every five days. Tumor volumes were measured by caliper. Data are expressed as mean ± SD. *p<0.05. E. Schematic of NSC59984-mediated p53 pathway restoration via the activation of p73 and mutant p53 degradation.
Discussion
Most small molecular weight compounds targeting mutant p53 in cancer therapy do so by either restoring the p53 pathway or by abolishing mutant p53 to remove GOF. Here, we report a small molecule NSC59984 that not only specifically restores the p53 pathway through p73 but also depletes mutant p53 protein in mutant p53-expressing cancer cells.
There are several thousand mutations of p53 that have been reported in human cancer (26). Most small molecules restoring the p53 pathway have been identified and tested against a certain mutation of p53. For example, identification of Phikan083 was based on the Y222C mutation (27), SCH529074 was based on N268R mutation (28), PRIMA-1 was based on the mutations at residues 273 and 175 (6) and NSC-319726 was based on R175H mutation (7). Therefore, it has remained a challenge to develop universal p53-restoring drugs.
In our study, we demonstrate that NSC59984 causes degradation of multiple mutant p53 proteins in a variety of human cancer cell lines (figure 3A), suggesting the versatility of NSC59984 to target various mutants. Importantly, NSC59984 specifically targets mutant p53 and the restoration of the p53 pathway in mutant p53-expressing cancer cells, although it appears to have p53 pathway-independent effects in other tumor cell lines, i.e. p21 upregulation, that may suppress their growth. We found that NSC59984 treatment leads to the degradation of mutant p53, but not wild-type p53 protein. Wild-type p53 protein was up-regulated by NSC59984 at high doses in normal cells and cancer cells (figure 3B and C). We documented a corresponding effect of NSC59984 to specifically restore the p53 pathway in mutant p53-expressing colorectal cancer cells, but not reactivation of the p53 pathway in tumor cells with wild-type p53 or restoration of the p53 pathway in those that are p53-null. This conclusion is based on our findings that: 1) NSC59984 increased p53-Luc reporter bioluminescence only in mutant p53-expressing cancer cells SW480 and DLD-1; 2) expression of the p53 target genes p21, Puma, DR5 and Noxa was significantly up-regulated at the mRNA and the protein levels by NSC59984 in mutant p53-expressing cancer cells SW480 and DLD-1 as compared to those in wild-type p53-expressing HCT116 and p53-null HCT116 (figure1); 3) NSC59984 significantly increased p53-responsive bioluminescence to much greater levels in p73-overexpressing SW480 cells than in p53-overexpressing SW480 cancer cells (figure 5A). These data suggest the specificity of NSC59984 for targeting p53 mutant in cancer. The specificity and versatility of NSC59984 for targeting mutant p53 indicates that NSC59984 is a promising small molecule drug candidate for further development through targeting restoration of the p53 pathway in part through degradation of the mutant p53 protein.
The mutant p53 protein level is high in tumor cells due to its stabilization (4). Stabilization of mutant p53 is mostly due to the inability of mutant p53 to interact with MDM2, an E3 ubiquitin ligase(3). Our data demonstrates that NSC59984 induces mutant p53 protein degradation via MDM2-mediated ubiquitination and proteasomal degradation (figure 4). Another compound, NSC319726, provided a model for the wild-type conformational change from mutant p53 to be sequentially degraded through MDM2-mediated ubiquitination (7). However, we did not find a wild-type p53 conformational change in tumor cells treated with NSC59984 (Supplementary figure S2) or down-regulation of wild-type p53 protein by NSC59984 treatment (figure 3B and C). These results suggest that NSC59984 mediates the down-regulation of p53 due to mutant p53 degradation rather than restoration of a less stable wild-type p53 protein.
Hsp90 and Hsp70 are two molecular chaperones that stabilize mutant p53 protein by affecting the MDM2-mediated turnover of mutant p53 (29). Thus, mutant p53 escapes from MDM2-mediated degradation, and there are lower levels of MDM2 in mutant p53-expressing cells due to reduced transactivation of the MDM2 gene by p53 (3). Inhibition of Hsp90 has been shown to destabilize mutant p53 through MDM2 activity (9, 10). It remained unclear whether NSC59984 induces mutant p53 protein degradation through disturbing the MDM2-Hsp chaperone axis. A conformational change of mutant p53 is another possible mechanism of mutant p53 degradation by NSC59984. For example, CP31398 induces a wild-type conformational change in mutant p53 by modifying the unfolded mutant p53 (5). PRIMA-1 causes a conformational change by forming adducts with thiols in the mutant p53 core domain (30). Although NSC59984 did not restore a wild-type p53 conformation, it is possible that NSC59984 converts the mutant p53 structure to one more amenable to the MDM2-mediated ubiquitination to regulate mutant p53 degradation. We found phosphorylation of mutant p53 at Thr55 and phosphorylation of MDM2 at Ser166 in SW480 cancer cells treated with NSC59984 (figure 4). Phosphorylation of p53 at Thr55 and phosphorylation of MDM2 at Ser166 have been reported to contribute to p53 protein ubiquitination (24, 25). Our results suggest a possibility that NSC59984 induces mutant p53 and MDM2 protein modifications which contribute to mutant p53 protein degradation. It remains unclear how mutant p53 and MDM2 are phosphorylated by signaling pathways stimulated by NSC59984 in mutant p53-expressing cancer cells.
Mutant p53 protein degradation is an effective means to remove its GOF, resulting in release of p73 and other factors from inhibitory complexes with mutant p53. However, many mutant p53 protein targeting small molecules, such as HDACI and 17AAG, do not restore the p53 pathway (10, 31). Unlike these mutant p53-targeting agents, NSC59984 not only degrades mutant p53 protein to release p73, but also induces p73-dependent p53 restoration (figure 1 and 5). It is possible that NSC59984 converts the released p73 to an active form or stabilizes p73. Active p73 functions as a transcription factor to up-regulate p53 target genes such as p21, puma and DR5. We found that NSC59984 significantly induces p73-dependent p53 restoration only in mutant p53-expressing cancer cells (figure 1 and 5), but not in p53-null or wild-type p53-expressing HCT116 cells that contain wild-type p73 (figure 1). These results suggest that restoration of the p53 pathway occurs, at least in part, through the release of p73 in mutant p53-expressing tumor cells. The fraction of released and active p73 could be increased by mutant p53 degradation. We did not find increased p73 protein levels in the mutant p53-expressing tumor cells treated with NSC59984 at 8 hours by Western blot assay using anti-p73 antibody (Bethyl Laboratories Inc.). It is possible that post-translational modifications of p73 protein are induced by NSC59984, and that such NSC59984-mediated modifications interrupt the anti-p73 antibody (Bethyl Laboratories Inc.) recognizing modified p73 protein. We found that NSC59984 significantly enhanced p73-dependent p53-responsive reporter bioluminescence in the p73-overexpressing cells (figure 5A), suggesting that active p73 is involved in NSC59984-mediated p53 restoration. p73 activity is regulated by complex post-translational modifications and protein-protein interactions (14). The mechanism by which NSC59984 activates the released p73 remains to be further elucidated. Nevertheless, we demonstrate that p73 is required for NSC59984 to induce cell death in addition to restoring p53 pathway. We found that knock-down of p73 not only rescues cells from apoptosis induced by NSC59984 in vitro (figure 6), but also prevents the NSC59984-mediated suppression of xenograft tumor growth in vivo (figure 7). Although mutant p53 deletion has been reported to be sufficient to induce cell death (10, 31), our findings suggest that NSC59984-induced cell death is p73-dependent in mutant p53-expressing cancer cells. Activation of p73 is an important step for p73-induced cell death (14). How active p73 regulates cell death in response to NSC59984 treatment will be further investigated in the future. Because p73 is required for both NSC59984-mediated p53 restoration and cell death, it is possible that NSC59984 induces cell death via p73-dependent restoration of the p53 pathway. Therefore, NSC59984 offers a rational bypass mechanism of p53 restoration via the activation of p73 to kill cancer cells. Based on our findings, we provide a model for NSC59984 working in cancer cells (figure7E). NSC59984 releases p73 from the inhibitory complex of mutant p53 by degrading mutant p53. The released p73 may be further converted to the active form by NSC59984. Active p73 then restores the p53 pathway. NSC59984-mediated p53 restoration and/or active p73 together with depletion of GOF may result in tumor suppression (figure 7E).
NSC59984 induces cell death in mutant p53-expressing cancer cells with minimal genotoxicity. Importantly, we did not find cytotoxicity of NSC59984 against normal cells at the doses tested (figure 2), suggesting the safety of NSC59984 administration for cancer therapy. Indeed, in vivo experiments demonstrate that i.p. injection of NSC59984 suppresses xenograft tumor growth (figure 7), but was not toxic toward animals, suggesting that NSC59984 may be further evaluated for clinical development. Mutant p53 protein degradation releases many factors, including p73, from the mutant p53 protein complex (4), which might result in p73 independent off-target effects. Because of the specificity of NSC59984 for mutant p53, the potential off-target effects may be limited in mutant p53-expressing cells and may not affect normal cells containing wild- type p53. However, the anti-tumor effect of NSC59984 on cancer cells is not limited in p53-mutant cancer cells (figure 2). We noted that NSC59984 induced cell apoptosis in wild-type p53-expressing as well as p53-null cancer cells, suggesting that the cell death induced by NSC59984 in these cell lines is p53-independent. It is possible there may be tumor suppressive effects due to NSC59984-mediated up-regulation of p21 protein in these two cell lines (figure 1). Our findings suggest that NSC59984 regulates p21 expression at the post-translational level in HCT116 and p53-null HCT116 cancer cells. p21 regulation at the post-translational level could be an off-target effect of NSC59984 in cancer cells, and may involve effects of NSC59984 on MDM2 in a p53-independent manner that remains to be further unraveled.
p73 is an important determinant of chemosensitivity. In response to cellular stresses and DNA damage, p73 is activated through different signaling pathways and enhances chemosensitivity (14, 15). However, mutant p53 inhibits p73 activation, resulting in drug resistance. Our finding that NSC59984 rescues p73 activity to restore the p53 pathway provides a potential application of NSC59984 to reduce chemoresistance. Indeed, NSC59984 synergizes with CPT11 to suppress colorectal cancer cell growth (figure 6E, supplementary table 2). Therefore, NSC59984 warrants further evaluation in combination therapy to reduce the dose of CPT11 required for growth suppression in colorectal cancer. Combinatorial treatment with NSC59984 may minimize the side effects of CPT11 chemotherapy and increase its anti-tumor effects in colorectal cancer patients. Taken together, these results demonstrate that NSC59984 is a candidate therapeutic as both a single agent or in combination with conventional chemotherapy. Based on the findings in this study, we conclude that NSC59984 is a promising drug candidate that specifically targets mutant p53 via a mechanism involving both mutant p53 depletion and p73-dependent p53 pathway restoration.
Supplementary Material
Acknowledgements
The work was presented in part at the AACR 104th annual meeting in Washington, DC (April, 2013). The work was supported in part by NIH grants (N01-CN43302-WA-17, N01-CN43302-WA-27) to W.S.E-D. W.S.E-D. is a Founder of p53-Therapeutics, Inc., a biotech company focused on developing small molecule anti-cancer therapies targeting mutant p53.
Footnotes
Conflict of Interest Disclosure:
W.S.E-D. is a Founder of p53-Therapeutics, Inc., a biotech company focused on developing small molecule anti-cancer therapies targeting mutant p53. Dr. El-Deiry has disclosed his relationship with p53-Therapeutics and potential conflict of interest to his academic institution/employer and is fully compliant with institutional policy that is managing this potential conflict of interest.
References
- 1.van Oijen MG, Slootweg PJ. Gain-of-function mutations in the tumor suppressor gene p53. Clin Cancer Res. 2000;6:2138–2145. [PubMed] [Google Scholar]
- 2.Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 3.Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol. 2010;2:a001107. doi: 10.1101/cshperspect.a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foster BA, Coffey HA, Morin MJ, Rastinejad F. Pharmacological rescue of mutant p53 conformation and function. Science. 1999;286:2507–2510. doi: 10.1126/science.286.5449.2507. [DOI] [PubMed] [Google Scholar]
- 6.Bykov VJ, Issaeva N, Shilov A, Hultcrantz M, Pugacheva E, Chumakov P, et al. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med. 2002;8:282–288. doi: 10.1038/nm0302-282. [DOI] [PubMed] [Google Scholar]
- 7.Yu X, Vazquez A, Levine AJ, Carpizo DR. Allele-specific p53 mutant reactivation. Cancer Cell. 2012;21:614–625. doi: 10.1016/j.ccr.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Zhao Q, Qi Q, Gu HY, Rong JJ, Mu R, et al. Gambogic acid-induced degradation of mutant p53 is mediated by proteasome and related to CHIP. J Cell Biochem. 2011;112:509–519. doi: 10.1002/jcb.22941. [DOI] [PubMed] [Google Scholar]
- 9.Li D, Marchenko ND, Moll UM. SAHA shows preferential cytotoxicity in mutant p53 cancer cells by destabilizing mutant p53 through inhibition of the HDAC6-Hsp90 chaperone axis. Cell Death Differ. 2011;18:1904–1913. doi: 10.1038/cdd.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li D, Marchenko ND, Schulz R, Fischer V, Velasco-Hernandez T, Talos F, et al. Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol Cancer Res. 2011;9:577–588. doi: 10.1158/1541-7786.MCR-10-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Yan W, Chen X. Mutant p53 disrupts MCF-10A cell polarity in three-dimensional culture via epithelial-to-mesenchymal transitions. J Biol Chem. 2011;286:16218–16228. doi: 10.1074/jbc.M110.214585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melino G, De Laurenzi V, Vousden KH. p73: Friend or foe in tumorigenesis. Nat Rev Cancer. 2002;2:605–615. doi: 10.1038/nrc861. [DOI] [PubMed] [Google Scholar]
- 13.Lunghi P, Costanzo A, Mazzera L, Rizzoli V, Levrero M, Bonati A. The p53 family protein p73 provides new insights into cancer chemosensitivity and targeting. Clin Cancer Res. 2009;15:6495–6502. doi: 10.1158/1078-0432.CCR-09-1229. [DOI] [PubMed] [Google Scholar]
- 14.Conforti F, Sayan AE, Sreekumar R, Sayan BS. Regulation of p73 activity by post-translational modifications. Cell Death Dis. 2012;3:e285. doi: 10.1038/cddis.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irwin MS, Kondo K, Marin MC, Cheng LS, Hahn WC, Kaelin WG., Jr Chemosensitivity linked to p73 function. Cancer Cell. 2003;3:403–410. doi: 10.1016/s1535-6108(03)00078-3. [DOI] [PubMed] [Google Scholar]
- 16.Bell HS, Dufes C, O'Prey J, Crighton D, Bergamaschi D, Lu X, et al. A p53-derived apoptotic peptide derepresses p73 to cause tumor regression in vivo. J Clin Invest. 2007;117:1008–1018. doi: 10.1172/JCI28920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kravchenko JE, Ilyinskaya GV, Komarov PG, Agapova LS, Kochetkov DV, Strom E, et al. Small-molecule RETRA suppresses mutant p53-bearing cancer cells through a p73-dependent salvage pathway. Proc Natl Acad Sci U S A. 2008;105:6302–6307. doi: 10.1073/pnas.0802091105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W, Kim SH, El-Deiry WS. Small-molecule modulators of p53 family signaling and antitumor effects in p53-deficient human colon tumor xenografts. Proc Natl Acad Sci U S A. 2006;103:11003–11008. doi: 10.1073/pnas.0604507103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W, El-Deiry WS. Bioluminescent molecular imaging of endogenous and exogenous p53-mediated transcription in vitro and in vivo using an HCT116 human colon carcinoma xenograft model. Cancer Biol Ther. 2003;2:196–202. doi: 10.4161/cbt.2.2.347. [DOI] [PubMed] [Google Scholar]
- 20.Huang C, Zhang XM, Tavaluc RT, Hart LS, Dicker DT, Wang W, et al. The combination of 5-fluorouracil plus p53 pathway restoration is associated with depletion of p53-deficient or mutant p53-expressing putative colon cancer stem cells. Cancer Biol Ther. 2009;8:2186–2193. doi: 10.4161/cbt.8.22.10446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1:2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 22.Kuo LJ, Yang LX. Gamma-H2AX - a novel biomarker for DNA double-strand breaks. In Vivo. 2008;22:305–309. [PubMed] [Google Scholar]
- 23.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li HH, Li AG, Sheppard HM, Liu X. Phosphorylation on Thr-55 by TAF1 mediates degradation of p53: a role for TAF1 in cell G1 progression. Mol Cell. 2004;13:867–878. doi: 10.1016/s1097-2765(04)00123-6. [DOI] [PubMed] [Google Scholar]
- 25.Meek DW, Knippschild U. Posttranslational modification of MDM2. Mol Cancer Res. 2003;1:1017–1026. [PubMed] [Google Scholar]
- 26.Olivier M, Hollstein M, Hainaut P. TP53 Mutations in Human Cancers: Origins, Consequences, and Clinical Use. Cold Spring Harb Perspect Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boeckler FM, Joerger AC, Jaggi G, Rutherford TJ, Veprintsev DB, Fersht AR. Targeted rescue of a destabilized mutant of p53 by an in silico screened drug. Proc Natl Acad Sci U S A. 2008;105:10360–10365. doi: 10.1073/pnas.0805326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Demma M, Maxwell E, Ramos R, Liang L, Li C, Hesk D, et al. SCH529074, a small molecule activator of mutant p53, which binds p53 DNA binding domain (DBD), restores growth-suppressive function to mutant p53 and interrupts HDM2-mediated ubiquitination of wild type p53. J Biol Chem. 2010;285:10198–10212. doi: 10.1074/jbc.M109.083469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng Y, Chen L, Li C, Lu W, Chen J. Inhibition of MDM2 by hsp90 contributes to mutant p53 stabilization. J Biol Chem. 2001;276:40583–40590. doi: 10.1074/jbc.M102817200. [DOI] [PubMed] [Google Scholar]
- 30.Lambert JM, Gorzov P, Veprintsev DB, Soderqvist M, Segerback D, Bergman J, et al. PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell. 2009;15:376–388. doi: 10.1016/j.ccr.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 31.Xu J, Reumers J, Couceiro JR, De Smet F, Gallardo R, Rudyak S, et al. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat Chem Biol. 2011;7:285–295. doi: 10.1038/nchembio.546. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.