SUMMARY
Alterations of IKZF1, encoding the lymphoid transcription factor IKAROS, are a hallmark of high risk acute lymphoblastic leukemia (ALL), however the role of IKZF1 alterations in ALL pathogenesis is poorly understood. Here we show that in mouse models of BCR-ABL1 leukemia, Ikzf1 and Arf alterations synergistically promote the development of an aggressive lymphoid leukemia. Ikzf1 alterations result in acquisition of stem cell-like features, including self-renewal and increased bone marrow stromal adhesion. Retinoid receptor agonists reversed this phenotype, partly by inducing expression of IKZF1, resulting in abrogation of adhesion and self-renewal, cell cycle arrest and attenuation of proliferation without direct cytotoxicity. Retinoids potentiated the activity of dasatinib in mouse and human BCR-ABL1 ALL, providing an additional therapeutic option in IKZF1-mutated ALL.
INTRODUCTION
ALL is the most common tumor and a leading cause of cancer death in the young (Stock, 2010; Inaba et al., 2013). B-progenitor ALL (B-ALL) is more common than T-lineage ALL, and comprises a number of subtypes characterized by constellations of chromosomal alterations, submicroscopic deletions and sequence mutations (Mullighan, 2013). Loss-of-function or dominant-negative deletions and mutations of lymphoid transcription factor genes including PAX5 (encoding paired box 5), IKZF1 (IKAROS), and EBF1 (early B-cell factor 1) are observed in the majority of B-ALL cases (Mullighan et al., 2007).
IKZF1 alterations are a hallmark of high-risk B-ALL, particularly BCR-ABL1 positive (Ph+) ALL (Mullighan et al., 2008) and Ph-like ALL, which is characterized by a range of genetic alterations driving cytokine receptor and kinase signaling (Den Boer et al., 2009; Mullighan et al., 2009; Roberts et al., 2012; Roberts et al., 2014). IKZF1 alterations include deletions that result in loss of expression of wild-type (WT) IKZF1 (IK1), and focal deletions or sequence mutations that alter IKZF1 function. A common deletion involving exons 4-7 results in expression of the IK6 isoform that lacks the N-terminal DNA-binding zinc fingers, but retains the C-terminal zinc fingers responsible for dimerization (Mullighan et al., 2008). IK6 has dominant negative effects, in part by mislocalizing WT IKZF1 from the nucleus to the cytoplasm. Sequence mutations commonly involve zinc finger residues that mediate DNA binding (Mullighan et al., 2009). IKZF1 alterations are also common in chronic myeloid leukemia (CML) at progression to lymphoid blast crisis, but are rare at chronic phase and progression to myeloid blast crisis, suggesting a central role in determining disease lineage and progression to acute leukemia (Mullighan et al., 2008). Additional genetic alterations are also observed in BCR-ABL1 lymphoid leukemia, most commonly deletion of CDKN2A/B (INK4/ARF) in approximately 50% of cases (Mullighan et al., 2008).
IKZF1 alterations are associated with poor outcome in Ph− positive ALL, despite the advent of TKI therapy (Martinelli et al., 2009; van der Veer et al., 2014) and Ph-negative B-ALL (Mullighan et al., 2009; Kuiper et al., 2010). Consequently, new therapeutic approaches to improve the outcome of IKZF1-mutated ALL are required. However, a detailed understanding of the relative roles of IKZF1 alterations and concomitant genomic alterations in lymphoid leukemogenesis and resistance to therapy is lacking. IKZF1 is required for the specification of the lymphoid lineage (Georgopoulos et al., 1994) by activating a lymphoid transcriptional network while repressing stem cell-, myeloid-, and erythroid-specific genes (Yoshida et al., 2010). Ikzf1 haploinsufficiency accelerates the onset of BCR-ABL1 lymphoid leukemia (Virely et al., 2010), and deletions of selected N-terminal zinc fingers in B cells results in stromal adhesion and progression to acute leukemia in mice (Schjerven et al., 2013; Joshi et al., 2014). Howevever, these studies do not fully recapitulate the genomic alterations in human IKZF1-mutated leukemia (including BCR-ABL1, expression of IK6 and CDKN2A/B deletion), nor do they directly model the role of IKZF1 alterations in determining disease lineage and responsiveness to TKI therapy.
Here we describe mouse models of Ikzf1-mutated, Ph+ ALL. We have used these models to examine the effects of different Ikzf1 alterations, including haploinsufficiency and/or expression of IK6, and Arf loss, on disease lineage and responsiveness to TKI therapy. We examine the role of Ikzf1 alterations on the acquisition of hematopoietic stem-cell like features, and have used these models as a platform of drug discovery to identify agents that enhance responsiveness to TKI therapy.
RESULTS
IKAROS Alterations in Human BCR-ABL1 B-ALL
We previously reported a prevalence of IKZF1 alterations in approximately 15% of childhood ALL, and over 80% of Ph+ ALL (Mullighan et al., 2008; Zhang et al., 2011). However, the prevalence of IKZF1 alterations has differed between studies. We examined IKZF1 status in large cohorts of childhood and adult Ph-positive and negative ALL (Figure 1A and Table S1) (Roberts et al., 2014). Twenty-five percent of childhood and 44% of young adult precursor B-cell ALL cases had alterations of IKZF1. Exon 4-7 (IK6) deletions were present in 22.9% of B-ALL cases, and biallelic alterations in 13.2% of cases. IKZF1 sequence mutations were observed in 2.6% of childhood and 3.4% of young adult ALL. Many sequence alterations were missense mutations in the N-terminal zinc fingers at or near residues known to be critical for DNA binding (Cobb et al., 2000), and are thus likely to be loss-of-function and/or exert dominant negative effects (Figure 1B).
Figure 1. IKZF1 alterations in human B-ALL.
(A) Distribution of IKZF1 alterations in human Ph+ (left, n=89) and Ph- (right, n=405) B-ALL. (B) Point mutations found in human B-ALL. Highlighted mutations were modeled in Arf−/− BCR ABL1-expressing pre-B cells studied in (C). (C) WT IKZF1, IK6, and six IKZF1 point mutants were expressed in Arf−/− BCR-ABL1-expressing pre-B cells (scale bars, 2 m).
See also Table S1.
To compare the effects of point mutations to IK6, we expressed six different IKZF1 point mutant alleles, as well as WT IKZF1 and IK6 in Arf−/− BCR-ABL1-expressing pre-B cells. These experiments were performed in Arf−/− cells as although Cdkn2a encodes both p19Arf and p16Ink4a, prior studies have loss of Arf but not Ink4a promotes the development of BCR-ABL1 ALL (Kamijo et al., 1997; Williams et al., 2006; Signer et al., 2010). Enforced expression of WT IKZF1 was not tolerated and resulted in cell death. In contrast, all Ikzf1 point mutant alleles resulted in perturbed subcellular localization of the protein (Figure 1C). Endogenous IKZF1 exhibited punctate nuclear staining, whereas IK6 or IKZF1 point mutant alleles exhibited cytoplasmic or perturbed nuclear localization (Figure 1C). Thus, like IK6 (Nishii et al., 2002), the IKZF1 sequence mutations observed in human ALL result in cellular mislocalization and may also perturb IKZF1 function.
Cooperativity of Ikzf1 and Arf Alterations in BCR-ABL1 lymphoid leukemogenesis
To examine the relative contributions of IKZF1 and Arf alterations in leukemogenesis, we used unfractionated, lineage negative (lin-) and pre-B cell retroviral bone marrow (BM) transplant models of BCR-ABL1 leukemia. Consistent with prior data (Virely et al., 2010), heterozygosity for an Ikzf1 null allele increased the penetrance and reduced latency of pre-B cell ALL in both whole BM and pre-B cell transplant models (Figures 2A-B). The leukemias were of pre-B cell lineage (CD43+, B220+, CD19+, BP-1+, and IgM−; Figure S1A). There was no expansion of lymphoid progenitors in Ikzf1+/− marrow (Figure S1B), and Ikzf1+/− mice did not develop leukemia in the absence of BCR-ABL1, suggesting that loss of IKZF1 does not promote leukemogenesis by expanding the pool of lymphoid progenitors susceptible to transformation.
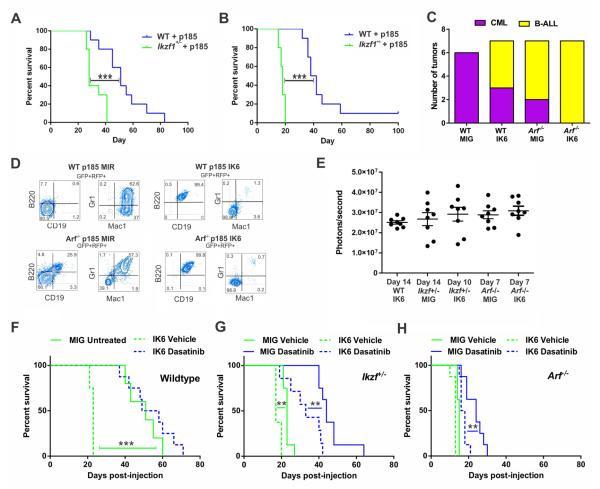
Figure 2. Perturbation of IKZF1 drives a lymphoid leukemia with less responsiveness to TKI therapy.
(A) Unmanipulated BM from WT or Ikzf1+/− mice transduced with p185 BCR-ABL1 expressing retrovirus and immediately transplanted into lethally irradiated recipients. Statistical significance was assessed by log rank Mantel Cox ***p<0.0005; n=10 mice per group. (B) Transplantation of in vitro-derived BCR-ABL1 transformed pre-B cells into sublethally irradiated recipients. Statistical significance was assessed by log rank Mantel Cox ***p<0.0005; n=10 mice per group. (C) Summarized leukemia lineage data from lin- BM transplant experiments (see also Figure S1) (Χ2 p=0.0029; n≥6 mice per group). (D) Representative flow cytometric analysis of tumors derived from p185 BCR-ABL1 transduced lin- BM cells to determine disease lineage. (E) In vivo quantification of luciferase activity, representing disease burden, in mice transplanted with preB cells of the defined genotypes (WT, Arf−/−, or Ikzf+/−) transduced with BCR-ABL1-ires-luciferase and empty vector (MIG) or IK6. Raw data points ± S.D. are plotted. Treatment was started when the average luminescence was greater than 2 × 107 photons/second, indicating a substantial and similar disease burden at the start of therapy between genotypic groups shown in (F-H) and Table 1. (F) Kaplan Meier survival curves of vehicle or dasatinib-treated mice inoculated with pre-B cells derived from WT BM described in (E). (G) Kaplan Meier survival curves of vehicle or dasatinib-treated mice inoculated with pre-B cells derived from Ikzf+/− BM described in (E). (H) Kaplan Meier survival curves of vehicle or dasatinib-treated mice inoculated with pre-B cells derived from Arf−/− BM described in (E). Statistical significance was access by log-rank Mantel-Cox test in F-H. **p < 0.005; n=8 mice per group.
Human Ph+ ALL tumors commonly harbor recurring DNA copy number alterations in addition to deletion of IKZF1, including deletions of CDKN2A/B, PAX5, EBF1 and MEF2C (Mullighan et al., 2008; Mullighan et al., 2009). Using a comparative genomic hybridization microarray with dense tiling of targets of genetic alteration in human B-ALL (Table S2), we identified deletions also observed in human B-ALL, including deletions of Ebf1 and Cdkn2a/b, but not Ikzf1 (Figure S1C). These data support the notion that IKZF1 haploinsufficiency promotes tumorigenesis, but this process involves the sequential acquisition of additional genetic alterations, including second hits disrupting transcriptional regulation of lymphoid development.
The lineage of BCR-ABL1 leukemia has been attributed to the cellular target of transformation (McLaughlin et al., 1987; Daley et al., 1990), but the importance of cooperating genetic alterations has not been formally examined. To examine the role of Ikzf1 and Arf alterations in determining disease lineage, we adopted a retroviral BM transplant model of BCRABL1 leukemia, in which expression of BCR-ABL1 in lineage-negative hematopoietic progenitors robustly induces myeloproliferative disease that recapitulates CML (Daley et al., 1990). As expected, WT BM expressing BCR-ABL1 resulted in a fully penetrant myeloid leukemia (Figure 2C; Figure S1D). In combination with IK6, BCR-ABL1 drove either myeloid or B-lymphoid disease (Figure 2C; Figure S1D). On an Arf−/− background, BCR-ABL1 resulted in 29% myeloid tumors and 71% B-lymphoid tumors; with IK6, BCR-ABL1 uniformly induced BALL (Figure 2C; Figure S1D). One recipient of Arf−/− BCR-ABL1 cells developed a bilineal tumor with both myeloid and lymphoid (Figures 2D and S1E). All other tumors were either myeloid or lymphoid by immunophenotype and morphology (Figures 2D and S1F), and were retransplantable maintaining immunophenotypic fidelity to the primary tumor (Figure S1E). These results indicate that both IK6 and loss of Arf shift differentiating hematopoietic precursors toward a lymphoid fate during BCR-ABL1-driven leukemogenesis.
Ikzf1 Alterations Reduce Responsiveness of BCR-ABL1 ALL to Dasatinib
We next examined the effects of Ikzf1 haploinsufficiency, IK6 and Arf loss on responsiveness to TKI therapy. To establish ALL models of each Arf/Ikzf1 genotype with equivalent tumor burden at the commencement of therapy, 2×105 pre-B cells from WT, Arf−/−, or Ikzf+/− mice expressing MSCV-BCR-ABL1-ires-luc and either MSCV-IK6-IRES-GFP (MIG-IK6) or empty vector (MIG) were inoculated into sublethally irradiated WT recipients. Dasatinib or vehicle was commenced at equivalent tumor burden as determined by bioluminescent imaging (Figure 2E). ALL lacking Ikzf1 or Arf alterations was not modeled due to incomplete penetrance and highly variable latency.
Ikzf1 haploinsufficiency and IK6 both reduced disease latency in a synergistic fashion (Ikzf1+/+ + IK6 vs Ikzf1+/− + IK6 p<0.0001; Table 1 and Figures 2F-2H). Expression of IK6 resulted in a reduced response to dasatinib treatment, also in a synergistic manner with Ikzf1 haploinsufficiency (Figure 2F-2H). The reduced effect of dasatinib in Ikzf1-altered leukemias was not due to impaired inhibition of ABL1 (as measured by STAT5 and CRKL phosphorylation, Figure S1G) or the acquisition of ABL1 tyrosine kinase domain mutations (data not shown).
Table 1.
| Genetic Combination |
Day Treatment Started |
Median Survival (post-injection) Untreated |
Median Survival (post-injection) Dasatinib |
|---|---|---|---|
| WT MIG | n/a | 51 | n/a |
| WT IK6 | 14 | 23 | 51.5 |
| Ikzf+/− MIG | 14 | 23 | 44 |
| Ikzf+/− IK6 | 10 | 17 | 31.5 |
| Arf−/− MIG | 7 | 15 | 24 |
| Arf−/− IK6 | 7 | 13 | 17 |
Increased Adhesion of Ikzf1-altered BCR-ABL1 pre-B Cells
We next examined the growth kinetics and phenotypic properties of BCR-ABL1 pre-B cells of various Ikzf1 and Arf genotypes (WT, Arf−/−, Ikzf1+/−, and Ikzf1+/−;Arf+/− with and without IK6). Both Ikzf1 haploinsufficiency and expression of IK6 reduced doubling times (Figure S2A).
We observed striking effects of all Ikzf1 alterations on intercellular adhesion (Figure 3A). Regardless of genetic background, BCR-ABL1 pre-B cells harboring Ikzf1 alterations exhibited marked changes in their morphology with adherence in sphere-like aggregates in vitro. This was observed for Ikzf1 haploinsufficiency, expression of IK6 or zinc finger point mutant alleles, and shRNA-knockdown of Ikzf1 (Figure S2B). Knockdown of Ikzf3 (AIOLOS), an IKAROS family member that with IKZF1 is part of the Mi-2/nucleosome remodeling and deacetylase (NuRD) complex (Zhang et al., 2012b), resulted in similar cellular aggregation (Figure S2B, panel XIV). Knockdown of Ikzf2 (HELIOS), that does not share a role in the NuRD complex and is not expressed in pre-B cells, had no effect.
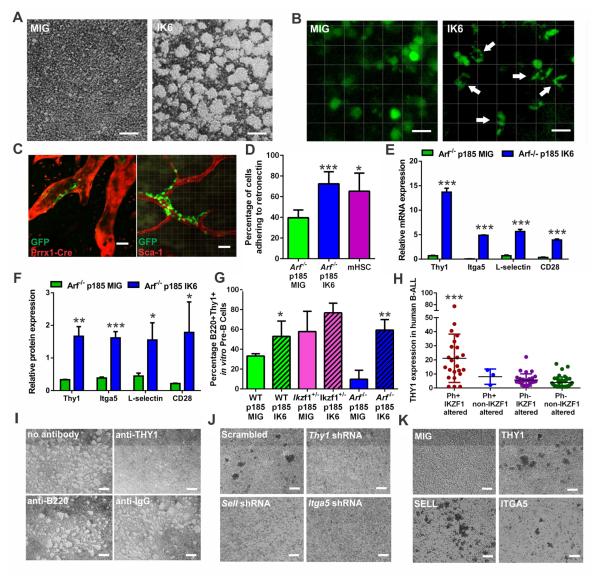
Figure 3. IKZF1 alterations induce aberrant adhesion in BCR-ABL1 leukemic cells.
(A) Suspension cultures of Arf−/− BCR-ABL1 (p185) pre-B leukemic cells showing adherent growth in IK6 expressing cells in vitro (scale bars, 100μM). (B) Calvarial imaging of mice transplanted with Arf−/− BCR-ABL1-transduced pre-B cells expressing MIG or IK6-GFP in vivo (white arrows indicate cells with marked change in pre-B cell morphology; scale bars, 20μM). (C) Calvarial imaging of Arf−/− BCR-ABL1 IK6-GFP leukemic cells in Prrx1-Cre;Ai9 tomato mice showing associations with the PRRX1+ perivascular stromal cells (left) and in vivo-labeled SCA-1+ arterioles (right) in the BM niche (scale bars, 20μM). (D) Arf−/− BCR-ABL1 IK6-expressing cells are more adherent to fibronectin monolayers in vitro, comparable to murine hematopoietic stem cells (mHSC). Data are means ± S.D.; *p<0.05, *** p<0.0005; n=3 biological replicates. (E) The adherent phenotype of IK6-expressing Arf−/− BCR-ABL1 pre-B cells is accompanied by increased expression of adhesion markers compared to empty vector controls as determined by mRNA-sequencing. Data are means ± S.D.; *** p<0.0005; n=4 biological replicates per group. (F) Proteomic analysis by tandem mass tag based mass spectrometry confirms increased protein expression of adhesion markers in IK6-expressing pre-B cells. Data are means ± S.D.; ***p<0.0005, **p<0.005, *p<0.05; n=3 biological replicates per group. (G) Flow cytometric analysis for THY1 in cultured WT, Ikzf1+/−, and Arf−/− BCR-ABL1-transduced cells expressing empty vector or IK6. Data are means ± S.D.; **p<0.005, *p<0.05; n=3 biological replicates. (H) Increased expression THY1 (CD90) by immunophenotypic analysis of human BCR-ABL1 (Ph+) and IKZF1-mutated leukemias. Data are means ± S.D.; **p<0.005, 98 human cases were analyzed, see also Table S5. (I) Cellular adherence of Arf−/− BCR-ABL1 IK6-expressing pre-B cells is abrogated in vitro by anti-THY1 antibody, but not B220 and IgG control antibodies (scale bars, 100μM). (J) Cell aggregation is also attenuated by shRNA knockdown of Thy1, Sell, or Itga5 (scale bars, 100μM). (K) Overexpression of THY1, SELL, or ITGA5 resulted in cellular aggregation (scale bars, 100μM).
To assess whether the increased adhesion of Ikzf1-altered BCR-ABL1 pre-B cells conferred an advantage in homing and engraftment in the BM niche, we transplanted 2×105 Arf−/− pre-B cells expressing BCR-ABL1 and either IK6-MIG or MIG and harvested calvaria at 24, 48, and 72 hours post-injection to track the infiltration of GFP-labeled cells in the BM cavities utilizing multiphoton microscopy. By 48 hours sufficient cells were visible in each group to enable accurate quantitation of engraftment (Figure S2C). No quantitative differences in homing were detected, however IK6-expressing cells adopted spindle-like morphology, with adherence to and infiltration of surrounding BM stroma (Figure 3B). To assess localization in the BM niche, we repeated this experiment using Prrx1-Cre;Ai9 recipient mice, which express tdTomato in mesenchymal stem cells, osteoblasts and CXCL12-abundant reticular cells (Greenbaum et al., 2013), followed by in vivo antibody-based marking of the BM vasculature. This showed localization of leukemic cells adjacent to perivascular mesenchymal cells and SCA-1+ arteriolar endothelial cells (Figure 3C). This stromal adherence in vivo was recapitulated in vitro by measuring adherence to the fibronectin, which showed increased adherence of IK6-expressing cells, comparable to purified mHSCs, after short-term incubation (Figure 3D).
Characterization of the Adhesive Phenotype of Ikzf1-altered Leukemia
We next performed transcriptomic and proteomic profiling of Arf−/− BCR-ABL1 pre-B cells with or without expression of IK6, and immunophenotyping of a large cohort of human Ph+ ALL cases of known IKZF1 genotype (Table S5). These analyses showed highly significant correlation of the transcriptomic and proteomic signatures of BCR-ABL1 IK6 v. non-IK6 leukemias, with gene set enrichment analysis p and FDR q values <0.0001 for enrichment of the expression signature in the proteomic signature. IK6-expressing leukemic cells showed overexpression of multiple adhesion molecules implicated in leukemic and stem cell adherence, including THY1 (thymocyte differentiation antigen 1, CD90), SELL (L-selectin), and the THY1 ligand ITGA5 (integrin alpha 5) (Saalbach et al., 2005) as well as other genes involved in THY1 integrin signaling, including PTK2 (protein kinase 2; or focal adhesion kinase, FAK) and PTK2B (Figures 3E-F and S2D-E; Tables S3 and S4). THY1 is a GPI-anchored glycoprotein involved in adhesion (Rege and Hagood, 2006) and integrin signaling (Barker and Hagood, 2009). THY1 is expressed by thymocytes, hematopoietic stem cells (Craig et al., 1993; Mayani and Lansdorp, 1994; Notta et al., 2011) and stem cell-enriched populations in non-BCR-ABL1 leukemia (Lamkin et al., 1994; Yamazaki et al., 2009), but not normal B-cells. THY1 and ITGA5 are both expressed at high levels in mouse and human HSCs (Figure S2F-G), and mouse and human HSCs were highly adherent to fibronectin in vitro (Figures 3D and S2H). Thy1 is a target of IKZF1 transcriptional repression (Zhang et al., 2012b). Analysis of existing chromatin immunoprecipitation (ChIP) sequencing data (Ferreiros-Vidal et al., 2013), and ChIP-PCR of BCR-ABL1 Arf−/− pre-B cells showed that Thy1, Sell, Cd28, and Itga5 are IKZF1 targets in primary mouse pre-B cells (Figures S2I-J).
All BCR-ABL1 pre-B cell lines harboring an Ikzf1 alteration displayed increased THY1 expression relative to controls (Figures 3G and S2K). IK6-expressing tumors cells from both lineage negative and pre-B cell experiments, and BCR-ABL1 cells following RNAi-mediated Ikzf1/3 knockdown showed increases of THY1 expression (Figures 3E-G and S2K-M). Immunophenotyping of 98 human ALL samples showed higher THY1 expression in IKZF1-altered BCR-ABL1 B-ALL cases compared to those lacking IKZF1 alterations and non-Ph+ cases (Figure 3H; Table S5).
To examine the role of adhesion molecule upregulation, we incubated Arf−/− BCR-ABL1 IK6 pre-B cells with a neutralizing anti-THY1 antibody, which abrogated the clustering phenotype, whereas an antibody directed to B220 and an IgG control had no effect (Figure 3I). shRNA-mediated knockdown of THY1, SELL, or ITGA5 also disrupted cellular aggregation in vitro (Figures 3J and S2N). Conversely, overexpression of THY1, SELL, or ITGA5 alone in Arf−/− BCR-ABL1-expressing pre-B cells was sufficient to induce clustering (Figure 3K and Figure S2O), demonstrating that THY1, SELL, and ITGA5 mediate adhesion of IKZF1-altered cells.
Ikzf1 Alterations Result in Acquisition of a Stem Cell-like Phenotype
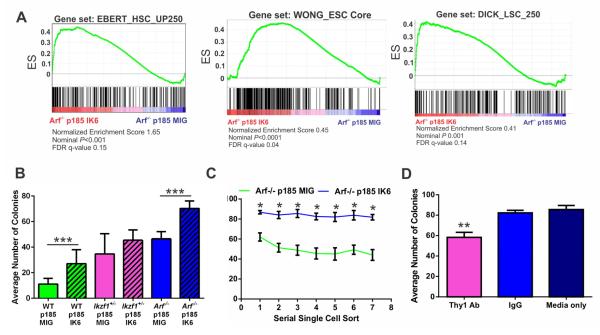
The gene expression profile of IK6-expressing BCR-ABL1 Arf−/− pre-B cells was significantly enriched for gene sets upregulated in human Ph+ B-ALL, transcriptional targets of IKZF1 and gene sets representative of human embryonic (Wong et al., 2008), hematopoietic (Doulatov et al., 2010; Novershtern et al., 2011) and leukemic (Zhang et al., 2012a) stem cells (Figure 4A). This is consistent with previous observations in human IKZF1-altered ALL (Mullighan et al., 2009), and suggests that IKZF1 alterations impair hematopoietic development and confer stem-cell like features. To investigate this, we examined single cell colony forming potential of WT, Ikzf1+/−, and Arf−/− BCR-ABL1 pre-B cell lines with and without IK6 as a measure of self-renewal. In all genetic backgrounds, Ikzf1-alterations conferred increased clonogenicity (Figure 4B). Serial re-plating of the Arf−/− groups demonstrated that IK6-expressing cells maintain a consistent level of clonogenicity over eight weeks, while cells expressing empty vector decrease the number of colonies formed over time (Figure 4C). WT cultures could not be maintained more than two serial platings due to loss of ARF. Addition of THY1-neutralizing antibody to the media prior to single cell sorting reduced the ability of IK6-expressing cells to form colonies (Figure 4D).
Figure 4. IKZF1 alterations induce stem cell-like features in BCR-ABL1 leukemic cells.
(A) GSEA analyses of Arf−/− BCR-ABL1 MIG vs IK6-expressing pre-B cells showing IK6 expression is associated with acquisition of a stem-like gene expression programs. A full list of significantly enriched gene sets is provided in Table S3. (B) IKZF1 perturbation and Arf loss synergistically result in enhanced self-renewal of BCR-ABL1-expressing pre-B cells, as shown by the ability of single cell cultures to form colonies. Data are means ± S.D.; ***p<0.0005, n=3 biological replicates performed in triplicate. (C) Single cell replatings were sustained over serial passages in the immortal Arf−/− lines. Data are means ± S.D.; *p<0.05; n=4 technical replicates (96 well plates). (D) A neutralizing THY1 antibody attenuates the colony-forming ability of Arf−/− BCR-ABL1 IK6 pre-B cells. Data are means ± S.D.; **p<0.005; n=3 biological replicates.
RXR Agonists Induce IKZF1 Expression and Ameliorate the Effects of Ikzf1 Alterations
As IKZF1 alterations induce stem cell and adhesive properties, and are associated with poor response to TKI therapy in human ALL, we performed a drug screen to identify agents that reverse this stem-cell-like phenotype and enhance TKI sensitivity. We used high-throughput microscopy to measure the clustering of IK6-expressing Arf−/− BCR-ABL1 pre-B cells with the aim of identifying compounds that abolish their sphere-forming capacity without directly affecting viability, although direct cytotoxicity was also measured (Figure S3A-C). Disruption of clustering suggested that a drug may reverse the stem-cell like characteristics induced by IKZF1 alteration, and may augment responsiveness to existing therapy.
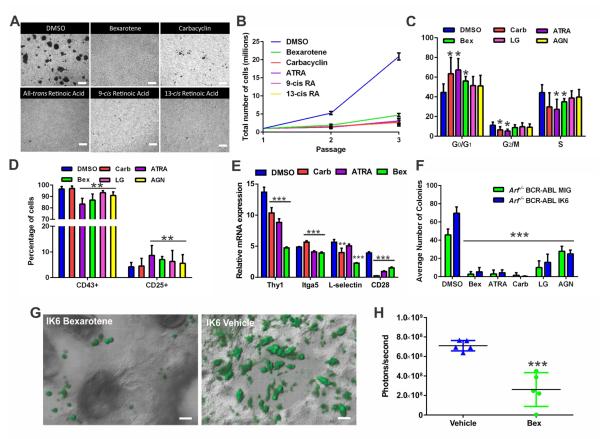
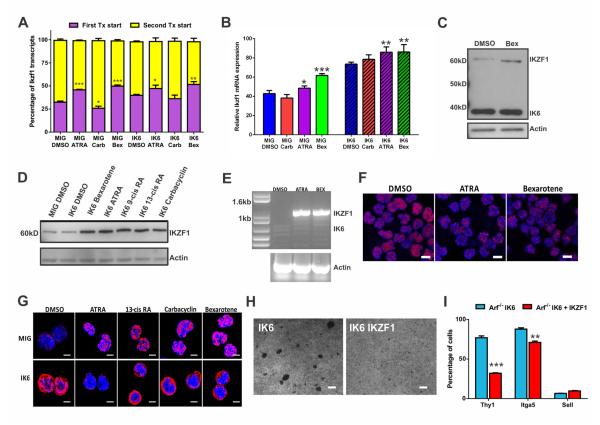
An initial screen of 356 compounds currently in clinical use or trials (Table S6) identified bexarotene as the agent that most potently inhibited cellular aggregation (Figure 5A). Bexarotene is a synthetic retinoid that activates retinoid X receptors (RXRs). A secondary screen using 127 nuclear hormone receptor effectors (Table S6) identified carbacyclin, all-trans retinoic acid (ATRA), 9-cis retinoic acid (RA), and 13-cis RA as potent inhibitors of cellular aggregation. Carbacyclin is a peroxisome proliferator-activated receptor (PPAR) agonist, whereas ATRA and 9/13-cis RA activate retinoic acid receptors (RARs), although all have promiscuous activity for multiple receptors. All compounds identified converge on the retinoid pathway, whereby RXRs, RARs, and PPARs can interchangeably heterodimerize to transcriptionally activate target genes containing appropriate response elements (Altucci et al., 2007). Rara, Rarg, Rarres, Rxra, Rxrb, Ppard, and Ppargc1b were expressed in Arf−/− BCR ABL1 pre-B cells, all of which harbor IKZF1 binding sites, with significant upregulation of Rxra, encoding retinoid X receptor alpha, in IK6 expressing cells (Figures S3D-E). As these agents exhibit cross reactivity for the different retinoid receptors, we also treated IK6-expressing Arf−/− BCR-ABL1 pre-B cells with RXR-specific agonist LG100268 (LG) and the RAR-specific agonist, AGN-195183 (AGN). LG abrogated the clustering phenotype in vitro, whereas AGN and the L-selectin inhibitor rivipansel had no effect on adhesion (Figure S3F). In addition, connectivity map analysis (Lamb et al., 2006) identified the gene expression signature of 13-cis-retinoic acid as negatively enriched in that of Arf−/− BCR-ABL1 MIG-IK6 v. MIG leukemia, further supporting the importance of this pathway in Ikzf1-altered ALL (P=0.0048, specificity=0.0079, Table S7).
Figure 5. Retinoids disrupt cellular aggregation and reverse the stem-cell like features of BCR-ABL1 leukemic cells.
(A) Suspension cultures of Arf−/− BCR-ABL1 IK6-expressing cells exposed to DMSO or retinoids at 1μM for 72 hours. Each drug abolishes adherent growth induced by IK6 (scale bars, 100μM). (B) In vitro growth kinetics of DMSO or retinoid-treated Arf−/− BCR-ABL1 IK6-expressing cells. Data are means ± S.D.; n=3 biological replicates. (C) Retinoid treatment arrests Arf−/− BCR-ABL1 IK6-expressing pre-B cells at the G0/G1 phase of the cell cycle. Data are means ± S.D.; *p<0.05; n=3 biological replicates performed in triplicate. (D) Flow cytometry of retinoid-treated Arf−/− BCR ABL1 IK6-expressing pre-B cells shows decreased CD43 expression and increased CD25 levels. Data are means ± S.D.; **p<0.005; n=3 biological replicates performed in triplicate. (E) Relative expression of aberrantly expressed adhesion molecules in Arf−/− BCR-ABL1 IK6-expressing pre-B cells following retinoid treatment, as determined by mRNA-sequencing. Data are means ± S.D.; ***p<0.0005, **p<0.005; n=4 biological replicates. (F) Retinoid treatment attenuates the self-renewal of Arf−/− BCR-ABL1 IK6-expressing pre-B cells as demonstrated by single cell colony forming ability. Data are means ± S.D.; ***p<0.0005; n= 3 biological replicates performed in triplicate 96 well plates. (G) In vivo bex treatment reversed the adherent phenotype of Arf−/− BCR-ABL1 IK6-GFP cells in the calvarial BM niche (scale bars, 20μM). (H) Luciferase quantification of vehicle or bex-treated mice 14 days after inoculation with 2×105 Arf−/− p185 IK6 leukemic cells. Daily vehicle or 45mg/kg oral bex treatment commenced at the time of transplant. Raw data points ± S.D. are plotted; ***p<0.0005; n=5 mice per group.
Each non-specific retinoid abrogated sphere formation of Arf−/− BCR-ABL1 IK6-expressing pre-B cells (Figure 5A), and arrested cell proliferation (Figure 5B) at the G0/G1 phase of the cell cycle (Figure 5C), with minimal apoptosis (Figure S3G). With the exception of carbacyclin, this was associated with partial maturation, with reduced CD43 and increased CD25 expression (Figure 5D), and on transcriptomic analysis, reversal of the hematopoietic stem cell state and induction of a B-cell differentiation program (Figure S3H). THY1 and CD28 expression was reduced after 72 hours of treatment in vitro, with drug-dependent effects on ITGA5 and SELL levels (Figure 5E). Expression of the downstream integrin signaling mediator PTK2B was also reduced by each compound (data not shown). All retinoids profoundly decreased the colony-forming potential of IK6-expressing pre-B cells (Figure 5F). In calvarial BM imaging experiments, in vivo treatment with bexarotene at 0, 24 and 48 hours post-transplant, restored the spherical, non-adherent phenotype of transplanted Arf−/− BCR-ABL1 IK6-expressing pre-B cells (Figure 5G). In addition, leukemic burden was significantly decreased in mice treated with bexarotene (Figure 5H).
IKZF1 has alternate non-coding first exons, and the promoter, 5′ untranslated region, and gene body of IKZF1 harbors multiple putative retinoic acid response elements (RARE; Figure S4A). Ikzf1 is a direct target of RXR signaling, as determined by ChIP-PCR in Arf−/− BCR ABL1 IK6 pre-B cells. No enrichment was observed for RAR binding at the Ikzf1 locus (Figure S4A). Retinoid treatment resulted in induction of WT IKZF1, but not IK6 expression from the first non-coding exon in both mouse and human leukemic cells as determined by RNA-seq (Figures 6A-B). Western blotting, RT-PCR and immunofluorescence confirmed selective induction of WT IKZF1 but not IK6 by retinoids, and partial relocalization of IKZF1 to the nucleus in IK6-expressing cells (Figures 6C-G). Moreover, 1745 (85%) of 2061 genes differentially expressed in Arf−/− BCR-ABL1 IK6 cells following bexarotene treatment were IKZF1 targets (Ferreiros-Vidal et al., 2013), compared to 40% of non-differentially expressed genes (P<2×10−16), supporting the notion that induction of IKZF1 directly contributes to the effects observed upon retinoid treatment. Multiple RAREs within the genomic region of IKZF1 deleted in the IK6 allele showed significant enrichment for RXR binding, particularly after treatment with the RXR-specific agonist LG100268 (Figure S4A). In addition, the IKZF1 locus has a putative intragenic enhancer within the region of IK6 deletion marked by H3K27 acetylation in K562 (BCR-ABL1+ IKZF1 WT cells, Figure S4B) that has been shown to interact with the IKZF1 promoter through Chromatin Interaction Analysis Paired-End Tags only in K562 among the 6 ENCODE cell lines (Li et al., 2012), suggesting that this region that is lost in IK6-deleted cases influences regulation of IKZF1 expression.
Figure 6. Retinoids induce IKZF1 expression, but not IK6, in BCR-ABL1 leukemic cells.
(A) Proportion of Ikzf1 transcripts from alternate transcriptional start sites as determined by mRNA-seq in Arf−/− p185-expressing pre-B cells. Data are means ± S.D.; ***p<0.0005, **p<0.005, *p<0.05; n=4 biological replicates. (B) Ikzf1 mRNA levels are increased after retinoid treatment in Arf−/− p185 MIG or IK6 pre-B cells. Data are means ± S.D.; ***p<0.0005, **P < 0.005, *p<0.05; n=4 biological replicates. (C) Western blot of retinoid-treated murine Arf−/− BCR-ABL1 IK6-expressing pre-B cells. (D) Western blot of retinoid-treated human CDKN2A-deleted (Arf-null) BCR-ABL1+ IK6-expressing B-ALL cells. (E) RT-PCR analysis of retinoid-treated human BCR-ABL1 IK6 B-ALL cells for IKZF1 and IK6 transcripts. (F) Immunofluorescence for IKZF1 in CDKN2A-deleted (Arf-null) BCR-ABL1+ IK6-expressing B-ALL cells (scale bars, 5 μm). (G) Immunofluorescence for IKZF1 in murine Arf−/− BCR-ABL1 MIG (top) and IK6-exressing (bottom) pre-B cells (scale bars, 2 μm). (H) Overexpression of IKZF1 in Arf−/− BCR-ABL1 IK6 pre-B cells abrogates cellular adhesion in vitro (scale bars, 100μm). (I) Overexpression of IKZF1 in Arf−/− BCR-ABL1 IK6 pre-B cells downregulates THY1 and ITGA5 (as determined by flow cytometry). Data are means ± S.D.; ***p<0.0005, **p<0.005; n=3 technical replicates.
See also Figure S4.
To test if directly increasing IKZF1 expression levels recapitulates the effects of retinoids, we enforced expression of IKZF1 in Arf−/− BCR-ABL1 pre-B cells with or without expression of IK6. Enforced expression of IKZF1 is not tolerated by Ikzf1 WT cells and results in apoptotic cell death (Figure 1C). In contrast, IKZF1 expression in IK6-expressing cells did not suppress proliferation or cause cell death (Figure S4C), but resulted in attenuation of aggregation accompanied by a decrease in THY1 and ITGA5 expression (Figures 6H and 6I). Therefore in IKZF1-altered cells, the induction of WT Ikzf1 by retinoids leads to abrogation of the adherent phenotype, but cell cycle arrest and differentiation are caused by retinoid-related, non-IKZF1-dependent mechanisms. In contrast, bexarotene did not significantly reduce proliferation in mouse BCR-ABL1 leukemic cells lacking Arf/Ikzf1 alterations (Figure S4D). Reduced proliferation and abrogation of clustering was observed in non-BCR-ABL1 human leukemic cells with IKZF1 alterations, including Ph-like ALL cells expressing PAG1-ABL2 or harboring EPOR rearrangements (Figure S4D-E), as has been reported for retinoids in non-BCR-ABL1 ALL cells (Zhang et al., 2002; Lin et al., 2007)
Retinoids Increase Responsiveness of Ikzf1-altered Leukemias to TKIs
Using in vitro viability assays with drug added at the time of cell plating, bexarotene significantly increased responsiveness to dasatinib in IK6-expressing Arf−/− BCR-ABL1 IK6 pre-B cells (data not shown). Response to dasatinib was also improved in Arf−/− BCR-ABL1 pre-B cells lacking IK6, consistent with retinoid induction of IKZF1 expression in non-IK6 expressing cells causing cell cycle arrest and subsequent death (Figures 1C, S4C).
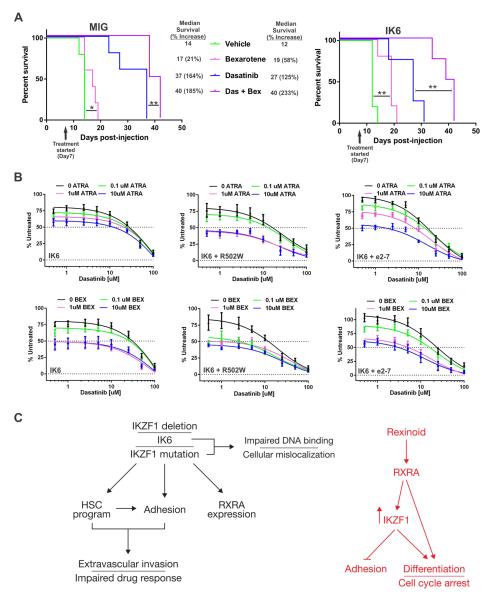
We examined the combinatorial and single-agent effects of bexarotene, dasatinib and additional cytotoxic agents used in ALL therapy, L-asparaginase and dexamethasone, in mice transplanted with Arf−/− BCR-ABL1 pre-B cells with or without IK6 expression. In contrast to L-asparaginase and dexamethasone (Boulos et al., 2011), bexarotene monotherapy resulted in significant benefit without detectable toxicity (Figure 7A and S5A). Dasatinib monotherapy increased survival with inferior responsiveness in the IK6 group (Figure 7A). The combination of dasatinib and bexarotene resulted in the greatest survival advantage, with a near-doubling of survival time in IK6 tumors compared to dasatinib monotherapy. Thus, bexarotene enhances efficacy of TKI therapy in Ph+ tumors regardless of Ikzf1 status, but with the greatest potentiation observed in Ikzf1-altered tumors.
Figure 7. Retinoids potentiate TKI therapy in BCR-ABL1 leukemia.
(A) WT C57Bl/6 mice were engrafted with Arf−/− BCR-ABL1 MIG-IK6 or MIG cells and randomized to vehicle, bex and/or dasatinib. On Kaplan-Meier analysis, bexarotene significantly increased survival time, with the greatest effect observed in mice treated with both drugs. **p<0.005, *p<0.05; n=5 mice per group. (B) Dose-response curves of human BCR-ABL1 CDKN2A/B-deleted (i.e., INK4/ARF−/−), IKZF1 Δ4-7 (IK6), IK6 + R502W mutation, or IK6 + Δ2-7 leukemic cells harvested from NSG mice and immediately treated ex vivo with increasing concentrations of bexarotene (Bex), or all-trans-retinoic acid (ATRA) at increasing concentrations of dasatinib. Each retinoid inhibited cell proliferation, even at very low concentrations of dasatinib, and reduced the TKI IC50. n=3 biological replicates performed in triplicate. (C) schematic summarizing effects of IKZF1 alterations and their reversal by retinoids.
See also Figure S5.
To test the activity of retinoids in human leukemic cells, we established xenografts of Ph+ ALL that faithfully recapitulate a range of IKZF1 genotypes. The tumors harbored homozygous deletion of CDKN2A/B (i.e. INK4/ARF−null), and had deletions or sequence mutations of IKZF1 (IKZF1 Δ4-7 (IK6), Δ2-7 or R502W). Xenografts were treated with dasatinib in combination with bexarotene, ATRA or the FAK inhibitors PF-562271, NVP-TAE226, and PF-573228 ex vivo. This resulted in significant potentiation of cell killing at increasing concentrations of the drugs (Figures 7B and S5B).
DISCUSSION
Here, we have examined large B-ALL cohorts to define the spectrum of IKZF1 alterations, and used this to inform the development of faithful mouse models of human Ph+ ALL. These models incorporate modeling of ARF loss, the second most common genetic alteration in Ph+ ALL, which has been shown to reduce the latency of experimental Ph+ ALL, but has not previously been co-modeled with IKZF1 alterations (Williams et al., 2006; Williams et al., 2007). Importantly, we have studied Ikzf1 haploinsufficiency and expression of IK6. In contrast to previously modeled Ikzf1 deletions (Joshi et al., 2014), IK6 lacks all 4 N-terminal zinc fingers required for normal DNA binding, and arises from the most common focal IKZF1 deletion observed in human ALL.
Using these models and complementary genomic and proteomic approaches, we have shown that perturbation of IKZF1 activity is a central event in the pathogenesis of BCR-ABL1 lymphoid leukemogenesis and reduced response to TKI therapy. We show that Ikzf1 alterations induce a hematopoietic stem cell-like gene expression program accompanied by upregulation of adhesion molecules and signaling pathways that mediate abnormal adherence. We provide evidence that Ikzf1 alterations confer stem cell like properties, as demonstrated by increased single cell colony formation and induction of adhesion molecules in mouse and human ALL.
Increased expression of THY1 (Yamazaki et al., 2009) and integrins (Hsieh et al., 2013; Miller et al., 2013) have been noted previously in ALL and Ikzf1-mutant mouse B-cells (Joshi et al., 2014) but not in Ph+ ALL. In gain- and loss-of-function studies, we have shown that upregulation of these genes is a consequence of IKZF1 alterations and directly contributes to enhanced adhesion and self-renewal properties in Ikzf1-altered BCR-ABL1 cells. Moreover, we have shown that each adhesion molecule is a direct transcriptional target of IKZF1, providing a mechanism for deregulation of expression in IKZF1-mutant leukemia.
The reversal of this phenotype with retinoids was striking and emphasizes the utility of drug screens interrogating phenotypes more complex than killing alone. Administration of retinoid agonists resulted in profound reversal of the stem cell transcriptional program, and abrogation of adherence and self-renewal, without direct cytotoxicity. The Ikzf1 locus has multiple retinoid receptor binding sites and strikingly, retinoids selectively induced expression of WT IKZF1 resulting in expression of IKZF1 target genes. Using adhesion assays and calvarial imaging, we showed that retinoids completely reversed the stromal adhesion of IKZF1-altered leukemic cells. Together with the observation that enforced IKZF1 expression also abrogated adhesion, these findings suggest that retinoid-mediated induction of IKZF1 directly contributes to the reversal of stem cell features and increased responsiveness to dasatinib in Ph+ ALL. The observed induction of IKZF1 expression suggests that retinoids may have less effect in the minority of ALL cases with biallelic alterations resulting in complete loss of IKZF1 expression. However, in contrast to enforced expression of IKZF1, retinoids induced differentiation and cell cycle arrest independent of IKZF1 induction, indicating that retinoids have both IKZF1 dependent and independent effects in Ph+ ALL (Figure 7C).
Retinoids are approved for use in several diseases, including all-trans-retinoic acid in acute promyelocytic leukemia (Park and Tallman, 2011), and bexarotene in cutaneous T-cell lymphoma (Connolly et al., 2013), but have not been previously systematically investigated in Ph+ ALL. Our findings provide a rationale for clinical evaluation of these agents in IKZF1-altered Ph+ ALL. Collectively, these results indicate that IKZF1 alterations have important roles in determining disease lineage, reduced TKI sensitivity, and induction of stem cell features in Ph+ B-ALL, and that disrupting these phenotypes by targeting the retinoid pathway, or downstream signaling pathways, represent important treatment opportunities that should be pursued in clinical trials.
EXPERIMENTAL PROCEDURES
Mouse modeling of BCR-ABL1 leukemia
For whole BM experiments, unmanipulated BM was transduced with retrovirus for three hours and 1 million unsorted cells were transplanted by tail vein injection into lethally irradiated WT recipients. To derive pre-B cells, BCR-ABL1-transduced BM was cultured in the absence of feeder layers or cytokines, cells were subsequently transduced with retroviruses on retronectin and 2×105 sorted cells were injected by tail vein into sublethally irradiated WT recipients. For lin-cell transplantation experiments, unmanipulated whole BM cells lacking lineage markers were isolated and transduced with retroviruses prior to sorting and injection into lethally irradiated WT recipients. Mice were housed in an American Association of Laboratory Animal Care (AALAC)-accredited facility and were treated on Institutional Animal Care and Use Committee (IACUC)-approved protocols in accordance with NIH guidelines.
Analysis of Human Leukemias
Previously reported cohorts (Mullighan et al., 2007; Mullighan et al., 2008; Mullighan et al., 2009; Roberts et al., 2012; Roberts et al., 2014) and ninety-eight adult B-progenitor ALL samples collected on the Eastern Cooperative Oncology Group E2993 (Rowe et al., 2005) and ClinicalTrials.gov identifier NCT00002514 and Alliance for Clinical Oncology C10403 (clinicaltrials.gov NCT00558519) studies were analyzed for IKZF1 deletions and mutations by SNP 6.0 microarrays (Affymetrix, Santa Clara, CA) and by genomic PCR and Sanger Sequencing as previously described (Mullighan et al., 2007; Mullighan et al., 2009; Mullighan et al., 2011). All samples were obtained with patient or parent/guardian provided informed consent under protocols approved by the Institutional Review Board at each COG and ECOG institution and St. Jude Children’s Research Hospital. All samples were de-identified prior to analysis.
Statistical Analyses
Data analyses were performed using GraphPad Prism Version 6.0 (GraphPad, La Jolla, CA). All data are presented as mean ± SD. Significance was determined using Student’s t test, ANOVA, or Mantel-Cox log rank test as appropriate. For survival studies, Kaplan-Meier curves were generated and Mantel-Cox p values were determined for pairwise comparisons of cohorts. A P value of less than 0.05 was considered significant.
Other Procedures
All methods are described in detail in the Supplemental Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
Ikzf1 alterations confer stem-like properties and increased adhesion in BCR-ABL1 ALL
IKZF1-altered BCR-ABL1+ leukemias display reduced responsiveness to dasatinib
Retinoids reverse the effects of Ikzf1 alterations, partially by induction of IKZF1
Retinoids potentiate the activity of dasatinb in human and mouse BCR-ABL1 ALL
SIGNIFICANCE.
The prognosis of high-risk acute lymphoblastic leukemia remains suboptimal despite contemporary chemotherapy and the advent of targeted therapeutic approaches. Deletions or mutations of IKZF1 are a hallmark of high-risk ALL, but an understanding of how IKZF1 alteration contribute to leukemia development are lacking. Here we show that IKZF1 alterations drive lymphoid lineage, a stem cell-like phenotype, abnormal bone marrow adhesion, and poor responsiveness to tyrosine kinase inhibitor (TKI) therapy. Using a high-content screen, we show that retinoids reverse this phenotype in part by inducing expression of wild-type IKZF1, and increase responsiveness to TKIs. These findings provide insight into the pathogenesis of high-risk ALL and potential therapeutic approaches.
ACKNOWLEDGMENTS
We thank K. Georgopolous and S. Nutt for providing Ikzf1+/− mice; and the St Jude Children’s Research Hospital Flow Cytometry and Cell Sorting Shared Resource, Cell Tissue and Imaging Center, and Small Animal Imaging Facility. We thank D Link for discussions regarding Prrx1-Cre;Ai9 mice. This work was supported by the American Lebanese Syrian Associated Charities of St Jude Children’s Research hospital; NCI Cancer Center Support Grant P30 CA021765, NCI grant R25 CA23944 (M.J.A.), a Stand Up to Cancer Innovative Research Grant (C.G.M.) the Pew Charitable Trusts (C.G.M.), American Association for Cancer Research/Aflac Career Development Award (C.G.M.), an American Society of Hematology Scholar Award (C.G.M.), and ECOG grants: U10 CA21115, U24-CA114737. This project has been funded in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views of policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Accession numbers
Array-based comparative genomic hybridization and RNA sequencing data have been deposited in the Gene Expression Omnibus, accessions number GSE54821 and GSE68391.
AUTHOR CONTRIBUTIONS
M.L.C., R.K.G, T.C. and C.G.M. designed experiments. M.L.C., J.L., E.M.P., L.H.K., Y.C., D.P.-T., M.J.A., W.C., L.L., K.G.R., K.M., I.I., J.P., V.E.C., K.K., V.P., and C.G.M. performed experiments. E.M.P., F.N., S.P.D, J.E.D., C.L.W., J.R. and S.L. provided clinical samples and data. Y.-L.Y., J.M, A.M., R.T.W. and R.A.D supplied reagents. M.L.C., J.L., C.Q., S.-C.C., J.M., G.S., M.R., D.M., M.E., P.G., Y.-D.W., B.F., J.C.P., S.B., L.J., J.P., V.P. and C.G.M. analyzed data. M.L.C. and C.G.M. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Altucci L, Leibowitz MD, Ogilvie KM, de Lera AR, Gronemeyer H. RAR and RXR modulation in cancer and metabolic disease. Nat Rev Drug Discov. 2007;6:793–810. doi: 10.1038/nrd2397. [DOI] [PubMed] [Google Scholar]
- Barker TH, Hagood JS. Getting a grip on Thy-1 signaling. Biochim Biophys Acta. 2009;1793:921–923. doi: 10.1016/j.bbamcr.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulos N, Mulder HL, Calabrese CR, Morrison JB, Rehg JE, Relling MV, Sherr CJ, Williams RT. Chemotherapeutic agents circumvent emergence of dasatinib-resistant BCR-ABL kinase mutations in a precise mouse model of Philadelphia chromosome-positive acute lymphoblastic leukemia. Blood. 2011;117:3585–3595. doi: 10.1182/blood-2010-08-301267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb BS, Morales-Alcelay S, Kleiger G, Brown KE, Fisher AG, Smale ST. Targeting of Ikaros to pericentromeric heterochromatin by direct DNA binding. Genes Dev. 2000;14:2146–2160. doi: 10.1101/gad.816400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly RM, Nguyen NK, Sukumar S. Molecular pathways: current role and future directions of the retinoic acid pathway in cancer prevention and treatment. Clin Cancer Res. 2013;19:1651–1659. doi: 10.1158/1078-0432.CCR-12-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig W, Kay R, Cutler RL, Lansdorp PM. Expression of Thy-1 on human hematopoietic progenitor cells. J Exp Med. 1993;177:1331–1342. doi: 10.1084/jem.177.5.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley GQ, Van Etten RA, Baltimore D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- Den Boer ML, van Slegtenhorst M, De Menezes RX, Cheok MH, Buijs-Gladdines JG, Peters ST, Van Zutven LJ, Beverloo HB, Van der Spek PJ, Escherich G, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 2009;10:125–134. doi: 10.1016/S1470-2045(08)70339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulatov S, Notta F, Eppert K, Nguyen LT, Ohashi PS, Dick JE. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nature immunology. 2010;11:585–593. doi: 10.1038/ni.1889. [DOI] [PubMed] [Google Scholar]
- Ferreiros-Vidal I, Carroll T, Taylor B, Terry A, Liang Z, Bruno L, Dharmalingam G, Khadayate S, Cobb BS, Smale ST, et al. Genome-wide identification of Ikaros targets elucidates its contribution to mouse B-cell lineage specification and pre-B-cell differentiation. Blood. 2013;121:1769–1782. doi: 10.1182/blood-2012-08-450114. [DOI] [PubMed] [Google Scholar]
- Georgopoulos K, Bigby M, Wang JH, Molnar A, Wu P, Winandy S, Sharpe A. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79:143–156. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- Greenbaum A, Hsu YM, Day RB, Schuettpelz LG, Christopher MJ, Borgerding JN, Nagasawa T, Link DC. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013;495:227–230. doi: 10.1038/nature11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh YT, Gang EJ, Geng H, Park E, Huantes S, Chudziak D, Dauber K, Schaefer P, Scharman C, Shimada H, et al. Integrin alpha4 blockade sensitizes drug resistant pre-B acute lymphoblastic leukemia to chemotherapy. Blood. 2013;121:1814–1818. doi: 10.1182/blood-2012-01-406272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba H, Greaves M, Mullighan CG. Acute lymphoblastic leukaemia. Lancet. 2013;381:1943–1955. doi: 10.1016/S0140-6736(12)62187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi I, Yoshida T, Jena N, Qi X, Zhang J, Van Etten RA, Georgopoulos K. Loss of Ikaros DNA-binding function confers integrin-dependent survival on pre-B cells and progression to acute lymphoblastic leukemia. Nat Immunol. 2014;15:294–304. doi: 10.1038/ni.2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Sherr CJ. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- Kuiper RP, Waanders E, van der Velden VH, van Reijmersdal SV, Venkatachalam R, Scheijen B, Sonneveld E, van Dongen JJ, Veerman AJ, van Leeuwen FN, et al. IKZF1 deletions predict relapse in uniformly treated pediatric precursor B-ALL. Leukemia. 2010;24:1258–1264. doi: 10.1038/leu.2010.87. [DOI] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Lamkin T, Brooks J, Annett G, Roberts W, Weinberg K. Immunophenotypic differences between putative hematopoietic stem cells and childhood B-cell precursor acute lymphoblastic leukemia cells. Leukemia. 1994;8:1871–1878. [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TL, Vala MS, Barber JP, Karp JE, Smith BD, Matsui W, Jones RJ. Induction of acute lymphocytic leukemia differentiation by maintenance therapy. Leukemia. 2007;21:1915–1920. doi: 10.1038/sj.leu.2404823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli G, Iacobucci I, Storlazzi CT, Vignetti M, Paoloni F, Cilloni D, Soverini S, Vitale A, Chiaretti S, Cimino G, et al. IKZF1 (Ikaros) deletions in BCR-ABL1-positive acute lymphoblastic leukemia are associated with short disease-free survival and high rate of cumulative incidence of relapse: a GIMEMA AL WP report. J Clin Oncol. 2009;27:5202–5207. doi: 10.1200/JCO.2008.21.6408. [DOI] [PubMed] [Google Scholar]
- Mayani H, Lansdorp PM. Thy-1 expression is linked to functional properties of primitive hematopoietic progenitor cells from human umbilical cord blood. Blood. 1994;83:2410–2417. [PubMed] [Google Scholar]
- McLaughlin J, Chianese E, Witte ON. In vitro transformation of immature hematopoietic cells by the P210 BCR/ABL oncogene product of the Philadelphia chromosome. Proc Natl Acad Sci U S A. 1987;84:6558–6562. doi: 10.1073/pnas.84.18.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller PG, Al-Shahrour F, Hartwell KA, Chu LP, Jaras M, Puram RV, Puissant A, Callahan KP, Ashton J, McConkey ME, et al. In Vivo RNAi screening identifies a leukemia-specific dependence on integrin beta 3 signaling. Cancer Cell. 2013;24:45–58. doi: 10.1016/j.ccr.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG. Genomic characterization of childhood acute lymphoblastic leukemia. Semin Hematol. 2013;50:314–324. doi: 10.1053/j.seminhematol.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, White D, Hughes TP, Le Beau MM, Pui CH, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, Ma J, Liu W, Cheng C, Schulman BA, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360:470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG. Single nucleotide polymorphism microarray analysis of genetic alterations in cancer. Methods in molecular biology. 2011;730:235–258. doi: 10.1007/978-1-61779-074-4_17. [DOI] [PubMed] [Google Scholar]
- Nishii K, Katayama N, Miwa H, Shikami M, Usui E, Masuya M, Araki H, Lorenzo F, Ogawa T, Kyo T, et al. Non-DNA-binding Ikaros isoform gene expressed in adult B-precursor acute lymphoblastic leukemia. Leukemia. 2002;16:1285–1292. doi: 10.1038/sj.leu.2402533. [DOI] [PubMed] [Google Scholar]
- Notta F, Doulatov S, Laurenti E, Poeppl A, Jurisica I, Dick JE. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. 2011;333:218–221. doi: 10.1126/science.1201219. [DOI] [PubMed] [Google Scholar]
- Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Tallman MS. Treatment of acute promyelocytic leukemia without cytotoxic chemotherapy. Oncology. 2011;25:733–741. [PubMed] [Google Scholar]
- Rege TA, Hagood JS. Thy-1, a versatile modulator of signaling affecting cellular adhesion, proliferation, survival, and cytokine/growth factor responses. Biochim Biophys Acta. 2006;1763:991–999. doi: 10.1016/j.bbamcr.2006.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Pei D, McCastlain K, Ding L, Lu C, Song G, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med. 2014;371:1005–1015. doi: 10.1056/NEJMoa1403088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KG, Morin RD, Zhang J, Hirst M, Zhao Y, Su X, Chen SC, Payne-Turner D, Churchman ML, Harvey RC, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22:153–166. doi: 10.1016/j.ccr.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe JM, Buck G, Burnett AK, Chopra R, Wiernik PH, Richards SM, Lazarus HM, Franklin IM, Litzow MR, Ciobanu N, et al. Induction therapy for adults with acute lymphoblastic leukemia: results of more than 1500 patients from the international ALL trial: MRC UKALL XII/ECOG E2993. Blood. 2005;106:3760–3767. doi: 10.1182/blood-2005-04-1623. [DOI] [PubMed] [Google Scholar]
- Saalbach A, Wetzel A, Haustein UF, Sticherling M, Simon JC, Anderegg U. Interaction of human Thy-1 (CD 90) with the integrin alphavbeta3 (CD51/CD61): an important mechanism mediating melanoma cell adhesion to activated endothelium. Oncogene. 2005;24:4710–4720. doi: 10.1038/sj.onc.1208559. [DOI] [PubMed] [Google Scholar]
- Schjerven H, McLaughlin J, Arenzana TL, Frietze S, Cheng D, Wadsworth SE, Lawson GW, Bensinger SJ, Farnham PJ, Witte ON, Smale ST. Selective regulation of lymphopoiesis and leukemogenesis by individual zinc fingers of Ikaros. Nat Immunol. 2013;14:1073–1083. doi: 10.1038/ni.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer RA, Montecino-Rodriguez E, Witte ON, Dorshkind K. Immature B-cell progenitors survive oncogenic stress and efficiently initiate Ph+ B-acute lymphoblastic leukemia. Blood. 2010;116:2522–2530. doi: 10.1182/blood-2010-01-264093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock W. Adolescents and young adults with acute lymphoblastic leukemia. Hematology / the Education Program of the American Society of Hematology American Society of Hematology Education Program. 2010;2010:21–29. doi: 10.1182/asheducation-2010.1.21. [DOI] [PubMed] [Google Scholar]
- van der Veer A, Zaliova M, Mottadelli F, De Lorenzo P, Te Kronnie G, Harrison CJ, Cave H, Trka J, Saha V, Schrappe M, et al. IKZF1 status as a prognostic feature in BCR-ABL1-positive childhood ALL. Blood. 2014;123:1691–1698. doi: 10.1182/blood-2013-06-509794. [DOI] [PubMed] [Google Scholar]
- Virely C, Moulin S, Cobaleda C, Lasgi C, Alberdi A, Soulier J, Sigaux F, Chan S, Kastner P, Ghysdael J. Haploinsufficiency of the IKZF1 (IKAROS) tumor suppressor gene cooperates with BCR-ABL in a transgenic model of acute lymphoblastic leukemia. Leukemia. 2010;24:1200–1204. doi: 10.1038/leu.2010.63. [DOI] [PubMed] [Google Scholar]
- Williams RT, den Besten W, Sherr CJ. Cytokine-dependent imatinib resistance in mouse BCR-ABL+, Arf-null lymphoblastic leukemia. Genes Dev. 2007;21:2283–2287. doi: 10.1101/gad.1588607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RT, Roussel MF, Sherr CJ. Arf gene loss enhances oncogenicity and limits imatinib response in mouse models of Bcr-Abl-induced acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2006;103:6688–6693. doi: 10.1073/pnas.0602030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Nishida H, Iwata S, Dang NH, Morimoto C. CD90 and CD110 correlate with cancer stem cell potentials in human T-acute lymphoblastic leukemia cells. Biochem Biophys Res Commun. 2009;383:172–177. doi: 10.1016/j.bbrc.2009.03.127. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Ng SY, Georgopoulos K. Awakening lineage potential by Ikaros-mediated transcriptional priming. Curr Opin Immunol. 2010;22:154–160. doi: 10.1016/j.coi.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012a;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Jackson AF, Naito T, Dose M, Seavitt J, Liu F, Heller EJ, Kashiwagi M, Yoshida T, Gounari F, et al. Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nat Immunol. 2012b;13:86–94. doi: 10.1038/ni.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Mullighan CG, Harvey RC, Wu G, Chen X, Edmonson M, Buetow KH, Carroll WL, Chen IM, Devidas M, et al. Key pathways are frequently mutated in high-risk childhood acute lymphoblastic leukemia: a report from the Children’s Oncology Group. Blood. 2011;118:3080–3087. doi: 10.1182/blood-2011-03-341412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Dawson MI, Mohammad R, Rishi AK, Farhana L, Feng KC, Leid M, Peterson V, Zhang XK, Edelstein M, et al. Induction of apoptosis of human BCLL and ALL cells by a novel retinoid and its nonretinoidal analog. Blood. 2002;100:2917–2925. doi: 10.1182/blood.V100.8.2917. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.