Abstract
Background
Aspergillus niger is widely used for enzyme production and achievement of high enzyme production depends on the comprehensive understanding of cell’s metabolic regulation mechanisms.
Results
In this paper, we investigate the metabolic differences and regulation mechanisms between a high glucoamylase-producing strain A. niger DS03043 and its wild-type parent strain A. niger CBS513.88 via an integrated isotope-assisted metabolomics and 13C metabolic flux analysis approach. We found that A. niger DS03043 had higher cell growth, glucose uptake, and glucoamylase production rates but lower oxalic acid and citric acid secretion rates. In response to above phenotype changes, A. niger DS03043 was characterized by an increased carbon flux directed to the oxidative pentose phosphate pathway in contrast to reduced flux through TCA cycle, which were confirmed by consistent changes in pool sizes of metabolites. A higher ratio of ATP over AMP in the high producing strain might contribute to the increase in the PP pathway flux as glucosephosphate isomerase was inhibited at higher ATP concentrations. A. niger CBS513.88, however, was in a higher redox state due to the imbalance of NADH regeneration and consumption, resulting in the secretion of oxalic acid and citric acid, as well as the accumulation of intracellular OAA and PEP, which may in turn result in the decrease in the glucose uptake rate.
Conclusions
The application of integrated metabolomics and 13C metabolic flux analysis highlights the regulation mechanisms of energy and redox metabolism on flux redistribution in A. niger.
Graphical abstract.
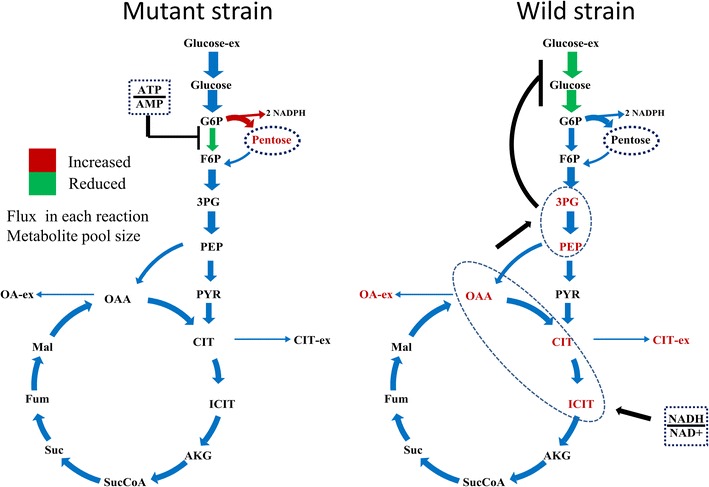
An integrated isotope-assisted metabolomics and 13C metabolic flux analysis was was firstly systematically performed in A. niger. In response to enzyme production, the metabolic flux in A. niger DS03043 (high-producing) was redistributed, characterized by an increased carbon flux directed to the oxidative pentose phosphate pathway as well as an increased pool size of pentose. The consistency in 13C metabolic flux analysis and metabolites quantification indicated that an imbalance of NADH formation and consumption led to the accumulation and secretion of organic acids in A. niger CBS513.88 (wild-type)
Electronic supplementary material
The online version of this article (doi:10.1186/s12934-015-0329-y) contains supplementary material, which is available to authorized users.
Keywords: Aspergillus niger, Glucoamylase, 13C metabolic flux analysis, Metabolomics, Cofactor metabolism
Background
Aspergillus niger, one of the most important filamentous fungi, has been widely used for the production of organic acids [1–3], such as citric and gluconic acid, and for the production of enzymes [4, 5] including glucoamylase, proteases, cellulase and glucose oxidase due to its excellent capacity in expression and secretion of proteins.
Despite the wide usage of A. niger for enzyme production, metabolic regulation mechanisms for high enzyme production by A. niger are still largely unclear. The complexity in metabolic regulation for high enzyme production calls for the introduction of multi-omics analysis. Metabolomics and fluxomics analysis, as an important complement to the commonly used genomics, transcriptomics and proteomics analysis, have undergone a quick development during the past years [6, 7]. With the aid of metabolomics analysis, the bottleneck for the target product synthesis can be identified [8], thus the addition of the pivotal limiting precursors can largely improve the strain performance. In the perturbed experiments, the profiles in dynamic changes of metabolites pool sizes can be used to assess the kinetic information of intracellular enzymes and finally estimate the limiting steps for the cell metabolism [9, 10]. Furthermore, the metabolomics play an important role in revealing the effects of energy metabolism and accumulation of polysaccharides on the strain physiology [11].
13C metabolic flux analysis has become a powerful tool to study the strain metabolic properties due to its capability of providing precise in vivo flux data [12–14]. For fructofuranosidase production by A. niger, Driouch et al. [15] reported that the production of recombinant enzyme led to a flux redistribution and in the recombinant strain the cytosolic pentose phosphate pathway (PPP) and mitochondrial malic enzyme was activated, indicating that the supply of NADPH is essential for efficient production of this protein. Klein et al. [16] observed that there is a positive relation between the lipid content and protein secretion by Schizosaccharomyces pombe. The latest 13C metabolic flux analysis also revealed that the imbalance in regeneration and utilization of cofactors result in the secretion of by-products [17].
To comprehensively understand the metabolic regulation mechanism for high glucoamylase production by A. niger, an integrated isotope-assisted metabolomics and 13C metabolic flux analysis was performed in this work. The integrative analysis of 13C MFA and metabolomics made then possible to identify the metabolic regulation mechanisms for high glucoamylase production by A. niger from the clear distinctions between the two strains in energy and redox metabolism.
Results
Physiology of A. niger DS03043 and CBS513.88
The two strains showed obviously different physiological characteristics (Fig. 1; Table 1). Consistent with the copy number of glucoamylase glaA gene, the specific glucoamylase production rate in A. niger DS03043 was about 8 times of that in A. niger CBS513.88. Whereas, the formation rate of oxalic and citric acid, as two main by-products, decreased significantly in A. niger DS03043. Though the specific growth and glucose uptake rate of A. niger DS03043 was about 40 and 25 % higher than those of A. niger CBS513.88, respectively, the two strains showed no statistically significant differences in the yields of biomass (YX/S) and CO2 (YCO2/S), initially indicating that mainly the reduction in by-products formation was compensated for the large increase in glucoamylase production by A. niger DS03043.
Fig. 1.

Physiological profiles of A. niger DS03043 (red line) and CBS513.88 (black line) in batch cultivations. The parameters including OUR (a), CER (b), DO (c), glucose concentration (d), biomass (e), enzyme activity (f), oxalate (g) and citrate (h) for. All parameters were measured based on at least triplicate measurements. The suddenly increased glucose concentration in d represent the pulsed addition of glucose during exponential phase
Table 1.
Kinetic parameters and carbon recovery for A. niger DS03043 and CBS513.88
| A. niger DS03043 | A. niger CBS513.88 | |
|---|---|---|
| A | ||
| μ (h−1) | 0.14 ± 0.01 | 0.10 ± 0.01 |
| qS (mmol/gDCW h) | 1.83 ± 0.01 | 1.46 ± 0.01 |
| qGA (μmol/gDCW h)a | 0.40 ± 0.03 | 0.05 ± 0.00 |
| qCIT (mmol/gDCW h) | 0.02 ± 0.00 | 0.11 ± 0.01 |
| qOA (mmol/gDCW h) | 0.11 ± 0.01 | 0.35 ± 0.02 |
| qO2 (mmol/gDCW h) | 2.94 ± 0.03 | 2.67 ± 0.02 |
| qCO2 (mmol/gDCW h) | 3.21 ± 0.02 | 2.79 ± 0.05 |
| B | ||
| YX/S (mmol/mmol) | 0.49 ± 0.03 | 0.47 ± 0.02 |
| YGA/S (mmol/mmol) | 0.12 ± 0.01 | 0.02 ± 0.00 |
| YCIT/S (mmol/mmol) | 0.01 ± 0.00 | 0.07 ± 0.01 |
| YOA/S (mmol/mmol) | 0.02 ± 0.00 | 0.07 ± 0.01 |
| YCO2/S (mmol/mmol) | 0.30 ± 0.02 | 0.32 ± 0.03 |
| Carbon recovery | 94 % | 95 % |
All parameters were measured based on at least triplicate measurements. The closeness to 100 % of carbon recovery indicates that almost all carbon compounds produced have been considered
aqGA: Specific production rate of glucoamylase. For the calculation of qGA, 1 mg glucoamylase equals to 50 units of enzyme activity
Estimation of intracellular metabolic flux and metabolites pool sizes
The achievement of metabolic and isotopic pseudo steady-state is a pre-requisite for stationary 13C metabolic flux analysis. During batch cultivations, the specific growth rate and yield coefficients were approximately similar (data not shown), indicating that the strains were in metabolic steady-state. The labelling information for most of the free amino acids had reached the isotopic steady-state at 3 h after the labeled carbon was added into the broth (Fig. 2). Based on the kinetic parameters (Table 1) and the labelling information of free amino acids (Additional file 1), the fluxes were estimated using INCA software [18]. The best-fit flux distribution is shown in Fig. 3. The best-fit fluxes and confidence intervals are presented in Additional file 2. The obtained minimal weighted residual from the parameter estimations is below the cut-off value determined form the χ2 at a 95 % confidence level (Additional file 2), indicating that the estimated fluxes are statistically acceptable. The measured and estimated labelling information of amino acids fragments for optimal flux distributions is listed in Additional file 1.
Fig. 2.
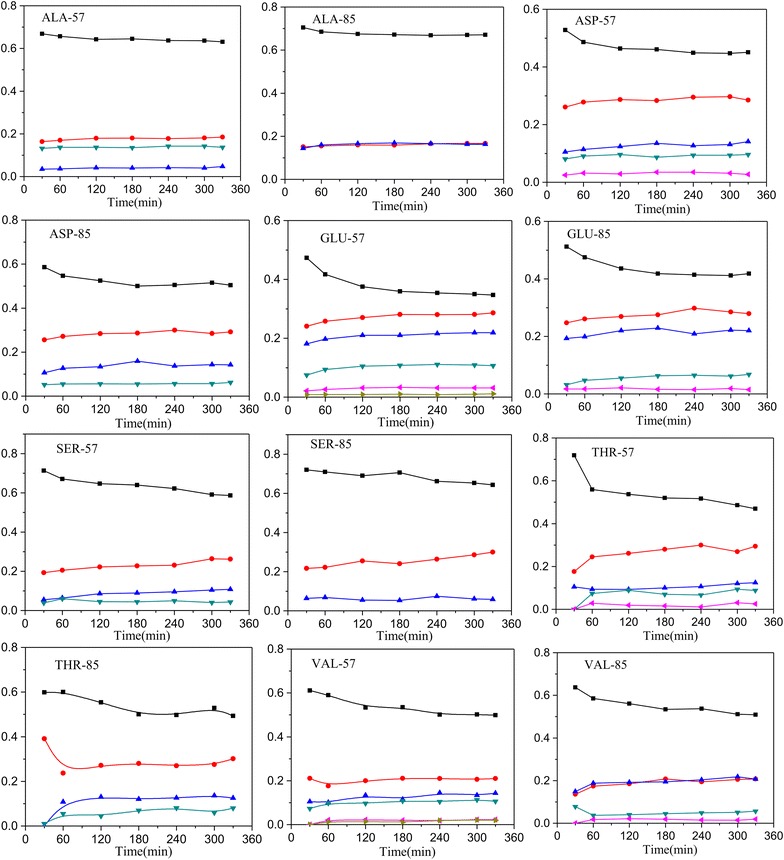
Profiles of enrichment for amino acid fragments in A. niger CBS513.88. The amino acid fragments including ALA-57, ALA-85, ASP-57, ASP-85, GLU-57, GLU-85, SER-57, SER-85, THR-57, THR-85, VAL-57 and VAL-85 after the labeled carbon source was fed into the broth
Fig. 3.

Relative flux (mmol/100 mmol glucose) of A. niger DS03043 (upper) and CBS513.88 (lower). The 68 and 95 % confidence intervals for each reaction and exchange flux values for reversible reactions can be found in Additional file 2. The detailed metabolic model of A. niger and the abbreviation of metabolites could be found in Additional file 4
For both strains, the pool sizes of central carbon metabolites, amino acids and cofactors were determined by isotope-assisted LC–MS and EnzyFluo™ Assay Kit (Fig. 4; Tables 2, 3). In the following, an integrated isotope-assisted metabolomics and 13C metabolic flux analysis of central metabolism of A. niger was conducted to comprehensively understand the metabolic regulation mechanisms for high glucoamylase production by A. niger.
Fig. 4.

The distribution of metabolites pools between A. niger DS03043 (left column) and CBS513.88 (right column). The bar graphs with black frames indicate the significant difference (P value <0.05) in pool size of metabolites for the two strains. All metabolite pool sizes were measured in at least triplicate measurements. The P values were obtained from two-tailed T test statistical analyses. The upper and lower numbers represent the absolute flux values (mmol/gDCW h) through each reactions in A. niger DS03043 and CBS513.88 respectively
Table 2.
Pool sizes of amino acids for A. niger DS03043 and CBS513.88
| Name | A. niger DS03043 | A. niger CBS513.88 | P value of T test | ||
|---|---|---|---|---|---|
| Value | SD | Value | SD | ||
| Gln | 56.23 | 3.05 | 51.20 | 4.81 | 0.73 |
| Lys | 52.36 | 3.88 | 46.50 | 6.50 | 0.66 |
| Glu | 63.85 | 3.71 | 44.00 | 4.68 | 0.01 |
| Ala | 25.56 | 1.39 | 13.74 | 2.36 | 6.36E−03 |
| Asp | 21.55 | 2.64 | 12.47 | 1.89 | 0.02 |
| Thr | 5.49 | 0.28 | 8.82 | 0.24 | 7.57E−05 |
| Asn | 6.94 | 0.45 | 8.61 | 0.25 | 4.45E−03 |
| Trp | 5.45 | 0.44 | 6.24 | 0.97 | 0.19 |
| Orn | 3.98 | 0.44 | 5.53 | 0.51 | 0.01 |
| Tyr | 3.63 | 0.18 | 5.36 | 0.94 | 0.07 |
| Ser | 5.78 | 0.41 | 5.03 | 0.70 | 0.43 |
| Aaa | 5.49 | 0.71 | 4.80 | 0.63 | 0.52 |
| Val | 2.46 | 0.07 | 3.54 | 0.43 | 0.04 |
| Arg | 4.11 | 0.33 | 3.44 | 0.34 | 0.24 |
| His | 2.07 | 0.08 | 1.61 | 0.19 | 0.62 |
| Ile | 0.86 | 0.03 | 0.98 | 0.13 | 0.19 |
| Leu | 0.72 | 0.01 | 0.87 | 0.14 | 0.14 |
| Pro | 0.84 | 0.07 | 0.58 | 0.09 | 0.04 |
| Phe | 0.64 | 0.02 | 0.44 | 0.08 | 0.06 |
| Met | 0.35 | 0.05 | 0.38 | 0.08 | 0.56 |
The unit is µmol/gDCW. All metabolite pool sizes were measured based on at least triplicate measurements. The P values were obtained based on two-tailed T test statistical analyses
Table 3.
Pool sizes of nucleotides and coenzymes in A. niger DS03043 and CBS513.88 during batch cultivations
| A. niger DS03043 | A. niger CBS513.88 | |||
|---|---|---|---|---|
| Value | SD | Value | SD | |
| GSH | 0.46 | 0.01 | 0.31 | 0.03 |
| GSSG | 2.59 | 0.02 | 1.61 | 0.05 |
| UTP | 1.62 | 0.29 | 2.78 | 0.56 |
| GTP | 1.27 | 0.43 | 0.79 | 0.10 |
| GDP | 0.57 | 0.05 | 0.62 | 0.03 |
| GMP | 0.38 | 0.02 | 0.46 | 0.01 |
| CMP | 0.10 | 0.02 | 0.13 | 0.02 |
| UMP | 0.06 | 0.04 | 0.08 | 0.01 |
| IMP | 0.13 | 0.07 | 0.14 | 0.01 |
| IDP | 0.36 | 0.05 | 0.39 | 0.01 |
| NADH | 1.12 | 0.23 | 0.86 | 0.17 |
| NAD+ | 10.31 | 0.72 | 5.33 | 0.70 |
| NADPH | 1.19 | 0.26 | 1.39 | 0.21 |
| NADP+ | 5.13 | 0.51 | 4.55 | 0.96 |
| ATP | 13.37 | 1.67 | 6.68 | 0.81 |
| ADP | 1.39 | 0.36 | 1.17 | 0.27 |
| AMP | 1.29 | 0.39 | 1.20 | 0.45 |
The unit is µmol/gDCW. All metabolites pool size was measured at least triplicate measurements
Central carbon metabolism
The central carbon metabolism, including EMP, PP and TCA cycle pathways, exhibited obvious differences between two strains. Firstly, consistent with a larger specific glucose uptake rate, most of the absolute fluxes in A. niger DS03043 were relatively higher than those of A. niger CBS513.88 (Fig. 4). In the following comparison, all fluxes are normalized to the specific glucose uptake rate (Fig. 3). The relative flux channeled into the PP pathway was obviously larger in A. niger DS03043 than in A. niger CBS513.88. Accordingly, the flux through EMP in A. niger DS03043 was decreased. Exceeding the demand for biomass synthesis, a fraction of pentose was recycled back into the EMP pathway in the form of F6P and GAP. As the main pathway for NADH formation, the flux through TCA cycle in A. niger DS03043 was only half of that in wild type strain (Fig. 3). Such results agree with previous studies involving Aspergilli [15, 19] and the detailed comparison could be found in Additional file 3.
The pool sizes of metabolites from the core carbon metabolic network showed a relationship with the flux distribution for both strains. Due to the increased flux from G6P to 6PG, the pool sizes of R5P and S7P in A. niger DS03043 were clearly larger (P value <0.05) than those in A. niger CBS513.88 (Fig. 4). In the upper EMP pathway, the pool sizes of G6P and F6P were nearly constant for the two strains, indicating that their pool sizes were strictly controlled. However, in the lower EMP pathway, except for PYR, the pool sizes of 3PG and PEP were largely increased in A. niger CBS513.88 (P value <0.05). It was also found that the intracellular OAA, CIT, isCIT and AKG pool sizes were obviously bigger in A. niger CBS513.88 than those in A. niger DS03043 (P value <0.05). On the contrary, in the TCA cycle of CBS513.88, the pool sizes of MAL and FUM were slightly smaller than A. niger DS03043. It could be speculated that there exists a close relationship between the accumulation of intermediates in the TCA cycle and the excretion of large amounts of oxalic acid and citric acid in A. niger CBS513.88.
Amino acids metabolism
The amino acids pool sizes for A. niger DS03043 and CBS513.88 are shown in Table 2. Although there was a similar distribution of amino acids, the total amino acid pools displayed a significant difference (235 ± 8 vs 213 ± 9 µm/gDCW for DS03043 and CBS513.88, respectively). Interestingly, the pool sizes of Gln, Lys, Glu, Ala and Asp were distinctively larger than those of His, Ile, Leu, Pro, Phe and Met (p value <0.05) in both strains. It also should be noted that the pool sizes of Ala, Asp, Glu and Pro were significantly larger in A. niger DS03043 compared to those in A. niger CBS513.88 (p value <0.05). According to the original sources, the average pool size of amino acids from the family of glutamic acid was significantly higher than those of other families (Fig. 5). By comparison, the pools of histidine and aromatic amino acids were significantly decreased.
Fig. 5.

The average values of pool size for amino acids from different amino acid families
Energy and redox metabolism
To investigate the balance of cofactor metabolism, the regeneration and consumption rates of NADPH, NADH and ATP were estimated. As displayed in Table 4, the total generation rate of NADPH for A. niger DS03043 and CBS513.88 were 2.06 and 1.44 mmol/gDCW h, respectively. For both strains, the amount of NADPH involved in biomass formation was remarkably higher than that in the glucoamylase production. There are mainly two sources for NADH production, including the EMP pathway and the TCA cycle. The total rates of NADH regeneration were 6.09 and 6.35 mmol/gDCW h in A. niger DS03043 and CBS513.88 respectively (Table 4). With a higher generation rate of NADH through the TCA cycle (4.06 mmol/gDCW h) in A. niger CBS513.88, the NADH from the TCA cycle was about twofold of that from EMP pathway (2.03 mmol/gDCW h). While for A. niger DS03043, the relatively small NADH formation rate through the TCA cycle flux (3.28 mmol/gDCW h) decreased the total generation rate of NADH. Clearly, the result further showed that in A. niger CBS513.88, the regeneration rate of NADH was exceeding its utilization rate.
Table 4.
Estimated production and consumption of NADPH, NADH, ATP ( mmol/gDCW h)
| Pathways | NADPH | NADH | ATP | |||
|---|---|---|---|---|---|---|
| A. niger DS03043 | A. niger CBS513.88 | A. niger DS03043 | A. niger CBS513.88 | A. niger DS03043 | A. niger CBS513.88 | |
| Glycolysis | 0 | 0 | 2.3 | 2.03 | 0.8 | 0.55 |
| PP pathway | 2.06 | 1.22 | 0 | 0 | 0 | 0 |
| TCA cycle | 0 | 0.22 | 3.28 | 4.06 | 0.49 | 0.79 |
| Amino acid synthesis | – | – | 0.42 | 0.24 | – | – |
| Oxidative phosphorylationa | 0 | 0 | −5.88 | −5.33 | 12.38 | 11.22 |
| AcCoA formation by Ac | – | – | – | – | −0.1 | −0.3 |
| GA formation | −0.37 | −0.06 | 0.09 | 0.02 | −1.3 | −0.17 |
| Biomass formation | −1.69 | −1.16 | −0.54 | −0.38 | −8.54 | −6.4 |
aThe P/O ratios were set to 2.64 for mitochondrial NADH, and 1.64 for succinate and cytosolic NADH
Among the three pathways to generate ATP, including EMP, TCA cycle and the oxidative phosphorylation, the oxidative phosphorylation was the main source of ATP in both strains and the total specific generation rate of ATP were 12.38 and 11.22 mmol ATP/gDCW h respectively (Table 4) in A. niger DS03043 and CBS513.88. As shown in Table 4, most ATP was used for the strain growth and the fractions of ATP for biomass formation were about 68 and 57 % of total ATP generated in A. niger DS03043 and CBS513.88. The specific rate of ATP being used for glucoamylase synthesis was increased from 0.17 mmol ATP/gDCW h in A. niger CBS513.88 to 1.3 mmol ATP/gDCW h in A. niger DS03043. The rest ATP was potentially consumed for the non-growth maintenance requirements, which was calculated to be 3.73 and 5.68 mmol ATP/gDCW h respectively in A. niger DS03043 and CBS 513.88.
As displayed in Table 3, the pool sizes of cofactors in A. niger DS03043 were strikingly higher than those in A. niger CBS513.88, especially for NAD+ and ATP. The higher ratio of NADH to NAD+ in A. niger CBS513.88 (Fig. 6b) was consistent with the surplus NADH supply (Table 4). By comparison, the ratio of NADPH to NADP+ was only slightly larger in A. niger CBS513.88 than DS03043 (Fig. 6c). In comparison with A. niger CBS513.88, the pool size of ATP within A. niger DS03043 was increased significantly. The energy charge in A. niger DS03043 and CBS513.88 were 0.88 ± 0.13 and 0.80 ± 0.11 respectively, consistent with the previous reported values (0.8–0.9) [20–22]. Nevertheless, the ratio of ATP to AMP was strikingly increased in A. niger DS03043 compared with A. niger CBS513.88 (Fig. 6a) and the mass spectrum profiles of ATP and AMP for two strains could be found in Additional file 4.
Fig. 6.

Ratios of ATP/AMP (a), NADH/NAD+ (b) and NADPH/NADP+ (c) in A. niger DS03043 and CBS513.88. The error bars were calculated from error propagation
Discussion
Amino acid metabolism
The pool sizes of intracellular free amino acids are influenced by multiple factors [11], including the energy availability, the flux distribution, as well as the synthesis and degradation of specific proteins. In this work, we found that there existed no direct relations between the pool sizes of amino acids and the synthesis of glucoamylase (data not shown), except for Ala, Asp, Glu and Pro (Table 2), which may be due to the fact that during the batch phase, the main part of carbon substrate (about 80 %) was used for strain growth and aerobic respiration. Similar to previous reports [11], the overall distribution of amino acid pools were slightly affected by the enzyme production levels, indicating the robustness of strain metabolism. It has been reported that [23], for Pichia pastoris, the single amino acid pool size was correlated with overall cell protein amino acid compositional variations. However, in this study, we found that the amino acids pool sizes were directly related to the precursor metabolites. The average pool size of amino acids from the families of aromatic and histidine amino acids were lower on the whole (Fig. 5). Therefore, we speculated that the synthesis of these specific amino acids might be the bottleneck for further improvement in enzyme production.
Mechanism for PP pathway regulation
To adapt to changes in the external environment or differences in genotype, the metabolic network should be flexible and, meanwhile, maintain robustness [24]. In this study, the consistent changes in metabolites pool sizes and metabolic flux demonstrate the flexibility in metabolic activities. According to Heyland et al. [25], there existed a direct correlation between the relative PP pathway flux and the yield of biomass. However, the PP pathway provides not only essential precursors for strain growth, but also the reducing power NADPH for protein synthesis. As reported previously [15, 17, 19], for strains with a high yield of protein, the flux directed into the PP pathway was always increased. A. niger DS03043 shows the same characteristics as the yield of glucoamylase (YP/S), rather than the yield of biomass (YX/S), was statistically significantly increased in A. niger DS03043 (Table 1). Besides the increased demand of NADPH from anabolic reaction [26], there may exist two reasons for the reinforcement of flux channeled into the PP pathway. Firstly, the key enzymes including glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase in the PP pathway may be up-regulated [27]. Secondly, the pivotal enzymes in the EMP pathway, such as glucose-6-phosphate isomerase, could be weakened to reinforce more carbon into the PP pathway [28]. Actually, the glucose-6-phosphate isomerase is inhibited by high ratio of ATP to AMP [29, 30]. In a related matter, we found that the pool size of ATP and the ratio of ATP/AMP were strikingly increased in A. niger DS03043 (Table 3; Fig. 6a). Thus, we hypothesize that there exists a positive correlation between the ratio of ATP/AMP and the flux through the PP pathway (Fig. 7a). However, additional evidences from multi-omics study are required to clarify the question whether the key enzymes for the EMP and PP pathways are transcriptionally regulated or not.
Fig. 7.
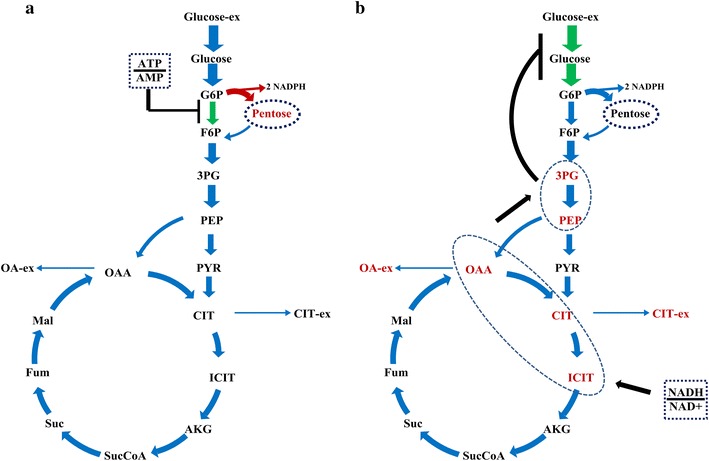
Speculated regulation mechanism revealed by integrated isotope-assisted metabolomics and 13C metabolic flux analysis. Possible effect of the ratio between ATP and AMP on flux redistribution in A. niger DS03043 (a) and regulation mechanism of ratio between NADH and NAD+ on the accumulation of OAA and PER pools, as well as on the specific glucose uptake rates in A. niger CBS513.88 (b). The red color in this graph represent increased flux or metabolites pool size and the green color represent weakened flux
TCA cycle pathway and energy metabolism
The TCA cycle pathway is the main source for NADH formation [31], as evident from the observation that the generation rate of NADH from the TCA cycle pathway was nearly twofold of that from the EMP pathway (Table 4). Via oxidative phosphorylation, NADH was transformed into ATP to meet the energy demand for maintenance, growth and product formation. For protein synthesis, the changes in the TCA cycle flux might depend on the strain. For Schizosaccharomyces pombe [16], it has been reported that to compensate for metabolic burden for heterologous protein synthesis, the flux through TCA cycle pathway was increased. As a result, there exists a positive correlation between the protein yield and the flux directed into the TCA cycle pathway. In contrast, for A. niger producing fructofuranosidase [15], the flux of the TCA cycle pathway dropped. Similarly, we found that this flux changed in the same direction in A. niger DS03043 (Fig. 3) and, more strikingly, the pool sizes of TCA cycle intermediates, like OAA, CIT, is CIT and AKG were also reduced (Fig. 4), which was accompanied with the reduction in by-products (oxalic acid and citric acid) formation (Table 1). The reported m_ATP for A. niger was 1.9 mmol ATP/gDCW h [32], while the calculated maintenance energy for both strains in this study were obviously higher, especially for A. niger CBS513.88. In general, the ATP cost for non-growth maintenance depends on product and by-products formation and the type of substrate used. It has been reported [33] that the cellular maintenance ATP consumption in engineered E. coli cells producing fatty acids was nearly twofold of that in control strain due to an increased osmotic stress resulting from product accumulation. Chung and co-workers [34] also reported that maintenance requirements for P. pastoris growing on glucose and glycerol–methanol mixed media were 2.3 and about 6 mmol ATP/gDCW h, respectively. Moreover, the transmembrane transport of organic acids was also accompanied with loss of energy [35]. Therefore, it could be concluded here that A. niger CBS513.88 needs more non-growth maintenance energy due to the accumulation and secretion of oxalic and citric acids, which, to some extent, decreased the utilization efficiency of energy in A. niger CBS513.88.
How metabolites modulate metabolic flux ?
Among thousands of metabolites, cofactors participate in 300 oxidation and reduction reactions [36] and play an important role in regulating the cell metabolic activities. Generally, the formation of by-products are correlated with the cofactors metabolism, for example, the glycerol synthesis could contribute to the recycle of NADH [37]. It has been reported that the substrate uptake rate was depressed under higher intracellular ratio of NADH/NAD+ [38]. However, the mechanisms for the modulation in fluxes via cofactors are generally not clear. The physiological parameters of A. niger CBS513.88 and DS03043, as found in this study, indicated that with large amounts of by-products formation the specific growth rate of the wild type strain was decreased. Interestingly, we found that the ratios of NADH/NAD+ and NADPH/NADP+ in A. niger CBS513.88 were higher than that in A. niger DS03043 (Fig. 6b, c), meaning that the A. niger CBS513.88 was in high redox condition. Accompanied with the high ratio of NADH/NAD+, the intermediates in the TCA cycle (CIT, is CIT, AKG and OAA) were accumulated in A. niger CBS513.88 (Fig. 4). The accumulation of organic acid from the TCA cycle also happened under oxygen limited conditions when the NADH recycle was hindered [39]. As reported by Heux et al. [38] and Hou et al. [40], decreasing the intracellular NADH level by introducing NADH oxidase or alternative oxidase lowered the by-products formation greatly. So it could be speculated that the higher ratio of NADH/NAD+ contributes to the increase in pool sizes of these intermediates from TCA cycle. As PEP was linked with the TCA cycle through anaplerotic reactions, the pool size of PEP in the cytoplasm was also obviously increased accordingly in A. niger CBS513.88 (Fig. 4).
According to Ogawa et al. [41], the accumulation of PEP could regulate the glucose uptake and glycolytic flux in cells by inhibiting glucokinase and glucose-6-phosphate isomerase. For mammalian cells culture, it was found that the high lactate level can exert an inhibitory effect on glycolysis enzyme phosphofructokinase, thus reducing the glycolytic flux [42]. So for A. niger CBS513.88 in this work, the lower absolute glycolytic flux (Fig. 4) might be caused by the inhibitory effect of organic acid, including PEP and 3PG, which in turn results in a lower specific growth rate (Table 1). As the accumulation of organic acids is related to the cell redox condition, the regulation of glycolytic flux actually seems to depend on the redox condition of the strain. In a comprehensive view, the possible regulation mechanism in flux and metabolites pools by the ratio of NADH/NAD+ could be summarized by Fig. 7b. With integrative analysis of metabolomics and 13C MFA, this work provides new clues to illustrate the interactive effects of flux and metabolites, which could be applied in other strains or process studies as well.
Conclusion
In this study, the combination of 13C metabolic flux analysis and quantitative metabolomics was adopted for the first time to investigate the metabolic changes in A. niger correlated with high glucoamylase production, initially revealing the metabolic regulation mechanisms between intracellular flux redistribution and metabolites pool sizes. The consistency in 13C metabolic flux analysis and metabolites quantification indicated that an imbalance of NADH formation and consumption led to the accumulation and secretion of organic acids in A. niger CBS513.88. The significantly reduced by-products formation rates resulted in an increased energy utilization efficiency and specific growth rate in A. niger DS03043. To enable a more efficient enzyme production and faster cell growth, the total amino acids pool size, the ratio of ATP to AMP and the flux channeled into the PP pathway were obviously improved in A. niger DS03043, which bring new insights on metabolic engineering targets for improved enzyme production using A. niger.
Methods
Strain and cultivations
Two A. niger strains with different glucoamylase production levels were kindly donated by DSM Corporation (Delft, the Netherlands). A. niger DS03043, a high glucoamylase-producing strain, contains seven copies of glucoamylase glaA gene, while A. niger CBS513.88, as a wild-type parent strain, contains only one glaA gene copy [43].
To obtain spores, Petri dishes containing PDA (Potato Dextrose Agar) medium were incubated with spores from a frozen stock (stored in 50 % glycerin at −80 °C). The seed cultivation was conducted in 500 mL shaking flasks with three baffles. Each flask was inoculated with 107 spores per 100 mL broth. The batch cultivations were conducted in a 5 L fermenter with an electronic balance, inoculated with the seed broth from the shake flasks after 24 h seed culture at 34 °C, with the speed of shaking table controlled at 150 rpm. The working volume for 5 L fermenter was 3.0 L. The agitation and aeration in the 5 L fermenter were kept at 375 rpm and 1 vvm respectively, with overpressure maintained at 0.05 MPa and temperature at 34 °C during the whole process. The pH was automatically maintained at pH 4.5 with 5 % (w/w) NH4·OH. The cultivations for each strain were performed in three independent cultivation replicates.
The medium for seed culture contained (g/L) glucose·H2O 22 and spray dried corn steep liquor 20. Before sterilization, the initial pH was adjusted to 6.5 using 3 mol/L NaOH. A chemically modified defined medium was used for all cultivations in this study, which contained (g/L) glucose·H2O 20, KH2PO4 3, NaH2PO4·H2O 1.5, (NH4)2SO4 3, MgSO4·7H2O 1.0, CaCl2·2H2O 0.1, ZnCl2 0.02, CuSO4·5H2O 0.015, CoCl2·6H2O 0.015, MnSO4·H2O 0.04, FeSO4·7H2O 0.3.
13C labeling experiments
Considering the high cost, the 13C labeling experiments were conducted in a reactor system consisting of a 5 L bioreactor and a 250 mL mini bioreactor equipped with pH and oxygen sensors. Before the formal experiment, the agitation and aeration in the mini bioreactor were carefully optimized to guarantee that the strain had similar physiological parameters in two scales of fermenters. The initial glucose concentration in the 5 L bioreactor was 10 g/L. When the glucose concentration decreased to approximately 5 g/L, about 170 mL broth was rapidly transferred from the 5 L bioreactor into the mini bioreactor. Then the labeled glucose including 0.56 g [U–13C]–Glc and 1.41 g [1–13C]–Glc (isotopic enrichment 98–99 %, Cambridge Isotope Laboratories, Inc.) were added as a pulse into the mini bioreactor.
Quantification of biomass and enzyme activity
10 mL broth was filtered through pre-dried and pre-weighed suction filter paper. Before filtering, the filter should be dried to a constant weight (24 h at 80 °C). To remove solutes, the samples were rinsed three times with deionized water. Then the wet filters with the biomass were put in the 80 °C oven and dried for 24 h. The dried filter was re-weighed immediately. A previous protocol was adopted to determine the enzyme activity in all samples [44].
Analysis of extracellular glucose and organic acids
A glucose analyzer (Shandong Academy of Sciences, China) was used for residual glucose concentration analysis. The amounts of extracellular organic acids (acetic, citric, oxalic, fumaric, malic, pyruvic, and succinic acids) were analyzed using a high-performance liquid chromatography (HPLC) system. An ion-exclusion column (Hi-Plex H, Agilent) was eluted at 50 °C with 10 mM H2SO4 at a flow rate of 0.5 mL/min and connected with an absorbance detector spectrophotometer at 210 nm. No acetic acid was detected under these conditions of cultivation.
Sampling and quantification of intracellular metabolites concentration
The protocol for quantification of intracellular metabolites pool size was adapted from the works of Douma et al. [45]. With a dedicated fast-sample equipment, 1–2 mL broth was rapidly withdrawn from sensor bioreactors in tubes with 10 mL precooled quench solution (−27.6 °C 40 % v/v methanol solution) at 30, 60, 90, 120, 180, 240 and 300 min since the start of 13C labelled experiments. The tubes were weighed before and after sampling procedure in order to estimate the exact amount of broth taken. Subsequently, fast filtration with a vacuum pump was conducted to remove the extracellular metabolites as soon as possible. Then, 120 mL precooled quench solution was used to wash the filter cake. To accurately determine the metabolites concentration, Isotope Dilution Mass Spectrometry (IDMS) [46] was adopted in this study. During the extraction process, the washed filter cake and 13C internal standard solution were together added into the pre-warmed 25 mL 75 % (v/v) ethanol solution and the extraction continued for 3 min at 95 °C. The metabolites pools were determined with the UPLC-MS/MS (Thermo Fisher Scientific Coporation).
For analysis of the intracellular pool size of NADH, NAD+, NADPH, and NADP+, the EnzyFluo™ Assay Kit (BioAssay Systems Coporation) was used. Firstly, 1 mL broth was quenched with 10 mL quench solution. The supernatant was discarded after centrifugation at −4 °C and 6000 rpm. The remaining processes were referred to the standard specifications (BioAssay Systems Coporation).
Analysis of amino acids enrichment with GC–MS
To obtain the enrichment of intracellular free amino acids during the 13C labelled experiments, the extracting solution was concentrated to 100 μL of supernatant before being transferred to a glass vial. After lyophilization, 80 μL acetonitrile and 80 μL of N-methyl-N-(tert-butyldimethylsilyl) trifluoroacetamide (MTBSTFA, Thermo Scientific) were added and the vial was incubated for 2 h at 60 °C. After that, the sample was centrifuged (10,000g, 2 min) and 160 μL of the supernatant was transferred to a GC glass vial with an insert. The sample was then analyzed by GC–MS instrument coupled to a 5975 C MSD single quadrupole mass spectrometer (Agilent, Santa Clara, CA, USA).
Calculation of metabolic fluxes based on 13C-labeled experiments
13C-MFA was performed using isotopomer Network Compartmental Analysis (INCA) [18], which is based on the elementary metabolite units (EMU) framework [47]. The A. niger network model used for flux calculations was described previously [15] and is given in Additional file 5. In brief, the model included all major reactions of the central carbon metabolism, amino acid biosynthesis, and lumped biomass formation. To determine the goodness-of- fit, the 13C-MFA fitting results were subjected to a χ2-statistical test [48]. The estimated fluxes were considered acceptable only when the obtained minimal weighted residual was below the χ2 at a 95 % confidence level. At convergence, accurate 68 and 95 % confidence intervals were computed for all estimated fluxes by evaluating the sensitivity of the minimized SSR to flux variations. The precision (or standard errors) of estimated fluxes were determined as follows [48]:
Authors’ contributions
HL carried out bioreactor cultivations, 13C labelled experiments, 13C flux calculation and metabolomics analysis. XL participated in the measurement of metabolites concentration. MH and JC participated in the design and coordination of the study. JX, YZ, SZ and NH helped to draft the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This work was financially supported by Royal DSM (Delft, the Netherlands) and partially supported by National Basic Research Program (973 Program 2013CB733600), NWO-MoST Joint Program (2013DFG32630), National High Technology Research and Development Program of China (863 Program 2012AA021201) and National Key Technology R&D Program (2012BAI44G01).
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Additional files
Additional file 1. The measured and estimated labelling information of amino acids fragments for optimal flux distributions.
Additional file 2. The best-fit flux distribution and minimal weighted residual from the parameter estimations.
Additional file 3. Overview of the current status of flux analysis in different Aspergillus species.
Additional file 4. Mass spectrum profiles of AXP for two strains and standard curves of AXP with 13C labelled metabolites as internal standards.
Additional file 5. Stoichiometric metabolic model of Aspergillus niger.
Contributor Information
Hongzhong Lu, Email: luhongzhong2006@126.com.
Xiaoyun Liu, Email: alice.liu@mail.ecust.edu.cn.
Mingzhi Huang, Email: huangmz@ecust.edu.cn.
Jianye Xia, Email: jyxia@ecust.edu.cn.
Ju Chu, Email: juchu@ecust.edu.cn.
Yingping Zhuang, Email: ypzhuang@ecust.edu.cn.
Siliang Zhang, Email: siliangz@ecust.edu.cn.
Henk Noorman, Email: henk.noorman@dsm.com.
References
- 1.Andersen MR, Nielsen J. Current status of systems biology in Aspergilli. Fungal Genet Biol. 2009;46:180–190. doi: 10.1016/j.fgb.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 2.Meyer V, Wu B, Ram AFJ. Aspergillus as a multi-purpose cell factory: current status and perspectives. Biotechnol Lett. 2011;33:469–476. doi: 10.1007/s10529-010-0473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knuf C, Nielsen J. Aspergilli: systems biology and industrial applications. Biotechnol J. 2012;7:1147–1155. doi: 10.1002/biot.201200169. [DOI] [PubMed] [Google Scholar]
- 4.Ward OP. Production of recombinant proteins by filamentous fungi. Biotechnol Adv. 2012;30:1119–1139. doi: 10.1016/j.biotechadv.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 5.Sun H, Zhao P, Ge X, Xia Y, Hao Z, Liu J, et al. Recent advances in microbial raw starch degrading enzymes. Appl Biochem Biotechnol. 2010;160:988–1003. doi: 10.1007/s12010-009-8579-y. [DOI] [PubMed] [Google Scholar]
- 6.Sévin DC, Kuehne A, Zamboni N, Sauer U. Biological insights through nontargeted metabolomics. Curr Opin Biotechnol. 2015;34:1–8. doi: 10.1016/j.copbio.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Ellis DI, Goodacre R. Metabolomics-assisted synthetic biology. Curr Opin Biotechnol. 2012;23:22–28. doi: 10.1016/j.copbio.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 8.Xia M, Huang D, Li S, Wen J, Jia X, Chen Y. Enhanced FK506 production in Streptomyces tsukubaensis by rational feeding strategies based on comparative metabolic profiling analysis. Biotechnol Bioeng. 2013;110:2717–2730. doi: 10.1002/bit.24941. [DOI] [PubMed] [Google Scholar]
- 9.Taymaz-Nikerel H, De Mey M, Baart G, Maertens J, Heijnen JJ, van Gulik W. Changes in substrate availability in Escherichia coli lead to rapid metabolite, flux and growth rate responses. Metab Eng. 2013;16:115–129. doi: 10.1016/j.ymben.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 10.de Jonge LP, Buijs NAA, ten Pierick A, Deshmukh A, Zhao Z, Kiel JAKW, et al. Scale-down of penicillin production in Penicillium chrysogenum. Biotechnol J. 2011;6:944–958. doi: 10.1002/biot.201000409. [DOI] [PubMed] [Google Scholar]
- 11.Jordà J, Rojas H, Carnicer M, Wahl A, Ferrer P, Albiol J. Quantitative metabolomics and instationary 13C-metabolic flux analysis reveals impact of recombinant protein production on trehalose and energy metabolism in Pichia pastoris. Metabolites. 2014;4:281–299. doi: 10.3390/metabo4020281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zamboni N, Fendt S-M, Ruhl M, Sauer U. 13C-based metabolic flux analysis. Nat Protoc. 2009;4:878–892. doi: 10.1038/nprot.2009.58. [DOI] [PubMed] [Google Scholar]
- 13.Antoniewicz MR. 13C metabolic flux analysis: optimal design of isotopic labeling experiments. Curr Opin Biotechnol. 2013;24:1116–1121. doi: 10.1016/j.copbio.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Niedenführ S, Wiechert W, Nöh K. How to measure metabolic fluxes: a taxonomic guide for 13C fluxomics. Curr Opin Biotechnol. 2015;34:82–90. doi: 10.1016/j.copbio.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Driouch H, Melzer G, Wittmann C. Integration of in vivo and in silico metabolic fluxes for improvement of recombinant protein production. Metab Eng. 2012;14:47–58. doi: 10.1016/j.ymben.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Klein T, Lange S, Wilhelm N, Bureik M, Yang T-H, Heinzle E, et al. Overcoming the metabolic burden of protein secretion in Schizosaccharomyces pombe—a quantitative approach using 13C-based metabolic flux analysis. Metab Eng. 2014;21:34–45. doi: 10.1016/j.ymben.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Nie Y, Huang M, Lu J, Qian J, Lin W, Chu J, et al. Impacts of high β-galactosidase expression on central metabolism of recombinant Pichia pastoris GS115 using glucose as sole carbon source via 13C metabolic flux analysis. J Biotechnol. 2014;187:124–134. doi: 10.1016/j.jbiotec.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 18.Young JD. INCA: a computational platform for isotopically non-stationary metabolic flux analysis. Bioinformatics. 2014;30:1333–1335. doi: 10.1093/bioinformatics/btu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pedersen H, Carlsen M, Nielsen J. Identification of enzymes and quantification of metabolic fluxes in the wild type and in a recombinant Aspergillus oryzae strain. Appl Environ Microbiol. 1999;65:11–19. doi: 10.1128/aem.65.1.11-19.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ball WJ, Atkinson DE. Adenylate energy charge in Saccharomyces cerevisiae during starvation. J Bacteriol. 1975;121:975–982. doi: 10.1128/jb.121.3.975-982.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chapman AG, Fall L, Atkinson DE. Adenylate energy charge in Escherichia coli during growth and starvation. J Bacteriol. 1971;108:1072–1086. doi: 10.1128/jb.108.3.1072-1086.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vrabl P, Mutschlechner W, Burgstaller W. Dynamics of energy charge and adenine nucleotides during uncoupling of catabolism and anabolism in Penicillium ochrochloron. Mycol Res. 2009;113:1422–1432. doi: 10.1016/j.mycres.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 23.Carnicer M, ten Pierick A, van Dam J, Heijnen JJ, Albiol J, van Gulik W, et al. Quantitative metabolomics analysis of amino acid metabolism in recombinant Pichia pastoris under different oxygen availability conditions. Microb Cell Fact. 2012;11:83–93. doi: 10.1186/1475-2859-11-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bordbar A, Monk JM, King ZA, Palsson BO. Constraint-based models predict metabolic and associated cellular functions. Nat Rev Genet. 2014;15:107–120. doi: 10.1038/nrg3643. [DOI] [PubMed] [Google Scholar]
- 25.Heyland J, Fu J, Blank LM, Schmid A. Carbon metabolism limits recombinant protein production in Pichia pastoris. Biotechnol Bioeng. 2011;108:1942–1953. doi: 10.1002/bit.23114. [DOI] [PubMed] [Google Scholar]
- 26.Wasylenko TM, Ahn WS, Stephanopoulos G. The oxidative pentose phosphate pathway is the primary source of NADPH for lipid overproduction from glucose in Yarrowia lipolytica. Metab Eng. 2015;30:27–39. doi: 10.1016/j.ymben.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 27.Chen H, Hao G, Wang L, Wang H, Gu Z, Liu L, et al. Identification of a critical determinant that enables efficient fatty acid synthesis in oleaginous fungi. Sci Rep. 2015;5:11247. doi: 10.1038/srep11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park SH, Kim HU, Kim TY, Park JS, Kim S-S, Lee SY. Metabolic engineering of Corynebacterium glutamicum for l-arginine production. Nat Commun. 2014;5:4618. doi: 10.1038/ncomms5618. [DOI] [PubMed] [Google Scholar]
- 29.Ghorbaniaghdam A, Henry O, Jolicoeur M. An in silico study of the regulation of CHO cells glycolysis. J Theor Biol. 2014;357:112–122. doi: 10.1016/j.jtbi.2014.04.035. [DOI] [PubMed] [Google Scholar]
- 30.Takama M, Nosoh Y. Effect of ATP on glucose-6-phosphate isomerase from Bacillus caldotenax. Biochim Biophys Acta. 1982;705:127–130. doi: 10.1016/0167-4838(82)90345-4. [DOI] [PubMed] [Google Scholar]
- 31.Zhou J, Liu L, Shi Z, Du G, Chen J. ATP in current biotechnology: regulation, applications and perspectives. Biotechnol Adv. 2009;27:94–101. doi: 10.1016/j.biotechadv.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 32.David H, Åkesson M, Nielsen J. Reconstruction of the central carbon metabolism of Aspergillus niger. Eur J Biochem. 2003;270:4243–4253. doi: 10.1046/j.1432-1033.2003.03798.x. [DOI] [PubMed] [Google Scholar]
- 33.He L, Xiao Y, Gebreselassie N, Zhang F, Antoniewicz MR, Tang YJ, et al. Central metabolic responses to the overproduction of fatty acids in Escherichia coli based on 13C-metabolic flux analysis. Biotechnol Bioeng. 2014;111:575–585. doi: 10.1002/bit.25124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung B, Selvarasu S, Camattari A, Ryu J, Lee H, Ahn J, et al. Genome-scale metabolic reconstruction and in silico analysis of methylotrophic yeast Pichia pastoris for strain improvement. Microb Cell Fact. 2010;9:50–65. doi: 10.1186/1475-2859-9-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Maris AJA, Winkler AA, Porro D, van Dijken JP, Pronk JT. Homofermentative lactate production cannot sustain anaerobic growth of engineered Saccharomyces cerevisiae: possible consequence of energy-dependent lactate export. Appl Environ Microbiol. 2004;70:2898–2905. doi: 10.1128/AEM.70.5.2898-2905.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Förster J, Famili I, Fu P, Palsson BØ, Nielsen J. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 2003;13:244–253. doi: 10.1101/gr.234503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rigoulet M, Aguilaniu H, Avéret N, Bunoust O, Camougrand N, Grandier-Vazeille X, et al. Organization and regulation of the cytosolic NADH metabolism in the yeast Saccharomyces cerevisiae. Mol Cell Biochem. 2004;256:73–81. doi: 10.1023/B:MCBI.0000009888.79484.fd. [DOI] [PubMed] [Google Scholar]
- 38.Heux S, Cachon R, Dequin S. Cofactor engineering in Saccharomyces cerevisiae: expression of a H2O-forming NADH oxidase and impact on redox metabolism. Metab Eng. 2006;8:303–314. doi: 10.1016/j.ymben.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 39.Diano A, Peeters J, Dynesen J, Nielsen J. Physiology of Aspergillus niger in oxygen-limited continuous cultures: influence of aeration, carbon source concentration and dilution rate. Biotechnol Bioeng. 2009;103:956–965. doi: 10.1002/bit.22329. [DOI] [PubMed] [Google Scholar]
- 40.Hou J, Lages NF, Oldiges M, Vemuri GN. Metabolic impact of redox cofactor perturbations in Saccharomyces cerevisiae. Metab Eng. 2009;11:253–261. doi: 10.1016/j.ymben.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 41.Ogawa T, Mori H, Tomita M, Yoshino M. Inhibitory effect of phosphoenolpyruvate on glycolytic enzymes in Escherichia coli. Res Microbiol. 2007;158:159–163. doi: 10.1016/j.resmic.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 42.Mulukutla BC, Gramer M, Hu W-S. On metabolic shift to lactate consumption in fed-batch culture of mammalian cells. Metab Eng. 2012;14:138–149. doi: 10.1016/j.ymben.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 43.van Dijck PWM, Selten GCM, Hempenius RA. On the safety of a new generation of DSM Aspergillus niger enzyme production strains. Regul Toxicol Pharm. 2003;38:27–35. doi: 10.1016/S0273-2300(03)00049-7. [DOI] [PubMed] [Google Scholar]
- 44.Lu H, Li C, Tang W, Wang Z, Xia J, Zhang S, et al. Dependence of fungal characteristics on seed morphology and shear stress in bioreactors. Bioprocess Biosyst Eng. 2015;38:917–928. doi: 10.1007/s00449-014-1337-8. [DOI] [PubMed] [Google Scholar]
- 45.Douma RD, de Jonge LP, Jonker CTH, Seifar RM, Heijnen JJ, van Gulik WM. Intracellular metabolite determination in the presence of extracellular abundance: application to the penicillin biosynthesis pathway in Penicillium chrysogenum. Biotechnol Bioeng. 2010;107:105–115. doi: 10.1002/bit.22786. [DOI] [PubMed] [Google Scholar]
- 46.Wu L, Mashego MR, van Dam JC, Proell AM, Vinke JL, Ras C, et al. Quantitative analysis of the microbial metabolome by isotope dilution mass spectrometry using uniformly C-13-labeled cell extracts as internal standards. Anal Biochem. 2005;336:164–171. doi: 10.1016/j.ab.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 47.Antoniewicz MR, Kelleher JK, Stephanopoulos G. Elementary metabolite units (EMU): a novel framework for modeling isotopic distributions. Metab Eng. 2007;9:68–86. doi: 10.1016/j.ymben.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Antoniewicz MR, Kelleher JK, Stephanopoulos G. Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab Eng. 2006;8:324–337. doi: 10.1016/j.ymben.2006.01.004. [DOI] [PubMed] [Google Scholar]


