Abstract
Background
Glutathione (GSH), a pivotal non-protein thiol, can be biosynthesized through three pathways in different organisms: (1) two consecutive enzymatic reactions catalyzed by γ-glutamylcysteine synthetase (Gsh1 or GshA) and glutathione synthetase (Gsh2 or GshB); (2) a bifunctional γ-glutamylcysteine synthetase/glutathione synthetase (GshF); (3) an alternative condensation of γ-glutamyl phosphate synthesized by γ-glutamyl kinase (Pro1 or ProB) with cysteine to form γ-glutamylcysteine which was further conjugated to glycine by glutathione synthetase. The Gsh1 and Gsh2 of conventional GSH biosynthetic pathway or the bifunctional GshF reported previously have been independently modulated for GSH production. This study developed a novel three-pathway combination method to improve GSH production in Saccharomyces cerevisiae.
Results
A bifunctional enzyme GshF of Actinobacillus pleuropneumoniae was functionally expressed in S. cerevisiae and Pro1 in proline biosynthetic pathway was exploited for improving GSH yield. Moreover, two fusion proteins Gsh2-Gsh1 and Pro1-GshB were constructed to increase the two-step coupling efficiency of GSH synthesis by mimicking the native domain fusion of GshF. The engineered strain W303-1b/FGP with three biosynthetic pathways presented the highest GSH concentration (216.50 mg/L) and GSH production of W303-1b/FGP was further improved by 61.37 % when amino acid precursors (5 mM glutamic acid, 5 mM cysteine and 5 mM glycine) were fed in shake flask cultures. In batch culture process, the recombinant strain W303-1b/FGP also kept high efficiency in GSH production and reached an intracellular GSH content of 2.27 % after 24-h fermentation.
Conclusions
The engineered strains harbouring three GSH pathways displayed higher GSH producing capacity than those with individually modulated pathways. Three-pathway combinatorial biosynthesis of GSH promises more effective industrial production of GSH using S. cerevisiae.
Electronic supplementary material
The online version of this article (doi:10.1186/s12934-015-0327-0) contains supplementary material, which is available to authorized users.
Keywords: Glutathione, Three-pathway combination, Combinatorial biosynthesis, Saccharomyces cerevisiae
Background
Glutathione (γ-l-glutamyl-l-cysteinylglycine, GSH) is an intracellular redox-active tripeptide thiol found in living organisms [1, 2]. The free sulfhydryl moiety of cysteine residue contributes to a wide variety of biological activities, such as anti-oxidization [3], detoxification [4, 5] and immune regulation [6], etc. GSH has been widely used in pharmaceutical, food and cosmetic industries.
GSH is primarily synthesized intracellularly through two consecutive reactions catalyzed by γ-glutamylcysteine synthetase (Gsh1 in eukaryotes and GshA in prokaryotes, encoded by GSH1 and gshA, respectively) and glutathione synthetase (Gsh2 in eukaryotes and GshB in prokaryotes, encoded by GSH2 and gshB, respectively). Gsh1 catalyzes the formation of γ-glutamylcysteine (γ-GC), which is subsequently conjugated to glycine by Gsh2 to form GSH. The activity of Gsh1 could be physiologically feedback-inhibited by GSH to avoid over-accumulation of GSH [7]. However, many organisms without Gsh1 or Gsh2 were reported to have the ability to produce GSH, which suggests that they may have a different pathway for generation of GSH. Recently, a novel bifunctional enzyme GshF (encoded by gshF) found in some Gram-positive bacteria such as: Listeria monocytogenes, Streptococcus agalactiae and Pasteurella multocida, was able to perform complete synthesis of GSH [8–11]. In other studies, compensatory pathway for GSH synthesis was characterized in yeast and Escherichia coli that lack Gsh1 and GshA [12, 13]. γ-glutamyl kinase (GK) encoded by PRO1 in eukaryotes or proB in prokaryotes, the first enzyme in proline biosynthetic pathway, catalyzes the formation of intermediate γ-glutamyl phosphate, which if accumulated could partially react with cysteine to generate γ-GC, and then Gsh2 adds glycine to γ-GC to form GSH. Different biosynthetic pathways of GSH are as shown in Fig. 1 and referred as G pathway, F pathway and P pathway.
Fig. 1.

Biosynthetic pathways of GSH. G pathway, Gsh1 catalyzes the reaction between glutamic acid and cysteine to form γ-GC, which is subsequently conjugated to glycine by Gsh2 to synthesize GSH. F pathway, GshF accounts for Gsh1 and Gsh2 activities and carries out complete synthesis of GSH. P pathway, Pro1 converts glutamate to γ-glutamyl phosphate, which reacts with cysteine to generate γ-GC, and then Gsh2 adds glycine to γ-GC to form GSH
To date, GSH is mainly produced via fermentation. Yeasts, usually S. cerevisiae and Candida utilis, are more commonly used in industrial scale GSH production. GSH contents of the wild type (wt) yeasts are typically in the range of 0.1–1 %. The most successful strategy toward obtaining GSH overproducing strains was classical selection which resulted in GSH contents of 3–5 % [14]. Along with the identification and characterization of genes involved in GSH metabolism, many genetic engineering strategy studies have been conducted in an effort to increase GSH production using various model microorganisms. The reported GSH contents in genetically engineering strains ranged from 1 to 2 % in the absence of amino acid precursors [15–22]. Genetic manipulation of the GSH biosynthetic pathway has been proved to be an effective method to produce GSH and provides a platform for further improvement. In previous reports, single modulation of G pathway and F pathway for GSH production could achieve GSH contents of 1.5–1.7 % [19, 22] (Table 1). Besides, many efforts have been focused on medium optimization and process control to enhance GSH production, in particular introduction of amino acid precursors during GSH fermentation [23, 24].
Table 1.
An overview of literature on the production of GSH without addition of amino acid precursors by microorganisms
| Host strain | Strategy | GSH content in mutant | GSH content in wild type | References |
|---|---|---|---|---|
| C. utilis | Classical selection | 3–5 % | 0.1–1 % | [14] |
| S. cerevisiae | ||||
| S. cerevisiae | pGSR2518-x containing gshA of E. coli B fused with a S. cerevisiae promoter fragment P8 was used to transform S. cerevisiae YNN27 | 1.54 % | 0.5 % | [17] |
| S. cerevisiae | A recombinant plasmid pGMF with GSH1 from S. cerevisiae was introduced into S. cerevisiae YSF-31 | 1.31 % | 0.87 % | [18] |
| S. cerevisiae | Plasmids, pδAUR-GCS and pδAUR-GS, containing GSH1 and GSH2 from S. cerevisiae were linearized and integrated at δ-sites with high copy numbers into the ribosomal DNA of S. cerevisiae YPH499 | 1.51 % | 1.03 % | [19] |
| Sulfate assimilation metabolism and GSH synthetic metabolism were combinatorially engineered in S. cerevisiae YPH499 | 1.83 % | |||
| S. cerevisiae | gshF derived from Streptococcus thermophilus was integrated at a high copy number into the ribosomal DNA of S. cerevisiae BY4741 | 54.9 μM/g DCW (1.69 %) | 11.7 μM/g DCW (0.36 %) | [22] |
| P. pastoris | An integrative expression vector, pGAPZHGSH, containing GSH1 and GSH2 from S. cerevisiae regulated by GAP promoter was transformed into P. pastoris GS115 | 0.92 g (GSH)/L | [16] | |
| 94.98 g (DCW)/L (0.97 %) | ||||
| P. pastoris | An integrative expression vector, pGAPZHGSH, containing GSH1 and GSH2 from S. cerevisiae regulated by GAP promoter was transformed into P. pastoris GS115 | <0.05 mM/g DCW (<1.54 %) | <0.02 mM/g DCW (<0.61 %) | [21] |
| An integrative expression vector, pGAPZH-Lmgsh, containing gshF from Listeria monocytogenes regulated by GAP promoter was transformed into P. pastoris GS115 | <0.04 mM/g DCW (<1.23 %) |
Although genetic manipulations of a single pathway have increased GSH production of engineered strains, discovering how to further improve GSH production is still a real challenge for us. In this study, we focused on direct modulation of the GSH biosynthetic pathway via a method of three-pathway combination. Two artificially recombinant enzymes glutathione synthetase-linker-γ-glutamylcysteine synthetase fusion protein (Gsh2-Gsh1) from S. cerevisiae, γ-glutamyl kinase-linker-glutathione synthetase hybrid (Pro1-GshB) from S. cerevisiae and E. coli and a resynthesized GshF of Actinobacillus pleuropneumoniae were introduced into S. cerevisiae genome via independent or combinatorial genetic integration. The overexpression of genes from different pathways and synthetic capacities of GSH of different engineered strains were investigated. The data demonstrated that three-pathway combination was superior to single-pathway for synthesis of GSH.
Results
Activity analysis of GshF from A. pleuropneumoniae expressed under the control of different promoters in S. cerevisiae
To examine the activity of codon-conformed A. pleuropneumoniae GshF in S. cerevisiae and evaluate effect of promoter strength on its activity, four δ DNA-mediated integrative expression vectors pδGAPg-gshF, pδGAP′g-gshF, pδGAL1g-gshF and pδPGK1g-gshF were constructed, in which the gshF gene was driven by glyceraldehyde-3-phosphate dehydrogenase promoter (GAP) of S. cerevisiae, GAP promoter from Pichia pastoris (GAP′), galactokinase promoter (GAL1) of S. cerevisiae and phosphoglycerate kinase promoter (PGK1) of S. cerevisiae, respectively. S. cerevisiae W303-1b cells were transformed with the linearized recombinant plasmids to obtain engineered strains W303-1b/F/GAP, W303-1b/F/GAP′, W303-1b/F/GAL1 and W303-1b/F/PGK1. The plasmids and strains constructed in the present study are listed in Table 2. GshF activities and promoter strengths could be determined by GSH concentrations of the engineered strains in shake flask cultures.
Table 2.
Strains and plasmids used in this study
| Strains or plasmids | Relevant properties | Source or references |
|---|---|---|
| Strains | ||
| E. coli Trans1-T1 | F− φ80(lacZ) ΔM15 ΔlacX74 hsdR(rK− mK+) ΔrecA1398 endA1 tonA | Our lab |
| S. cerevisiae W303-1b | MATα ade2-1 leu2-3,112 his3-11,15 ura3-1 trp1-1 | Our lab |
| W303-1b/Δgsh1 | W303-1b derivative with pΔgsh1h, GSH1::HygB | This study |
| W303-1b/Δgsh1/pro1 | W303-1b derivative with pΔgsh1h-pro1, GSH1::P gap-PRO1-T pgk1 | This study |
| W303-1b/Δgsh1/proB | W303-1b derivative with pΔgsh1h-proB, GSH1::P gap-proB-T pgk1 | This study |
| W303-1b/F/GAP (W303-1b/F)a | W303-1b derivative with pδGAPg-gshF | This study |
| W303-1b/F/GAP′ | W303-1b derivative with pδGAP′g-gshF | This study |
| W303-1b/F/PGK1 | W303-1b derivative with pδPGK1g-gshF | This study |
| W303-1b/F/GAL1 | W303-1b derivative with pδGAL1g-gshF | This study |
| W303-1b/Δgsh1/F | W303-1b/F derivative with pΔgsh1h, GSH1::HygB | This study |
| W303-1b/G | W303-1b derivative with pδGAPh-gsh2gsh1 | This study |
| W303-1b/P | W303-1b derivative with pδGAPg-pro1gshB | This study |
| W303-1b/P′ | W303-1b derivative with pδGAPg-proBgshB | This study |
| W303-1b/FF | W303-1b/F derivative with pδGAPg-gshF | This study |
| W303-1b/FG | W303-1b/F derivative with pδGAPh-gsh2gsh1 | This study |
| W303-1b/FP | W303-1b/F derivative with pδGAPg-pro1gshB | This study |
| W303-1b/FGP | W303-1b/FG derivative with pδGAPg-pro1gshB | This study |
| Plasmids | ||
| pG | pBluescript II KS(+) derivative with homologous region of GSH1 | Our lab |
| pΔgsh1h | pBluescript II KS(+) derivative with homologous region of GSH1, HygBr | This study |
| pΔgsh1h-pro1 | pG derivative with P gap, PRO1, T pgk1, HygBr | This study |
| pΔgsh1h-proB | pG derivative with P gap, proB, T pgk1, HygBr | This study |
| pδGAPg | pBluescript II KS(+) derivative with homologous δ region, P gap, T pgk1, G418r | Our lab |
| pδGAP′g | pBluescript II KS(+) derivative with homologous δ region, P gap′, T pgk1, G418r | Our lab |
| pδPGK1g | pBluescript II KS(+) derivative with homologous δ region, P pgk1, T pgk1, G418r | Our lab |
| pδGAL1g | pBluescript II KS(+) derivative with homologous δ region, P gal1, T pgk1, G418r | Our lab |
| pδGAPh | pBluescript II KS(+) derivative with homologous δ region, P gap, T pgk1, HygBr | Our lab |
| pδGAPg-gshF | pδGAPg derivative with A. pleuropneumoniae gshF, G418r | This study |
| pδGAP′g-gshF | pδGAP′g derivative with A. pleuropneumoniae gshF, G418r | This study |
| pδPGK1g-gshF | pδPGK1 g derivative with A. pleuropneumoniae gshF, G418r | This study |
| pδGAL1g-gshF | pδGAL1 g derivative with A. pleuropneumoniae gshF, G418r | This study |
| pδGAPh-gsh2gsh1 | pδGAPh derivative with GSH2-GSH1 from S. cerevisiae, HygBr | This study |
| pδGAPg-pro1gshB | pδGAPg derivative with PRO1-gshB from S. cerevisiae and E. coli, respectively, G418r | This study |
| pδGAPg-proBgshB | pδGAPg derivative with proB-gshB from E. coli, G418r | This study |
aW303-1b/F/GAP and W303-1b/F represent the same engineered strain in this study
GSH production in the engineered strains together with host strain W303-1b was compared as shown in Fig. 2. The four engineered strains conferred higher GSH concentrations than the parental strain. Among them, W303-1b/F/GAP (abbreviated to W303-1b/F in the following sections) produced higher intracellular GSH levels; the highest concentration was achieved after fermentation for 48–60 h. This result indicated that A. pleuropneumoniae GshF was active for GSH synthesis in S. cerevisiae. In addition, differences in synthetic capacities of GSH were considered to be directly related to the strengths of different promoters, thus GAP promoter of S. cerevisiae was utilized to keep uniform expression of each construct in this work.
Fig. 2.
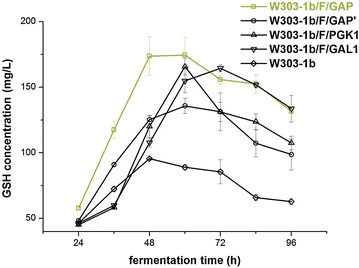
Evaluation of synthetic capacities of GSH of engineered S. cerevisiae strains harbouring gshF gene under the control of four different promoters. Intracellular GSH amount was monitored after 24-h fermentation and measured every 12 h. The values are presented as the means, and the error bars show the SD (n = 3)
Active GK for GSH biosynthesis in S. cerevisiae W303-1b
In the cells lacking Gsh1-dependent biosynthetic pathway of GSH, it was found that γ-glutamyl phosphate is a common precursor shared by proline and GSH biosynthetic pathways. Thus, biosynthesis of proline could be partially diverted toward GSH production via γ-glutamyl phosphate (Fig. 1) and GK may play a central role in the alternative pathway.
Preliminary experiments had proved in vitro activity of GK for formation of intermediate γ-GC (data not shown). For the purpose of exploiting an active GK for biosynthesis of GSH in S. cerevisiae W303-1b, plasmids pΔgsh1h, pΔgsh1h-pro1 and pΔgsh1h-proB were constructed. With the homologous integration of the linearized plasmids, GSH1 gene of wt W303-1b was deleted or replaced by recombinant DNA fragment of S. cerevisiaePRO1 or E. coli proB, and the resulting strains were W303-1b/Δgsh1 (GSH1 deletion), W303-1b/Δgsh1/pro1 (GSH1::Pgap-PRO1-Tpgk1) and W303-1b/Δgsh1/proB (GSH1::Pgap-proB-Tpgk1) (Table 2). Patch assay on YPD containing the given amounts of H2O2 was carried out with wt W303-1b, W303-1b/Δgsh1, W303-1b/Δgsh1/pro1 and W303-1b/Δgsh1/proB to detect their H2O2 tolerances that normally require GSH. As for W303-1b/Δgsh1/pro1 or W303-1b/Δgsh1/proB, the overexpression of introduced Pro1 or ProB under the control of strong constitutive GAP promoter might allow a small amount of GSH in the host strains conferring tolerance to oxidative stress. As shown in Fig. 3, all transformants grew on the plates with the concentration of H2O2 below 1.0 mM. However, their growth rates were apparently slower than wt strain (data not shown). W303-1b/Δgsh1 and W303-1b/Δgsh1/proB failed to grow when addition of H2O2 was more than 1.5 mM, while W303-1b/Δgsh1/pro1 with Pro1 overexpression construct afforded to form colonies on YPD containing 3.0 mM H2O2. The presence of GSH in W303-1b/Δgsh1/pro1 was about 4 % of wt level (Fig. 4c). This data showed that overexpression of Pro1 could produce more γ-glutamyl phosphate involved in an alternative pathway for a small amount of GSH formation in S. cerevisiae W303-1b.
Fig. 3.
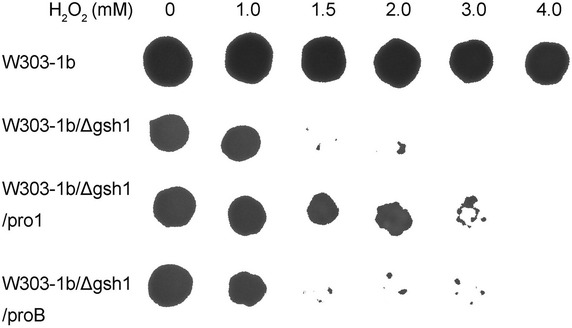
Patch assay on YPD containing the indicated amounts of H2O2. The tolerances to H2O2 of W303-1b (wt), W303-1b/Δgsh1 (GSH1 deletion), W303-1b/Δgsh1/pro1 (GSH1::P gap-PRO1-T pgk1) and W303-1b/Δgsh1/proB (GSH1::P gap-proB-T pgk1) were evaluated
Fig. 4.

Single modulation of G pathway, F pathway and P pathway for GSH biosynthesis in S. cerevisiae. a GSH concentration (mg/L). b Biomass (DCW). c GSH content (%). G (F or FF, P or P′) represents improvement of GSH amount conferred by the overexpression of G (F, P or P′) pathway and H represents GSH amount of host strain W303-1b. DCW is calculated from OD600 values (OD600 = 10 corresponds to 1.19 g/L DCW). The values are presented as the means, and the error bars show the SD (n = 3)
Overexpression of enzymes involved in GSH biosynthesis
In earlier studies, Gsh1 and Gsh2 (GshA and GshB) derived from yeast or E. coli were independently applied to various model microorganisms to generate genetically engineered strains with higher GSH productive capacities [15–19]. GshF from bacteria was also expressed in E. coli and yeast [20–22]. Here, A. pleuropneumoniae GshF was successfully expressed in S. cerevisiae as mentioned above. However, the recombinant constructs of Gsh2-Gsh1 and Pro1-GshB with a flexible linker have not been exploited for improving intracellular GSH synthesis until now.
To evaluate the potential of different pathways geared toward improving GSH synthesis, several δ DNA-mediated integrative expression vectors pδGAPh-gsh2gsh1, pδGAPg-pro1gshB or pδGAPg-proBgshB harbouring different pathway genes were constructed. Recombinant S. cerevisiae strains W303-1b/G, W303-1b/P or W303-1b/P′ (Table 2) were then generated by transforming different linearized plasmids into S. cerevisiae W303-1b as constructed W303-1b/F. The pathway genes with strong constitutive GAP promoters were introduced into the genome of W303-1b via homologous integration at δ-sites. To further compare GSH synthetic capacities of different pathways, GSH1 gene of engineered strain W303-1b/F was deleted to eliminate synthesis of GSH by endogenous enzymes. The resulting strain W303-1b/Δgsh1/F (F pathway) together with W303-1b/G (G pathway) and W303-1b/Δgsh1/pro1 (P pathway) represent different pathway strains (Table 2). No dramatic differences in biomass between W303-1b/G, W303-1b/F, W303-1b/Δgsh1/F, W303-1b/P or W303-1b/P′ and original strain W303-1b were observed during shake flask cultures (Fig. 4). Three engineered strains W303-1b/G, W303-1b/F and W303-1b/P displayed higher activities for synthesizing GSH than the parental strain. The highest intracellular concentration of GSH was 186.77 mg/L produced by strain W303-1b/F, and most of the synthesized GSH could be attributed to the function of the introduced GshF as strain W303-1b/Δgsh1/F yielded a similar amount (about 90 %) GSH to W303-1b/F. Other strains W303-1b/G, W303-1b/P and W303-1b produced 145.72, 111.82 and 98.75 mg/L of GSH, respectively. Results of the synthetic capacities of different pathways (ranked in descending order) are as follows: F pathway, G pathway and P pathway.
Since W303-1b/F synthesized the highest GSH concentration among tested strains, it was chosen to examine the effect of gene dosage on GSH production. The antibiotic-resistant gene was removed from the genomic DNA of W303-1b/F using the Cre-loxP system, then W303-1b/F cells were further transformed with the linearized recombinant plasmid pδGAPg-gshF to obtain engineered strain W303-1b/FF (Table 2). Three recombinant strains harbouring two copies of gshF gene relative to that of W303-1b/F (Additional file 1: Table S8) were obtained and compared for their GSH production with W303-1b/F. However, no further improvement was observed (Fig. 4). This result suggests that increasing copy number was shown to be effective up to a certain limit of gene copy number.
Three biosynthetic pathways combined to increase GSH production
To obtain a higher GSH concentration, three biosynthetic pathways of GSH were combinatorially expressed in S. cerevisiae W303-1b. Strain W303-1b/F with higher synthetic capacity of GSH was chosen as the starting strain for further introduction of other biosynthetic pathways. The two-pathway strains W303-1b/FG and W303-1b/FP were obtained by the integration of linearized pδGAPh-gsh2gsh1 and pδGAPg-pro1gshB, respectively. Further integration of linearized pδGAPg-pro1gshB into the engineered W303-1b/FG generated the three-pathway strain W303-1b/FGP (Table 2).
As expected, strains W303-1b/FG, W303-1b/FP and W303-1b/FGP yielded 206.70, 190.57 and 216.50 mg/L of GSH in shake flask cultures, indicating that introduction of a second pathway could enhance the synthetic capacity of GSH. Among all the engineered strains, the strain W303-1b/FGP with three biosynthetic pathways showed the highest GSH producing capacity and improved the intracellular GSH concentration by 2.19-fold compared with W303-1b (Table 3; Fig. 5).
Table 3.
Biomass, GSH concentration, GSH content and molar ratio of γ-GC to GSH of various strains in flask cultures with or without addition of amino acid precursors (AA)
| Strains | Biomass (DCW)a | GSH concentration (mg/L) | GSH content (%) | Molar ratio of γ-GC to GSH (%) |
|---|---|---|---|---|
| W303-1b | 9.14 ± 0.48 | 98.75 ± 7.30 | 1.09 ± 0.14 | 6.74 ± 0.24 |
| W303-1b/F | 9.09 ± 0.13 | 186.77 ± 3.19 | 2.05 ± 0.06 | 10.29 ± 0.87 |
| W303-1b/G | 9.56 ± 0.26 | 145.72 ± 14.87 | 1.52 ± 0.14 | 9.72 ± 0.93 |
| W303-1b/P | 9.23 ± 0.32 | 111.82 ± 11.34 | 1.21 ± 0.09 | 8.58 ± 2.53 |
| W303-1b/P′ | 8.74 ± 0.44 | 104.24 ± 9.86 | 1.19 ± 0.08 | 7.34 ± 0.42 |
| W303-1b/FG | 9.35 ± 0.38 | 206.70 ± 7.23 | 2.21 ± 0.02 | 11.26 ± 0.67 |
| W303-1b/FP | 9.04 ± 0.20 | 190.57 ± 1.56 | 2.11 ± 0.05 | 10.92 ± 0.93 |
| W303-1b/FGP | 9.26 ± 0.07 | 216.50 ± 11.46 | 2.34 ± 0.13 | 9.97 ± 0.40 |
| W303-1b/F (AA) | 8.58 ± 0.09 | 252.33 ± 28.62 | 2.94 ± 0.36 | 9.65 ± 0.51 |
| W303-1b/FGP (AA) | 8.38 ± 0.37 | 316.77 ± 45.69 | 3.77 ± 0.43 | 9.50 ± 1.43 |
aDCW is calculated from OD600 values (OD600 = 10 corresponds to 1.19 g/L DCW). The values given are mean values ± standard deviation
Fig. 5.

GSH production of single-pathway, two-pathway and three-pathway engineered strains. a GSH concentration (mg/L). b Biomass (DCW). c GSH content (%). G (F, P or P′) represents improvement of GSH amount conferred by the overexpression of G (F, P or P′) pathway and H represents GSH amount of host strain W303-1b. DCW is calculated from OD600 values (OD600 = 10 corresponds to 1.19 g/L DCW). The values are presented as the means, and the error bars show the SD (n = 3)
Shake flask culture with addition of amino acid precursors
Genetic manipulation and medium optimization together with the addition of amino acid precursors are required to increase GSH production. In order to further investigate the potential of three-pathway combinatorial biosynthesis of GSH, we compared utilization and conversion efficiency of amino acid precursors of engineered strain W303-1b/FGP with that of strain W303-1b/F in shake flask cultures. Figure 6 showed that the biomass of both W303-1b/FGP and W303-1b/F dropped by around 10 % when three amino acid precursors were added, and GSH contents of W303-1b/FGP and W303-1b/F were 3.77 and 2.94 %, respectively. After addition of amino acid precursors, GSH production of W303-1b/FGP was improved by 61.37 %, while that of W303-1b/F was 43.17 %. This result indicated that the engineered strain W303-1b/FGP held a higher utilization and conversion efficiency of amino acid precursors. That is to say, advantages of W303-1b/FGP over W303-1b/F include higher GSH yield and utilization and conversion efficiency of amino acid precursors.
Fig. 6.
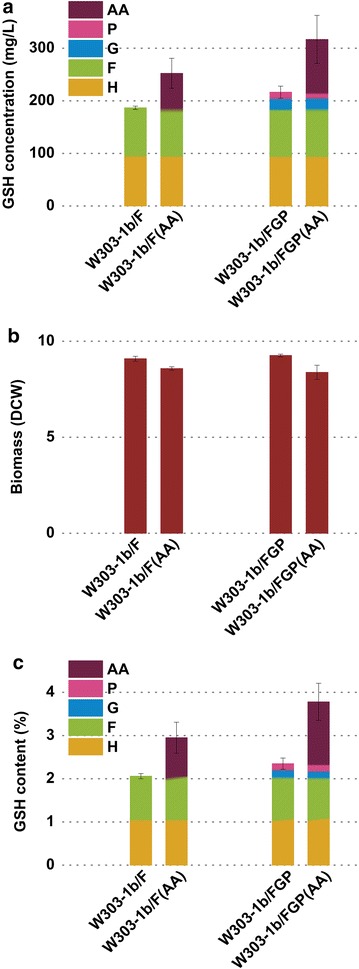
Evaluation of synthetic capacities of GSH of engineered S. cerevisiae W303-1b/F and W303-1b/FGP in shake flask cultures with or without addition of three amino acid precursors. a GSH concentration (mg/L). b Biomass (DCW). c GSH content (%). G (F or P) represents improvement of GSH content conferred by overexpression of G (F or P) pathway, H represents GSH content of host strain W303-1b and AA indicates improvement of GSH amount conferred by addition of amino acid precursors. DCW is calculated from OD600 values (OD600 = 10 corresponds to 1.19 g/L DCW). The values are presented as the means, and the error bars show the SD (n = 3)
Batch culture of engineered strain with three biosynthetic pathways
To examine the GSH productive capacity of engineered strain W303-1b/FGP in a larger scale, batch cultivation for GSH production was carried out in a 10-L fermentor. The time course of batch culture with W303-1b/FGP was shown in Fig. 7. Both GSH concentration and biomass were higher than that of shake flask culture. The GSH concentration increased to the highest point of 272.52 mg/L after fermentation for 24 h, and the GSH content was 2.27 %. It was observed that the GSH productive capacity measured by GSH content showed no obvious change after scale-up fermentation.
Fig. 7.
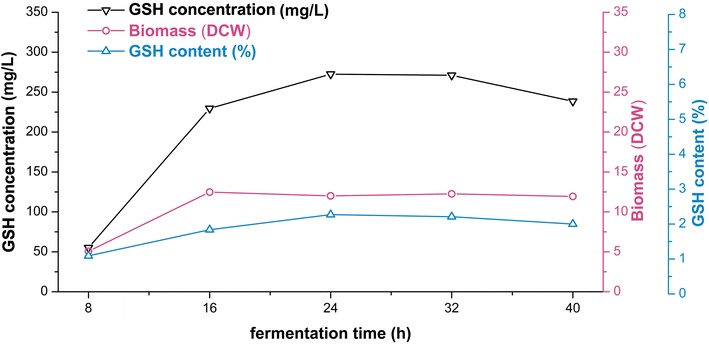
Batch culture of S. cerevisiae W303-1b/FGP in 10-L fermentor. The working volume was 4 L. Culture temperature and DO were controlled at 30 °C and 25 %, and pH was uncontrolled. Intracellular GSH amount was measured every 8 h
Discussion
Recently, a novel bifunctional enzyme GshF has aroused concerns about GSH productive capacity as most reported GshFs are insensitive to feedback inhibition of GSH except that of P. multocida and L. monocytogenes [8–11, 20]. In this study, the putative gshF from A. pleuropneumoniae resynthesized according to codon usage of S. cerevisiae was firstly functionally expressed in S. cerevisiae, and then the relationship between GSH production and the strengths of three different glucose-based promoters, including GAP and PGK1 promoters of S. cerevisiae and GAP promoter from P. pastoris (GAP′), and a galactose-based GAL1 promoter of S. cerevisiae was analyzed. Figure 2 showed that the expressed GshF under the control of constitutive GAP promoter of S. cerevisiae produced the highest concentration of GSH. Thus it is presumed that the expression of GshF driven by GAP promoter of S. cerevisiae functioned well for GSH synthesis. Through the comparison of the productive capacity of GSH of engineered strain W303-1b/F with that of the reported GS115/LmgshF with L. monocytogenes GshF, an observed variance was that F pathway presented higher synthetic capacity of GSH than G pathway (Fig. 4) [21]. This conflicting result may be attributed to the different sensitivities of GshFs from L. monocytogenes and A. pleuropneumoniae to feedback inhibition of GSH [8, 11]. Since single F pathway could produce higher amounts of GSH compared with other pathways, we tried to augment the copies of F pathway by further integration of gshF to improve the productive capacity of GSH of engineered strain W303-1b/F. However, the gshF copy number increase didn’t further improve GSH concentration in W303-1b/F, indicating that a reasonable rather than high copy number of gshF gene driven by GAP promoter is sufficient for the highest expression levels and synthetic capacity of GSH through one pathway is limited.
It has been reported that proline biosynthesis could contribute to GSH synthesis. γ-glutamyl phosphate is required as the common intermediate for the crosstalk between proline biosynthetic pathway and conventional Gsh1/Gsh2 pathway. Veeravalli et al. reported that, different from in vitro assay, as l-cysteine is present in micromolar concentrations in the intracellular environment, the activity of γ-glutamyl phosphate reductase (GPR) encoded by PRO2 in eukaryotes or proA in prokaryotes must be shut off to allow γ-GC formation from γ-glutamyl phosphate [12]. In this study, as amino acid precursors could be fed to improve concentrations of intracellular free l-cysteine, we resorted to overexpression of GK instead of loss-of-function mutations in GPR for accumulation of γ-glutamyl phosphate to strengthen the redirection of γ-glutamyl phosphate to GSH synthesis. It was indirectly confirmed by the result of patch assay that W303-1b/Δgsh1/pro1 with Pro1 overexpression construct performed a relatively stronger H2O2 tolerance than W303-1b/Δgsh1/proB and W303-1b/Δgsh1 (Fig. 3). Overexpressed Pro1 mediated accumulation of γ-glutamyl phosphate which was partially diverted toward GSH production, while heterologous ProB (optimized according to codon usage of S. cerevisiae) did not work well. Consistent with the result of patch assay, W303-1b/P provided a slightly higher GSH production than W303-1b/P′ and strain W303-1b in shake flask cultures (Fig. 4). When P pathway was upregulated, the accumulated γ-glutamyl phosphate could improve utilization of extra l-cysteine by addition of amino acid precursors for improvement of GSH synthesis. Thus, P pathway could be modulated for improving intracellular GSH synthesis.
Previous studies have reported individual modulation of G pathway and F pathway for GSH production [15–22]. In this study, P pathway was exploited for GSH production for the first time. Combining G, F and P three pathways is a novel method for GSH production. Genes of three pathways were uniformly regulated and expressed by exactly the same strategy to evaluate synthetic capacities of engineered strains with different combinations of biosynthetic pathways of GSH. As expected, the engineered strain W303-1b/FGP presented the highest yield of GSH. Moreover, the result of shake flask cultures with addition of amino acid precursors (Fig. 6) further proved the advantages of W303-1b/FGP. In batch culture process, the recombinant strain W303-1b/FGP also afforded high efficiency in GSH production and reached an intracellular GSH content of 2.27 % after 24-h fermentation. Generally, a GSH content of 3–5 % could be obtained using strains selected via classical selection strategy, but the exact mechanisms underlying higher GSH accumulation in these mutants has remained rather obscure. Nisamedtinov et al. reported that GSH over-accumulation in S. cerevisiae mutants selected via random mutagenesis is caused by the combined effect of higher cysteine synthesis and increased flux through Gsh1 reaction [25]. These genetic manipulations could confer a GSH content of 1–2 % on the engineered strains. The GSH synthetic capacity of engineered strain W303-1b/FGP constructed in this study was comparable to that of reported genetically engineered strains and higher than that of engineered strains with single modulation of G pathway or F pathway (Table 1). Increase in GSH content of W303-1b/FGP may be attributed to the fact that the three pathways form a complex network of GSH biosynthesis producing comprehensive effects. Here, it should be noted that the balance of two consecutive enzymatic reactions especially in the complex network of GSH biosynthesis is very important. Previous studies have reported that overexpression of conventional Gsh1/Gsh2 pathway improved production of intracellular GSH along with accumulation of intermediate γ-GC which increased significantly when amino acid precursors were added [21]. In our study, as the intracellular γ-GC level was only increased slightly compared with that of original strain W303-1b when F pathway was introduced, we artificially introduced a flexible six-glycine linker between the two enzymes of G pathway as well as P pathway to improve the coupling efficiency by mimicking the native domain fusion of GshF. As expected, intracellular γ-GC amounts of W303-1b/FGP were still maintained at low levels even when amino acid precursors were added (Table 3).
Conclusions
In summary, a three-pathway combinatorial method of GSH biosynthesis presents higher GSH yield and utilization efficiency of amino acid precursors than that of a single pathway. This strategy is an important development toward improving industrial production of GSH using S. cerevisiae.
Methods
Strains
E. coli Trans1-T1 (TransGen, Beijing, China) was used for propagation and manipulation of the recombinant DNA. S. cerevisiae W303-1b (MATα ade2-1 leu2-3,112 his3-11,15 ura3-1 trp1-1) was used as host strain for DNA transformation and expression of recombinant genes. Yeast transformants were grown at 30 °C in YPD medium (10 g/L yeast extract, 20 g/L tryptone and 20 g/L glucose).
Plasmid construction and yeast transformation
All plasmids were constructed using conventional restriction enzyme-mediated cloning methods. Based on the nucleotide sequences of the target genes, development of primer sets were designed and used to amplify gene fragments by PCR (Additional file 1: Tables S1–S6). δ DNA-mediated integrative expression vectors as described in Ref. [26], and plasmids for deletion of GSH1 gene were constructed as shown in Additional file 1: Figures S1–S4. The resulting plasmids were linearized by digestion with restriction enzyme NotI and transformed into S. cerevisiae using lithium acetate method. The positive transformants were selected on YPD plates with addition of antibiotics geneticin (G418, 4 mg/mL) or hygromycin B (HygB, 1 mg/mL) and verified by DNA sequencing after the integrated gene fragments were amplified by PCR using the corresponding primers and the genomic DNA templates. The copy numbers of integrated genes were verified using real-time PCR (primers GshF-RT1/GshF-RT2, Additional file 1: Table S7) and calculated by a comparative CT method [27]. For each transformation, three positive colonies were picked randomly for further investigation. The repeated use of the marker genes were performed via a loxP-marker-loxP gene disruption cassette according to Ref. [28].
Patch assay
Single colonies of strains W303-1b, W303-1b/Δgsh1, W303-1b/Δgsh1/pro1 and W303-1b/Δgsh1/proB were diluted in distilled H2O and patched (about 500 cells/μL) on YPD plates containing various concentrations of H2O2. Plates were incubated for about 72 h at 30 °C to compare H2O2 tolerances by watching their growth.
GSH production of the engineered S. cerevisiae strains in shake flasks
Three positive colonies of each engineered S. cerevisiae strain were cultured at 30 °C in shake flasks containing 20 mL of liquid YPD medium with agitation at 250 rpm for 18–24 h for primary culture. An adequate volume of fresh cultures were then inoculated into a set of 250-mL flasks containing 50 mL of liquid YPD medium to keep an initial optical density at 600 nm (OD600) value of 0.2. The cultures were incubated at 30 °C with agitation at 250 rpm for additional 48 h. In order to further increase the production of GSH, the addition of amino acid precursors (5 mM glutamic acid, 5 mM cysteine and 5 mM glycine) was single shot after 36-h cultivation for feeding assay.
Batch culture of the engineered strain W303-1b/FGP for GSH production
The batch fermentation was operated in a 10-L fermentor (China Beauty). Liquid YPD medium for batch culture was sterilized by autoclave at 121 °C for 20 min. Engineered strain W303-1b/FGP grown on YPD agar plate was inoculated into liquid YPD medium and cultured at 30 °C with agitation at 250 rpm for 18–24 h. 10 % (v/v) seed cultures were transferred into the fermentor. The dissolved oxygen (DO) was maintained at 25 % through adjustment of stirrer speed (100–500 rpm) and the aeration rate (1–5 L/h). The temperature was set to 30 °C and the pH was uncontrolled.
Analytical methods
Grown cultures were removed from the incubator at a given time and biomass was measured as optical density at 600 nm (OD600) and/or the dry cell weight (DCW). OD600 was determined after the sample was appropriately diluted. DCW (g/L) was determined after yeast cells were centrifuged at 4000 rpm for 10 min and washed twice with distilled water before drying at 105 °C to constant weight. OD600 was converted to DCW using calibration coefficient (OD600 = 10, equivalent to 1.19 g/L DCW in this study). To monitor the intracellular GSH and γ-GC amounts, cells were harvested and washed with distilled water. Intracellular GSH and γ-GC were extracted from the cells by 40 % ethanol for 2 h at 30 °C. The GSH and γ-GC concentrations (mg/L) of the sample were measured using the 4-fluoro-7-aminosulfonylbenzofurazan (ABD-F) derivatization method (Additional file 1: Figure S5) according to a previous report [29] with the reduced GSH as the standard. The GSH content (%) was calculated from the intracellular GSH concentration (mg/L) divided by DCW.
Authors’ contributions
LT, WW, WZ and KC performed the experiments. LT, WW, YY and ML analyzed the data, and LT drafted the manuscript. KC commented on the manuscript. WW conceived and coordinated the study and revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This work was financially supported by the National Science Foundation of China (Grant No. 30772677, 81072562), the Fundamental Research Funds for the Central Universities (Grant No. 2012N06), and the National Science and Technology Project of China (Grant No. 2012ZX09301002-001).
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Additional file
Additional file 1. Table S1. Primers used for cloning of A. pleuropneumoniae gshF gene optimized according to codon usage of S. cerevisiae. Table S2. Primers used for cloning of S. cerevisiae GSH2 gene. Table S3. Primers used for cloning of S. cerevisiae GSH1 gene. Table S4. Primers used for cloning of S. cerevisiae PRO1 gene. Table S5. Primers used for cloning of E. coli proB mutant optimized according to codon usage of S. cerevisiae. Table S6. Primers used for cloning of E. coli gshB mutant optimized according to codon usage of S. cerevisiae. Table S7. Primers used for real-time PCR to verify copy number of gshF gene. Table S8. Relative quantitation of copy number of gshF gene. Figure S1. Construction of integrative expression vectors pδGAPg-gshF, pδGAP′g-gshF, pδGAL1g-gshF and pδPGK1g-gshF used to evaluate effects of different promoter strengths on the activity of GshF. Figure S2. Construction of integrative expression vector pδGAPh-gsh2gsh1 for overexpression of Gsh2-Gsh1 fusion protein. Figure S3. Construction of integrative expression vectors pδGAPg-pro1gshB and pδGAPg-proBgshB for overexpression of Pro1-GshB and ProB-GshB fusion proteins. Figure S4. Construction of plasmids pΔgsh1h-pro1, pΔgsh1h-proB and pΔgsh1h used for deletion of GSH1 gene. Figure S5. Chromatograms of standard thiols and samples labelled with ABD-F.
Footnotes
Liang Tang and Weiwei Wang contributed equally to this work
Contributor Information
Liang Tang, Email: tangl@imm.ac.cn.
Weiwei Wang, Email: wangweiwei_fly@126.com.
Wenlong Zhou, Email: zhouwl@imm.ac.cn.
Kai Cheng, Email: 492793582@qq.com.
Yan Yang, Email: yangyan@imm.ac.cn.
Minzhi Liu, Email: lmzluoxue@imm.ac.cn.
Kedi Cheng, Email: chengkd@imm.ac.cn.
Wei Wang, Email: wwang@imm.ac.cn.
References
- 1.Meister A, Anderson ME. Glutathione. Annu Rev Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 2.Penninckx MJ, Elskens MT. Metabolism and functions of glutathione in micro-organisms. Adv Microb Physiol. 1993;34:239–301. doi: 10.1016/S0065-2911(08)60031-4. [DOI] [PubMed] [Google Scholar]
- 3.Jones DP. Redefining oxidative stress. Antioxid Redox Signal. 2006;8:1865–1879. doi: 10.1089/ars.2006.8.1865. [DOI] [PubMed] [Google Scholar]
- 4.Bock KW, Lilienblum W, Fischer G, Schirmer G, Bock-Hennig BS. The role of conjugation reactions in detoxication. Arch Toxicol. 1987;60:22–29. doi: 10.1007/BF00296941. [DOI] [PubMed] [Google Scholar]
- 5.Ketterer B, Coles B, Meyer DJ. The role of glutathione in detoxication. Environ Health Perspect. 1983;49:59–69. doi: 10.1289/ehp.834959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dröge W, Breitkreutz R. Glutathione and immune function. Proc Nutr Soc. 2000;59:595–600. doi: 10.1017/S0029665100000847. [DOI] [PubMed] [Google Scholar]
- 7.Richman PG, Meister A. Regulation of gamma-glutamyl–cysteine synthetase by nonallosteric feedback inhibition by glutathione. J Biol Chem. 1975;250:1422–1426. [PubMed] [Google Scholar]
- 8.Gopal S, Borovok I, Ofer A, Yanku M, Cohen G, Goebel W, et al. A multidomain fusion protein in Listeria monocytogenes catalyzes the two primary activities for glutathione biosynthesis. J Bacteriol. 2005;187:3839–3847. doi: 10.1128/JB.187.11.3839-3847.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janowiak BE, Griffith OW. Glutathione synthesis in Streptococcus agalactiae one protein accounts for γ-glutamylcysteine synthetase and glutathione synthetase activities. J Biol Chem. 2005;280:11829–11839. doi: 10.1074/jbc.M414326200. [DOI] [PubMed] [Google Scholar]
- 10.Vergauwen B, De Vos D, Van Beeumen JJ. Characterization of the bifunctional γ-glutamate–cysteine ligase/glutathione synthetase (GshF) of Pasteurella multocida. J Biol Chem. 2006;281:4380–4394. doi: 10.1074/jbc.M509517200. [DOI] [PubMed] [Google Scholar]
- 11.Yang J. Cloning, expression, characteristics and application of novel bifunctional glutathione synthetase. Master’s Thesis, East China University of Science and Technology, CN. 2012. (In Chinese).
- 12.Veeravalli K, Boyd D, Iverson BL, Beckwith J, Georgiou G. Laboratory evolution of glutathione biosynthesis reveals natural compensatory pathways. Nat Chem Biol. 2011;7:101–105. doi: 10.1038/nchembio.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spector D, Labarre J, Toledano MB. A genetic investigation of the essential role of glutathione mutations in the proline biosynthesis pathway are the only suppressors of glutathione auxotrophy in yeast. J Biol Chem. 2001;276:7011–7016. doi: 10.1074/jbc.M009814200. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Wei G, Chen J. Glutathione: a review on biotechnological production. Appl Microbiol Biotechnol. 2004;66:233–242. doi: 10.1007/s00253-004-1751-y. [DOI] [PubMed] [Google Scholar]
- 15.Liao XY, Shen W, Chen J, Li Y, Du GC. Improved glutathione production by gene expression in Escherichia coli. Lett Appl Microbiol. 2006;43:211–214. doi: 10.1111/j.1472-765X.2006.01927.x. [DOI] [PubMed] [Google Scholar]
- 16.Fei L, Wang Y, Chen S. Improved glutathione production by gene expression in Pichia pastoris. Bioprocess Biosyst Eng. 2009;32:729–735. doi: 10.1007/s00449-009-0297-x. [DOI] [PubMed] [Google Scholar]
- 17.Ohtake Y, Watanabe K, Tezuka H, Ogata T, Yabuuchi S, Murata K, et al. The expression of the γ-glutamylcysteine synthetase gene of Escherichia coli B in Saccharomyces cerevisiae. Agric Biol Chem. 1988;52:2753–2762. doi: 10.1271/bbb1961.52.2753. [DOI] [Google Scholar]
- 18.Fan X, He X, Guo X, Qu N, Wang C, Zhang B. Increasing glutathione formation by functional expression of the γ-glutamylcysteine synthetase gene in Saccharomyces cerevisiae. Biotechnol Lett. 2004;26:415–417. doi: 10.1023/B:BILE.0000018261.14427.e8. [DOI] [PubMed] [Google Scholar]
- 19.Hara KY, Kiriyama K, Inagaki A, Nakayama H, Kondo A. Improvement of glutathione production by metabolic engineering the sulfate assimilation pathway of Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2012;94:1313–1319. doi: 10.1007/s00253-011-3841-y. [DOI] [PubMed] [Google Scholar]
- 20.Li W, Li Z, Yang J, Ye Q. Production of glutathione using a bifunctional enzyme encoded by gshF from Streptococcus thermophilus expressed in Escherichia coli. J Biotechnol. 2011;154:261–268. doi: 10.1016/j.jbiotec.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 21.Ge S, Zhu T, Li Y. Expression of bacterial GshF in Pichia pastoris for Glutathione Production. Appl Environ Microbiol. 2012;78:5435–5439. doi: 10.1128/AEM.00509-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu ZQ, Deng ZJ, Tan HM, Zhou SN, Cao LX. Engineering the robustness of Saccharomyces cerevisiae by introducing bifunctional glutathione synthase gene. J Ind Microbiol Biotechnol. 2015;42:537–542. doi: 10.1007/s10295-014-1573-6. [DOI] [PubMed] [Google Scholar]
- 23.Alfafara C, Miura K, Shimizu H, Shioya S, Suga KI. Cysteine addition strategy for maximum glutathione production in fed-batch culture of Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 1992;37:141–146. [Google Scholar]
- 24.Wen S, Zhang T, Tan T. Utilization of amino acids to enhance glutathione production in Saccharomyces cerevisiae. Enzyme Microb Technol. 2004;35:501–507. doi: 10.1016/j.enzmictec.2004.08.003. [DOI] [Google Scholar]
- 25.Nisamedtinov I, Kevvai K, Orumets K, Arike L, Sarand I, Korhola M, et al. Metabolic changes underlying the higher accumulation of glutathione in Saccharomyces cerevisiae mutants. Appl Microbiol Biotechnol. 2011;89:1029–1037. doi: 10.1007/s00253-010-2946-z. [DOI] [PubMed] [Google Scholar]
- 26.Lee FWF, Da Silva NA. Improved efficiency and stability of multiple cloned gene insertions at the δ sequences of Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 1997;48:339–345. doi: 10.1007/s002530051059. [DOI] [PubMed] [Google Scholar]
- 27.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 28.Güldener U, Heck S, Fiedler T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steele ML, Ooi L, Münch G. Development of a high-performance liquid chromatography method for the simultaneous quantitation of glutathione and related thiols. Anal Biochem. 2012;429:45–52. doi: 10.1016/j.ab.2012.06.023. [DOI] [PubMed] [Google Scholar]


