SUMMARY
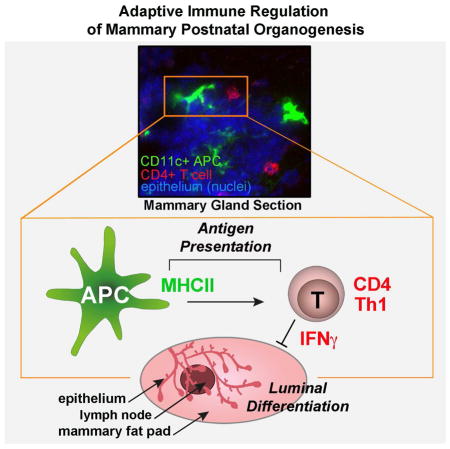
Postnatal organogenesis occurs in an immune competent environment and is tightly controlled by interplay between positive and negative regulators. Innate immune cells have beneficial roles in postnatal tissue remodeling, but roles for the adaptive immune system are currently unexplored. Here we show that adaptive immune responses participate in the normal postnatal development of a non-lymphoid epithelial tissue. Since the mammary gland (MG) is the only organ developing predominantly after birth, we utilized it as a powerful system to study adaptive immune regulation of organogenesis. We found that antigen-mediated interactions between mammary antigen-presenting cells and interferon-γ (IFNγ)-producing CD4+ T helper 1 cells participate in MG postnatal organogenesis as negative regulators, locally orchestrating epithelial rearrangement. IFNγ then affects luminal lineage differentiation. This function of adaptive immune responses regulating normal development changes the paradigm for studying players of postnatal organogenesis and provides insights into immune surveillance and cancer transformation.
Keywords: mammary postnatal organogenesis, adaptive immune system, antigen-presenting cells, T cells
Graphical Abstract
INTRODUCTION
Prenatal organ development takes place in the presence of an immature immune system, and continues after birth as the immune system reaches its full potential. In epithelial organs such as lung, breast and intestine, postnatal organogenesis is regulated by communication between epithelial cells and innate immune cells (Reed and Schwertfeger, 2010; Renz et al., 2012). During this period, the previously immature adaptive immune system is exposed to antigens and rapidly develops. Therefore, postnatal development of various epithelial tissues occurs in an immune competent environment. However, whether the adaptive immune system regulates normal organ development in non-pathologic conditions has yet to be explored. While other epithelial organs have relatively minor postnatal developmental phases, the mammary gland (MG) is the only organ that develops predominantly after birth, during puberty (Wiseman and Werb, 2002). At birth, the MG consists of an epithelial rudimentary tree. In mice, mammary postnatal organogenesis starts with onset of puberty, at 3–4 weeks of age, and is characterized by ductal invasion until the growing and branching epithelial ducts reach the fat pad limits by 8–9 weeks of age (Sternlicht et al., 2006) (Fig. S1A). A secondary expansion occurs with each estrus or menstrual cycle in rodents and humans, respectively, as well as during pregnancy and lactation. This secondary expansion regresses during the process of involution (Watson et al., 2011). Although steroid and peptide hormones are major initiators and regulators of mammary postnatal organogenesis, it is also tightly regulated by intracellular and extracellular signals between epithelial cells and the surrounding microenvironment or stroma, which includes extracellular matrix, fibroblasts, adipocytes and immune cells (Lu and Werb, 2008).
Innate immune cells as CSF1-dependent macrophages, eotaxin-dependent eosinophils (Gouon-Evans et al., 2002) and mast cells (Lilla and Werb, 2010) are involved in the positive regulation of pubertal mammary ductal expansion. Interestingly, based on the immune cells involved, postnatal MG development has been compared to a wound healing process (Reed and Schwertfeger, 2010). During wound healing, antigen-presenting cells (APCs) become activated and recruit other immune cells, including CD4+ T helper cells, to create an environment for tissue remodeling. However, to date, the presence and role of APCs and T cells in the development of the mammary organ has not been assessed.
Since in the mouse the MG develops from a rudimentary epithelial tree to a fully developed organ within the few weeks of puberty (Sternlicht et al., 2006), it is a powerful model system to explore whether antigen-mediated interactions between APCs and tissue T cells contribute to the normal postnatal development of non-lymphoid epithelial tissues. To explore roles for the adaptive immune system in local regulation of mammary postnatal organ development, we utilized transgenic mouse models that allowed for genetic ablation or overexpression of immune cell subsets and explored ductal invasion in whole mount MGs. In parallel, we also performed live imaging and functional analyses in primary three-dimensional (3D) organotypic mammary branching assays (i.e., organoids).
RESULTS
Epithelial-associated mammary CD11c+ cells negatively regulate branching morphogenesis
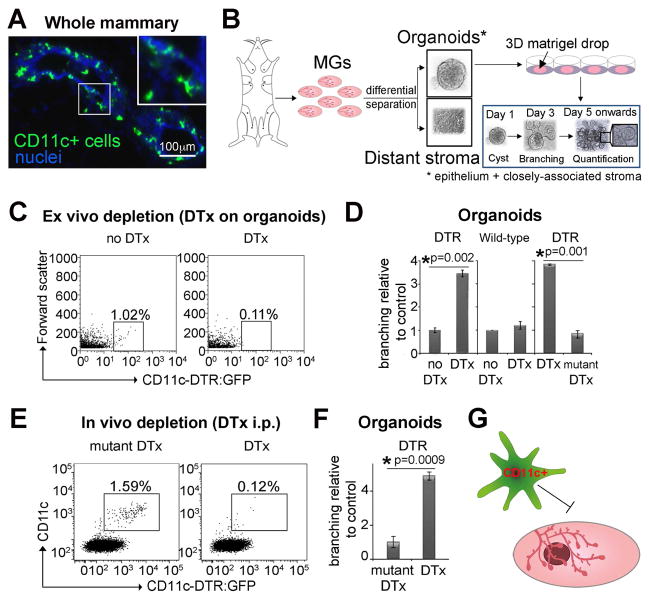
CD11c is a selective marker for dendritic cells but also for some macrophages (Engelhardt et al., 2012; Vallon-Eberhard et al., 2006), both of which have antigen presentation capacities (Gautier et al., 2012; Miller et al., 2012). We found CD11c+ cells, which have dendritic-like features, closely associated with the mammary epithelium (Fig. 1A, Fig. S1B, Movie 1). To study the role of these CD11c+ cells in mammary postnatal organogenesis, we utilized organoids cultured in FGF2 (Fig. 1B), which are well characterized ex vivo branching models to study postnatal mammary organogenesis (Ewald et al., 2008). These surrogate assays not only reflect the ductal elongation aspect of epithelial branching, which depends on cell proliferation and epithelial surface expansion (Zhang et al., 2014), but also allow the elimination of any organ non-specific or hormone-dependent effects. To assess whether these CD11c+ cells influenced MG organogenesis, we used CD11c-DTR:GFP mice (Jung et al., 2002), which express the diphtheria toxin receptor under the CD11c promoter. Utilizing organoids from CD11c-DTR:GFP MGs, we found that CD11c+ cells are closely associated with the mammary epithelium and then depleted them by diphtheria toxin (DTx) administration either ex vivo during organotypic culture (Fig. 1C–D), or in vivo before organoid preparation (Fig. 1E–F). In both cases, CD11c+ cell depletion accelerated epithelial branching (Fig. 1D, 1F, Fig. S1C–E). These data suggest an inhibitory role for CD11c+ cells in the morphogenesis of pubertal MGs (Fig. 1G).
Figure 1. Epithelial-associated Mammary CD11c+ Cells Negatively Regulate Branching Morphogenesis.
(A) Immunostaining of CD11c+ cells in MGs of CD11c-DTR:GFP mice shows co-localization of these cells to the mammary epithelium (Movie 1).
(B) Experimental design of differential separation, embedding in Matrigel, culture and quantification of epithelial branching in 3D primary mammary epithelial organotypic cultures (organoids). Organoids initiate as cysts (day 1), which start branching on day 3 of culture. Quantification of branching was performed on day 5 unless indicated otherwise.
(C) Flow cytometry of CD11c-DTR:GFP organoids 24 h after culture with DTx. Note that organoids were retrieved from Matrigel so number of cells and autofluorescence are a challenge.
(D) Branching of CD11c-DTR:GFP organoids cultured with DTx. Controls were DTx on wild-type and mutated DTx on CD11c-DTR:GFP organoids (n=8, 3 and 3 experiments, respectively).
(E) Flow cytometry of CD11c-DTR:GFP epithelial-associated APCs, 48 h after mDTx or DTx injections.
(F) Branching of CD11c-DTR:GFP organoids cultured from MGs harvested 48 h after DTx injection (n=3 experiments).
(G) Schematic depicting mammary CD11c+ cells as negative regulators of branching.
Data in (D) and (E) are represented as mean ± SEM. See also Figure S1, Movies S1.
Epithelial-associated mammary CD11c+ cells have characteristics of APCs
We next characterized the epithelial-associated mammary CD11c+ cells. Interrogation of molecular markers using surface stains and transgenic reporters (See Supplementary Experimental Procedures, qPCR Primers and Function of Gene Targeted) revealed that these CD11c+ cells express high levels of CX3CR1 (Fig. 2A), colony stimulating factor-1 receptor (CSF-1R, using the c-fms transgene) (Fig. 2B) and F4/80 (Fig. 2C). Most interestingly, they express high levels of major histocompatibility complex (MHC) II (Fig. 2D), which is essential for antigen presentation, as well as intermediate levels of CD11b (Fig. 2E). The absence of Siglec-F expression (Fig. S2A) suggested that these CD11c+ cells are APCs of the monocytic lineage, rather than eosinophils (Gautier et al., 2012; Gouon-Evans et al., 2000; Miller et al., 2012). In addition, we observed a macrophage-type population associated with the organoids, which is F4/80+, high for CD11b and low for CD11c and MHCII (Fig. 2C, E).
Figure 2. Epithelial-associated Mammary CD11c+ Cells Respond to Epithelial Branching and Present APC Characteristics.

(A) Flow cytometry of epithelial-associated CD11c+ cells indicated almost all are CX3CR1+. Data obtained using CX3CR1-GFP/− transgenic mice and gated on single live cells.
(B) Flow cytometry of epithelial-associated CD11c+ cells indicated they are CSF-1R+. Data obtained using c-fms- GFP transgenic mice (c-fms is transgene for CSF-1R) and gated on single live cells.
(C) Flow cytometry of epithelial-associated CD11c+ cells, gated on single live cells, shows they are F4/80 high.
(D) Flow cytometry of epithelial-associated CD11c+ cells, gated on single live cells, shows they are MHCII high.
(E) Flow cytometry of epithelial-associated CD11c+ cells, gated on single live cells, shows they are CD11b intermediate. These cells are also MHCII high and F4/80 high. Another population of CD11b high cells are low for MHCII, CD11c and F4/80+.
(F) Time-lapse microscopy of CX3CR1-GFP/+ organoids (vast majority of CX3CR1+ cells are CD11c+), from culture days 3–15 (Movies 2, 3). Arrows, proliferating APC (epithelium marked by Cell Tracker red).
(G) Bar graph quantifies APC proliferation in (f), n= 12 fields.
(H) Arrows point to APC taking up a cellular particle (Movie 4).
See also Figure S2, Movies S2–4 and see Supplementary Experimental Procedures, qPCR Primers and Function of Gene Targeted.
We examined the interactions of these CD11c+ APCs with the mammary epithelium during branching morphogenesis. Because the fluorescence of CD11c+ cells from the CD11c-DTR:GFP model is too dim for time-lapse confocal microscopy and since most CD11c+ cells are CX3CR1+, we used organoids from CX3CR1-GFP/+ reporter mice, in which the GFP signal is considerably brighter (Jung et al., 2000). These experiments revealed that the CX3CR1+ APCs actively interacted with the mammary epithelium and proliferated during branching of organoids (Fig. 2F, G, Movies 2, 3) and mammary ductal invasion in vivo (Fig. S2B). Interestingly, several studies report the in situ proliferation of both macrophages and dendritic cells (Davies et al., 2013; Veres et al., 2013; Ginhoux et al., 2009). CSF-1 expressed by mammary epithelial cells (Fig. S2C) located at the neck of terminal end buds (Coussens and Pollard, 2011), could contribute to the proliferation of mammary APCs, which express CSF-1R (Fig. 2B). During branching, these APCs also engulfed cellular particles (Fig. 2H, Movie 4), a prerequisite for antigen presentation by monocyte-derived APCs.
T cells are involved in the negative regulation of mammary organogenesis
Activation of the adaptive immune system requires the presence of APCs (Hoebe et al., 2004). APCs link the innate and adaptive immune systems by capturing antigens in tissues and presenting them to T cells in lymph nodes. After activation, T cells home to non-lymphoid tissues, where they exert their effector functions (Hoebe et al., 2004). We therefore asked whether T cells are also involved, along with CD11c+ APCs, in the negative regulatory loop of mammary postnatal organogenesis (Fig. 3A). Significantly, we found a reduction of mammary CD4+ and CD8+ T cell numbers after depletion of APCs, both in vivo (Fig. 3B, Fig. S1C) and in organoids (Fig. S3A). These data suggest that APCs may regulate epithelial morphogenesis by not only attracting T cells to and/or retaining T cells in MGs, but also by preventing their death. We reasoned that in order to regulate mammary postnatal organogenesis, T cells would need to closely interact with the epithelium, and indeed we found CD4+ and CD8+ T cells closely associated with the mammary ducts (Fig. 3C, Fig. S3B, C). Finally, to test whether T cells contribute to MG organogenesis, we examined TCRα-deficient (Tcra−/−) mice, which lack CD4+ and CD8+ T cells. We observed accelerated ductal outgrowth in Tcrα−/− MGs (Fig. 3D, E) and increased branching in Tcra−/− organoids (Fig. 3F), supporting an inhibitory role for CD4+ and/or CD8+ T cells in MG organogenesis.
Figure 3. T cells are Involved in the Negative Regulation of Mammary Organogenesis.
(A) Schematic depicting the question of whether APC may negatively regulate MG postnatal organogenesis via interaction with T cells.
(B) Flow cytometry of CD4+ and CD8+ T cells (gated on CD3+), 48 h post PBS/DTx injections to CD11c-DTR (n=6 MGs).
(C) Immunohistochemistry shows co-localization of CD4+ and CD8+ T cells to mammary epithelium in intact glands.
(D) Comparison of epithelial ductal outgrowths from carmine red-stained MG whole mounts from wild-type (WT) and Tcra−/− mice.
(E) Quantification of ductal outgrowths in (d), (n=8 WT and 4 Tcra−/− MGs).
(F) Increased branching in Tcra−/− organoids (n=3 experiments).
Data in (B), (E) and (F) are represented as mean ± SEM. See also Figure S3.
Antigen presentation via MHCII to CD4+ T cells negatively regulates mammary organogenesis
We next sought to determine which T cell type was involved in the regulation of MG morphogenesis. For this purpose we used organoid cultures that, similar to intact MGs, also contained T cells (Fig. 4A). In addition to their role in the lymph node, APCs may reactivate T cells within non-lymphoid tissues (Broz et al., 2014; Honda et al., 2014). To determine whether APCs regulate mammary organogenesis through T cells, we inhibited MHC-mediated APC-T cell activation with blocking antibodies (Banchereau and Steinman, 1998). Inhibiting MHCII-mediated CD4+ T cell stimulation resulted in accelerated branching (Fig. 4B), whereas inhibiting MHCI-mediated CD8+ T cell stimulation had no effect. These data suggest that antigen presentation to CD4+ T cells may be involved in the negative regulation of mammary organogenesis. Antigen presentation requires the close association of APCs and T cells. Indeed, CD11c+ APCs, and CD4+ T cells were in close proximity and co-localized to the mammary epithelium in sections of pubertal MGs (Fig. 4C). To confirm that epithelial-associated mammary APCs were capable of T cell activation via antigen presentation, we measured T cell proliferation by co-culturing organoids with ovalbumin (OVA)-specific (OT-II) CD4+ T cells. In this system, only presentation of their cognate peptide antigen (pOVA) by APCs will cause the activation and proliferation of these OT-II naïve CD4+ T cells. Indeed, upon the addition of pOVA to the culture media, OT-II T cells proliferated extensively (Fig. 4D, Fig. S4A, B). To further test whether the presence and activation of CD4+ T cells is important for MG organogenesis in vivo, we utilized MHCIIflox/− (fl/−) ×CD11c-cre mice, which are devoid of CD4+, but not CD8+ T cells, due to lack of MHCII expression on CD11c+ APCs (Fig. S4C). MGs from these mice showed accelerated ductal outgrowth (Fig. 4E, F) and increased organoid branching (Fig. 4G). Taken together, these results suggest a negative regulatory role for CD4+ T cell activation in MG organogenesis (Fig. 4H).
Figure 4. Antigen Presentation to CD4+ T cells Negatively Regulates Mammary Organogenesis.

(A) Flow cytometry of MHCII+ CD11c+ APCs and CD3+ T cells (which marks both CD4+ and CD8+ T cells) associated with organoids.
(B) Branching of organoids cultured with blocking antibodies to MHCI, MHCII or CD4 quantified relative to respective isotype controls (red line, n=5 experiments).
(C) Immunohistochemistry shows co-localization of CD11c+ APCs, CD4+ T cells and mammary epithelium in intact glands. Nuclei, DAPI.
(D) Naïve eFluor 670-labeled OVA-specific CD4+ (OT-II) T cells proliferate when co-cultured with organoids with pOVA (n=5 experiments, a representative histogram is presented. Cell counts: OT-II no pOVA: 87; OT-II +pOVA: 1656).
(E) Comparison of epithelial ductal outgrowths from MHCIIfl/− control versus MHCIIfl/−×CD11c-cre MG whole mounts.
(F) Quantification of outgrowths in (e), (n=4 control and 8 MHCIIfl/−×CD11c-cre MGs).
(G) Increased branching in MHCIIfl/−×CD11c-cre organoids (n=3 experiments).
(H) Schematic depicting APC and CD4+ T cells interactions via MHCII mediating mammary postnatal organogenesis.
Data in (B), (F) and (G) are represented as mean ± SEM. See also Figure S4.
CD4+ Th1 cells negatively regulate mammary organogenesis
Since CD4+ effector T cells function as distinct subsets (e.g., Th1, 2, 17, and T regulatory cells) specialized for specific adaptive immune responses (Zhu et al., 2010), we next asked which subset regulates MG organogenesis. By flow cytometric analysis, we observed predominantly the Th1 phenotype within MGs (Fig. 5A, Fig. S5A). We therefore determined whether activation of CD4+ Th1 cells could inhibit mammary epithelial morphogenesis, by adding these cells to the organoid cultures and stimulating the cultures with the pOVA peptide. Even though CD4+ Th2 cells are very rare in pubertal MGs, we included these cells in our analysis as they often counteract the effects of CD4+ Th1 and therefore may yield opposing results. Using time-lapse confocal microcopy, we noticed that Th1 and Th2 cells both interacted with MG APCs (Fig. S5B, Movies S5, 6) and dye-dilution experiments showed that antigen-presentation induced their proliferation (Fig. 5B). Interestingly, while Th1-polarized OT-II cells inhibited branching, the presence of Th2 cells increased branching (Fig. 5C, Fig. S5C). Remarkably, only the addition of pOVA to the OVA-specific effector T cell-organoid co-cultures caused the significant decrease or increase in branching, suggesting that T cell activation by mammary APCs is required in situ in order to regulate MG organogenesis. However, since culturing the Th1 or Th2 cells in the presence of interleukin (IL) 2 likely promoted the production of their effector cytokines (Zhou et al., 2003), this could explain the respective initial negative or positive effect they still had on branching even without pOVA (Fig. 5C). Interestingly, depletion of APCs from the T cell-organoid co-cultures rescued the effect that OVA-specific Th1 (and Th2) cells had on branching (Fig. S5D), validating a role for in situ antigen presentation. The reduction in T cells numbers after depletion (Fig. 3B and Fig. S3A) may further suggest that restimulation from local tissue APCs is required for T cell survival. Moreover, in support of this notion, we observed that mammary organoids from OT-II mice that are naturally enriched with OVA specific Th1 cells exhibited decreased branching when cultured with pOVA (Fig. 5D). Given that Th1 effector cells are the predominant CD4+ T cells within MGs, the accelerated branching of WT organoids after blockade of MHCII (Fig. 4B) also suggested that in situ antigen presentation to epithelial-associated CD4+ Th1 cells plays a role in organoid branching.
Figure 5. Antigen Presentation to CD4+Th1 Effector Cells Negatively Regulates Mammary Organogenesis.
(A) CD4+ T cell subsets in pubertal MGs based on flow cytometry data. Other CD4+ cells are naïve/non-activated cells.
(B) In vitro differentiated Th1 and Th2-polarized OVA-specific CD4+ (OT-II) T cells proliferate when co-cultured with organoids and pOVA (n=3 experiments) (Movies 5, 6). Figure shows representative histograms. Counts: Th1 no pOVA: 165; +pOVA: 1798. Counts: Th2 no pOVA: 158; +pOVA: 451.
(C) In vitro differentiated Th1 or Th2-polarized OT-II cells decreased and increased branching, respectively, when co-cultured with organoids. The addition of pOVA alone did not affect branching as compared to IL2 control. However, the addition of pOVA to the OVA-specific effector T cell-organoid co-culture caused the significant decrease or increase in branching, suggesting that antigen presentation by MHCII+ CD11c+ APCs (the only MHCII high cells associated with organoids) plays a significant role in mediating the effect of these effector T cells on organogenesis (n=3 experiments).
(D) The addition of pOVA to organoids from OTII×CD2-RFP mice (bearing only OVA-specific Th1 cells) inhibited branching, suggesting that the effector OTII cells associated with the organoids were reactivated with the addition of their cognate peptide antigen (n=3 experiments).
(E) Flow cytometry of Th1 cells.
(F) Analysis of CD4+ T cell subpopulations in control and β8fl/fl×CD11c-cre mice (n=4 control n=8 β8fl/fl×CD11c-cre MGs).
(G) Comparison of epithelial ductal MG outgrowths from control versus β8fl/fl×CD11c-cre mice.
(H) Quantification of ductal outgrowths from (G), (n=6 MGs each).
(I) Decreased branching in β8fl/fl×CD11c-cre organoids (n=3 experiments).
(J) Comparison of epithelial ductal MG outgrowths from SCID control versus SCID β8fl/fl×CD11c-cre mice.
(K) Quantification of ductal outgrowths from (j), (n=4 control n=6 β8fl/fl×CD11c-cre MGs).
(L) Comparable branching of SCID control and β8fl/fl×CD11c-cre organoids (n=3 experiments).
(M) Decreased branching in β8fl/fl×CD11c-cre MGs persists beyond puberty. These MGs showed inhibited postnatal organogenesis (versus accelerated organogenesis in all other models examined here), with blunted ductal outgrowths persisting beyond the pubertal organogenesis phase, as examined at 9 weeks of age (n=4 MGs each).
(N) Schematic depicting antigen presentation to CD4+ Th1 cells negatively regulating mammary organogenesis.
Data in (C), (D), (F), (H), (I), (K) and (L) are represented as mean ± SEM. See also Figure S5 and Movies S5, 6.
We next examined the role of CD4+ Th1 cells in mammary postnatal organogenesis in vivo. Given the MG Th1 dominance, the MHCIIfl/−×CD11c-cre mice lacking CD4+ T cells (Fig. 4E–G) effectively served as a model devoid of mammary Th1 cells. For a mouse model with an overrepresentation of CD4+ Th1 cells within MGs, we exploited the integrin β8flox/flox (fl/fl) ×CD11c-cre mice, which have increased frequency and number of Th1 cells, while all other MG CD4+ T cell subsets are diminished (Fig. 5F, Fig. S5E, F). Importantly, these mice show no change in CD11c+ cell number or phenotype (Travis et al., 2007; Melton et al., 2010). The β8fl/fl×CD11c-cre mice demonstrated significantly impaired MG ductal invasion, reinforcing the role of Th1 cells as the relevant negative regulatory subset within MGs (Fig. 5G, H). We also observed blunted branching in organoids from these Th1-enhanced β8fl/fl×CD11c-cre MGs (Fig. 5I, Fig. S5G, I). To confirm that the impaired organogenesis of β8fl/fl×CD11c-cre mice depends on increased frequency and numbers of Th1 cells rather than defective APCs, we crossed β8fl/fl×CD11c-cre mice to severe combined immunodeficient (SCID) mice, which lack T (and B) cells, and found that levels of MG ductal growth and branching were similar to those of SCID littermates (Fig. 5J–L). Moreover, since the β8fl/fl×CD11c-cre is the only model thus far that exhibited inhibited branching, we examined the MGs from older mice and found that the blunted branching of β8fl/fl×CD11c-cre MGs persisted beyond puberty (Fig. 5M). IFNγ secreted by Th1 cells participates in driving Th1 differentiation downstream of IL-12 from naïve T cells in a positive feedback loop (Magram et al., 1996). In the absence of IFNγ, most CD4+ T cells fail to differentiate into the Th1 effector phenotype (Ishii et al., 2013). We therefore crossed the β8fl/fl×CD11c-cre mice into the interferon-γ (IFNγ) null background. MGs from Ifng−/− β8fl/fl×CD11c-cre mice revealed comparable ductal invasion and organoid branching to Ifng−/− controls (Fig. S5I–K). These data further suggest that the excess of Th1 cells in β8fl/fl×CD11c-cre MGs led to their impaired development. To this end, our data suggest that tissue antigen presentation by mammary CD11c+ APCs and reactivation of CD4+ Th1 cells play roles in the negative regulation of MG organogenesis (Fig. 5N).
IFNγ mediates the inhibitory effect of CD4+ Th1 cells on mammary organogenesis by affecting luminal differentiation
IFNγ is the primary effector cytokine secreted by Th1 cells (Schoenborn and Wilson, 2007; Zhu et al., 2010) and is also a characteristic of mammary CD4+ Th1 cells (Fig. 5A, Fig. S5A). Morphological changes of branching epithelial organs are governed by a meticulous local regulation of epithelial rearrangement (Ewald et al., 2008). To determine whether IFNγ can mediate the effects of Th1 cells on mammary organogenesis we first tested whether it has a direct effect on mammary epithelial cells. The mammary epithelium consists of luminal and basal lineages, which are characterized by specific surface markers and cytokeratins (Rios et al., 2014; Shehata et al., 2012) (Fig. S6A). Luminal cells express cytokeratin (Krt) 8 and basal cells express Krt14. In addition, in mouse MGs, both lineages are positive for the surface marker epithelial cell adhesion molecule (EpCAM), although basal cells express lower levels of this marker but are high for CD49f. We incubated mammary organoids with IFNγ and observed a transient nuclear translocation of phosphorylated STAT1 (pSTAT1), a hallmark of IFNγ receptor (IFNγR) signaling (Fig. 6A, Fig. S6B). The nuclei of mammary luminal cells (marked by Krt8) were positive for pSTAT1 in confirmation of previous findings (Khalkhali-Ellis et al., 2008; Zhu et al., 2010) (Fig. 6B). Corroborating the activation of IFNγR signaling in luminal cells, we also observed enriched expression of IFNγR1 in the luminal fraction of organoids (Fig. S6C). This is in keeping with previous studies showing IFNγR1 and IFNγR2 expression in cultured normal human mammary epithelial cells, which represent a mixture of luminal and basal cell populations based on their cytokeratin expression (Khalkhali-Ellis et al., 2008), while proteomic profiling revealed that IFNγR1 is present only on a subset of luminal progenitors in vivo (Ji et al., 2011). Functionally, we found that incubation of organoids with IFNγ inhibited branching (Fig 6C), confirming an inhibitory role for IFNγ in mammary morphogenesis
Figure 6. IFNγ Mediates the Inhibitory Effect of CD4+Th1 Cells on Mammary Organogenesis by Affecting Luminal Differentiation.
(A) Transient nuclear STAT1 phosphorylation (pSTAT1), 30 min after IFNγ exposure, that disappears after 2 h. Insets showing high magnifications of the immunostained organoids without the nuclear staining. Borders of nuclei (Dapi) are designated.
(B) pSTAT1 is localized to nuclei of luminal cells [Krt8+] in organoids. Magnification of boxed area (right) without the nuclear staining.
(C) Reduced branching in organoids incubated with IFNγ (n=4 experiments).
(D) Quantitative PCR on luminal cells sorted from organoids incubated with IFNγ (n= 7 experiments).
(E) IFNγ blocking antibodies increase branching in β8fl/fl×CD11c-cre organoids (n=3 experiments).
(F) Accelerated branching in IFNγR1−/− organoids (n=3 experiments).
(G) Schematic depicting CD4+ Th1 cells as inhibitors of mammary organogenesis via IFNγ by affecting luminal differentiation.
Data in (C) – (F) are represented as mean ± SEM. See also Figure S6.
Given that IFNγ signaling affects mammary epithelial branching, we examined the effects of IFNγ on mammary epithelial lineage determination. Indeed, gene profiling of mammary organoids after incubation with IFNγ and sorting into the luminal and basal cell populations (Fig. S6C) confirmed that IFNγ affects the luminal lineage by downregulating the expression of some mature and progenitor luminal cell markers (Fig. 6D). We found a significant reduction in the expression of the luminal progenitor marker CD49b (Shehata et al., 2012), as well as CD14. We also found a significant decrease in expression of the mature luminal markers progesterone receptor (PR), Gata3 and Muc1. The expression of the luminal progenitor marker Elf5 did not change. Lmo4 and Mfge8, two other luminal markers also did not significantly change in expression following incubation with IFNγ (Shehata et al., 2012). Although Krt8 decreased, the change in its expression was not statistically significant, similar to Ifngr1. These results imply that branching in organoids cultured in FGF2, which reflects ductal elongation and is governed by proper luminal differentiation, is disrupted by IFNγ. Even though we detected no significant change in the overall frequency of luminal cells (Fig. S6C), we found increases in both proliferation and apoptosis of cells located within the lumens of IFNγ-treated organoids (Fig. S6D, E), reinforcing that changes occur within the luminal population. The effects on the basal lineage were not significant for the progenitor and basal markers tested (Fig. S6F).
Lastly, we investigated how significant is IFNγ secreted by CD4+ Th1 cells to mammary epithelial branching and whether CD4+ Th1 cells could regulate the mammary epithelium through other factors than IFNγ. For this purpose we added IFNγ-neutralizing antibodies to organoid cultures with excessive numbers of CD4+ Th1 cells, and found that these cultures showed significantly accelerated branching (Fig. 6E). Similarly, organoids from Ifngr1−/− mice, which similar to Ifng−/− mice also do not have properly functioning Th1 cells, showed accelerated branching (Fig. 6F). Taken together, our data suggest that Th1 cells inhibit mammary organogenesis by locally inhibiting luminal differentiation through IFNγ secretion (Fig. 6G).
DISCUSSION
In this study we have investigated whether the adaptive immune system locally contributes to postnatal mammary organogenesis. By using the postnatal MG as a model of immune-competent organ development, we have demonstrated a role for the adaptive immune system in regulating organogenesis of a non-lymphoid epithelial tissue. We provide compelling evidence that antigen-mediated interactions between mammary CD11c+ antigen-presenting cells (APCs) and CD4+ IFNγ+ Th1 cells provide signals that negatively regulate ductal invasion. We show that mammary epithelial cells respond directly to IFNγ. These effects are reinforced by enriched expression of IFNγR1 on mammary luminal epithelial cells and activation of downstream signaling pathways upon IFNγ stimulation. This response participates in orchestrating epithelial rearrangement by locally affecting luminal epithelial differentiation.
Although the nature of the antigen(s) involved remains to be discovered, the significance of our findings is several folds. First, we recognized an engagement and/or reactivation of the adaptive immune system in the regulation of normal epithelial organ architecture, under non-pathologic conditions. Second, these results have implications for understanding immune surveillance. Third, the data raise interesting questions regarding possible roles for T cell dysfunction in pathologic organ remodeling as in early stages of breast cancer transformation.
APCs versus Other Innate Immune Cells Implicated in Mammary Organogenesis
Several seminal papers over the last two decades have demonstrated roles for mast cells, eosinophils, and macrophages in terminal end bud elongation during postnatal MG organogenesis (Coussens and Pollard, 2011; Gouon-Evans et al., 2000; Lilla and Werb, 2010; Van Nguyen and Pollard, 2002). Macrophages also have a role in regulating epithelial cell death during postpartum MG involution (O’Brien et al., 2012).
Csf1op/op mice, as well as Csf1−/− mice, both lack F4/80+ CSF-1R+ macrophages and exhibit decreased terminal end bud numbers, reduced mammary duct length and branching during puberty (Dai et al., 2002; Gouon-Evans et al., 2000; Van Nguyen and Pollard, 2002). This phenotype is consistent with CSF-1R+ cells positively regulating ductal elongation. The mammary CD11c+ APCs that we describe here are also F4/80+ and CSF1-R+, yet these cells negatively regulate MG development. However, we also found a larger population of F4/80+ cells closely associated with the mammary epithelium, which expresses higher levels of the macrophage marker CD11b, but is low for CD11c and MHCII (Fig. 2E). These potentially immature/non-activated macrophages could be part of those macrophages previously described as positive regulators of mammary pubertal development (Gouon-Evans et al., 2000).
Since the Csf1op/op study does not characterize epithelial-associated cells differentially, but rather examines the MG stroma as a whole, there may potentially be additional F4/80+ and CSF1-R+ populations located in distant stroma. Indeed, F4/80+ eosinophils, which are also lacking in Csf1op/op mice and have been identified as positive regulators of mammary ductal morphogenesis (Gouon-Evans et al., 2000), are almost exclusively located in the distant stroma fraction (Fig. S2A). Therefore, it seems that our study identified a discrete population of CSF-1R+ antigen-presenting cells within the mammary stromal milieu, with opposing regulatory potential and/or location to those cell types identified in the studies on Csf1op/op MGs. The fact that very little is known about the negative regulators of mammary postnatal organogenesis further adds to the value of this study. Since positive and negative regulators need to act in concert to orchestrate organogenesis, it will be interesting to examine how these cells strike the balance for proper MG postnatal organogenesis and whether antigen-mediated interactions participate in other stages of mammary morphogenesis such as pregnancy, lactation and involution.
Downstream Effects of the Th1 Effector IFNγ on Mammary Epithelial Cells
In the present study, we focused on IFNγ, which is the cytokine characteristic of Th1 effector T cells, the predominant CD4+ effector T cell subset in the developing MG. We showed that IFNγ mediates the effect of Th1 cells on postnatal mammary organogenesis and acts directly on epithelial cells, as demonstrated by the transient nuclear translocation of pSTAT1 after IFNγ stimulation. Gene profiling of mammary organoids revealed changes in mature and progenitor luminal cell markers, most of which exhibited decreased expression after incubation with IFNγ. Our results are reinforced by a recent study, demonstrating a role for STAT1, the downstream target of IFNγ, in MG development (Chan et al., 2014). This study showed that there is a significant increase in the number of alveolar buds in Stat1−/− mice as compared with WT mice and that excessive bud formation is observed in mature Stat1−/− mice. Although no T cell data or treatment with IFNγ was examined, the authors suggest that signaling via STAT1 negatively regulates luminal progenitor cells, as loss of STAT1 results in an expansion of CD61+ luminal progenitor cells, similar to the effects of loss of IFNγ in our study. Specifically, we show that organoids exposed to IFNγ exhibit significantly reduced expression of CD49b, a marker for mammary luminal progenitor cells. This CD49b+ population also includes IFNγR1-expressing CD61+ cells (Ji et al., 2011; Shehata et al., 2012).
Furthermore, in our experiments we observed that exposing organoids to activated Th2 cells induces accelerated branching (Fig. 5C). This is in line with data showing that epithelial-secreted Th2 type cytokines IL-4 and IL-13, along with their downstream activators STAT6 and GATA3, promote luminal differentiation during alveolar expansion throughout gestation (Khaled et al., 2007). Together, this supports the idea that T cell effector cytokines directly regulate mammary epithelial cells. While we did not find a role for CD8+ T cells in our study, these cells also secret IFNγ upon activation (Schoenborn and Wilson, 2007), and thus may still have roles in mammary organogenesis. We do not rule out the potential existence of pathways alternative to the one we have delineated in this study.
As a side note, our study does not argue that disrupted adaptive immune regulation would necessarily have fatal affects on mammary ductal invasion, but rather suggests a role for adaptive immune regulation as an important homeostatic modulator of normal epithelial development. Since mice have ten MGs, they could potentially still nurse pups even if the epithelia in some/all glands is partially impaired or fail to cover the entire fat pad.
Implications for Cancer Progression
Immune surveillance is an important factor in preventing tumor growth, and mechanisms of normal organogenesis are frequently disrupted during cancer progression. Our data suggest that a Th1 bias in normal MGs restricts epithelial branching. By interacting with IFNγ+ Th1 cells, APCs may be part of a positive feedback loop as IFNγ induces MHCII expression that further drives local Th1 responses (Biswas and Mantovani, 2010; Strutt et al., 2010). In contrast, breast tumor models are characterized by a Th2-bias (Czarneski et al., 2001; Jensen et al., 2003). Our observation of increased branching in Th2-organoid co-cultures echoes the observation of Th2 cells dominating mammary adenocarcinomas, and promoting invasion as well as pulmonary metastasis (DeNardo et al., 2009). In fact, numerous findings in cancer research imply that survival and differentiation of mammary epithelial cells that form tumors are indeed regulated by a delicate balance between Th1/Th2 signaling:
A Th1 bias is more effective in tumor rejection. It has been shown that the combined action of the Th1 cell cytokines, IFNγ and tumor necrosis factor (TNF) induces permanent growth arrest and drives breast and other cancers into senescence (Braumuller et al., 2013). This cytokine-induced senescence strictly requires STAT1 and TNFR1 signaling, respectively. Moreover, in an estrogen receptor (ER) α+ luminal type human breast cancer model which is heavily infiltrated by innate immune cells and T lymphocytes (Bos et al., 2013), IFNγ and CD4+ T cells (but not NK or CD8+ T cells) show anti-tumor activity and inhibit metastatic tumor progression via extensive apoptotic tumor cell death. Other studies imply that immune escape mechanisms in ERα+ breast cancer may be facilitated through an ERα suppressive mechanism on IFNγ signaling (Mostafa et al., 2014). Indeed, in a mouse model of human epidermal growth factor receptor (HER) 2-positive breast cancers, tumor cells control the outcome of tumor immune surveillance through modulation of IFNγR1 expression on tumor cells. Those that express low levels of IFNγR1 fail to be eliminated by IFNγ producing CD8+ T cells, and remain dormant and quiescent until they conceal themselves from the adaptive immune system by losing the tumor antigen, Neu (Kmieciak et al., 2013). Finally, using a Stat1 floxed model that permits tissue-specific disruption of STAT1 in mammary epithelium and a model of HER2-positive breast cancer (Neu/ERBB2-induced) allowed the investigators to identify a role for STAT1 in mammary tumor suppression separate from its role in the immune system, (Klover et al., 2010). This is also in line with the findings that Stat1-deficient mice develop ERα+ mammary tumors (Chan et al., 2014).
Our work warrants future studies examining how the adaptive immune system modulates the fate of normal epithelial and cancer cells. A follow-up study could then focus on the transition between positive and negative regulation during the different cycles of MG expansion and involution, and during early mammary carcinogenesis. Understanding how this switch occurs and what impact it has on the epithelium could identify potential targets for intervention.
EXPERIMENTAL PROCEDURES
Experimental Animal Models
Experiments were approved by the UCSF Institutional Animal Use and Care Committee (IACUC). CX3CR1-GFP/+, CD11c-DTR:GFP, integrin β8fl/fl×CD11c-cre (also crossed to SCID or Ifng−/−), Tcra−/− and MHCIIfl/−×CD11c-cre mice were on the C57BL/6 background.
In vivo Depletion of CD11c+ Antigen Presenting Cells (APCs)
CD11c-DTR:GFP mice were depleted of APC by injecting diphtheria toxin (DTx, 4 ng/g weight; Sigma) i.p. and harvested 48 h post injection (see scheme in Fig. S1C). Mutant DTX (mDTx, 4 ng/g weight) was injected as a control for possible DTx-activated EGF signaling.
Mammary Whole Mount Carmine Staining and Analysis of Ductal Invasion
Inguinal (#4) MGs excised at 7 weeks (no significant fat-pad size difference between littermates) were stained with Carmine Red and manually imaged with a Leica dissection scope using Nikon ACT-1 software. Data were quantified in a blind fashion.
Immunostaining of Intact Mammary Glands
MGs were fixed and frozen sections were stained with anti-CD4 (eBiosciences, 16-0041-82), anti-CD8a (eBiosciences, 16-0081-82) antibodies (1:500) and anti-GFP (CD11c-DTR:GFP, Abcam, ab5450, 1:200) and mounted with Vectashield + DAPI (Vector Laboratories) for nuclear staining.
3D Organotypic Culture and Analysis
MGs (#3, 4, 5 without lymph nodes) were digested in collagenase solution consisting of DMEM/F12, 2 μg/ml trypsin (GIBCO-BRL), 2 μg/ml collagenase IV (Sigma), 10%FBS, 0.5% insulin and 0.1% gentamicin (UCSF Cell Culture Facility) and then treated with DNase (2U/μl) (Sigma). Organoids were epithelial-enriched by differential centrifugations (pulsed to 1500 rpm), plated (50–100 organoids/well, 3–4 wells/condition) with Growth Factor Reduced Matrigel (BD Biosciences) and cultured in DMEM/F12 with 1% Pen/Strep, 1% insulin, transferrin, sodium selenite, and supplemented with 2.5 nM FGF2 (Sigma). Ex vivo depletion of CD11c+ APCs is achieved by mixing organoids with DTx (1 μg/ml) before plating. mDTx (1 μg/ml) was added to control for DTx activated EGF signaling. For some experiments, blocking antibodies were added to organoid cultures: 0.5 mg/ml anti-MHCII (14-5321-82, eBioscience), Rat IgG2b K Isotype Control (16-4031-81, eBiosciences), and 1 mg/ml anti-MHCI (16-5999-82, eBioscience), anti-CD4 (16-0041-82, eBioscience) and Mouse IgG2a K Isotype Control (16-4724-81, eBioscience). For IFNγ blocking, 0.2 mg/ml of anti IFNγ (Clone XMG1.2, BioXCell) was used. IFNγ stimulation experiments were performed by adding IFNγ (PeproTech, #315-05, 5 ng/ml) on day 3 of culture. Organoids were graded by direct manual examination of the culture wells on days 5, 6 and 7. Branching quantifications are shown for day 5 of culture, unless otherwise indicated. Images were taken on a ZeissAxio Observer A1 inverted microscope, using DFC400 camera and Leica application suite Version 3 software (see additional description in Fig. 1B).
Fixation and Staining of Organotypic Cultures
Matrigel drops were fixed and embedded in O.C.T. Sections (20 μm) were stained with antibodies against pSTAT1 (Tyr701) (Cell Signaling, 9167), GFP (for CD11c-DTR:GFP, Abcam, ab5450, 1:200), CD4 (eBiosciences Cat#: 16-0041-82, 1:200), cytokeratin 8 (Troma 1, Developmental Studies Hybridoma Bank, Iowa, 1:50), phospho-histone H3 (Cell Signaling Cat#: 9701,1:200), cleaved caspase 3 (Cell Signaling Cat#: 9661,1:200), imaged with a spinning disk confocal microscope and quantified using ImageJ.
Flow Cytometry
Single cells were isolated from MGs with collagenase solution similar to organoids but without trypsin in the digest. Differential centrifugations were used to isolate stroma versus epithelial-enriched fractions. For non-T cell immune populations, samples were trypsinized, and then incubated with RBC lysis buffer for red blood cell removal. Stainings were for CD11b, CD3, CD8, CD4, F4/80, MHCII, Siglec-F (eBioscience). Intracellular cytokine analysis was performed as previously described (Melton et al., 2010). In brief, cells were incubated for 4 h at 37°C in complete RPMI containing 50 ng/ml PMA (Sigma-Aldrich), 1 μM ionomycin (Sigma-Aldrich), and 2 μM monensin (eBioscience). Cells were blocked with PBS containing 2 μg/ml α-CD16/32 (UCSF Hybridoma Core) and 10% heat-inactivated normal rat serum (Sigma-Aldrich), and incubated with cell-surface antibodies and Aqua Live/Dead Fixable Dead cell stain (Invitrogen). Cells were fixed and permeabilized with Fix/Perm solution (eBioscience). Intracellular cytokines were labeled with antibodies in permeabilization buffer (eBioscience). Data was acquired using BD LSRII, and analyzed with FlowJo. The following antibodies (BD Biosciences or eBioscience) were used: anti-CD4 (clone RM4-5), anti-CD8 (clone 53-6.7), anti–IL17 (Th17+ CD4+ T cells, clone eBio17B7), anti–IFN-γ (Th1 effector CD4+ T cells, clone XMG1.2), anti–Foxp3 (T regulatory cells, clone FJK-16s), and anti–IL13 (Th2 effector CD4+ T cells, clone eBio13A).
In vivo proliferation was assessed by BrdU incorporation. 100 μl BrdU (10 μg/ml; BD Pharmingen) was injected i.p. Single cells from MGs were isolated as described above 2 h post-injection, stained for extracellular surface markers, fixed and permeabilized using the BD (Becton Dickinson) Cytofix/Cytoperm buffer (15 min at RT). The next day, intracellular BrdU was stained overnight using the FITC BrdU Flow Kit Staining Protocol (BD Pharmingen, Cat No. 559619). Data were acquired using BD LSRII, and analyzed with FlowJo.
For flow cytometry on organoid cultures, Matrigel was digested in 1U of dispase (Becton Dickinson, San Diego, CA) to release the organoids, followed by 0.05% trypsin and strained through a 70 μm filter. Dead cells and lineage-specific cells (CD31+, Ter119+, CD45+, as described previously (Plaks et al., 2013) were excluded. Luminal and basal populations were sorted using antibodies against EpCAM and CD49f (Shehata et al., 2012). To examine T cells after depletion of APCs in organoids, Matrigel drops were collected in medium and left on ice before the dispase and trypsin digestion. Dead cells were excluded and antibodies against CD11c versus CD90.2, CD4 and CD8 were used to quantify APCs versus T cells, respectively.
Th1/2 Polarization, T Cell Proliferation Assay and Co-Culture with Organoids
Lymph node cells or splenocytes from OT-II or OT-II x CD2-RFP x 4get mice were cultured in RPMI (Life Sciences/GIBCO) with 10% FCS (Hyclone, Thermo Scientific). Culture for Th1 differentiation: 0.5 μg/ml anti-CD28 (clone PV-1), 10 μg/ml anti-IL-4 (clone 11B11), 10 ng/ml IL-12, 2.5 U/ml IL-2 (Peprotech) and 1 μg/ml OVA peptide 323–336 (GenScript). For Th2 differentiation: 0.5 μg/ml anti-CD28, 10 μg/ml anti-IFNγ (clone XMG1.2), 20 ng/ml IL-4 (Peprotech), 2.5 U/ml IL-2 and 1 μg/ml OVA peptide. Th1/2 polarization was confirmed by intracellular Th1/2 cytokine stains (as in for Fig. S5A) or by GFP expression indicative of Th2 cytokines, using the 4get IL-4 reporter mice. Th1/2 helper cells were enriched using the EasySep Mouse CD4 Enrichment kit (StemCell Tech.) and 1 μM eFluor670 cell proliferation dye was used for helper T proliferation assays. 105 naïve or activated OT-II cells per well (stimulated with pOVA, 1μg/ml, as indicated in Fig. S4B) were co-cultured in organoid medium containing 10 U/ml human IL2.
Spinning Disk Time-Lapse Confocal Microscopy
Imaging of 3D organotypic culture was performed as described previously (Ewald et al., 2008). CellTracker Red or Blue (Life Technologies) stained the mammary epithelium. Quantification was done using ImageJ.
RNA Preparation and Quantitative RT-PCR Analysis
Basal and luminal mammary epithelial cells sorted from organoids were extracted for RNA using TRIzol Reagent (Life technologies). cDNA was generated using iScript Reverse Transciption Supermix of RT-qPCR (Biorad). RT-PCR reaction was performed with iTaq Universal SYBR Green Supermix (Biorad) using ABI 7900 Real Time PCR system. Analysis was normalized to the housekeeping gene Ppia. Primer sequences used are listed in Supplementary Experimental Procedures, qPCR Primers and Function of Gene Targeted.
Statistics
Two-tailed unpaired Student’s t-test was performed. For comparisons between more than two groups, one-way ANOVA was used. Statistical significance was considered when P < 0.05. Experiments were repeated at least 3 times, as indicated in the figure legends. Pooled data are represented as mean ± SEM.
Supplementary Material
Acknowledgments
We thank Dr. Steffen Jung for the MHCIIfl/−×CD11c-cre mice and for critical advice, Dr. Samantha Bailey-Bucktrout for help with the TCRα−/− mice, Dr. John Engelhardt for help with the T cell co-cultures, Dr. Miranda Broz and Michael Kissner for assistance with flow cytometry, Eline C. Van Kappel, Joanne Dai, Charlotte D. Koopman, Karren Wong, Carrie Maynard and Ana Marija Plavec for technical help, and Ying Yu and Elena Atamaniuc for mouse husbandry and genotyping. This work was supported by grants from the National Institutes of Health (CA057621, ES019458 and AI053194 to Z.W., AI077439 and HL53949 to D.S., and CA141451 and HL024136 to M.F.K.) and by a Department of Defense postdoctoral fellowship to VP (W81XWH-11-01-0139) and the Weizmann Institute of Science-National Postdoctoral Award Program for Advancing Women in Science (to V.P.).
Footnotes
Supplemental Information includes 6 Figures, Supplemental Experimental Procedures and References. It also includes 6 movies.
AUTHOR CONTRIBUTIONS
V.P. conceived the study. V.P., with the help of B. B. and A.C.M., designed the study. V.P. and K.K. performed APC depletion experiments in organoids, produced and analyzed live-imaging data. V.P. and A.C.M. preformed the mammary CD4+ effector T cell characterization. V.P. and Y.W. performed the experiments with mice deficient of CD4+ T cells. V.P. and J.R.L. performed whole mount analyses and supporting organoid experiments with all the other transgenic mouse models. V.P. and B.B. designed and performed the organoid and T cell co-culture experiments as well as analyzed the flow cytometry data throughout the study. N.H.N. performed qPCR experiments and analyses. N.H.N and N.K. performed the organoid experiments examining the effect of IFNγ on mammary epithelium. N.K. and R.J.E.v.d.B performed the organoid experiments examining Csf-1 expression and T cell presence after APC depletion. A.J.C. designed and performed the APC in vivo proliferation experiments. V.P. wrote the manuscript, and with N.K., N.H.N. and R.J.E.v.d.B designed figures and schematics. D.S., A.C.M., M.F.K. and Z.W. guided the overall approach, experimental design, and trajectory of the study as well as assisted in manuscript completion.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11:889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- Bos PD, Plitas G, Rudra D, Lee SY, Rudensky AY. Transient regulatory T cell ablation deters oncogene-driven breast cancer and enhances radiotherapy. J Exp Med. 2013;210:2435–2466. doi: 10.1084/jem.20130762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braumuller H, Wieder T, Brenner E, Assmann S, Hahn M, Alkhaled M, Schilbach K, Essmann F, Kneilling M, Griessinger C, et al. T-helper-1-cell cytokines drive cancer into senescence. Nature. 2013;494:361–365. doi: 10.1038/nature11824. [DOI] [PubMed] [Google Scholar]
- Broz ML, Binnewies M, Boldajipour B, Nelson AE, Pollack JL, Erle DJ, Barczak A, Rosenblum MD, Daud A, Barber DL, et al. Dissecting the tumor myeloid compartment reveals rare activating antigen-presenting cells critical for T cell immunity. Cancer Cell. 2014;26:638–652. doi: 10.1016/j.ccell.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SR, Rickert CG, Vermi W, Sheehan KC, Arthur C, Allen JA, White JM, Archambault J, Lonardi S, McDevitt TM, et al. Dysregulated STAT1-SOCS1 control of JAK2 promotes mammary luminal progenitor cell survival and drives ERalpha(+) tumorigenesis. Cell Death Diff. 2014;21:234–246. doi: 10.1038/cdd.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens LM, Pollard JW. Leukocytes in mammary development and cancer. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a003285. pii: a003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarneski J, Meyers J, Peng T, Abraham V, Mick R, Ross SR. Interleukin-4 up-regulates mouse mammary tumor virus expression yet is not required for in vivo virus spread. J Virol. 2001;75:11886–11890. doi: 10.1128/JVI.75.23.11886-11890.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai XM, Ryan GR, Hapel AJ, Dominguez MG, Russell RG, Kapp S, Sylvestre V, Stanley ER. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood. 2002;99:111–120. doi: 10.1182/blood.v99.1.111. [DOI] [PubMed] [Google Scholar]
- Davies LC, Rosas M, Jenkins SJ, Liao CT, Scurr MJ, Brombacher F, Fraser DJ, Allen JE, Jones SA, Taylor PR. Distinct bone marrow-derived and tissue-resident macrophage lineages proliferate at key stages during inflammation. Nat Comm. 2013;4:1886. doi: 10.1038/ncomms2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N, Coussens LM. CD4+ T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell. 2009;16:91–102. doi: 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt JJ, Boldajipour B, Beemiller P, Pandurangi P, Sorensen C, Werb Z, Egeblad M, Krummel MF. Marginating dendritic cells of the tumor microenvironment cross-present tumor antigens and stably engage tumor-specific T cells. Cancer Cell. 2012;21:402–417. doi: 10.1016/j.ccr.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald AJ, Brenot A, Duong M, Chan BS, Werb Z. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Developmental cell. 2008;14:570–581. doi: 10.1016/j.devcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Liu K, Helft J, Bogunovic M, Greter M, Hashimoto D, Price J, Yin N, Bromberg J, Lira SA, et al. The origin and development of nonlymphoid tissue CD103+ DCs. J Exp Med. 2009;206:3115–3130. doi: 10.1084/jem.20091756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouon-Evans V, Lin EY, Pollard JW. Requirement of macrophages and eosinophils and their cytokines/chemokines for mammary gland development. Breast Cancer Res. 2002;4:155. doi: 10.1186/bcr441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouon-Evans V, Rothenberg ME, Pollard JW. Postnatal mammary gland development requires macrophages and eosinophils. Development. 2000;127:2269–2282. doi: 10.1242/dev.127.11.2269. [DOI] [PubMed] [Google Scholar]
- Hoebe K, Janssen E, Beutler B. The interface between innate and adaptive immunity. Nat Immunol. 2004;5:971–974. doi: 10.1038/ni1004-971. [DOI] [PubMed] [Google Scholar]
- Honda T, Egen JG, Lammermann T, Kastenmuller W, Torabi-Parizi P, Germain RN. Tuning of antigen sensitivity by T cell receptor-dependent negative feedback controls T cell effector function in inflamed tissues. Immunity. 2014;40:235–247. doi: 10.1016/j.immuni.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii H, Tanabe S, Ueno M, Kubo T, Kayama H, Serada S, Fujimoto M, Takeda K, Naka T, Yamashita T. ifn-gamma-dependent secretion of IL-10 from Th1 cells and microglia/macrophages contributes to functional recovery after spinal cord injury. Cell Death Disease. 2013;4:e710. doi: 10.1038/cddis.2013.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen SM, Meijer SL, Kurt RA, Urba WJ, Hu HM, Fox BA. Regression of a mammary adenocarcinoma in STAT6−/− mice is dependent on the presence of STAT6-reactive T cells. J Immunol. 2003;170:2014–2021. doi: 10.4049/jimmunol.170.4.2014. [DOI] [PubMed] [Google Scholar]
- Ji H, Goode RJA, Vaillant F, Mathivanan S, Kapp EA, Mathias RA, Lindeman GJ, Visvader JE, Simpson RJ. Proteomic profiling of secretome and adherent plasma membranes from distinct mammary epithelial cell subpopulations. Proteomics. 2011;11:4029–4039. doi: 10.1002/pmic.201100102. [DOI] [PubMed] [Google Scholar]
- Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, Littman DR. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol. 2000;20:4106–4114. doi: 10.1128/mcb.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Unutmaz D, Wong P, Sano GI, De los Santos K, Sparwasser T, Wu S, Vuthoori S, Ko K, Zavala F, et al. In Vivo Depletion of CD11c+ Dendritic Cells Abrogates Priming of CD8+ T Cells by Exogenous Cell-Associated Antigens. Immunity. 2002;17:211–220. doi: 10.1016/s1074-7613(02)00365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaled WT, Read EKC, Nicholson SE, Baxter FO, Brennan AJ, Came PJ, Sprigg N, McKenzie ANJ, Watson CJ. The IL-4/IL-13/Stat6 signalling pathway promotes luminal mammary epithelial cell development. Development. 2007;134:2739–2750. doi: 10.1242/dev.003194. [DOI] [PubMed] [Google Scholar]
- Khalkhali-Ellis Z, Abbott DE, Bailey CM, Goossens W, Margaryan NV, Gluck SL, Reuveni M, Hendrix MJ. IFN-gamma regulation of vacuolar pH, cathepsin D processing and autophagy in mammary epithelial cells. J Cell Biochem. 2008;105:208–218. doi: 10.1002/jcb.21814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klover PJ, Muller WJ, Robinson GW, Pfeiffer RM, Yamaji D, Hennighausen L. Loss of STAT1 from mouse mammary epithelium results in an increased Neu-induced tumor burden. Neoplasia. 2010;12:899–905. doi: 10.1593/neo.10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmieciak M, Payne KK, Wang XY, Manjili MH. IFN-gamma Ralpha is a key determinant of CD8+ T cell-mediated tumor elimination or tumor escape and relapse in FVB mouse. PLoS One. 2013;8:e82544. doi: 10.1371/journal.pone.0082544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilla JN, Werb Z. Mast cells contribute to the stromal microenvironment in mammary gland branching morphogenesis. Dev Biol. 2010;337:124–133. doi: 10.1016/j.ydbio.2009.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Werb Z. Patterning mechanisms of branched organs. Science. 2008;322:1506–1509. doi: 10.1126/science.1162783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magram J, Connaughton SE, Warrier RR, Carvajal DM, Wu CY, Ferrante J, Stewart C, Sarmiento U, Faherty DA, Gately MK. IL-12-deficient mice are defective in IFN gamma production and type 1 cytokine responses. Immunity. 1996;4:471–481. doi: 10.1016/s1074-7613(00)80413-6. [DOI] [PubMed] [Google Scholar]
- Melton AC, Bailey-Bucktrout SL, Travis MA, Fife BT, Bluestone JA, Sheppard D. Expression of αvβ8 integrin on dendritic cells regulates Th17 cell development and experimental autoimmune encephalomyelitis in mice. J Clin Invest. 2010;120:4436–4444. doi: 10.1172/JCI43786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Brown BD, Shay T, Gautier EL, Jojic V, Cohain A, Pandey G, Leboeuf M, Elpek KG, Helft J, et al. Deciphering the transcriptional network of the dendritic cell lineage. Nat Immunol. 2012;13:888–899. doi: 10.1038/ni.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafa AA, Codner D, Hirasawa K, Komatsu Y, Young MN, Steimle V, Drover S. Activation of ERalpha signaling differentially modulates IFN-gamma induced HLA-class II expression in breast cancer cells. PLoS One. 2014;9:e87377. doi: 10.1371/journal.pone.0087377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien J, Martinson H, Durand-Rougely C, Schedin P. Macrophages are crucial for epithelial cell death and adipocyte repopulation during mammary gland involution. Development. 2012;139:269–275. doi: 10.1242/dev.071696. [DOI] [PubMed] [Google Scholar]
- Plaks V, Brenot A, Lawson DA, Linnemann JR, Van Kappel EC, Wong KC, de Sauvage F, Klein OD, Werb Z. Lgr5-expressing cells are sufficient and necessary for postnatal mammary gland organogenesis. Cell Rep. 2013;3:70–78. doi: 10.1016/j.celrep.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JR, Schwertfeger KL. Immune cell location and function during post-natal mammary gland development. J Mammary Gland Biol Neoplasia. 2010;15:329–339. doi: 10.1007/s10911-010-9188-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renz H, Brandtzaeg P, Hornef M. The impact of perinatal immune development on mucosal homeostasis and chronic inflammation. Nat Rev Immunol. 2012;12:9–23. doi: 10.1038/nri3112. [DOI] [PubMed] [Google Scholar]
- Rios AC, Fu NY, Lindeman GJ, Visvader JE. In situ identification of bipotent stem cells in the mammary gland. Nature. 2014;506:322–327. doi: 10.1038/nature12948. [DOI] [PubMed] [Google Scholar]
- Schoenborn JR, Wilson CB. Regulation of interferon-gamma during innate and adaptive immune responses. Adv Immunol. 2007;96:41–101. doi: 10.1016/S0065-2776(07)96002-2. [DOI] [PubMed] [Google Scholar]
- Shehata M, Teschendorff A, Sharp G, Novcic N, Russell IA, Avril S, Prater M, Eirew P, Caldas C, Watson CJ, et al. Phenotypic and functional characterisation of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 2012;14:R134. doi: 10.1186/bcr3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternlicht MD, Kouros-Mehr H, Lu P, Werb Z. Hormonal and local control of mammary branching morphogenesis. Differentiation. 2006;74:365–381. doi: 10.1111/j.1432-0436.2006.00105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt TM, McKinstry KK, Dibble JP, Winchell C, Kuang Y, Curtis JD, Huston G, Dutton RW, Swain SL. Memory CD4+ T cells induce innate responses independently of pathogen. Nat Med. 2010;16:558–564. doi: 10.1038/nm.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis MA, Reizis B, Melton AC, Masteller E, Tang Q, Proctor JM, Wang Y, Bernstein X, Huang X, Reichardt LF, et al. Loss of integrin αvβ8 on dendritic cells causes autoimmunity and colitis in mice. Nature. 2007;449:361–365. doi: 10.1038/nature06110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallon-Eberhard A, Landsman L, Yogev N, Verrier B, Jung S. Transepithelial pathogen uptake into the small intestinal lamina propria. J Immunolo. 2006;176:2465–2469. doi: 10.4049/jimmunol.176.4.2465. [DOI] [PubMed] [Google Scholar]
- Van Nguyen A, Pollard JW. Colony stimulating factor-1 is required to recruit macrophages into the mammary gland to facilitate mammary ductal outgrowth. Dev Biol. 2002;247:11–25. doi: 10.1006/dbio.2002.0669. [DOI] [PubMed] [Google Scholar]
- Veres TZ, Voedisch S, Spies E, Valtonen J, Prenzler F, Braun A. Aeroallergen challenge promotes dendritic cell proliferation in the airways. J Immunol. 2013;190:897–903. doi: 10.4049/jimmunol.1200220. [DOI] [PubMed] [Google Scholar]
- Watson CJ, Oliver CH, Khaled WT. Cytokine signalling in mammary gland development. J Reprod Immunol. 2011;88:124–129. doi: 10.1016/j.jri.2010.11.006. [DOI] [PubMed] [Google Scholar]
- Wiseman BS, Werb Z. Stromal effects on mammary gland development and breast cancer. Science. 2002;296:1046–1049. doi: 10.1126/science.1067431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Martinez D, Koledova Z, Qiao G, Streuli CH, Lu P. FGF ligands of the postnatal mammary stroma regulate distinct aspects of epithelial morphogenesis. Development. 2014;141:3352–3362. doi: 10.1242/dev.106732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Zhang F, Aune TM. Either IL-2 or IL-12 is sufficient to direct Th1 differentiation by nonobese diabetic T cells. J Immunol. 2003;170:735–740. doi: 10.4049/jimmunol.170.2.735. [DOI] [PubMed] [Google Scholar]
- Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.