Abstract
Purpose
To determine the relationship between the apparent diffusion coefficient (ADC, from diffusion weighted (DW) MRI), the extravascular, extracellular volume fraction (ve, from dynamic contrast enhanced (DCE) MRI), and histological measurement of the extracellular space fraction.
Methods
Athymic nude mice were injected with either human epidermal growth factor receptor 2 positive (HER2+) BT474 (n = 15) or triple negative MDA-MB-231 (n = 20) breast cancer cells, treated with either Herceptin (n = 8), Abraxane (low dose n = 7, high dose n =6), or saline (n = 7 for each cell line), and imaged using DW- and DCE-MRI before, during, and after treatment. After the final imaging acquisition, the tissue was resected and evaluated by histological analysis. H&E stained central slices were scanned using a digital brightfield microscope and evaluated with thresholding techniques to calculate the extracellular space.
Results
For both BT474 and MDA-MB-231, the median ADC of the central slice exhibited a significantly positive correlation with the corresponding central slice extracellular space as measured by H&E (p = 0.03, p < 0.01, respectively). Median ve calculated from the central slice showed differing results between the two cell lines. For BT474, a significant correlation between ve and extracellular space was calculated (p = 0.02), while MDA-MB-231 tumors did not demonstrate a significant correlation (p = 0.64). Additionally, there was no correlation discovered between ADC and ve with either whole tumor analysis or central slice analysis (p > 0.05).
Conclusion
While ADC correlates well with the histologically determined fraction of extracellular space, this data adds to the growing body of literature which suggests that ve derived from DCE-MRI is not a reliable biomarker of extracellular space for a range of physiological conditions.
Keywords: extracellular space, quantitative imaging, extravascular extracellular space, apparent diffusion coefficient
Introduction
Dynamic contrast enhanced-magnetic resonance imaging (DCE-MRI) involves the serial acquisition of heavily T1-weighted images to measure the perfusion and extravasation of an injected contrast agent into a tissue of interest (1). Typical contrast agents used in DCE-MRI are gadolinium-based chelates that alter the MR signal primarily by decreasing the native longitudinal relaxation rate R1 (≡ 1/T1) of tissue water protons to a degree dependent upon the local contrast agent concentration. By acquiring MR images before, during, and after an intravenous injection of a contrast agent, the resulting signal intensity time course can be fit to an appropriate model to extract various pharmacokinetic properties of the tissue including, for example, the volume transfer constant (Ktrans) and the extravascular, extracellular volume fraction (ve). Both pre-clinical (2-5) and clinical (6-10) data have shown these parameters change in response to chemotherapeutic and radiation treatments. This study is particularly focused on ve, which provides a measure of tissue cellularity.
The microscopic thermally-induced random motion of molecules is referred to as self-diffusion or Brownian motion (11). Diffusion-weighted MRI (DW-MRI) techniques have been developed which are sensitive to this motion of water molecules, and thus allow an evaluation of the relative movement of the molecules within a tissue of interest. Though cellular matrix and other components may affect diffusion, the predominant barrier to motion in tissue is cellular membranes, and therefore the apparent diffusion coefficient (ADC) can be related to the cell density of the tissue. ADC values obtained from DW-MRI have been shown to correlate inversely with tumor cellularity (12-18) and have been used to assess treatment response to both chemotherapy and radiotherapy in pre-clinical (2,4,19,20) and clinical (21-25) applications.
The goal of this study was to investigate the relationship between ADC and ve in two common breast tumor models before and after drug treatment. We reasoned that, in response to successful cytotoxic treatment, tumor cells would die, thereby increasing the extravascular, extracellular space and causing a concomitant increase in both ADC and ve. However, previous studies have indicated that ve decreases after treatment (22,26). Other studies examining both ADC and ve in glioblastomas (27) and breast tumors (28) yielded conflicting results as well. In particular, both the human breast tumors and glioblastomas revealed no correlations between the two parameters. While both ADC and ve have been interpreted as metrics of the extravascular, extracellular space, to the best of our knowledge, no direct relationship between ADC and ve has been shown. Here, DCE- and DW-MRI data were obtained simultaneously from two different murine models of breast cancer and used to examine the relationship between in vivo measurements of the ADC and ve, and the actual extracellular space as quantified by the gold-standard of ex vivo histology. This preclinical work serves as an experimental validation of our previous clinical research, which indicated that there was no correlation between ADC and ve in human breast cancer (28). By translating the work to the preclinical setting, we are able to extensively evaluate the relationship between the imaging parameters and histology in a manner which cannot be achieved as thoroughly in clinical analysis.
Methods
Tumor Model
Breast cancer cells (BT474 or MDA-MB-231) were cultured to 80-90% confluency prior to passaging and cell numbers were determined with a hemocytometer and trypan blue dye exclusion. BT474 cells were cultured in improved minimal essential medium (IMEM, Invitrogen, Carlsbad, CA) supplemented with 10% FBS, 1% penicillin/streptomycin, and 1% insulin at 37 ºC with 5% CO2. MDA-MB-231 cells were grown in Dulbecco’s Modified Eagle’s Minimal Essential Medium (Invitrogen, Carlsbad, CA) containing 10% FBS, 1% penicillin/streptomycin, also at 37 ºC with 5% CO2. Either 1 × 107 BT474 cells or 1 × 107 MDA-MB-231 cells suspended in 100 μL of medium plus 20-30% matrigel were injected subcutaneously on the right flank of each mouse. Tumors were grown to approximately 200-250 mm3 at which point the animals were entered into the study. The time between cell implantation and the baseline imaging session was approximately 6 to 8 weeks.
Animal Model
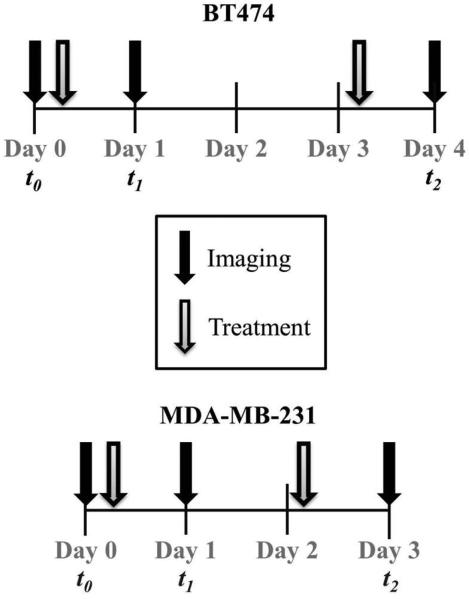
Adult female fox nu/nu mice (n = 35) were purchased from Charles River Laboratories (Raleigh, NC). One to two days prior to imaging, a 26-gauge jugular catheter was surgically implanted to allow for delivery of the contrast agent during DCE-MRI studies. All animals were imaged prior to any treatment to obtain DCE- and DW-MRI data at baseline (t0), one time point during treatment (t1), and on the endpoint day prior to sacrificing for histology (t2). The timing of the imaging and treatment protocols are shown in Figure 1. BT474 tumor-bearing mice were imaged on the following days: t0 = 0, t1 = 1, and t2 = 4. MDA-MB-231 tumor-bearing mice were imaged on: t0 = 0, t1 = 1, and t2 = 3. Catheters were flushed with heparin saline (10 USP/mL) on days on which imaging did not occur in order to maintain patency of the catheters. After the final imaging session (t2), the mice were humanely sacrificed for histological analysis of tumor tissues.
Figure 1.
Timeline of imaging and treatment regimens for BT474 and MDA-MB-231 tumors. Tumors were imaged at baseline (t0), during treatment (t1), and following treatment (t2). BT474 tumors were treated with Herceptin on days 0 and 3, and MDA-MB-231 tumors were treated with Abraxane on days 0 and 2.
Treatment
Two therapies were employed: Herceptin (Genentech, San Francisco, CA) for BT474 cells, and Abraxane (Celegene, Summit, NJ) for MDA-MB-231 cells. Herceptin (10 mg/kg, n = 8) was administered on days 0 (following imaging) and 3 via intraperitoneal (i.p.) injection. Abraxane treatment (15 mg/kg (n = 7) or 25 mg/kg (n = 6)) occurred on days 0 (following imaging) and day 2 via i.p. injection (Figure 1). Injections were prepared in 200 μL. In addition, control animals for each cell line (BT474 (n = 7) and MDA-MB-231 (n = 7)) were imaged and received i.p injections of the same volume of saline on the corresponding days of therapeutic injections.
Data Acquisition
All procedures in the experiment were reviewed and approved by our Institutional Animal Care and Use Committee. Animals were imaged on a 7T MR scanner (Agilent Technologies (formerly Varian), Palo Alto, CA) with a 38-mm quadrature coil (Doty Scientific, Columbia, SC). During all imaging procedures, the mice were anesthetized using a 2%/98% isoflurane/oxygen mixture. Body temperature was maintained via a flow of warm air through the magnet bore, and temperature and respiratory rate were monitored throughout the entire experiment. Each animal was placed in a custom built restraint, and the tumor region was first localized via 3D gradient echo scout images. T2-weighted images covering the entire tumor volume were acquired using a fast spin echo pulse sequence with the following parameters: TR = 5500 ms, effective TE = 35.6 ms, 15 slices, 1 mm slice thickness, and an acquisition matrix of 128 × 128 over a 28 × 28 × 15 mm3 field of view (FOV) yielding a voxel size of 0.22 × 0.22 × 1 mm3.
DW-MRI
Once the tumor region was located, diffusion data was collected via a respiratory gated and navigated (29) diffusion-weighted pulsed gradient spin echo sequence. To avoid slice cross-excitation artifacts, the 15 slices were collected with a 1 mm gap; after acquisition of each b value, the domain was offset by 1 mm in the z direction and acquired at the same b value in order to collect DW-MRI data over the entire tumor volume. The 15 slices of DW-MRI data that correspond to the DCE acquisition were used for the subsequent analysis. Diffusion-weighted images were acquired with three b values (150, 500, and 800 s/mm2) and gradients applied simultaneously along three orthogonal directions (x, y, and z). Scan acquisition parameters were: TR/TE = 2000/30 ms, gradient duration δ = 3 ms, gradient interval Δ = 20 ms, two signal excitations (NEX = 2), 15 slices, 1 mm slice thickness, and an acquisition matrix of 64 × 64 over a 28 × 28 × 15 mm3 FOV yielding a voxel size of 0.44 × 0.44 × 1 mm3. A 1 mL conical tube filled with water was placed near the hind limb (taped to the animal holder) and was used to verify the integrity of ADC measurements at each imaging time point.
DCE-MRI
After DW-MRI, pre-contrast T1 maps were obtained using a multi-slice inversion recovery snapshot FLASH (Fast Low Angle SHot) gradient echo sequence with an adiabatic inversion pulse over seven inversion times (TI) ranging from 250 – 10,000 ms (250, 450, 830, 1,500, 2,700, 5,000, 10,000 ms). Scan acquisition parameters were as follows: TR/TE = 100/2.1 ms, α = 25°, NEX = 2, 15 slices, 1 mm slice thickness, and an acquisition matrix of 64 × 64 over a 28 × 28 × 15 mm3 FOV yielding a voxel size of 0.44 × 0.44 × 1 mm3. Dynamic T1-weighted images were acquired using a spoiled gradient echo sequence at a temporal resolution of 12.8 seconds for approximately 20 minutes with the following parameters: TR/TE = 100 ms/2.1 ms, α = 25°, NEX = 2, and the same acquisition matrix and FOV as the pre-contrast T1 map, and over the same anatomical volume. Baseline images were acquired for approximately 2.5 min before a bolus of 0.05 mmol/kg of Magnevist (Bayer, Whippany, NJ) was administered in the jugular catheter over 3 seconds using an automated syringe pump (Harvard Appartatus, Holliston, MS) at a rate of 2.4 mL/min.
Data Analysis
Regions of interest (ROIs) were drawn around the tumor in all tumor-containing slices. The acquisition matrix, FOV, and animal position were maintained throughout the imaging protocol such that voxels within the DW-MRI and DCE-MRI acquisitions were inherently co-registered. Thus, once the ROI was drawn for each slice, that ROI could be used for both the DW-MRI analysis and the DCE-MRI analysis. Additionally, corresponding voxels could be selected from DCE-MRI and DW-MRI to perform voxel-wise analysis.
DW-MRI
The signal (S) collected at each b value at each voxel was fit to Eq. [1]:
| [1] |
where S0 and S(b) are the signal intensities before and after application of the diffusion gradients. Data fitting was performed using Matlab 2013a (The Mathworks, Natick, MA) to extract the ADC values for every ROI and voxel location. In order to help verify the integrity of tumor ADC, data were filtered to include only slices that had a median water phantom ADC value in the range of ± 15% of that reported for free water at 37 ºC (30).
DCE-MRI
Pre-contrast T1 maps from the same ROIs used in the DW-MRI analyses were computed by fitting data collected from the inversion recovery snapshot sequence using a nonlinear least squares method to Eq. [2]:
| [2] |
where S0 and S are the signal intensities at equilibrium and the inversion time (TI), respectively.
The description of the signal intensity measured from a spoiled gradient echo acquisition is given by:
| [3] |
where S0 is a constant describing the scanner gain and proton density, α is the flip angle, and TR is the repetition time and we have assumed that TE ≪ T2*. In this work, the fast exchange limit (FXL) model (31) was assumed, such that the longitudinal relaxation time is described by:
| [4] |
where r is the relaxivity for gadopentetate at 7T (measured at 4.7 mM−1s−1 (32)), Ct(t) is the dynamic concentration of the contrast in the tissue, and T10 is the previously established pre-contrast T1 value. In conjunction with a population-derived vascular input function, Cp(t) (32), the dynamic data can then be fit using a nonlinear least squares approach to the standard Tofts-Kety model:
| [5] |
to extract the volume transfer coefficient (Ktrans) and the extravascular, extracellular volume fraction (ve) for each voxel. For each animal, data were verified to include only slices that had physiological values of ve for the muscle (i.e., 0 < ve < 0.12). This was accomplished by selecting a muscle ROI near the femur for each slice at the same time the tumor ROI was selected, and subsequently calculating the median ve value within the muscle ROI.
Histology
On the final day of imaging (i.e., t2), animals were sacrificed and the tumor tissue was excised. Tumors were sliced in half in an orientation as closely matched to the imaging plane as possible, placed into tissue cassettes, and soaked in 10% Formalin (Fisher, Pittsburgh, PA) for 48-72 hours. The tissue was then stored in 70% ethanol. Samples were paraffin embedded and central slices were sectioned at 5 μm thickness, floated onto charged glass slides, and dried overnight. Three hematoxylin and eosin (H&E) stained sections were obtained from the tissue block and utilized for this analysis.
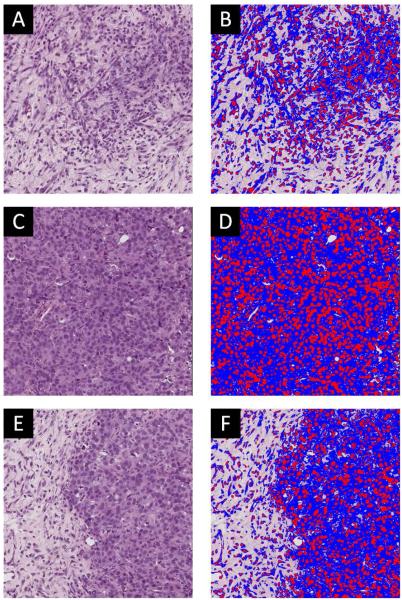
Slides were digitally scanned in high resolution (20× magnification) brightfield with a Leica (Leica Microsystems Inc, Ariol, Buffalo Grove, IL) SCN400 Slide Scanner. The software associated with the Leica SCN400 package was utilized for unbiased, automated image analysis and quantification of immunostaining in brightfield. This software was used to generate maps of intra- and extracellular regions based on intensity thresholding. Briefly, a region was drawn to encompass the entire histology slice and images were thresholded into three categories using automated selection: positive staining (i.e., hemotoxylin, nuclei) colored with red, counterstaining (i.e., eosin, cytoplasm) colored with blue, and no staining (i.e., extracellular regions). The thresholding protocol utilized to calculate the extracellular space is demonstrated in Figure 2. Figure 2 displays three representative histological slices revealing low cellular density (panel A), high cellular density (panel C), and variable cell density (panel E) and the corresponding color maps (panels B, D, and F, respectively). The total area (AROI) and total stained area (Astain) were used to determine the percentage of extracellular (EC) space as given by Eq. [6]:
| [6] |
This value was calculated for three central H&E slices for each animal. The average percent extracellular space was then calculated, and that average value was utilized for comparison to the imaging parameters.
Figure 2.
Panel A presents a H&E stain of a tumor with low cellularity, while panel B shows the color coded segmentation of the tissue into red (positive hemotoxylin staining), blue (eosin counterstaining), and no staining (extracellular regions). Panels C and D display similar data for a tumor with high cellularity, while panels E and F correspond to a more heterogeneous tumor.
Statistical Analysis
ADC-ve Voxel Correlation
The DW- and DCE-MRI image sets were inherently registered and thus were amenable to voxel-wise correlation analysis. The previously defined ROIs were utilized to select voxels within the tumor for each animal on the baseline (t0) scans. The correlation coefficient was then calculated for each animal between the ADC and ve values. The Pearson correlation coefficients were calculated for two sets of voxels. The first analysis consisted of all voxels within the ROI (ADC > 0 and ve > 0.01). The second analysis consisted only of physiologically relevant voxels (i.e., ADC > 0 and 1 ≥ ve > 0.01). These calculations were performed only for the baseline time point (t0), prior to any treatment, allowing analysis of the correlation between the two imaging parameters without the potentially confounding effects of treatment.
ADC-ve ROI Correlation
For each of the three time points, the median ADC and ve values were calculated from the total tumor ROI (voxels from all slices) for each animal. Voxels were not removed from this particular analysis for magnitudes outside of the physiological range (i.e. ve > 1) in order to maintain consistency with the central slice ADC-ve-EC analysis which employed a whole tissue section calculation. The median value parameter pairs for each mouse were then plotted against each other and linear regression was used to characterize the relationship between the two parameters at each time point. The Pearson correlation coefficient was calculated between data sets to assess the significance of the relationship.
Central Slice ve-EC, ADC-EC, and ADC-ve Correlation
Imaging data that was obtained at t2 was visually examined to find a single slice that displayed the most correspondence (based on shape and features) to the three sections of tissue that were stained using H&E. Care was taken to obtain the central region of the tumor as assessed by both imaging and histology, and to select the imaging slice which most closely represented the histology slices in structure and features. The central slice ROI from the imaging and histology data were then compared to evaluate the relationship between ADC, ve, and EC. As for the ROI analysis, parameters from all voxels were retained in the central slice analysis even if the parameter values were unphysiological, and median parameter values were calculated within the ROI. Linear regression parameters and Pearson correlation coefficients were determined for the central slice analysis for comparisons between 1) ve and EC, 2) ADC and EC, and 3) ADC and ve.
For all analyses, statistical analysis was completed using GraphPad Prism version 6.00 for Windows (GraphPad Software Inc, La Jolla, CA) and a p-value of less than 0.05 was considered significant.
Results
Representative Data
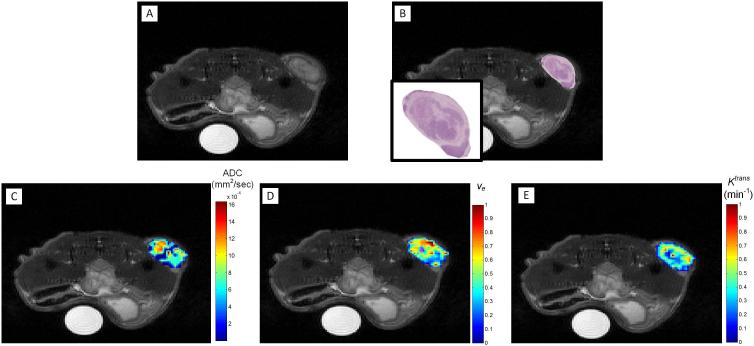
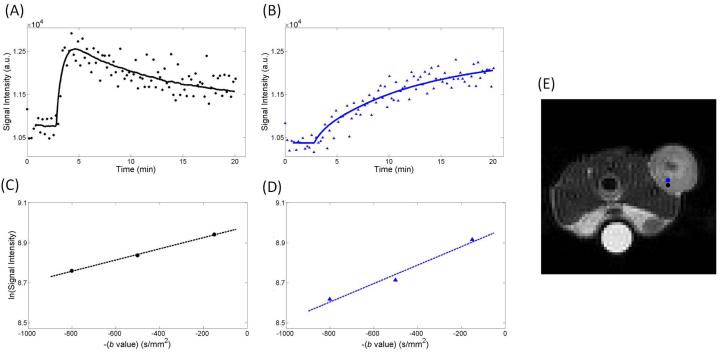
Figure 3 shows a representative anatomical grayscale image of the central slice of a mouse at t2 (panel A) and one of the corresponding H&E histology slices manually overlaid using visual alignment on the anatomical image (panel B). Also shown are representative parameter maps for ADC (panel C), ve (panel D), and Ktrans (panel E). Visual inspection of the spatial variations in these regions reveals that areas of high cellular density (shown by positive hemotoxylin staining in panel B) qualitatively correspond with regions of a lower ADC (panel C); this qualitative correspondence is not as obvious between ve (panel D) and the H&E staining. The Ktrans map (panel E) reveals increased blood flow in the periphery compared to the central region. Magenta-colored voxels (that are not utilized on the color bar range) were used to represent areas with a ve greater than 1 (panel D). Figure 4 shows representative voxel signal intensity data for the DCE acquisition (panels A and B) as well as the corresponding DW-MRI acquisition (panels C and D) for two voxels from a single slice of an individual animal that are in close proximity. (We return to this important point in the Discussion section.) In panels A and B, the circle and triangle markers represent the dynamic signal intensity data while the lines show the standard Tofts model fit to the signal intensity data (Eq. [3]). The representative data shows signal intensity curves for (panel A) a voxel exhibiting contrast agent delivery and wash-out (Ktrans = 0.22 min−1, ve = 0.29), compared to (panel B) a voxel that has an extremely high ve of 2.66 (and Ktrans = 0.024 min−1). Similarly, in panels C and D, the circle and triangle markers show the natural log values of the signal intensity data for the three b values and the dashed lines represent the fit obtained using Eq. [1]. Panel E shows the corresponding location of these voxels.
Figure 3.
Representative data displaying a grayscale, T2-weighted anatomical MR image (A), a H&E histological section (with zoomed view in the inset) overlaid on the anatomical image of panel A, and the corresponding ADC (C), ve (D), and Ktrans (E) parametric maps at t2. The ADC map shows higher values (red) in regions where H&E shows lower cellular densities (high extracellular space) as indicated by the lighter pink regions.
Figure 4.
Panel A displays an example DCE-MRI signal intensity curves for a voxel exhibiting rapid contrast agent delivery and washout (Ktrans = 0.22 min−1, ve of 0.29), compared to a voxel that has an extremely high (and unphysiological) ve of 2.66 (Ktrans = 0.024 min−1) in panel B. In panel B, a slow, steady increase of contrast agent is observed; a pattern more indicative of diffusion than perfusion. Panels C and D show the natural log of the signal intensity curves as a function of b value from DW-MRI data that correspond to the same voxels in panels A and B, respectively (the black voxel corresponds to graph A and C, while the blue voxel corresponds to graph B and D). The anatomical gray scale image E indicates the location of the voxels which are color coded to match the curves in panels A-D. This demonstrates the potential variability of ve, and consistency of ADC, within a region-of-interest.
BT474 Tumor Bearing Mice
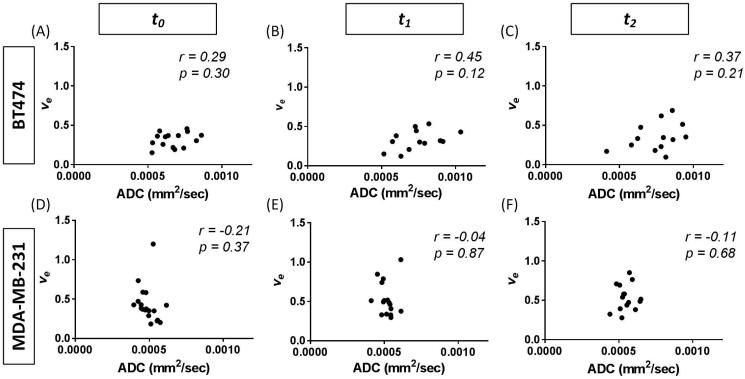
As the DCE- and DW-MRI were acquired sequentially (and with identical slices) in the same imaging session, we were able to make direct comparisons between ADC and ve both at the voxel level and at the ROI level. One BT474 control mouse was not able to be used for voxel-wise comparisons due to movement in the animal between the DW- and DCE-MRI scans. The results for the voxel-wise analysis at baseline are given in Table 1. The Pearson correlation values are shown for each mouse, with an average value (±SE) of 0.22 (±0.049) and a range of −0.05 to 0.51, when analyzing all voxels. Using only those voxels that returned physiological ve values, the Pearson correlation values had a mean value of 0.20 (±0.057) with a range from −0.1 to 0.59. In considering the whole tumor ROI analysis, the median values of ADC and ve showed no significant correlation at any of the three time points, with Pearson correlation coefficients of 0.29 (p = 0.30) at t0, 0.45 (p = 0.12) at t1, and 0.37 (p = 0.21) at t2 as seen in Figure 5 (panels A-C). The mean ADC and ve values, respectively, at each time point for all of the mice were 6.72×10−4 mm2/s and 0.32 at t0, 7.93×10−4 mm2/s and 0.35 at t1, and 7.59×10−4 mm2/s and 0.34 at t2.
Table 1.
Voxel-wise correlation values between ADC and ve for BT474 tumors at t0.
| Mouse Number |
Treatment | Correlation Coefficient | |
|---|---|---|---|
| All Voxels | ve≤1a | ||
| 1 | Control | −0.051 | −0.035 |
| 2 | Control | 0.28 | −0.042 |
| 3 | Control | 0.51 | 0.59 |
| 4 | Control | 0.34 | 0.31 |
| 5 | Control | −0.0058 | 0.34 |
| 6 | Trastuzumab | −0.015 | 0.48 |
| 7 | Trastuzumab | 0.20 | 0.029 |
| 8 | Trastuzumab | 0.50 | 0.17 |
| 9 | Trastuzumab | 0.17 | 0.25 |
| 10 | Trastuzumab | 0.25 | 0.20 |
| 11 | Trastuzumab | 0.33 | −0.10 |
| 12 | Trastuzumab | 0.048 | 0.21 |
| 13 | Trastuzumab | 0.11 | −0.029 |
| 14 | Trastuzumab | 0.36 | 0.37 |
The correlation between ADC and ve in this column was calculated using only voxels within the tumor ROI which had a physiological value of ve (i.e. less than or equal to 1).
Figure 5.
3D quantitative analysis correlation of ADC (mm2/sec) and ve over the whole tumor at baseline (t0), during treatment (t1), and following treatment (t2). In both the BT474 (panels A-C) and MDA-MB-231 (panels D-F) tumor lines, there is no significant correlation between ADC and ve prior to, during, or following treatment.
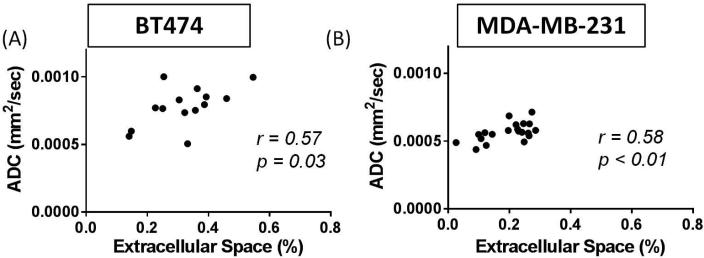
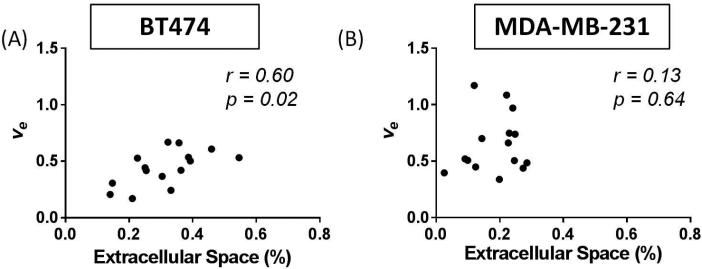
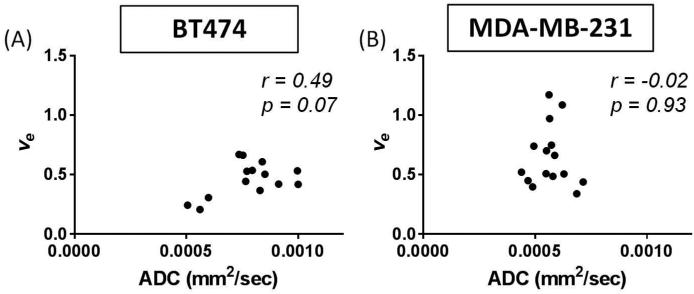
When comparing the central slice of the MR images to the EC calculated from histological analysis of the H&E slide at t2, a significant correlation was found between ADC and EC (r = 0.57, p = 0.03) as seen in Figure 6A. A similar comparison between ve and EC also demonstrated a significant correlation (r = 0.60, p = 0.02), as seen in Figure 7A. However, there was no significant correlation (r = 0.49, p = 0.07) between ADC and ve in the central slice at t2, as seen in Figure 8A. The group mean values of ADC, ve, and %EC were 7.80×10−4 mm2/s, 0.44, and 31%, respectively. In all evaluations, the number of data points for the comparisons at t1 and t2 is lower than for the comparisons at t0 due to either failed contrast agent injections (likely caused by a non-patent catheter), which did not allow for quantification of ve, or unusable ADC values (i.e. a water phantom ADC outside of the acceptable range).
Figure 6.
Analysis of the central slice of the tumor allowed direct comparisons of histology to DW-MRI at t2 through the correlation of percent of extracellular space (EC) and ADC (mm2/sec). Both the BT474 (A) and MDA-MB-231 (B) tumors show significant correlations between these parameters (p = 0.03, p < 0.01, respectively). The range of extracellular space values allows for a direct correlation between this noninvasive imaging parameter and the gold standard (histology).
Figure 7.
Analysis of the central slice of the tumor allowed comparisons of histology to DCE-MRI at t2 through the correlation of percent of extracellular space (EC) and ve. The BT474 tumors (A) revealed a significant correlation between these parameters (p = 0.02), while the MDA-MB-231 tumors (B) did not (p = 0.64). This data presents evidence that the standard model of DCE-MRI analysis provides a poor characterization of the extracellular space.
Figure 8.
Analysis of central slice of the tumor allowed comparisons of ADC (mm2/sec) and ve at t2. Both the BT474 (A) and MDA-MB-231 (B) tumors revealed no significant correlations between these parameters (p = 0.07, p = 0.93, respectively). As these parameters are believed to provide a measure of comparable biological features, it is reasonable to hypothesize a positive correlation between them. However, the data obtained in these tumor models does not support that hypothesis as no significant correlation was observed.
MDA-MB-231 Tumor Bearing Mice
As was the case in the BT474 tumor-bearing mice, there was no correlation in the voxel-wise analysis of the imaging parameters (Table 2). In Table 2, the Pearson correlation values are shown for each mouse, with an average value (±SE) of 0.002 (±0.028) and a range of −0.14 to 0.14, when analyzing all voxels. Using only those voxels that returned physiological ve values, the Pearson correlation values had a mean value of 0.16 (±0.050) with a range from −0.12 to 0.33. Additionally, there was no significant correlation between the median ADC and ve values for the whole tumor ROI at any of the imaging time points in the MDA-MB-231 tumor-bearing mice. The Pearson correlation coefficient was −0.21 (p = 0.37) at t0, −0.04 (p = 0.87) at t1, and 0.11 (p = 0.68) at t2, as shown in Figure 5 (panels D-F). The mean ADC and ve values, respectively, at each time point for all of the mice were 4.91×10−4 mm2/s and 0.43 at t0, 5.18×10−4 mm2/s and 0.53 at t1, and 5.49×10−4 mm2/s and 0.53 at t2.
Table 2.
Voxel-wise correlation values between ADC and ve for MDA-MB-231 tumors at t0.
| Mouse Number |
Treatment | Correlation Coefficient | |
|---|---|---|---|
| All Voxels | ve≤1a | ||
| 1 | Control | 0.14 | 0.12 |
| 2 | Control | 0.079 | 0.24 |
| 3 | Control | −0.0044 | 0.12 |
| 4 | Control | 0.013 | 0.25 |
| 5 | Control | 0.071 | 0.24 |
| 6 | Control | 0.053 | 0.20 |
| 7 | Control | 0.0029 | 0.19 |
| 8 | 15 mg/kg Abxb | −0.069 | 0.25 |
| 9 | 15 mg/kg Abx | 0.069 | 0.12 |
| 10 | 15 mg/kg Abx | −0.033 | −0.12 |
| 11 | 15 mg/kg Abx | −0.060 | 0.050 |
| 12 | 15 mg/kg Abx | −0.068 | 0.031 |
| 13 | 15 mg/kg Abx | −0.14 | −0.0049 |
| 14 | 15 mg/kg Abx | −0.061 | 0.33 |
| 15 | 25 mg/kg Abx | 0.028 | 0.11 |
| 16 | 25 mg/kg Abx | 0.032 | 0.23 |
| 17 | 25 mg/kg Abx | −0.052 | 0.049 |
| 18 | 25 mg/kg Abx | 0.002 | 0.27 |
| 19 | 25 mg/kg Abx | 0.042 | 0.26 |
| 20 | 25 mg/kg Abx | −0.008 | 0.25 |
The correlation between ADC and ve in this column was calculated using only voxels within the tumor ROI which had a physiological value of ve (i.e. less than or equal to 1).
Abx = Abraxane
When considering the central slice MR data relative to the histological calculation of EC at t2, a significant correlation was shown between ADC and EC (r = 0.58, p < 0.01), as seen in Figure 6B. However, ve did not significantly correlate to EC (r = 0.13, p = 0.64), as shown in Figure 7B. Additionally, there was no significant correlation (r = −0.02, p = 0.93) between ADC and ve in the central slice at t2, as seen in Figure 8B. The group mean values of ADC, ve, and %EC were 5.67×10−4 mm2/s, 0.72, and 19.7%, respectively. As was the case for the BT474 tumors, the number of data points for the comparisons at t1 and t2 is lower than for the comparisons at t0 due to either failed contrast agent injections, which did not allow for quantification of ve, or unusable ADC values.
Combined BT474 and MDA-MB-231 Data Sets
When grouping the tumors from both models together, there was a highly significant correlation between EC and ADC (r = 0.76, p < 0.0001) in the central slice analysis at t2, as seen in Figure 9A. However, ve did not significantly correlate to EC, with a correlation of −0.07 (p = 0.57), as seen in Figure 9B. Additionally, there was no significant correlation between ve and ADC for the central slice (r = −0.16, p = 0.41), as seen in Figure 9C.
Figure 9.
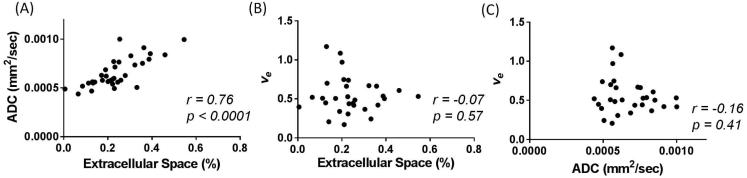
Imaging and histology comparisons of all tumors (n = 35) are shown: (A) EC and ADC, (B) EC and ve, and (C) ADC and ve. ADC demonstrates a strong significant correlation to EC (p < 0.0001), while ve shows no significant correlation with EC (p = 0.57). Additionally, ADC and ve show no correlation with each other (p = 0.41).
Physiological ve Voxels
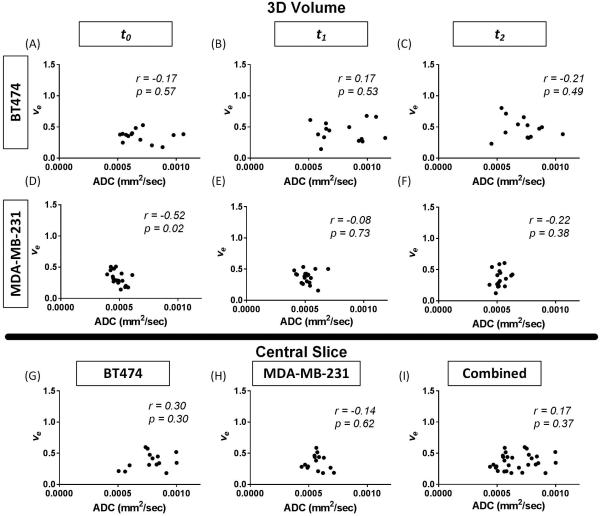
For completeness, the correlations between the imaging-derived parameter values were calculated utilizing only the voxels in which ve is physiologically relevant (i.e., 0.01 ≤ ve ≤ 1, 0 < ADC < 3×10−3 mm2/s). The results are shown in Figure 10. When considering the BT474 tumors, the ROI analysis yielded a correlation value of −0.17 (p = 0.57, panel A) between ADC and ve at t0, 0.17 (p = 0.53, panel B) at t1, and −0.21 (p = 0.49, panel C) at t2. For the MDA-MB-231 tumors, the ROI analysis of the physiologically relevant voxels resulted in a correlation coefficient of −0.52 (p = 0.02, panel D) at t0, −0.08 (p = 0.73, panel E) at t1, and −0.22 (p = 0.38, panel F) at t2.
Figure 10.
Correlations between ADC (mm2/sec) and physiologically relevant ve voxels (0.01 ≤ ve ≤ 1) are shown. 3D quantitative analysis correlation of ADC and ve over the whole tumor at baseline (t0), during treatment (t1), and following treatment (t2) in both the BT474 (panels A-C) and MDA-MB-231 (panels D-F) tumor lines. Central slice analysis is shown in the BT474 (G), MDA-MB-231 (H) and combination (I) cell lines. No consistent correlations are seen in these comparisons.
The central slice analysis of the physiologically relevant voxels is shown in panels G, H, and I of Figure 10. For the BT474 tumors, the correlation between ADC and ve at t2 resulted in a coefficient of 0.30 (p = 0.30, panel G). The MDA-MB-231 tumors had a central slice correlation of −0.14 (p = 0.62). When grouping the tumors from both models together, the correlation between ADC and ve in the center slice is 0.17 (p = 0.37).
Discussion
The use of biomarkers available from quantitative DCE- and DW-MRI to assess tumors and their response to therapy has shown considerable potential (21,33-39). As the Institute of Medicine notes, assessment of the performance of any biomarker requires specific measurements in a controlled setting which results in direct relation to an outcome (40). Thus, successful application of DCE- and DW-MRI derived parameters demands that they be properly qualified. Given their definitions and common physiological interpretations, proper qualification of ADC and ve requires that they correlate with each other, as well as with independent (gold-standard) assessments of cell density (12,41).
ADC is a reflection of how freely water can diffuse, so in areas of increased cellular boundaries (i.e., low EC), a decrease in ADC is anticipated. And thus, reciprocally, when there is a decrease in cell density, ADC should increase. The present study supports that hypothesis as ADC has a statistically significant positive correlation with the amount of extracellular space as measured by the gold-standard of H&E. These trends were seen in both the BT474 and MDA-MB-231 models (Figure 6). Furthermore, when the data from the two models are grouped together, a highly significant correlation is observed (Figure 9A). This represents a robust method for analyzing extracellular space in breast cancer pre-clinical data, which provides support for its clinical application. Similarly, an increase in extracellular space should, by definition, yield an increase in the DCE-MRI parameter ve. However, our data indicates a complicated relationship between EC and ve (as seen in Figure 7 and 9B). In the BT474 tumors, a significant relationship was calculated between EC and ve, while in the MDA-MB-231 tumors, and in the combined group, there was not a significant correlation between the two parameters. Thus, while both ve and ADC should correlate with cell density, it seems that there are other factors that influence these measurements in practice, particularly in regard to ve. While ADC has been shown to be highly sensitive to cellular changes during treatment (16,42), ve has revealed mixed results while assessing the differences in extravascular, extracellular space in various cancers (5,18,43,44). Utilizing two different breast cancer models, with identical approaches for data acquisition and analysis, there were differences in the correlation between ve and EC. This provides further evidence that ve should not universally be applied as a biomarker of cellularity; however, it may still explain some aspect of cellularity within the tumor (as revealed with a BT474 model showing a significant correlation between ve and EC in the central slice analysis). Since the DW-MRI and DCE-MRI image sets were inherently registered, we were also able to examine if correlation existed for individual animals on a voxel level between ADC and ve at the pretreatment time point. No significant correlations were observed on a voxel level between the image-derived parameters for either cell line. This lack of correlation between ADC and ve before the presence of any potential treatment-initiated changes further suggests an inherent difference in sensitivity of these two measures of cellularity to other factors in the context of the tumor model presented here. Finally, we also evaluated the correlation between the imaging parameters (ADC and ve) for only the physiologically relevant voxels (i.e., ve ≤ 1) for both the central slice and the ROI. The results highlight the lack of a consistent relationship between the parameters in both tumor lines.
The lack of a universal correlation between ve and histology could potentially be attributed to various aspects of the modeling approach commonly used in DCE-MRI. For example, analysis of DCE-MRI data with the Tofts models (i.e., Eq. [5] and its common extensions) often results in values of ve that are misestimated when compared to histological assessment (43,44). One explanation for this phenomenon is that the standard Tofts model accounts only for active delivery of the contrast agent (via Ktrans), but does not account for passive delivery of the contrast agent through diffusion. Specifically, consider the signal intensity time curve from the two voxels highlighted in Figure 4. Figure 4A shows a voxel with significant influx and efflux (Ktrans = 0.22 min−1, ve = 0.29), while Figure 4B presents a voxel that has an extremely high (and unphysiological) ve. The ve value shown in Figure 4B is 2.66 and there is a notable absence of the typical rapid uptake peak. The gradual uptake of the contrast agent (Ktrans = 0.024 min−1) suggests that it may be diffusing into the tumor from surrounding tissue, which the Tofts model does not explicitly consider. For this study, spatially dependent diffusion was not incorporated into the two compartment model, but others have incorporated this component into the quantification of contrast enhancement and have shown that diffusion has an effect on parameterization utilizing the standard model (45-48). The data presented here indicates that building and validating a model incorporating the effects of contrast agent diffusion should be a central goal of the DCE-MRI community. Regardless of the underlying mechanism, the slow, persistent uptake cannot be accurately described by the Tofts model, which assumes that ve reflects the volume of the extravascular, extracellular space into which the agent leaks. Since the assignment of ve is dependent on delivery to the voxel, regions that are not well perfused (or necrotic) misrepresent the extravascular, extracellular volume invoked in the model. Additionally, the representative neighboring voxels (location visualized in Figure 4E) reveal a high variability of contrast uptake between voxels that are within the same region of the tumor as shown by the signal intensity curves in the DCE-MRI data; however, the signal intensity from the DW-MRI data for these voxels reveal similar quality and trends (Figure 4C, 4D). That is, quantitative DCE-MRI analysis is based on an indirect method of quantifying the pharmacokinetics of a contrast agent in tissue; thus, physiology that affects the flow and/or distribution of the contrast agent (e.g., passive diffusion or active delivery of the contrast agent) will inherently confound the ve measurement. However, the ADC measurement is not as readily affected by changes in perfusion, especially at higher b values (as utilized in this particular DW-MRI acquisition). This distinction is further emphasized by the voxel-wise correlation between ADC and ve shown in Table 1. Specifically, in the MDA-MB-231 tumors, there is a poor correlation between the two parameters when all voxels are considered (first column). However, it is interesting to note that in most mice, the correlation coefficient increases when only the physiologically relevant voxel values of ve are included (second column). This provides further support for the argument that the standard pharmacokinetic modeling approaches are failing in the DCE-MRI analysis for a fraction of the voxels, thus resulting in ve values which do not accurately quantify cellularity in certain regions.
Another potential source of error in the ve measurement is the arterial input function (AIF). In this work, a population AIF was utilized, which, in previous efforts, was shown to result in parameters that correlated well to those obtained using an individual AIF (32). Although the population AIF is a reasonable approach for DCE-MRI analysis, and returns appropriate values of ve in contralateral muscle tissue, the potential for parameterization error is still present on the individual basis.
Given the preclinical results, the correlation of ADC and EC should be tested clinically in a variety of breast cancer patients to confirm ADC as a qualified biomarker for measuring overall patient response and prognosis. Matsubayashi et al. has shown that ADC values correlated to stroma features in invasive ductal breast carcinomas patients (49). Additionally, it was demonstrated that cellular density is inversely correlated to ADC values (49), which substantiates our results exhibiting a positive correlation between EC and ADC. Another group also demonstrated a correlation between cellular fraction and ADC, however it was only significant in the slow diffusion component of their model (50). ADC can also be shown to potentially predict the histological grade or aggressiveness of small hepatocellular carcinomas (51) and breast cancer (52). Pathologic validation and relationship with patient outcome is necessary to move forward with this quantitative parameter to monitor treatment response in the clinical setting.
Conclusions
This study suggests that ADC is influenced by cellular density. However, the lack of correlation between ADC and ve and the misestimation of ve compared to histology suggests that ve (as estimated from the standard two compartment model) is not a reliable biomarker of the extravascular, extracellular space. Conversely, the ADC appears to be a reliable method for noninvasively evaluating extracellular space in pre-clinical breast cancer models.
Acknowledgements
We thank the National Institutes of Health through funding mechanisms NCI R01 CA138599, NCI P50 CA098131, NCI P30 CA68485, NCI R25 CA092043, NCI 5T32 CA093240 and NCI U01 CA174706. We thank the Kleberg Foundation for the generous support of our imaging program.
Abbreviations
- ADC
apparent diffusion coefficient
- AIF
arterial input function
- DW-MRI
diffusion-weighted magnetic resonance imaging
- DCE-MRI
dynamic contrast-enhanced magnetic resonance imaging
- EC
extracellular space
- FOV
field of view
- HER2+
human epidermal growth factor receptor 2 positive
- H&E
hematoxylin and eosin
- FXL
fast exchange limit
- NEX
number of excitations
- ROI
region of interest
References
- 1.Yankeelov TE, Gore JC. Dynamic contrast enhanced magnetic resonance imaging in oncology: theory, data acquisition, analysis, and examples. Current Medical Imaging Reviews. 2007;3:91–107. doi: 10.2174/157340507780619179. 10.2174/157340507780619179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaustad JV, Pozdniakova V, Hompland T, Simonsen TG, Rofstad EK. Magnetic resonance imaging identifies early effects of sunitinib treatment in human melanoma xenografts. Journal of Experimental and Clinical Cancer Research. 2013;32:93. doi: 10.1186/1756-9966-32-93. 10.1186/1756-9966-32-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ovrebo KM, Gulliksrud K, Mathiesen B, Rofstad EK. Assessment of tumor radioresponsiveness and metastatic potential by dynamic contrast-enhanced magnetic resonance imaging. International Journal of Radiation Oncology Biology Physics. 2011;81:255–261. doi: 10.1016/j.ijrobp.2011.04.008. 10.1016/j.ijrobp.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 4.Roe K, Kakar M, Seierstad T, Ree AH, Olsen DR. Early prediction of response to radiotherapy and androgen-deprivation therapy in prostate cancer by repeated functional MRI: a preclinical study. Radiation oncology. 2011;6:65. doi: 10.1186/1748-717X-6-65. 10.1186/1748-717X-6-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whisenant J, Sorace A, McIntyre J, Hakmook K, Sanchez V, Loveless M, Yankeelov T. Evaluating treatment response using DW-MRI and DCE-MRI in trastuzumab responsive and resistant HER2-overexpressing human breast cancer xenografts. Translational Oncology. 2014;7:768–779. doi: 10.1016/j.tranon.2014.09.011. 10.1016/j.tranon.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang Y-C, Yu C-J, Chen C-M, Hu F-C, Hsu H-H, Tseng W-YI, Ting-Fang Shih T, Yang P-C, Chih-Hsin Yang J. Dynamic contrast-enhanced MRI in advanced nonsmall-cell lung cancer patients treated with first-line bevacizumab, gemcitabine, and cisplatin. Journal of Magnetic Resonance Imaging. 2012;36:387–396. doi: 10.1002/jmri.23660. 10.1002/jmri.23660. [DOI] [PubMed] [Google Scholar]
- 7.Guo J, Reddick WE, Glass JO, Ji Q, Billups CA, Wu J, Hoffer FA, Kaste SC, Jenkins JJ, Ortega Flores XC, Quintana J, Villarroel M, Daw NC. Dynamic contrast-enhanced magnetic resonance imaging as a prognostic factor in predicting event-free and overall survival in pediatric patients with osteosarcoma. Cancer. 2012;118:3776–3785. doi: 10.1002/cncr.26701. 10.1002/cncr.26701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chikui T, Kitamoto E, Kawano S, Sugiura T, Obara M, Simonetti AW, Hatakenaka M, Matsuo Y, Koga S, Ohga M, Nakamura K, Yoshiura K. Pharmacokinetic analysis based on dynamic contrast-enhanced MRI for evaluating tumor response to preoperative therapy for oral cancer. Journal of Magnetic Resonance Imaging. 2012;36:589–597. doi: 10.1002/jmri.23704. 10.1002/jmri.23704. [DOI] [PubMed] [Google Scholar]
- 9.Park JJ, Kim CK, Park SY, Simonetti AW, Kim E, Park BK, Huh SJ. Assessment of early response to concurrent chemoradiotherapy in cervical cancer: value of diffusion-weighted and dynamic contrast-enhanced MR imaging. Magnetic Resonance Imaging. 2014;32:993–1000. doi: 10.1016/j.mri.2014.05.009. 10.1016/j.mri.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Li X, Abramson RG, Arlinghaus LR, Hakmook K, Chakravarthy AB, Abramson VG, Farley J, Mayer IA, Kelley MC, Meszoely IM, Means-Powell J, Grau AM, Sanders M, Yankeelov TE. Multiparametric magnetic resonance imaging for predicting pathological response after the first cycle of neoadjuvant chemotherapy in breast cancer. Investigative Radiology. 2015;50:195–204. doi: 10.1097/RLI.0000000000000100. 10.1097/RLI.0000000000000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Einstein A. Über einen die Erzeugung und Verwandlung des Lichtes betreffenden heuristischen Gesichtspunkt. Annalen der Physik. 1905;322:132–148. 10.1002/andp.19053220607. [Google Scholar]
- 12.Padhani AR, Liu G, Koh DM, Chenevert TL, Thoeny HC, Takahara T, Dzik-Jurasz A, Ross BD, Van Cauteren M, Collins D, Hammoud DA, Rustin GJ, Taouli B, Choyke PL. Diffusion-weighted magnetic resonance imaging as a cancer biomarker: consensus and recommendations. Neoplasia. 2009;11:102–125. doi: 10.1593/neo.81328. 10.1593/neo.81328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stejskal EO, Tanner JE. Spin diffusion measurements: spin echoes in the presence of a time dependent field gradient. Journal of Chemical Physics. 1965;42:288–292. 10.1063/1.1695690. [Google Scholar]
- 14.Le Bihan D, Breton E, Lallemand D, Aubin ML, Vignaud J, Laval-Jeantet M. Separation of diffusion and perfusion in intravoxel incoherent motion MR imaging. Radiology. 1988;168:497–505. doi: 10.1148/radiology.168.2.3393671. [DOI] [PubMed] [Google Scholar]
- 15.Chenevert TL, Brunberg JA, Pipe JG. Anisotropic diffusion in human white matter: demonstration with MR techniques in vivo. Radiology. 1990;177:401–405. doi: 10.1148/radiology.177.2.2217776. [DOI] [PubMed] [Google Scholar]
- 16.Patterson DM, Padhani AR, Collins DJ. Technology insight: water diffusion MRI--a potential new biomarker of response to cancer therapy. Nature Clinical Practice Oncology. 2008;5:220–233. doi: 10.1038/ncponc1073. 10.1038/ncponc1073. [DOI] [PubMed] [Google Scholar]
- 17.Guo Y, Cai YQ, Cai ZL, Gao YG, An NY, Ma L, Mahankali S, Gao JH. Differentiation of clinically benign and malignant breast lesions using diffusion-weighted imaging. Journal of Magnetic Resonance Imaging. 2002;16:172–178. doi: 10.1002/jmri.10140. 10.1002/jmri.10140. [DOI] [PubMed] [Google Scholar]
- 18.Aryal MP, Nagaraja TN, Keenan KA, Bagher-Ebadian H, Panda S, Brown SL, Cabral G, Fenstermacher JD, Ewing JR. Dynamic contrast enhanced MRI parameters and tumor cellularity in a rat model of cerebral glioma at 7 T. Magnetic Resonance in Medicine. 2014;71:2206–2214. doi: 10.1002/mrm.24873. 10.1002/mrm.24873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moestue SA, Huuse EM, Lindholm EM, Bofin A, Engebraaten O, Maelandsmo GM, Akslen LA, Gribbestad IS. Low-molecular contrast agent dynamic contrast-enhanced (DCE)-MRI and diffusion-weighted (DW)-MRI in early assessment of bevacizumab treatment in breast cancer xenografts. Journal of Magnetic Resonance Imaging. 2013;38:1043–1053. doi: 10.1002/jmri.24079. 10.1002/jmri.24079. [DOI] [PubMed] [Google Scholar]
- 20.Graham TJ, Box G, Tunariu N, Crespo M, Spinks TJ, Miranda S, Attard G, de Bono J, Eccles SA, Davies FE, Robinson SP. Preclinical evaluation of imaging biomarkers for prostate cancer bone metastasis and response to cabozantinib. Journal of the National Cancer Institute. 2014;106:dju033. doi: 10.1093/jnci/dju033. 10.1093/jnci/dju033. [DOI] [PubMed] [Google Scholar]
- 21.Sahani DV, Jiang T, Hayano K, Duda DG, Catalano OA, Ancukiewicz M, Jain RK, Zhu AX. Magnetic resonance imaging biomarkers in hepatocellular carcinoma: association with response and circulating biomarkers after sunitinib therapy. Journal of Hematology and Oncology. 2013;6:51. doi: 10.1186/1756-8722-6-51. 10.1186/1756-8722-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pickles M, Lowry M, Manton D, Gibbs P, Turnbull L. Role of dynamic contrast enhanced MRI in monitoring early response of locally advanced breast cancer to neoadjuvant chemotherapy. Breast Cancer Research and Treatment. 2005;91:1–10. doi: 10.1007/s10549-004-5819-2. 10.1007/s10549-004-5819-2. [DOI] [PubMed] [Google Scholar]
- 23.Sinha S, Lucas-Quesada FA, Sinha U, DeBruhl N, Bassett LW. In vivo diffusion-weighted MRI of the breast: potential for lesion characterization. Journal of Magnetic Resonance Imaging. 2002;15:693–704. doi: 10.1002/jmri.10116. 10.1002/jmri.10116. [DOI] [PubMed] [Google Scholar]
- 24.Li X, Kang H, Arlinghaus LR, Abramson RG, Chakravarthy AB, Abramson VG, Farley J, Sanders M, Yankeelov TE. Analyzing spatial heterogeneity in DCE- and DW-MRI parametric maps to optimize prediction of pathologic response to neoadjuvant chemotherapy in breast cancer. Translational Oncology. 2014;7:14–22. doi: 10.1593/tlo.13748. 10.1593/tlo.13748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reischauer C, Froehlich JM, Pless M, Binkert CA, Koh DM, Gutzeit A. Early treatment response in non-small cell lung cancer patients using diffusion-weighted imaging and functional diffusion maps - a feasibility study. PloS one. 2014;9:e108052. doi: 10.1371/journal.pone.0108052. 10.1371/journal.pone.0108052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wedam SB, Low JA, Yang SX, Chow CK, Choyke P, Danforth D, Hewitt SM, Berman A, Steinberg SM, Liewehr DJ, Plehn J, Doshi A, Thomasson D, McCarthy N, Koeppen H, Sherman M, Zujewski J, Camphausen K, Chen H, Swain SM. Antiangiogenic and antitumor effects of bevacizumab in patients with inflammatory and locally advanced breast cancer. Journal of Clinical Oncology. 2006;24:769–777. doi: 10.1200/JCO.2005.03.4645. [DOI] [PubMed] [Google Scholar]
- 27.Mills SJ, Soh C, Rose CJ, Cheung S, Zhao S, Parker GJ, Jackson A. Candidate biomarkers of extravascular extracellular space: a direct comparison of apparent diffusion coefficient and dynamic contrast-enhanced MR imaging--derived measurement of the volume of the extravascular extracellular space in glioblastoma multiforme. American Journal of Neuroradiology. 2010;31:549–553. doi: 10.3174/ajnr.A1844. 10.3174/ajnr.A1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arlinghaus LR, Li X, Rahman AR, Welch EB, Xu L, Gore JC, Yankeelov TE. On the relationship between the apparent diffusion coefficient and extravascular extracellular volume fraction in human breast cancer. Magnetic Resonance Imaging. 2011;29:630–638. doi: 10.1016/j.mri.2011.02.004. 10.1016/j.mri.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson AW, Gore JC. Analysis and correction of motion artifacts in diffusion weighted imaging. Magnetic Resonance in Medicine. 1994;32:379–387. doi: 10.1002/mrm.1910320313. 10.1002/mrm.1910320313. [DOI] [PubMed] [Google Scholar]
- 30.Mills R. Self-diffusion in normal and heavy water in the range 1-45 degrees. The Journal of Physical Chemistry. 1973;77:685–688. 10.1021/j100624a025. [Google Scholar]
- 31.Landis CS, Li X, Telang FW, Coderre JA, Micca PL, Rooney WD, Latour LL, Vetek G, Palyka I, Springer CS., Jr Determination of the MRI contrast agent concentration time course in vivo following bolus injection: effect of equilibrium transcytolemmal water exchange. Magnetic Resonance in Medicine. 2000;44:563–574. doi: 10.1002/1522-2594(200010)44:4<563::aid-mrm10>3.0.co;2-#. 10.1002/1522-2594(200010)44:4<563::AID-MRM10>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 32.Loveless ME, Halliday J, Liess C, Xu L, Dortch RD, Whisenant J, Waterton JC, Gore JC, Yankeelov TE. A quantitative comparison of the influence of individual versus population-derived vascular input functions on dynamic contrast enhanced-MRI in small animals. Magnetic Resonance in Medicine. 2012;67:226–236. doi: 10.1002/mrm.22988. 10.1002/mrm.22988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abramson RG, Arlinghaus LR, Weis JA, Li X, Dula AN, Chekmenev EY, Smith SA, Miga MI, Abramson VG, Yankeelov TE. Current and emerging quantitative magnetic resonance imaging methods for assessing and predicting the response of breast cancer to neoadjuvant therapy. Breast cancer : targets and therapy. 2012;2012:139–154. doi: 10.2147/BCTT.S35882. 10.2147/BCTT.S35882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yankeelov TE, Abramson RG, Quarles CC. Quantitative multimodality imaging in cancer research and therapy. Nature Reviews Clinical Oncology. 2014 doi: 10.1038/nrclinonc.2014.134. 10.1038/nrclinonc.2014.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chung MP, Margolis D, Mesko S, Wang J, Kupelian P, Kamrava M. Correlation of quantitative diffusion-weighted and dynamic contrast-enhanced MRI parameters with prognostic factors in prostate cancer. Journal of medical imaging and radiation oncology. 2014 doi: 10.1111/1754-9485.12230. 10.1111/1754-9485.12230. [DOI] [PubMed] [Google Scholar]
- 36.Punwani S, Prakash V, Bainbridge A, Taylor SA, Bandula S, Olsen OE, Hain SF, Shankar A, Daw S, Humphries P. Quantitative diffusion weighted MRI: a functional biomarker of nodal disease in Hodgkin lymphoma? Cancer Biomarkers. 2010;7:249–259. doi: 10.3233/CBM-2010-0197. 10.3233/CBM-2010-0197. [DOI] [PubMed] [Google Scholar]
- 37.Hayashida Y, Yakushiji T, Awai K, Katahira K, Nakayama Y, Shimomura O, Kitajima M, Hirai T, Yamashita Y, Mizuta H. Monitoring therapeutic responses of primary bone tumors by diffusion-weighted image: Initial results. European radiology. 2006;16:2637–2643. doi: 10.1007/s00330-006-0342-y. 10.1007/s00330-006-0342-y. [DOI] [PubMed] [Google Scholar]
- 38.Xu J, Li K, Smith RA, Waterton JC, Zhao P, Chen H, Does MD, Manning HC, Gore JC. Characterizing tumor response to chemotherapy at various length scales using temporal diffusion spectroscopy. PloS one. 2012;7:e41714. doi: 10.1371/journal.pone.0041714. 10.1371/journal.pone.0041714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dudeck O, Zeile M, Pink D, Pech M, Tunn PU, Reichardt P, Ludwig WD, Hamm B. Diffusion-weighted magnetic resonance imaging allows monitoring of anticancer treatment effects in patients with soft-tissue sarcomas. Journal of Magnetic Resonance Imaging. 2008;27:1109–1113. doi: 10.1002/jmri.21358. 10.1002/jmri.21358. [DOI] [PubMed] [Google Scholar]
- 40.Institute of Medicine (U.S.). Forum on Drug Discovery Development and Translation. Olson S, Robinson S, Giffin RB. Accelerating the development of biomarkers for drug safety: workshop summary. National Academies Press; Washington, D.C.: 2009. p. 83. [Google Scholar]
- 41.Vaupel P, Harrison L. Tumor hypoxia: causative factors, compensatory mechanisms, and cellular response. Oncologist. 2004;9(Suppl 5):4–9. doi: 10.1634/theoncologist.9-90005-4. 10.1634/theoncologist.9-90005-4. [DOI] [PubMed] [Google Scholar]
- 42.Thoeny HC, De Keyzer F. Extracranial applications of diffusion-weighted magnetic resonance imaging. European Radiology. 2007;17:1385–1393. doi: 10.1007/s00330-006-0547-0. 10.1007/s00330-006-0547-0. [DOI] [PubMed] [Google Scholar]
- 43.Yao WW, Zhang H, Ding B, Fu T, Jia H, Pang L, Song L, Xu W, Song Q, Chen K, Pan Z. Rectal cancer: 3D dynamic contrast-enhanced MRI; correlation with microvascular density and clinicopathological features. Radiologia Medica. 2011;116:366–374. doi: 10.1007/s11547-011-0628-2. 10.1007/s11547-011-0628-2. [DOI] [PubMed] [Google Scholar]
- 44.Wu L, Lv P, Zhang H, Fu C, Yao X, Wang C, Zeng M, Li Y, Wang X. Dynamic contrast-enhanced (DCE) MRI assessment of microvascular characteristics in the murine orthotopic pancreatic cancer model. Magnetic Resonance Imaging. 2015;33:737–760. doi: 10.1016/j.mri.2014.08.014. 10.1016/j.mri.2014.08.014. [DOI] [PubMed] [Google Scholar]
- 45.Barnes SL, Quarles CC, Yankeelov TE. Modeling the effect of intra-voxel diffusion of contrast agent on the quantitative analysis of dynamic contrast enhanced magnetic resonance imaging. PloS one. 2014;9 doi: 10.1371/journal.pone.0108726. 0.1371/journal.pone.0108726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fluckiger JU, Loveless ME, Barnes SL, Lepage M, Yankeelov TE. A diffusion-compensated model for the analysis of DCE-MRI data: theory, simulations and experimental results. Phys Med Biol. 2013;58:1983–1998. doi: 10.1088/0031-9155/58/6/1983. 10.1088/0031-9155/58/6/1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pellerin M, Yankeelov TE, Lepage M. Incorporating contrast agent diffusion into the analysis of DCE-MRI data. Magnetic Resonance in Medicine. 2007;58:1124–1134. doi: 10.1002/mrm.21400. 10.1002/mrm.21400. [DOI] [PubMed] [Google Scholar]
- 48.Jia G, O'Dell C, Heverhagen JT, Yang X, Liang J, Jacko RV, Sammet S, Pellas T, Cole P, Knopp MV. Colorectal liver metastases: contrast agent diffusion coefficient for quantification of contrast enhancement heterogeneity at MR imaging. Radiology. 2008;248:901–909. doi: 10.1148/radiol.2491071936. 10.1148/radiol.2491071936. [DOI] [PubMed] [Google Scholar]
- 49.Matsubayashi RN, Fujii T, Yasumori K, Muranaka T, Momosaki S. Apparent diffusion coefficient in invasive ductal breast carcinoma: correlation with detailed histologic features and the enhancement ratio on dynamic contrast-enhanced MR images. Journal of oncology. 2010:2010. doi: 10.1155/2010/821048. 10.1155/2010/821048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamura T, Usui S, Murakami S, Arihiro K, Fujimoto T, Yamada T, Naito K, Akiyama M. Comparisons of multi b-value DWI signal analysis with pathological specimen of breast cancer. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2012;68:890–897. doi: 10.1002/mrm.23277. 10.1002/mrm.23277. [DOI] [PubMed] [Google Scholar]
- 51.Le Moigne F, Boussel L, Haquin A, Bancel B, Ducerf C, Berthezene Y, Rode A. Grading of small hepatocellular carcinomas (</=2 cm): correlation between histology, T2 and diffusion-weighted imaging. The British journal of radiology. 2014;87:20130763. doi: 10.1259/bjr.20130763. 10.1259/bjr.20130763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Costantini M, Belli P, Rinaldi P, Bufi E, Giardina G, Franceschini G, Petrone G, Bonomo L. Diffusion-weighted imaging in breast cancer: relationship between apparent diffusion coefficient and tumour aggressiveness. Clinical radiology. 2010;65:1005–1012. doi: 10.1016/j.crad.2010.07.008. 10.1016/j.crad.2010.07.008. [DOI] [PubMed] [Google Scholar]