Abstract
Heparanase is the only enzyme in mammals capable of cleaving heparan sulfate, an activity implicated in tumor inflammation, angiogenesis and metastasis. Heparanase is secreted as a latent enzyme that is internalized and subjected to proteolytic processing and activation in lysosomes. Its role under normal conditions has yet to be understood. Here we provide evidence that heparanase resides within autophagosomes where studies in heparanase-deficient or transgenic mice established its contributions to autophagy. The pro-tumorigenic properties of heparanase were found to be mediated in part by its pro-autophagic function, as demonstrated in tumor xenograft models of human cancer and through use of inhibitors of the lysosome (chloroquine) and heparanase (PG545), both alone and in combination. Notably, heparanase-overexpressing cells were more resistant to stress and chemotherapy in a manner associated with increased autophagy, effects that were reversed by chloroquine treatment. Collectively, our results establish a role for heparanase in modulating autophagy in normal and malignant cells, thereby conferring growth advantages under stress as well as resistance to chemotherapy.
Keywords: Heparanase, lysosome, autophagy, drug-resistance, tumorigenicity
Introduction
Heparanase is an endo-β-glucuronidase that cleaves heparan sulfate (HS) side chains presumably at sites of low sulfation. Enzymatic degradation of HS leads to disassembly of the ECM and is therefore involved in fundamental biological phenomena associated with tissue remodeling and cell invasion, including inflammation, angiogenesis and metastasis (1–4). In addition, the multitude of polypeptides sequestered and regulated by HS and the ability of heparanase to convert these into bio-available molecules (5, 6) require that these activities will be kept tightly regulated. We have shown previously that following secretion as a latent 65 kDa enzyme, heparanase rapidly interacts with cell membrane HS proteoglycans (HSPG; i.e., syndecans) (7–10), followed by internalization and processing into a highly active 50 kDa enzyme (7, 11). Notably, heparanase was shown to reside primarily within endocytic vesicles, assuming a polar, peri-nuclear localization and co-localizing with lysosomal markers (7, 8, 10, 12). In addition, it has been demonstrated that incubation with endosomal/lysosomal fraction, but not membrane or cytosolic preparations, leads to heparanase processing and activation (13). Likewise, heparanase processing was blocked by chloroquine and bafilomycin A1 which inhibit lysosomal proteases by raising the lysosome pH (11). Subsequent studies employing site-directed mutagenesis, gene silencing, and pharmacological inhibitors have identified cathepsin L as the primary lysosomal protease responsible for heparanase processing and activation (14–16). In spite of its localization in a highly active protein degradation environment such as the lysosome, heparanase appears stable (12) and exhibits a half-life of about 30 hours (17), relatively long compared with a t1/2 of 2–6 hours, and 25 minutes of transmembrane and GPI-anchored HSPGs, respectively (18). Residence and accumulation of heparanase in lysosomes may indicate that the enzyme functions in the normal physiology of this organelle, but such a function has not been described yet. Autophagy is an evolutionarily conserved catabolic pathway through which cytoplasmic components, including macromolecules such as proteins and lipids as well as whole organelles, are sequestered into double-membrane vesicles called autophagosomes. Autophagosomes are subsequently fused with lysosomes, where the intracellular material is degraded and recycled. This process occurs in every cell at a basal level and is required to remove unfolded proteins and damaged organelles, thus maintaining cellular homeostasis. Autophagy is induced significantly by starvation and stress, promoting cancer cells survival by providing their metabolic needs (19, 20). Here, we provide evidence that heparanase enhances autophagy. Moreover, we show that enhanced tumor growth and chemo-resistance exerted by heparanase are mediated in part by augmenting autophagy.
Materials and methods
Cells and cell culture
Control (Mock) and heparanase over expressing human FaDu pharyngeal carcinoma, U87 glioma and rat C6 glioma cells have been described previously (8, 21–23). SIHN-013 laryngeal carcinoma cells were kindly provided by Sue Eccles (Institute of Cancer Research, Sutton, Surrey, UK) (24). Cells were grown in Dulbecco’s modified Eagle’s medium (Biological Industries, Beit Haemek, Israel) supplemented with 10% fetal bovine serum and antibiotics. Mouse embryonic fibroblasts (MEF) of control and heparanase knockout (KO) mice have been described elsewhere (25). Cells were passed in culture no more than 2 months after being thawed from authentic stocks.
Antibodies and reagents
Anti-phopsho-p70S6-kinase, anti-p70S6-kinase, anti-TSC2, anti-PTEN, and anti-mTOR antibodies were purchased from Cell Signaling (Beverly, MA). Anti-LC3 and anti-actin monoclonal antibodies were purchased from Sigma (St. Louis, MO). The heparanase inhibitor PG545 was kindly provided by Progen Pharmaceuticals (Brisbane, Australia) (26). Cisplatin and doxorubicin were obtained from the Oncology Department, Rambam Health Care Campus (Haifa, Israel). LysoTracker was purchased from Molecular Probes (Life Technologies, Grand Island, NY); Torin was purchased from Tocris Bioscience (Bristol, UK).
Cell lysates and immunoblotting
Preparation of cell and tissue extracts and immunoblotting were carried out essentially as described (10). Following induction of autophagy, the cytosolic form of LC3 (LC3-I) is conjugated to phosphatidylethanolamine (LC3-II) which is recruited to autophagosomal membranes, serving as a most acceptable mean to measure autophagy levels. Immunoblots were subjected to densitometry analyses and the relative intensity of bands (i.e., fold change) is presented underneath the gel.
Immunocytochemistry
Immunofluorescent staining of methanol-fixed cells and analysis by confocal microscopy was carried out essentially as described (10). In order to better detect LC3 by immunofluorescence or electron microscopy (EM), autophagy was initiated by depriving the cells of amino acids (AA) for 3 h, in the presence of chloroquine (Chl, 50 μg/ml; Sigma).
Colony formation in soft agar
Dulbecco’s modified Eagle’s medium (DMEM) (3 ml) containing 0.5% low-melt agarose (Bio-Rad) and 10% FCS was poured into 60-mm Petri dishes. The layer was covered with cell suspension (2×103 cells) in 1.5 ml DMEM containing 0.3% low-melt agarose and 10% FCS, followed by addition of 2 ml DMEM containing 10% FCS without or with PG545 (25 μg/ml), chloroquine (50 μg/ml), or both. Medium was exchanged every 3 days. Colonies were visualized and counted under a microscope 2–5 weeks after seeding, as described previously (27).
MTT assay
The number of viable cells was evaluated by thiazolyl blue tetrazolium bromide (MTT; Sigma) that measures the activity of cellular enzymes that reduce the tetrazolium dye, MTT, to its insoluble formazan, yielding a purple color. Cells (5×103/well) were grown in 96 wells plate and subjected to various treatments as indicated. MTT (100 μg) was then added to each well for 2–3 hours, followed by centrifugation. The cell pellet was re-suspended in 150 μl of isopropanol and absorbance was measured at 570 nm using an ELISA plate reader, as described (27).
Tumorigenicity and immunohistochemistry
U87 cells were detached with trypsin/EDTA, washed with PBS, and brought to a concentration of 5×07 cells/ml. Cell suspension (5×106/0.1 ml) was inoculated subcutaneously at the right flank of 6-weeks-old female SCID mice (n=7). Mice were administrated with the heparanase inhibitor PG545 (20 mg/kg; once weekly), chloroquine (50 mg/kg; every day) or both and xenografts size was determined by externally measuring tumors in 2 dimensions using a caliper. At the end of the experiment, mice were sacrificed; tumor xenografts were removed, weighed and fixed in formalin. Paraffin-embedded 5μm sections were subjected to immunostaining applying anti-LC3, anti-phospho-p70S6K or anti-glial fibrillary acidic protein (GFAP; Dako) antibodies using the Envision kit according to the manufacturer’s (Dako) instructions, as described previously (28).
Statistics
Data are presented as means ± SE. Statistical significance was analyzed by 2-tailed Student’s t test. Values of P < 0.05 were considered significant. Data sets passed D’Agostino-Pearson normality (GraphPad Prism 5 utility software). All experiments were repeated at least 3 times with similar results.
Results
Heparanase augments autophagy
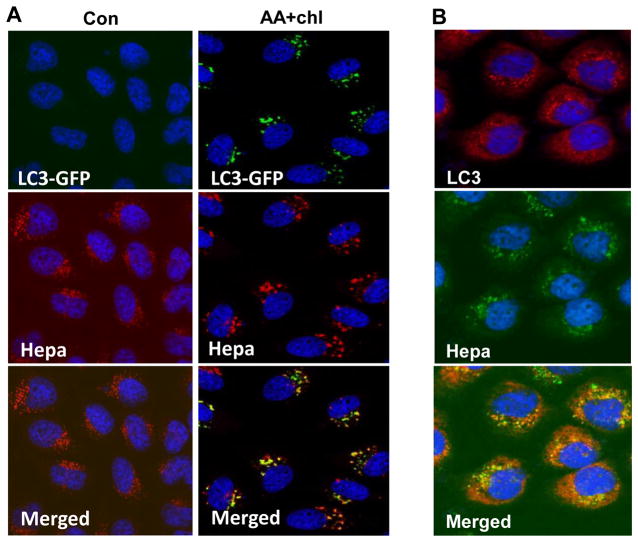
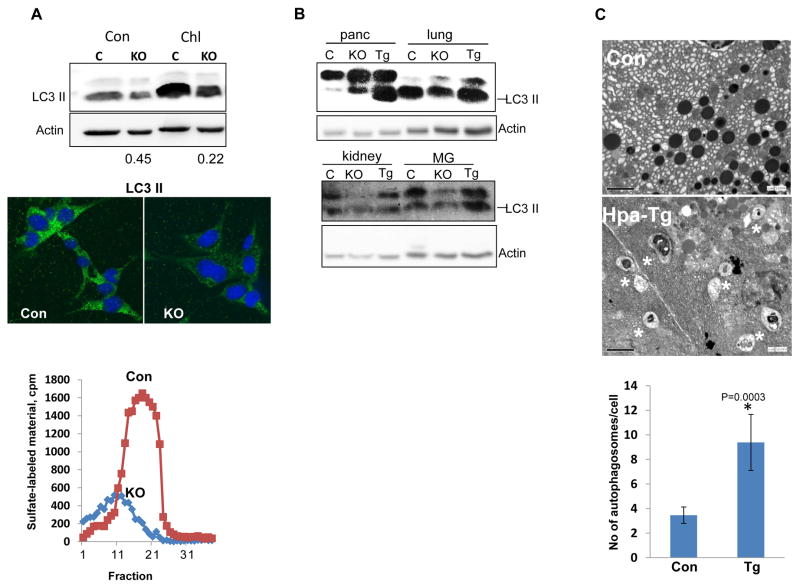
We first examined the possibility that heparanase is localized within autophagosome. To this end, latent heparanase (1 μg/ml) was added exogenously to HeLa cells stably expressing LC3-GFP gene construct and cells were left untreated (Con) or were deprived of amino acids (AA) in the presence of chloroquine. The latter is a lysosome inhibitor which allows the formation and accumulation of autophagosomes, but prevents their activity. LC3-GFP fluorescence was evident only in conditions that stimulate autophagy (AA+Chl; Fig. 1A, upper panels, green), co-localizing with heparanase (Fig. 1A right lower panel, yellow). A similar co-localization of endogenous LC3 and heparanase was noted in SIHN-013 laryngeal carcinoma cells over expressing heparanase following autophagy stimulation (Fig. 1B). We further found that LC3-II level was reduced in MEF derived from Hpa-KO mice that lack heparanase activity (25) (Fig. 2A, lower panel) vs. control MEF (Fig. 2A, upper panel, Con) and even greater reduction in LC3-II level was quantified after treatment with chloroquine (Fig. 2A, upper panel, Chl). Reduced LC3-II level in Hpa-KO MEF was also evident by immunofluorescent staining (Fig. 2A, middle panels). Similarly, we observed reduced autophagy levels (i.e., LC3-II) in the lung, kidney and mammary gland (MG) of Hpa-KO mice (Fig. 2B, KO), while increased level of autophagy was observed in the lung, MG and pancreas tissues of transgenic mice (Hpa-Tg) (29) over expressing heparanase (Fig. 2B, Tg). Increased autophagy in the pancreas of Hpa-Tg mice vs. control was further evident by electron microscopy (EM; Fig. 2C, asterisks; p=0.0003). Collectively, these results suggest that heparanase is localized within autophagosomes and is involved in the regulation of autophagy.
Figure 1.
Heparanase co-localizes with LC3-II. A. GFP-LC3. Heparanase (1 μg/ml) was added exogenously to HeLa cells stably expressing a GFP-LC3 gene construct for 24h. Cells were then deprived of amino acids in the presence of chloroquine (50 μg/ml; AA+Chl) for 3 h or were incubated under serum-free conditions as control (Con). Cells were then fixed with methanol and subjected to immunofluorescent staining applying anti-heparanase (middle panels, red) antibody. Co-localization of heparanase and GFP-LC3 appears yellow (lower right panel). B. SIHN-013 laryngeal carcinoma cells overexpressing heparanase were starved for amino acids for 3h in the presence of chloroquine. Cells were then fixed with methanol and subjected to immunofluorescent staining utilizing anti-heparanase (middle panel, green) and anti-LC3 (upper panel, red) antibodies. Note co-localization (lower panel, yellow) of heparanase and endogenous LC3 in autophagosomes. Original magnifications: ×60.
Figure 2.
Endogenous heparanase levels reflect autophagy extent. A. MEF. Mouse embryonic fibroblasts (MEF) were isolated from control (Con) and Hpa-KO (KO) mice and examined for heparanase activity (lower panel) and LC3 levels by immunofluorescent staining (green, middle panel). Nuclei counterstaining is shown in blue. Original magnifications: ×60. Control (C) and KO MEF were incubated under serum-free conditions without (Con) or with chloroquine (Chl; 50 μg/ml) for 3h. Cell lysates were then subjected to immunoblotting applying anti-LC3 (upper panel) and anti-actin (second panel) antibodies. Note, decreased levels of LC3 in Hpa-KO MEF. The extent of LC3-II decrease in Hpa-KO MEF was quantified (bottom numbers) by densitometry analyses compared with control MEF in each culture conditions.
B. Autophagy levels in Hpa-Tg and Hpa-KO mouse tissues. Extracts were prepared from the indicated tissues of control (C), Hpa-Tg (Tg) and Hpa-KO (KO) mice over expressing or lacking heparanase, respectively and lysate samples were subjected to immunoblotting applying anti-LC3 and anti-actin antibodies. C. Electron microscopy. Pancreas tissues from control (Con) and Hpa-Tg mice were fixed in glutaraldehyde and processed for EM. Shown are representative images at ×10,000 magnification. Quantification of autophagosomes (asterisks) is shown graphically in the lower panel. p=0.0003 for Con vs. Hpa-Tg.
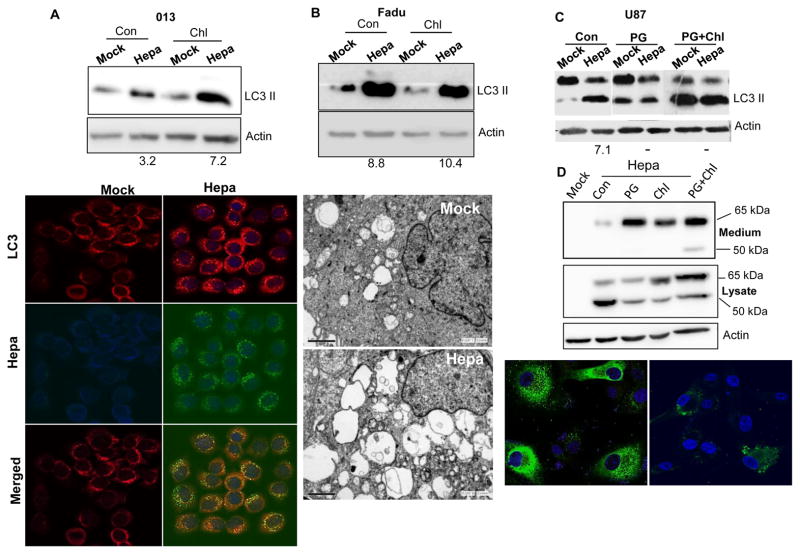
Even higher increase of autophagy was observed following heparanase over expression in tumor derived cells. Autophagy (i.e., LC3-II) was markedly increased under resting conditions (Con; Fig. 3A–C, upper panels), and further increase in LC3-II levels was noted following chloroquine treatment (Fig. 3A, B; Chl, upper panels), evident by immunoblotting (Fig. 3A–C, upper panels) and immunofluorescent staining (Fig. 3A, lower panels) of heparanase over-expressing SIHN-013 laryngeal carcinoma (Fig. 3A), FaDu pharyngeal carcinoma (Fig. 3B), and rat C6 glioma (Suppl. Fig. 1A) cells. Increased autophagy in FaDu cells over expressing heparanase is further demonstrated by EM of unstimulated (Suppl. Fig. 1B) and amino acids-starved (Fig. 3B, lower panels) cells, increase that was statistically highly significant (p=0.00001 and 0.01 for Mock vs. Hepa under resting (Con) and AA+Chl conditions, respectively; Suppl. Fig. 1B, right panel). LC3-II induction comparable in magnitude was evident also by U87 glioma cells over expressing heparanase (Hepa; Fig. 3C, Con); notably, autophagy induction was prevented by the heparanase inhibitor PG545 (PG; Fig. 3C) and was associated with accumulation of heparanase in the culture medium of U87-Hepa cells treated with this compound (PG; Fig. 3D, upper panel), thus mimicking the effect of heparin/HS (7). Unexpectedly, even higher accumulation of the latent (65 kDa) and active (50 kDa) forms of heparanase was detected in the culture medium of cells treated with PG545 and chloroquine (PG+Chl; Fig. 3D, upper panel). In PG545-, and to a lesser extent PG545 plus chloroquine-treated cells, accumulation of heparanase in the culture medium was accompanied by decreased levels of intracellular heparanase (Fig. 3D, second and bottom panels; Suppl. Fig. 1C), implying that PG545 attenuates autophagy also by decreasing the content of intracellular heparanase.
Figure 3.
Heparanase over expression enhances autophagy. A–B. Head and neck carcinoma-derived cells. Control (Mock) and heparanase over expressing (Hepa) SIHN-013 laryngeal carcinoma (A) and FaDu pharyngeal carcinoma (B) cells were cultured without (Con) or with chloroquine (Chl) for 3h. Lysate samples were then subjected to immunoblotting applying anti-LC3 (upper panels) and anti-actin (second panels) antibodies. Control (Mock) and heparanase over expressing SIHN-013 cells were subjected to immunofluorescent staining applying anti-LC3 (third panels, red) and anti-heparanase (fourth panel, green) antibodies. Merged images are shown in the lower panels. EM. Control (Mock) and heparanase over expressing Fadu cells were deprived of amino acids for 3h in the presence of chloroquine, fixed with glutaraldeyde, and processed for EM. Shown are representative images at ×10,000 magnification (B, ;lower panels). Note increased abundance and size of autophagic vacuoles following heparanase over expression. C,D. PG545 treatment. C. Control (Mock) and heparanase over expressing (Hepa) U87 glioma cells were left untreated (Con) or were incubated with the heparanase inhibitor PG545 without (PG; 25 μg/ml) or with chloroquine (PG+Chl) for 20 h and lysate samples were subjected to immunoblotting for LC3 and actin. Note a marked increase in LC3-II levels by heparanase over expression that is prevented by PG545. The extent of increased LC3-II was quantified by densitometry analyses of heparanase over expressing cells (Hepa) compared with control cells in each culture conditions (bottom numbers). D. Accumulation of heparanase in the culture medium of cells treated with PG545. Control (Mock) and heparanase over expressing (Hepa) U87 cells were cultured without (Con) or with PG545 (PG; 25 μg/ml), chloroquine (Chl; 50 μg/ml) or both for 20 h. Cell conditioned medium (upper panel) and cell lysates (second panel) were subjected to immunoblotting applying anti-heparanase and anti-actin (third panel) antibodies. U87-Hepa cells cultured without (Con) or with PG545 (PG) were similarly subjected to immunofluorescent staining with anti-heparanase antibody (lower panels). Original magnifications: ×60.
Pro-tumorigenic function of heparanase involves induction of autophagy
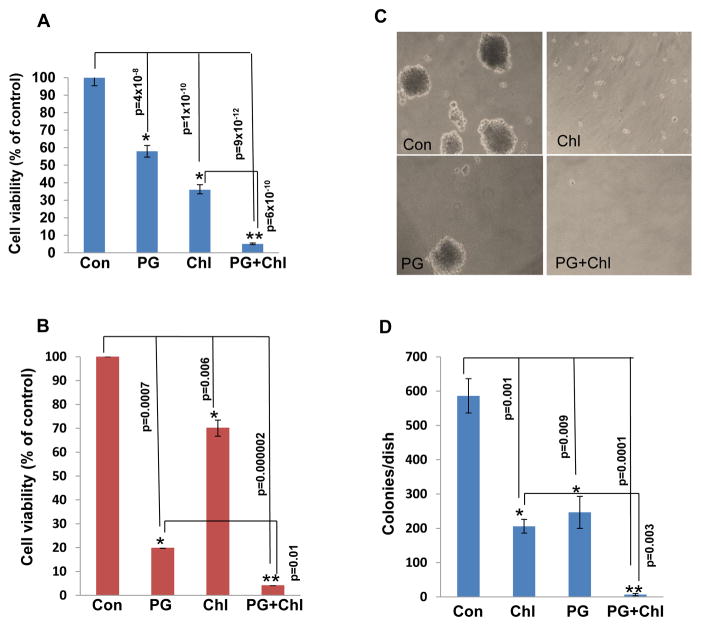
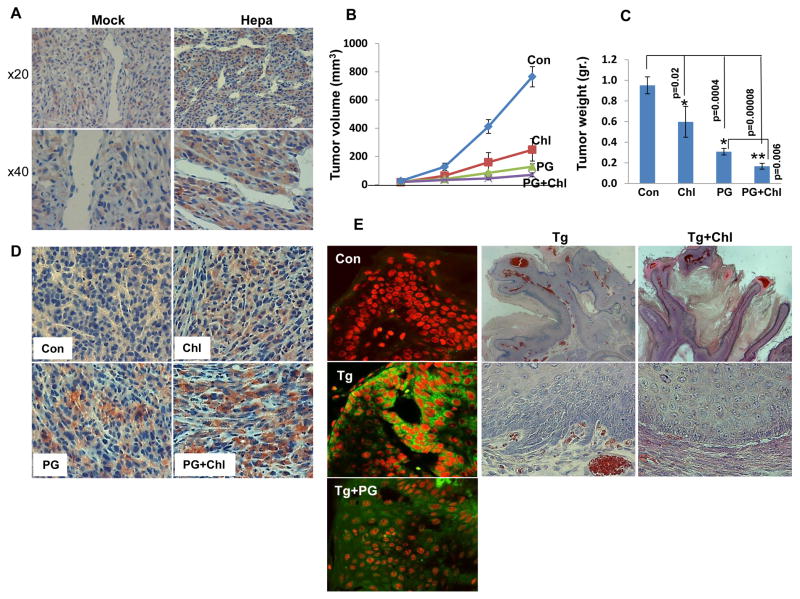
We next examined whether induction of autophagy underlies the pro-tumorigenic function of heparanase. We have shown previously that U87 glioma cells over expressing heparanase are endowed with higher proliferation rate and form more and bigger colonies in soft agar (21, 22, 30). Applying the MTT assay we found that chloroquine treatment reduces U87 cell viability compared with control untreated cells (Fig. 4A, Chl), decrease that was most pronounced when combined with pharmacologically relevant concentrations (8 μM) (31) of the heparanase inhibitor PG545 (Fig. 4A, Chl+PG; **p=6.2×10−10 for Chl vs. PG+Chl). A similar synergistic effect of chloroquine and PG545 was observed with C6-Hepa rat glioma cells (Fig. 4B; **p=0.01 for PG vs. PG+Chl). Notably, colony formation in soft agar was markedly reduced by chloroquine treatment (Fig. 4C, Chl), synergizing with PG545 in term of colony number (Fig. 4C, D; **p=0.003 for Chl vs. PG+Chl) and size (Fig. 4C). We have reported previously that over expression of heparanase in U87 cells enhanced the growth of tumor xenografts (21, 30). Immunohistochemistry revealed that the growth advantage of these tumor xenografts is associated with increased staining of LC3 (Fig. 5A, Hepa). In order to further reveal the role of autophagy in this system, mice were inoculated subcutaneously with U87 cells and treated with chloroquine (50 mg/kg, every day), PG545 (20 mg/kg; once a week), or both. Tumor growth was attenuated by chloroquine (p=0.02 for Con vs. Chl) and PG545 (p=0.0004 for Con vs. PG) as single agents and was most evident when the two compounds are combined (Fig. 5B, C; **p=0.006 for PG vs. PG+Chl and p=0.003 for Chl vs. PG+Chl), resembling the in vitro results (Fig. 4A, C, D). In addition, we found that U87 tumor xenografts from treated mice exhibit higher levels of GFAP immunoreactivity (Fig. 5D) suggesting that these lesions are not only smaller but also assume higher level of differentiation.
Figure 4.
Autophagy induction by heparanase is associated with increased cell survival and colony number. Human U87 (A) and rat C6-Hepa (B) glioma cells were left untreated (Con) or were incubated with chloroquine (Chl; 50 μg/ml), PG545 (PG; 25 μg/ml) or both (PG+Chl) for three days and cell survival was evaluated by MTT assay as described under ‘Materials and Methods’. U87 cells were cultured in soft agar and were left untreated (Con) or were incubated with chloroquine (Chl; 50 μg/ml), PG545 (PG; 25 μg/ml) or both (PG+Chl). After three weeks dishes were photographed (C) and the number of colonies in each dish was counted (D).
Figure 5.
Inhibition of tumor growth by autophagy- and heparanase-inhibitors. A. Five micron sections of tumor xenografts produced by control (Mock) and heparanase over expressing (Hepa) U87 cells were subjected to immunostaining applying anti-LC3 antibody. Note, increased autophagy (LC3) in tumor xenografts with high levels of heparanase. Original magnification: upper panels ×20, lower panels ×40. B. Tumor growth inhibition. U87 cells (5×106) were inoculated subcutaneously and mice were administered with PBS (Con; n=7), chloroquine (Chl; 50 mg/kg, every day; n=7), PG545 (PG; 20 mg/kg, once weekly; n=8) or both (PG +Chl; n=8) and tumor volume was inspected over time by external measurements of tumor lesions. At termination, tumors were resected, weighted (C), fixed in 4% PFA and five micron sections of paraffin embedded tumors were subjected to immunostaining applying anti-GFAP antibody (D). E. DMBA-TPA skin cancer model. Sections of tumor lesions from control and Hpa-Tg mice exposed to DMBA-TPA two-steps skin carcinogenesis model (22) untreated (Tg) or treated with PG545 (Tg+PG) were subjected to immunofluorescent staining applying anti-LC3 antibody (green). Nuclei counterstaining (PI) is shown in red. Original magnifications: ×40. Hpa-Tg mice were similarly exposed to DMBA-TPA regimen. After 15 weeks, when tumor lesions start to appear, mice were divided randomly into two groups and chloroquine was administrated every day at 50 mg/kg. At termination, tumors were collected, weighted and fixed in PFA. Shown are representative hematoxylin and eosin staining of tumor sections of Hpa-Tg mice untreated (Tg) or treated with chloroquine (Tg+Chl). Note reduced downward proliferation and invasion in Hpa-Tg mice treated with chloroquine. Original magnifications: upper panels ×10, lower panels × 40.
In the skin, LC3-II levels are undetectable under normal conditions in control, Hpa-Tg, or Hpa-KO mice (not shown). Exposing the skin tissue to two-step DMBA/TPA skin carcinogenesis model resulted in marked increase in the number and size of tumor lesions in Hpa-Tg compared with control mice (32). Notably, the increased tumorigenicity in Hpa-Tg mice was associated with a noticeable increase in autophagy revealed by LC3 staining that was reduced by PG545 (Fig. 5E, left panels), thus providing another indication for the involvement of autophagy in heparanase-mediated tumor expansion. In order to explore this notion further, we exposed control and Hpa-Tg mice to DMBA-TPA protocol. By 15 weeks, when small tumor lesions started to appear, Hpa-Tg mice were divided into two groups and chloroquine was administrated every day (50 mg/kg; ip) for four weeks. Chloroquine had no effect on the number or size of tumor lesions developed (not shown). Pathological examination revealed, however, that chloroquine treatment resulted in exophytic tumor growth, exhibiting less downward epithelial proliferation (Tg+Chl; Fig. 5E, rightmost panels) compared with untreated lesions that displayed endophytic, irregular growth and invaded the underlying stroma (Tg; Fig. 5E, middle panels), thus signifying more aggressive tumors compared with tumors developed in chloroquine treated mice.
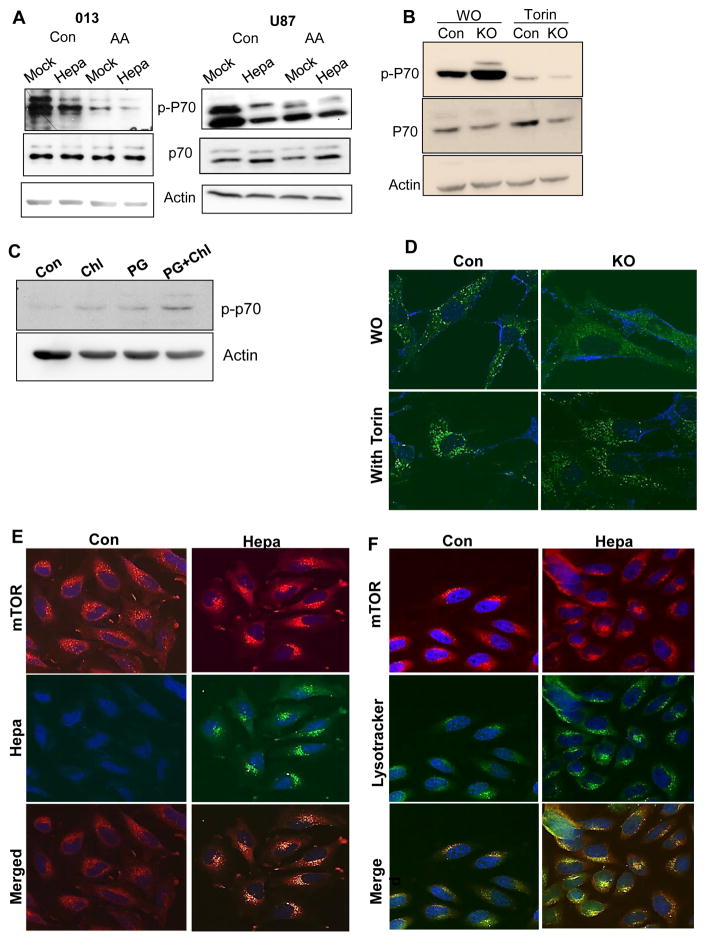
Mechanistically, autophagy induction by heparanase appears to involve the mammalian target of rapamycin complex 1 (mTOR1). This nutrient sensing kinase acts as a master negative regulator of autophagy because during starvation, mTOR1 is inhibited and this induces autophagy (33). mTOR1 activity can be assessed by the phosphorylation status of p70 S6-kinase, a specific downstream substrate of mTOR1. In cells over expressing heparanase we observed reduced phospho-p70 S6-kinase levels (Fig 6A, Con, upper panels), in agreement with increased autophagy (Fig. 3). Moreover, a substantial increase in p70 S6-kinase phosphorylation level was observed in Hpa-KO MEF vs control (WO; Fig. 6B), in agreement with reduced autophagy in these cells (Fig. 2A). In accordance with these results we observed increased p70 S6-kinase phosphorylation levels in tumor xenografts following treatment with inhibitors of heparanase (PG545) or autophagy (chloroquine) (Fig. 6C, Suppl. Fig. 1D). The alterations in p70 S6-kinase phosphorylation in relation to heparanase levels were associated with the cellular localization of mTOR1. In control MEF, mTOR1 was localized within scattered vesicles but appeared diffusely distributed in Hpa-KO MEF (Fig. 6D, WO). p70 S6-kinase phosphorylation was reduced markedly in MEF treated with mTOR1 inhibitor (Torin; Fig. 6B), resulting in accumulation of mTOR1 peri-nuclearly (Fig. 6D). We also found that mTOR1 assumes peri-nuclear localization in cells with high content of heparanase (Hepa; Fig. 6E, right) compared with a more diffused distribution in control cells (Con; Fig. 6E, left) and noted that mTOR1 co-localizes with heparanase (Fig. 6E) and LysoTracker that labels acidic lysosomal vesicles (Fig. 6F). Notably, Hpa-KO MEF were more sensitive to the mTOR1 inhibitor, resulting in decreased p70 S6-kinase phosphorylation to an extent 7-fold lower than in control MEF (Suppl. Fig. 2C).
Figure 6.
Heparanase modulates p70S6-kinase phosphorylation. A. Immunoblotting. Control (Mock) and heparanase (Hepa) over expressing SIHN-013 (left) and U87 (right) cells were cultured under serum-free conditions without (Con) or were deprived of amino acids (AA) for 3h. Cell lysates were subjected to immunoblotting applying anti-phospho-p70 S6-kinase (p-P70; upper panels), p70 S6-kinase (P70; second panels), and anti-actin (lower panels) antibodies. B. MEF obtained from control (Con) and heparanase-KO mice were incubated without (WO) or with Torin (mTOR inhibitor; 500 μM, 3h). Cell lysate samples were then subjected to immunoblotting applying anti-phospho-p70 S6-kinase (p-P70; upper panels), p70 S6-kinase (P70; second panels), and anti-actin (lower panels) antibodies. C. Mice were inoculated with U87 cells and were administered with PBS (Con) or with chloroquine (Chl, 50 μg/ml), PG545 (PG, 20 μg/ml) or both. At termination, tumor lysates were subjected to immunoblotting applying anti phospho-p70 (upper panel) and anti-actin (lower panel) antibodies. D. MEF obtained from control (Con) and heparanase-KO mice were incubated without (WO) or with Torin (500 μM, 3h) and were then subjected to immunofluorescent staining applying anti-mTOR1 antibody. E–F. mTOR localization. E. Control (Mock) and heparanase (Hepa) over expressing U87 cells were subjected to immunofluorescent staining applying anti-mTOR (upper panel, red) and anti-heparanase (middle panels, green) antibodies. Merged images are shown in the lower panels. Note diffused, scattered, distribution of mTOR in control cells compared with peri-nuclear staining that co-localized with heparanase (lower right) in transfected cells. F. Cells were similarly stained for mTOR1 (upper panel, red) and LysoTracker (middle panel, green). Nuclei counterstaining appear in blue. Merged images are shown in the lower panels.
Autophagy induction by heparanase endowed cells with chemo-resistance
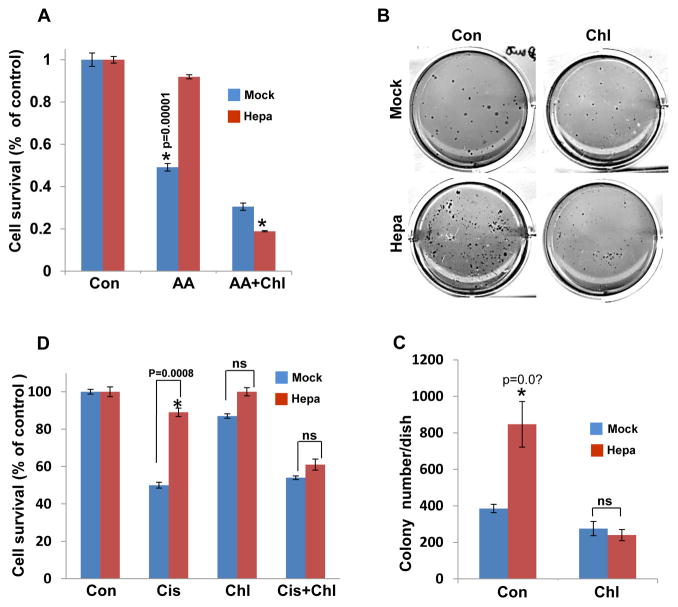
While the pro-tumorigenic properties of heparanase are well documented, its function in chemo-resistance has not been so far explored. In order to examine this aspect we first exposed control (Mock) and heparanase-over expressing (Hepa) U87 glioma cells to amino acid starvation without (AA, Fig. 7A) or with chloroquine (AA+Chl; Fig. 7A) and cell viability was evaluated by MTT assay. Amino acid starvation reduced the viability of control (Mock) cells to half compared with untreated cells (Con) but did not reduce the viability of heparanase over expressing cells (Hepa; Fig. 7A, AA), also evident by cell morphology (Suppl. Fig. 1E), differences that are statistically highly significant (p=0.00001 for Mock vs. Hepa). Notably, the resistance of heparanase over-expressing cells was reversed by combining amino acid starvation with chloroquine (Fig. 7A, AA+Chl), suggesting that resistance to starvation stress is mediated by induction of autophagy in U87-Hepa cells (Fig. 3C) and can be prevented by autophagy inhibitor. This was further evident by the increased number of colonies formed by cells over expressing heparanase vs. control cells (Mock, Fig. 7B, C; p= 0.01), growth advantage that was reversed by the inclusion of chloroquine (Fig. 7B, C). More importantly, heparanase over-expressing U87 glioma and FaDu pharyngeal carcinoma cells were more resistant to cisplatin treatment at concentrations of 0.1 μg/ml and above (Suppl. Fig. 2A), and this tolerance was similarly prevented by combining cisplatin and chloroquine (Fig. 7D). This implies that heparanase provides cells with chemo-resistance that is mediated, in part, by increased autophagy.
Figure 7.
Enhanced autophagy underlies heparanase-mediated stress-, and chemo-resistance. A. Stress-resistance. Control (Mock) and heparanase over expressing (Hepa) U87 cells were cultured under serum free conditions (Con) or were deprive of amino acids without (AA) or with chloroquine (AA+Chl) for 24h and cell survival was evaluated by MTT assay. Note, decreased cell survival of Mock, but not heparanase (Hepa) cells subjected to amino acids starvation, which is reversed by addition of chloroquine. B,C. Colony formation. Control (Mock) and heparanase over expressing (Hepa) C6 glioma cells were cultured in soft agar without (Con) or with chloroquine (50 μg/ml) for three weeks. Dishes were then photographed (B) and colonies were counted (C). Note increased colony number by heparanase over expressing cells which is prevented by chloroquine. D. Resistance to chemotherapy. Control (Mock) and heparanase over expressing (Hepa) U87 cells were cultured without (Con) or with cisplatin (Cis; 30 ug/ml), chloroquine (Chl; 50 μg/ml) or both for 24 h and cell survival was evaluated by MTT assay. Note resistance of heparanase over expressing cells to cisplatin that is prevented by chloroquine.
Discussion
Heparanase expression is increased in many types of tumors and this elevation is often associated with more aggressive disease and poor prognosis (1, 2, 34), but the role of heparanase under normal conditions has not been resolved thus far. Here, we describe for the first time a role of lysosomal heparanase in modulating autophagy, an evolutionary conserved mechanism that delivers intracellular proteins, lipids, and organelles to the lysosomal compartment for degradation and recycling (20).
Residing for relatively long period of time in the lysosome led us to hypothesize that heparanase may be found within autophagosomes. Indeed, double immunofluorescent staining clearly shows that heparanase co-localizes with GFP-labeled and endogenous LC3-II (Fig. 1). LC3-II is a convenient and widely used marker of autophagy because it forms and remains associated with the autophagosome even after fusion with lysosomes. Moreover, LC3-II is the only known protein that specifically associates with autophagosomes and not with other vesicles and thus its level reflects the amount of autophagic vacuoles (35). Our results indicate that heparanase does not only reside in autophagosomes but also modulates autophagy. This is concluded because reduced LC3-II levels are found in MEF and tissues obtained from heparanase KO mice (Fig. 2B). Likewise, LC3-II level was enhanced in transgenic mice that over express heparanase (Fig. 2B, Tg) (29). Even higher induction of autophagy was evident in head and neck carcinoma (SIHN-013, FaDu) and glioma (U87, C6) cells over expressing heparanase (Fig. 3A–C). These cell systems were preferred because previous results provided a strong pre-clinical and clinical significance of heparanase in the progression of these malignancies (21–23, 30, 36–38). A noticeable, 3–8 fold increase in LC3-II levels was observed under control, serum-free conditions (Fig. 3, Con) and following treatment with chloroquine (Fig. 3, Chl), revealed by biochemical measures and EM. Interestingly, EM analyses of FaDu cells over expressing heparanase revealed not only a higher number of autophagic vacuoles (Suppl. Fig. 1B, lower panels, asterisks), but also abundant release of vesicles, possibly exosomes, from the cell surface (Suppl. Fig. 1B, lower panel, arrow). This supports the notion that heparanase enhances exosome secretion that contributes to tumor growth (39, 40).
The mechanism underlying autophagy induction by heparanase is not entirely clear, but likely involves mTOR1 that plays a pivotal role in nutrient-sensing and autophagy regulation (41). mTOR1 activity inhibits autophagy but under starvation, its activity is repressed, leading to autophagy induction. Similarly, mTOR1 inhibitors such as rapamycin induce autophagy (42). Interestingly, we found that heparanase over expression associates with reduced mTOR1 activity, evident by decreased levels of p70 S6-kinase phosphorylation, an mTOR1 substrate (Fig. 6A, upper panels; Suppl. Fig. 2B), while heparanase-KO MEF are endowed with increased p70 S6-kinase phosphorylation levels (Fig. 6B). Similarly, heparanase inhibition by PG545 results in increased mTOR1 activity and p70 S6-kinase phosphorylation (Fig. 6C, Suppl. Fig. 1D). The reason for mTOR1 inhibition by heparanase may be related to alteration of mTOR1 localization within the cell evident in three independent experimental settings. First, vesicular mTOR1 appeared scattered in control MEF but accumulate at perinuclear regions following treatment with mTOR inhibitor (Torin, Fig. 6D, left). Similarly, mTOR1 appeared more diffusely scattered in control cells, whereas in cells with high content of heparanase, mTOR1 is found mostly in peri-nuclear regions, co-localizing with heparanase (Fig. 6E, Hepa) and LysoTracker (Fig. 6F, Hepa) that labels acidic lysosomal vesicles. Thirdly, in heparanase-KO MEF mTOR1 do not reside in vesicles but rather diffusely distributed in the cytoplasm (Fig. 6D), associated with increased p70 S6-kinase phosphorylation (Fig. 6B, WO). This agrees with the notion that activation of mTOR1 by nutrients is associated with peripheral lysosomes, whereas starvation leads to peri-nuclear clustering of lysosomes and decreased mTOR1 activity (43). Thus, Torin, and to a lesser degree heparanase, result in accumulation of mTOR1 at peri-nuclear regions, leading to reduced mTOR1 activity and increased autophagy (43, 44). Localization of mTOR1 to LysoTracker-positive acidic vesicles appeared unique because no such co-localization was evident for PTEN or TSC2 (Suppl. Fig. 3), intrinsic components of the PI3-K signaling pathway. This is not surprising because we employed nutrient starvation to induce autophagy which, unlike growth factors, is not regulated by the PTEN/Akt/TCS pathway but rather involves as yet uncharacterized cytoplasmic sensor (45, 46).
Compelling evidence has shown that heparanase is up regulated in various primary solid tumors (i.e., carcinomas and sarcomas) and hematological malignancies (2, 34, 47). The consequence of heparanase induction most often associates with disease progression and bad prognosis, thus encouraging the development of heparanase inhibitors (26, 48, 49). The molecular mechanism(s) exerted by heparanase to promote tumor initiation and progression is still incompletely understood. The results of the current study imply that autophagy induction contributes to the pro-tumorigenic function of heparanase. This emerges from in vitro and in vivo experiments utilizing inhibitors of autophagy (chloroquine) and heparanase (PG545) alone or in combination. The heparanase inhibitor PG545 significantly reduces U87 cell survival and colony number (Fig. 4A, C), resembling the effect of PG545 on pancreatic carcinoma cells (31). Notably, chloroquine treatment resulted in a comparable effect, and even more effective inhibition is obtained by combining PG545 and chloroquine in vitro (Fig. 4, PG+Chl) and, moreover, in a tumor xenograft model in vivo, resulting in significantly smaller and more differentiated tumors (Fig. 5A–D). Increased cell differentiation (i.e., elevation of E-cadherin) was also noted in pancreatic carcinoma cells treated with PG545 (31), suggesting that heparanase activity drives cancer cell de-differentiation as part of its pro-tumorigenic properties.
Immunostaining further revealed that enhanced tumor growth in Hpa-Tg mice exposed to two-steps skin carcinogenesis model (DMBA-TPA) (32) is associated with increased LC3 staining which is reduced substantially by PG545 (Fig. 5E, left), along with decreased tumor burden (32) thus adding another route by which this compound attenuates tumor growth (50). In a subsequent experiment, the number and size of tumor lesions developed by Hpa-Tg mice exposed to the DMBA-TPA regimen were not affected by chloroquine (not shown). Pathological examination revealed, nonetheless, that chloroquine treatment resulted in less aggressive tumors indicated by exophytic growth pattern and less invasive capacity towards the underlying skin tissue compared with un-treated lesions (Fig. 5E, middle and right panels), suggesting that autophagy dictates skin cancer severity.
Equally important is the ability of heparanase over expression to confer resistance to stress and chemotherapy (Fig. 7D; Suppl. Fig. 2A) mediated, at least in part, by enhancing autophagy (Fig. 7D). Accordingly, heparanase-KO MEF were more sensitive to mTOR inhibitor than control MEF (Torin; Fig. 6B, Suppl. Fig. 2C). Moreover, in cell invasion assay, heparanase-KO MEF exhibit lower invasion capacity and were more sensitive to chloroquine than control MEF (Suppl. Fig. 2D). This add mechanistic view to previous results demonstrating resistance of heparanase over expressing cells to inhibitors of EGFR (22, 51) and the established role of autophagy as a survival pathway. Indeed, diverse classes of anticancer drugs induce autophagy, thus attenuating tumor cell elimination, while autophagy inhibitors overcome chemo-resistance (42, 52). Based on this concept, chloroquine is currently evaluated in several clinical trials including head and neck carcinoma and glioma in combination with different classes of chemotherapeutic agents (42). Interestingly, bacterial heparinase III was noted to enhances autophagy in endothelial cells, and autophagy was further enhanced when heparinase III was combined with oxidized-low density lipoprotein, a mechanism thought to allow endothelial cell survival during stress (53). Heparanase is an endoglucuronidase that cleaves HS in a far more selective manner than heparinase III, yet cleavage and/or clustering of HS within lysosomes may still exert a pro-autophagy effect by heparanase, although non-enzymatic function (1, 30) cannot be excluded.
In addition to neutralizing heparanase enzymatic activity, PG545 has recently been shown to inhibit signaling properties of heparanase, attenuating Akt phosphorylation (32). PG545 similarly prevented autophagy induction by heparanase (Fig. 3C, Con vs. PG), associating with accumulation of heparanase extracellularly and decreased intracellular content of heparanase (Fig. 3D). Thus, PG545 emerges as a broad spectrum heparanase inhibitor, neutralizing its enzymatic, signaling, and autophagic properties. Combining PG545 with inhibitors of autophagy such as chloroquine may yield even higher efficacy in cases that more strictly depend on autophagy for tumor growth.
Supplementary Material
Acknowledgments
We acknowledge the devoted help of Svetlana Vesnovaty (Rambam Health Care Campus) in assisting with the electron microscopy analyses. This study was supported by research grants awarded to I.V. by the Israel Science Foundation (grant 601/14); National Cancer Institute, NIH (grant CA106456); the United States-Israel Binational Science Foundation (BSF); the Israel Cancer Research Fund (ICRF); and the Rappaport Family Institute Fund. I. Vlodavsky is a Research Professor of the ICRF.
Footnotes
Conflict of interest: Edward Hammond is employed by Progen Pharmaceuticals, Brisbane, Queensland, Australia. All other authors have no potential conflict of interest to declare.
Authors’ contribution:
Conception and design: IV, NI, AS
Development of methodology: AS, IB, NI
Acquisition of data: AS, MG, IB, NI
Analysis and interpretation of data: AS, IB, NI, IV, IN, ID
Writing, review, and/or revision of the manuscript: AS, NI, IV, EH
Administrative, technical, or material support: EH, ID
Study supervision: IV
References
- 1.Barash U, Cohen-Kaplan V, Dowek I, Sanderson RD, Ilan N, Vlodavsky I. Proteoglycans in health and disease: new concepts for heparanase function in tumor progression and metastasis. FEBS J. 2010;277:3890–903. doi: 10.1111/j.1742-4658.2010.07799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ilan N, Elkin M, Vlodavsky I. Regulation, function and clinical significance of heparanase in cancer metastasis and angiogenesis. Intl J Biochem & Cell Biol. 2006;38:2018–39. doi: 10.1016/j.biocel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Parish CR, Freeman C, Hulett MD. Heparanase: a key enzyme involved in cell invasion. Biochim Biophys Acta. 2001;1471:M99–108. doi: 10.1016/s0304-419x(01)00017-8. [DOI] [PubMed] [Google Scholar]
- 4.Vlodavsky I, Friedmann Y. Molecular properties and involvement of heparanase in cancer metastasis and angiogenesis. J Clin Invest. 2001;108:341–7. doi: 10.1172/JCI13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elkin M, Ilan N, Ishai-Michaeli R, Friedmann Y, Papo O, Pecker I, et al. Heparanase as mediator of angiogenesis: mode of action. FASEB J. 2001;15:1661–3. doi: 10.1096/fj.00-0895fje. [DOI] [PubMed] [Google Scholar]
- 6.Folkman J, Klagsbrun M, Sasse J, Wadzinski M, Ingber D, Vlodavsky I. A heparin-binding angiogenic protein--basic fibroblast growth factor--is stored within basement membrane. Am J Pathol. 1988;130:393–400. [PMC free article] [PubMed] [Google Scholar]
- 7.Gingis-Velitski S, Zetser A, Kaplan V, Ben-Zaken O, Cohen E, Levy-Adam F, et al. Heparanase uptake is mediated by cell membrane heparan sulfate proteoglycans. J Biol Chem. 2004;279:44084–92. doi: 10.1074/jbc.M402131200. [DOI] [PubMed] [Google Scholar]
- 8.Levy-Adam F, Abboud-Jarrous G, Guerrini M, Beccati D, Vlodavsky I, Ilan N. Identification and characterization of heparin/heparan sulfate binding domains of the endoglycosidase heparanase. J Biol Chem. 2005;280:20457–66. doi: 10.1074/jbc.M414546200. [DOI] [PubMed] [Google Scholar]
- 9.Nadav L, Eldor A, Yacoby-Zeevi O, Zamir E, Pecker I, Ilan N, et al. Activation, processing and trafficking of extracellular heparanase by primary human fibroblasts. J Cell Sci. 2002;115:2179–87. doi: 10.1242/jcs.115.10.2179. [DOI] [PubMed] [Google Scholar]
- 10.Shteingauz A, Ilan N, Vlodavsky I. Processing of heparanase is mediated by syndecan-1 cytoplasmic domain and involves syntenin and alpha-actinin. Cell Mol Life Sci. 2014;71:4457–70. doi: 10.1007/s00018-014-1629-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zetser A, Levy-Adam F, Kaplan V, Gingis-Velitski S, Bashenko Y, Schubert S, et al. Processing and activation of latent heparanase occurs in lysosomes. J Cell Sci. 2004;117:2249–58. doi: 10.1242/jcs.01068. [DOI] [PubMed] [Google Scholar]
- 12.Goldshmidt O, Nadav L, Aingorn H, Irit C, Feinstein N, Ilan N, et al. Human heparanase is localized within lysosomes in a stable form. Exp Cell Res. 2002;281:50–62. doi: 10.1006/excr.2002.5651. [DOI] [PubMed] [Google Scholar]
- 13.Cohen E, Atzmon R, Vlodavsky I, Ilan N. Heparanase processing by lysosomal/endosomal protein preparation. FEBS Lett. 2005;579:2334–8. doi: 10.1016/j.febslet.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 14.Abboud-Jarrous G, Atzmon R, Peretz T, Palermo C, Gadea BB, Joyce JA, et al. Cathepsin L is responsible for processing and activation of proheparanase through multiple cleavages of a linker segment. J Biol Chem. 2008;283:18167–76. doi: 10.1074/jbc.M801327200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abboud-Jarrous G, Rangini-Guetta Z, Aingorn H, Atzmon R, Elgavish S, Peretz T, et al. Site-directed mutagenesis, proteolytic cleavage, and activation of human proheparanase. J Biol Chem. 2005;280:13568–75. doi: 10.1074/jbc.M413370200. [DOI] [PubMed] [Google Scholar]
- 16.Arvatz G, Shafat I, Levy-Adam F, Ilan N, Vlodavsky I. The heparanase system and tumor metastasis: is heparanase the seed and soil? Cancer Metastasis Rev. 2011;30:253–68. doi: 10.1007/s10555-011-9288-x. [DOI] [PubMed] [Google Scholar]
- 17.Gingis-Velitski S, Zetser A, Flugelman MY, Vlodavsky I, Ilan N. Heparanase induces endothelial cell migration via protein kinase B/Akt activation. J Biol Chem. 2004;279:23536–41. doi: 10.1074/jbc.M400554200. [DOI] [PubMed] [Google Scholar]
- 18.Egeberg M, Kjeken R, Kolset SO, Berg T, Prydz K. Internalization and stepwise degradation of heparan sulfate proteoglycans in rat hepatocytes. Biochim Biophys Acta. 2001;1541:135–49. doi: 10.1016/s0167-4889(01)00132-x. [DOI] [PubMed] [Google Scholar]
- 19.Rosenfeldt MT, Ryan KM. The multiple roles of autophagy in cancer. Carcinogenesis. 2011;32:955–63. doi: 10.1093/carcin/bgr031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–10. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arvatz G, Barash U, Nativ O, Ilan N, Vlodavsky I. Post-transcriptional regulation of heparanase gene expression by a 3′ AU-rich element. FASEB J. 2011;24:4969–76. doi: 10.1096/fj.10-156372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen-Kaplan V, Doweck I, Naroditsky I, Vlodavsky I, Ilan N. Heparanase augments epidermal growth factor receptor phosphorylation: correlation with head and neck tumor progression. Cancer Res. 2008;68:10077–85. doi: 10.1158/0008-5472.CAN-08-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen-Kaplan V, Jrbashyan J, Yanir Y, Naroditsky I, Ben-Izhak O, Ilan N, et al. Heparanase Induces Signal Transducer and Activator of Transcription (STAT) Protein Phosphorylation: Preclinical and clinical significance in head and neck cancer. J Biol Chem. 2012;287:6668–78. doi: 10.1074/jbc.M111.271346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luangdilok S, Box C, Patterson L, Court W, Harrington K, Pitkin L, et al. Syk tyrosine kinase is linked to cell motility and progression in squamous cell carcinomas of the head and neck. Cancer Res. 2007;67:7907–16. doi: 10.1158/0008-5472.CAN-07-0331. [DOI] [PubMed] [Google Scholar]
- 25.Zcharia E, Jia J, Zhang X, Baraz L, Lindahl U, Peretz T, et al. Newly generated heparanase knock-out mice unravel co-regulation of heparanase and matrix metalloproteinases. PloS one. 2009;4:e5181. doi: 10.1371/journal.pone.0005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dredge K, Hammond E, Handley P, Gonda TJ, Smith MT, Vincent C, et al. PG545, a dual heparanase and angiogenesis inhibitor, induces potent anti-tumour and anti-metastatic efficacy in preclinical models. Br J Cancer. 2011;104:635–42. doi: 10.1038/bjc.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barash U, Zohar Y, Wildbaum G, Beider K, Nagler A, Karin N, et al. Heparanase enhances myeloma progression via CXCL10 downregulation. Leukemia. 2014;28:2178–87. doi: 10.1038/leu.2014.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barash U, Cohen-Kaplan V, Arvatz G, Gingis-Velitski S, Levy-Adam F, Nativ O, et al. A novel human heparanase splice variant, T5, endowed with protumorigenic characteristics. FASEB J. 2010;24:1239–48. doi: 10.1096/fj.09-147074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zcharia E, Metzger S, Chajek-ShaulL T, Aingorn H, Elikn M, Friedmann Y, et al. Transgenic expression of mammalian heparanase uncovers physiological functions of heparan sulfate in tissue morphogenesis, vascularization, and feeding behavior. FASEB J. 2004;18:252–63. doi: 10.1096/fj.03-0572com. [DOI] [PubMed] [Google Scholar]
- 30.Fux L, Feibish N, Cohen-Kaplan V, Gingis-Velitski S, Feld S, Geffen C, et al. Structure-function approach identifies a COOH-terminal domain that mediates heparanase signaling. Cancer Res. 2009;69:1758–67. doi: 10.1158/0008-5472.CAN-08-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ostapoff KT, Awasthi N, Cenik BK, Hinz S, Dredge K, Schwarz RE, et al. PG545, an angiogenesis and heparanase inhibitor, reduces primary tumor growth and metastasis in experimental pancreatic cancer. Mol Cancer Ther. 2013;12:1190–201. doi: 10.1158/1535-7163.MCT-12-1123. [DOI] [PubMed] [Google Scholar]
- 32.Boyango I, Barash U, Naroditsky I, Li JP, Hammond E, Ilan N, et al. Heparanase cooperates with Ras to drive breast and skin tumorigenesis. Cancer Res. 2014;74:4504–14. doi: 10.1158/0008-5472.CAN-13-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bar-Peled L, Sabatini DM. Regulation of mTORC1 by amino acids. Trends Cell Biol. 2014;24:400–6. doi: 10.1016/j.tcb.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vreys V, David G. Mammalian heparanase: what is the message? J Cell Mol Med. 2007;11:427–52. doi: 10.1111/j.1582-4934.2007.00039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ravikumar B, Futter M, Jahreiss L, Korolchuk VI, Lichtenberg M, Luo S, et al. Mammalian macroautophagy at a glance. J Cell Sci. 2009;122:1707–11. doi: 10.1242/jcs.031773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cohen-Kaplan V, Naroditsky I, Zetser A, Ilan N, Vlodavsky I, Doweck I. Heparanase induces VEGF C and facilitates tumor lymphangiogenesis. Intl J cancer. 2008;123:2566–73. doi: 10.1002/ijc.23898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doweck I, Kaplan-Cohen V, Naroditsky I, Sabo E, Ilan N, Vlodavsky I. Heparanase localization and expression by head and neck cancer: correlation with tumor progression and patient survival. Neoplasia. 2006;8:1055–61. doi: 10.1593/neo.06577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zetser A, Bashenko Y, Edovitsky E, Levy-Adam F, Vlodavsky I, Ilan N. Heparanase induces vascular endothelial growth factor expression: correlation with p38 phosphorylation levels and Src activation. Cancer Res. 2006;66:1455–63. doi: 10.1158/0008-5472.CAN-05-1811. [DOI] [PubMed] [Google Scholar]
- 39.Roucourt B, Meeussen S, Bao J, Zimmermann P, David G. Heparanase activates the syndecan-syntenin-ALIX exosome pathway. Cell Research. 2015;25:412–28. doi: 10.1038/cr.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson CA, Purushothaman A, Ramani VC, Vlodavsky I, Sanderson RD. Heparanase regulates secretion, composition, and function of tumor cell-derived exosomes. J Biol Chem. 2013;288:10093–9. doi: 10.1074/jbc.C112.444562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunlop EA, Tee AR. mTOR and autophagy: A dynamic relationship governed by nutrients and energy. Semin Cell Dev Biol. 2014;36:121–9. doi: 10.1016/j.semcdb.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 42.Levy JM, Thorburn A. Targeting autophagy during cancer therapy to improve clinical outcomes. Pharmacol Ther. 2011;131:130–41. doi: 10.1016/j.pharmthera.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korolchuk VI, Saiki S, Lichtenberg M, Siddiqi FH, Roberts EA, Imarisio S, et al. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol. 2011;13:453–60. doi: 10.1038/ncb2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korolchuk VI, Rubinsztein DC. Regulation of autophagy by lysosomal positioning. Autophagy. 2011;7:927–8. doi: 10.4161/auto.7.8.15862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Efeyan A, Comb WC, Sabatini DM. Nutrient-sensing mechanisms and pathways. Nature. 2015;517:302–10. doi: 10.1038/nature14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Efeyan A, Sabatini DM. Nutrients and growth factors in mTORC1 activation. Biochem Soc Trans. 2013;41:902–5. doi: 10.1042/BST20130063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanderson RD, Yang Y, Kelly T, Macleod V, Dai Y, Theus A. Enzymatic remodeling of heparan sulfate proteoglycans within the tumor microenvironment: Growth regulation and the prospect of new cancer therapies. J Cell Biochem. 2005;96:897–905. doi: 10.1002/jcb.20602. [DOI] [PubMed] [Google Scholar]
- 48.Miao HQ, Liu H, Navarro E, Kussie P, Zhu Z. Development of heparanase inhibitors for anti-cancer therapy. Curr Med Chem. 2006;13:2101–11. doi: 10.2174/092986706777935230. [DOI] [PubMed] [Google Scholar]
- 49.Vlodavsky I, Ilan N, Naggi A, Casu B. Heparanase: Structure, Biological Functions, and Inhibition by Heparin-Derived Mimetics of Heparan Sulfate. Curr Pharm Des. 2007;13:2057–73. doi: 10.2174/138161207781039742. [DOI] [PubMed] [Google Scholar]
- 50.Ferro V, Liu L, Johnstone KD, Wimmer N, Karoli T, Handley P, et al. Discovery of PG545: a highly potent and simultaneous inhibitor of angiogenesis, tumor growth, and metastasis. J Med Chem. 2012;55:3804–13. doi: 10.1021/jm201708h. [DOI] [PubMed] [Google Scholar]
- 51.Zhang L, Ngo JA, Wetzel MD, Marchetti D. Heparanase mediates a novel mechanism in lapatinib-resistant brain metastatic breast cancer. Neoplasia. 2015;17:101–13. doi: 10.1016/j.neo.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu WK, Coffelt SB, Cho CH, Wang XJ, Lee CW, Chan FK, et al. The autophagic paradox in cancer therapy. Oncogene. 2012;31:939–53. doi: 10.1038/onc.2011.295. [DOI] [PubMed] [Google Scholar]
- 53.Ding Z, Wang X, Khaidakov M, Liu S, Dai Y, Mehta JL. Degradation of heparan sulfate proteoglycans enhances oxidized-LDL-mediated autophagy and apoptosis in human endothelial cells. Biochem Biophys Res Commun. 2012;426:106–11. doi: 10.1016/j.bbrc.2012.08.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.