Abstract
Background
Malaria cases were estimated to 207 million in 2013. One of the problems of malaria control is the emergence and spread of Plasmodium falciparum strains that become resistant to almost all drugs available. Monitoring drug resistance is essential for early detection and subsequent prevention of the spread of drug resistance by timely changes of treatment policy. This review was performed to gather all data available on P. falciparum molecular resistance in DR Congo, as baseline for future assessments.
Methods
The search for this review was undertaken using the electronic databases PubMed and Google Scholar using the terms “malaria”, “Congo”, “resistance”, “molecular”, “antimalarial”, “efficacy”. Articles were classified based on year of collecting, year of publication, sample size and characteristics, molecular markers analysed and polymorphisms detected.
Results
Thirteen articles were included and five genes have been analysed in these studies: pfcrt, pfdhps, pfdhfr, pfmdr1 and K13-propeller. The majority of studies included were not representative of the whole country.
Conclusion
This systematic review demonstrates the lack of molecular resistance studies in DRC. Only 13 studies were identified in almost 15 years. The MOH must implement a national surveillance system for monitoring malaria drug resistance and this surveillance should be conducted frequently and country-representative.
Keywords: Malaria, DR Congo, Molecular, Drug resistance
Background
Malaria cases were estimated to 207 million in 2013, among with 584,000 deaths, 90 % found in Africa, Democratic Republic of Congo (DRC) and Nigeria together accounting for 40 % of the estimated global total [1]. The absence to date of an effective vaccine leaves us only chemotherapy to fight against Plasmodium falciparum infection, the most virulent Plasmodium species that infects humans. One of the problems of malaria control is the emergence and spread of P. falciparum strains that become resistant to almost all drugs available.
Chloroquine (CQ) was one of the most used molecules in the fight against malaria because of its cost-effectiveness, but just some years after being placed on the market, the first cases of chloroquine resistance emerged in Southeast Asia, then in Latin America before spreading to all endemic areas [2]. Later, this phenomenon was repeated for the other available drugs (proguanil, sulfadoxine–pyrimethamine, halofantrine, mefloquine). To prevent, or at least delay the onset of new resistant strains, WHO recommended in 2001 the use of drugs association and that one of the drugs be an artemisinin derivative [3]. At that time, no artemisinin resistance had been identified yet and many hopes were placed onto artemisinins. Unfortunately, in 2008 Noedl et al. reported evidence of artemisinin resistance in Western Cambodia [4]. To date, artemisinin resistance has spread to Thailand, Myanmar, Laos and Vietnam [5–7]. In 2014, Ariey et al. identified a molecular marker for artemisinin resistance [8].
Monitoring drug resistance is essential for early detection and subsequent prevention of the spread of drug resistance by timely changes of treatment policy. There are three possibilities to study drug resistance: in vitro tests, in vivo tests and study of molecular markers [9]. In vivo tests are the gold standard, as involving the three factors (human host, parasite and anti-malarial molecule), but they are difficult to implement and very expensive due to the heavy organizational machinery that must be in place (drugs, monitoring of patients for 24 or 48 days, repeated biological tests, an experienced staff).
In vitro tests allow studying the inhibition of parasite growth in culture subjected to different concentrations of an anti-malarial. Their results do not reflect a real situation where parasites can survive in vivo in presence of an optimal serum concentration of the drug, as they do not consider host immunity or drug pharmacokinetics. These tests are also financially heavy and require live parasites, qualified persons for its implementation and laboratory facilities for in vitro culture. Finally, the study of molecular markers is a good alternative although the factors related to the host and the drugs are not taken into account. They can explore a wide range of markers (previously identified as related to resistance) onto large samples and in punctual manner. So the analysis of molecular markers gives a snapshot of the situation at a given time [10]. In DRC, which is the second largest African country, CQ was widely used for years till its withdrawal in 2002, first replaced by sulfadoxine–pyrimethamine (SP), then by artemisinin-based combination therapy (ACT) [11]. Due to its size, studying malaria resistance in DRC by in vitro or in vivo methods would be very expensive and time-consuming; analysing molecular markers would be a better alternative.
This systematic review was performed to gather all data available on P. falciparum molecular resistance in DRC, as baseline for future assessments.
Methods
The search for this review was conducted during November 2014 and was undertaken using the electronic databases Pub Med and Google scholar. The following search terms “malaria”, “Congo”, “resistance”, “molecular”, “antimalarial”, “efficacy” were used. Additional results were obtained from the references in the articles identified through the search. Selection criteria were: (a) samples related to malaria infection in DRC; (b) study comprising analysis of one or more malaria resistance molecular marker; (c) original articles, short reports but no review articles. Full-text articles were read to check for selection criteria. An overall of 34 articles has been identified among which 16 were non-molecular studies and 5 were review articles or duplicates (based on same samples). Thirteen articles were finally included. Articles were classified based on year of collecting, year of publication, sample size and characteristics, molecular markers analyzed and polymorphisms detected. Figure 1 shows the search strategy used.
Fig. 1.
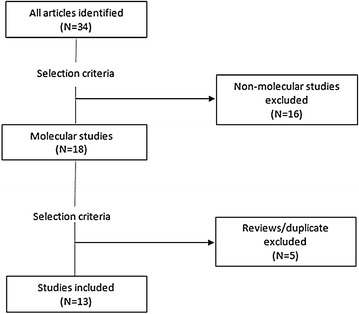
Search strategy. Twenty-one studies have been excluded after application of selection criteria
Results
The overall of the articles included in this review were of samples collected from 1999 to 2014 [12–24]. Some of them concerned non-Congolese people but supposed to have been infected in DRC. Five genes have been analysed in these studies: the Plasmodium falciparum chloroquine resistance transporter gene (pfcrt), the dihydropteroate synthase gene (pfdhps), the dihydrofolate reductase gene (pfdhfr), the Plasmodium falciparum multidrug resistance 1 gene (pfmdr1) and the K13-propeller gene (k13).
The most analysed gene was pfcrt, linked to CQ and amodiaquine (AMQ) resistance [25, 26], and it has been studied in seven articles (53.8 %). Pfdhps, known to be related to sulfadoxine resistance [27] and pfdhfr for which evidence of point mutations conferring resistance to pyrimethamine has been made [28], have been respectively analysed in five (38.4 %) and four articles (30.7 %). Analysis concerning K13-propeller and the pfmdr1 genes have been found, respectively in two (15.3 %) and one (7.6 %) studies. Pfmdr1 is correlated to resistance to many drugs as mefloquine (MQ), lumefantrine (LMF), AMQ and may be to artemisinin [29–32] and k13 is correlated to artemisinin resistance [8]. Table 1 provides a summary of the studies included in the review.
Table 1.
Summary of the studies included in the review
| Authors | Year of collecting | Year of publication | Sample sizea | Age | Molecular markers | |
|---|---|---|---|---|---|---|
| 1. | Andriantsoanirina et al. | 1999, 2002, 2005 | 2010 | 3 | – | pfcrt |
| 2. | Severini et al. | 2000 | 2005 | 32 | – | pfcrt |
| 3. | Wilson et al. | 2002 | 2005 | 56 | 6–58 months | pfcrt |
| 4. | Alker et al. | 2002 | 2008 | 249 | 6–59 months | pfdhfr, pfdhps |
| 5. | Cohuet et al. | 2003–2004 | 2006 | 458 | – | pfdhfr, pfdhps |
| 6. | Swarthout et al. | 2004 | 2006 | 168 | 6–59 months | pfdhfr, pfdhps |
| 7. | Taylor et al. | 2007 | 2013 | 179 | >15 years | dhps |
| 8. | Antonia et al. | 2007 | 2014 | 180 | >15 years | pfcrt |
| 9. | Taylor et al. | 2007 | 2014 | 151 | >15 years | K13 |
| 10. | Mobula et al. | 2008 | 2009 | 142 | 1–10 years | pfcrt, pfmdr1, pfdhfr, pfdhps |
| 11. | Juliao et al. | 2010 | 2013 | 12 | 19–55 years | pfcrt |
| 12. | Mvumbi et al. | 2010 | 2013 | 145 | 6–59 months | pfcrt |
| 13. | Kamau et al. | 2013–2014 | 2014 | 82 | >6 years | K13 |
aConsidering only samples from DRC, not the total sample size of the study
pfcrt gene
On the seven studies involving the K76T mutation, four have also explored the haplotypes defined by specific mutations at amino acid positions 72–76 (related to AMQ resistance). The main findings of these articles are given in Table 2.
Table 2.
Main findings of studies that analysed the pfcrt gene
| Authors | Year of collecting | Haplotypes found | Mutant 76T | ||
|---|---|---|---|---|---|
| 1. | Andriantsoanirina et al. | 1999, 2002, 2005 | _ | 3/3 | 100 % |
| 2. | Severini et al. | 2000 | CVIET, CVMNT, SVIET | 27/27 | 100 % |
| 3. | Wilson et al. | 2002 | _ | 52/56 | 93 % |
| 4. | Antonia et al. | 2007 | CVMNK, CVIET, CVMNT, CVMDK | 92/166 | 55.4 % |
| 5. | Mobula et al. | 2008 | _ | 88/105 | 83.8 % |
| 6. | Juliao et al. | 2010 | CVMNK, CVIET | 1/12 | 8 % |
| 7. | Mvumbi et al. | 2010 | CVMNK, CVIET | 148/198 | 73.2 % |
pfdhfr and pfdhps genes
Both genes are generally analysed together as they are related to SP combination susceptibility. Only one study analysed the dhps gene alone. Most point mutations analysed were on positions 51, 59 and 108 on dhfr and on positions 437 and 540 on dhps. Prevalence of point mutations found in these studies is presented in Table 3.
Table 3.
Prevalence of point mutations found in dhfr/dhps genes from included studies
| Authors | Year of collecting | Dhps mutations | Dhfr mutations | Quint. mutant (dhfr and dhps) | Wild | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A437G | K540E | Double mutants | N51I | C59R | S108N | Triple mutants | – | dhps | dhfr | both | |||
| 1. | Alker et al. | 2002 | 72 % | 67 % | – | >95 % | 66 % | >95 % | – | 43 % | – | – | 1 % |
| 2. | Cohuet et al. | 2003–2004 | – | – | <20 % | – | – | – | >50 % | 0.9–12.8 % | 29.4–75.6 % | 1–3.9 % | – |
| 3. | Swarthout et al. | 2004 | 0.6 % | – | 45.2 % | – | – | 3.2 % | 46.2 % | 27.1 % | 30 % | 7.6 % | 3 % |
| 4 | Taylor et al. | 2007 | – | – | 14.6 % | – | – | – | – | – | 35.1 % | – | – |
| 5. | Mobula et al. | 2008 | 93.1 % | 9.5 % | – | 97.9 % | 80.7 % | 99.1 % | – | – | – | – | – |
K13-propeller gene
Recently linked to artemisinin resistance, this gene has been analysed twice in sample from DRC. The older samples are about 2007 and the recent one is of 2013–2014. None of mutations related to artemisinin resistance discovered in South-East Asia were found in DRC to date. All mutations found are shown in Table 4.
Table 4.
Mutations found on k13 in DRC samples
| Authors | Year of collecting | Mutations found | |
|---|---|---|---|
| 1. | Kamau et al. | 2013–2014 | T493T, A578S |
| 2. | Taylor et al. | 2007 | R471R, G496G, R513R, V520A, A557S, V581V, A617T, V637A |
pfmdr1 gene
Only one study analysed this gene in Kinshasa. Five point mutations were explored on codons positions 86, 184, 1034, 1042 and 1046. The N86Y mutation was present in 66.7 % of samples studied [20].
Discussion
During almost 15 years, only 13 molecular studies were conducted based on samples from the DRC. These studies were of samples from 3 to 458 individuals (mean = 142) and that usually were focalized to one or two geographical sites. However, three studies [18, 19, 21] were based on samples collected in the whole country as part of the Demographic and Health Survey (DHS), funded mostly by the international community [33]. Considering the size of the DRC (second largest African country with 2,345,000 km2) and the different patterns of malaria from one corner to another, results of most of these studies are not generalizable to the nationwide, but must be considered in their local context.
Sometimes for the same molecular markers in the same study, differences are significant in two different sites. The study of Cohuet et al. [16] illustrates well this fact: the prevalence of dhps double mutants (resistance to sulfadoxine) was estimated at less than 1 % in a site (Basankusu) whereas it was close to 20 % in another (Kisangani). This would like to suggest that the molecule remains active in one place while it should be replaced by another. As WHO recommends withdrawal of a drug if the prevalence of resistant is above 10 %, this suggests in that case that the drug should be replaced in one place while it should be maintained in another. This underlines the fact that making national decision about treatment policy could not be based only on “localized” studies. The Ministry of Health should initiate, regularly, anti-malarial drugs efficacy monitoring studies but with a representative sampling. This scarcity of representative studies across the country is a real handicap. Treatment policies are applied in light of the available study (and thus the geographical location concerned). Thus, when changing the SP to ACT in the DRC, one of the studies available to support the selection of the combination AS + AQ compared with AS + SP were localized to one site, Shabunda (East of the DRC) [17]. Maybe if another study was conducted simultaneously in the West of the country, the data would have been different. The main hurdle is generally the high cost of surveillance studies, in particular for molecular studies that require equipment and reagents often unavailable locally. Thus, none of molecular analysis performed for these 13 studies have been performed in a Congolese laboratory. All of the molecular analysis was carried out in Europe or in USA. Another implication of this high cost is that making these studies in a repetitive manner is very difficult. Yet the WHO experts recommend that the surveillance of the resistance must be carried out every 2 years in endemic countries [34].
The various data existing on the pfcrt gene confirm the decline of CQ resistant strains after its withdrawal. The latest available figures (which only concern Kinshasa) revealed a prevalence of 73.2 % [23], whereas previous studies before chloroquine replacement in 2002 were giving values beyond 90 % [12–14]. Unfortunately, as mentioned above, it is difficult to correctly compare these studies because of the differences in the applied methodology (essentially the sampling and the study location). It is important to notice that none of the studies concerning the pfcrt gene have detected the presence of the SVMNT haplotype (linked to AMQ resistance) in the DRC. As this haplotype was recently found in Angola [35] and Tanzania [36], two countries bordering the DRC, it would be prudent that a continuous and regular monitoring of the occurrence of this haplotype be conducted.
The SP was quickly removed for malaria first-line-treatment in malaria endemic areas when many reports showed strong resistance rates. Studies conducted in the DRC have also confirmed it, but with important inter-sites and inter-studies variations [15–18]. It is not easy to say that SP resistance is declining or increasing because there were not two repetitive studies conducted in the same place, with the same methodology and at different time. Besides, it appears that SP resistance is more pronounced in the East than the West of the DRC. SP is nevertheless still used for intermittent preventive treatment in pregnant women (IPTp) and none of these studies assessed the impact of the use of this molecule in high resistance area.
At the time of the use of ACT, it is very important to monitor the emergence of artemisinin resistant strains in Africa. Indeed, this is the first time that a resistance molecular marker is identified before the resistant strains spreads to all malaria endemic regions. Actually, two studies were related to DRC samples on the k13-propeller gene [21, 24]. None of polymorphisms identified in Southeast Asia have been found in the DRC but new mutations on k13 have been discovered in Africa. Further studies should be conducted to assess the impact of these new mutations on susceptibility to artemisinin.
Conclusion
Only 13 studies were identified in this review and the majority of them were not representative of the whole country. So their results are not generalizable. DRC is a very large country and malaria responses to drugs are very different from one place to another. Ministry of Health must implement a national surveillance system for monitoring malaria drug resistance and this surveillance should be conducted frequently, in many sentinel sites and this especially to react as fast as the occurrence of artemisinin-resistant or AMQ-resistant strains in a part of the DRC.
Authors’ contributions
DMM and TB make the web search. DMM and HS drafted the manuscript. JK, CN and GM read full-text articles and reviewed manuscript. MPH and PDM reviewed manuscript. All authors read and approved the final manuscript.
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Contributor Information
Dieudonné Makaba Mvumbi, Email: didimvumbi@gmail.com.
Jean-Marie Kayembe, Email: dr12jmkayembe@yahoo.com.
Hippolyte Situakibanza, Email: hsitua@gmail.com.
Thierry L. Bobanga, Email: tbobanga@gmail.com
Célestin N. Nsibu, Email: celnsibu@hotmail.fr
Georges L. Mvumbi, Email: mvumbilelo@yahoo.fr
Pierrette Melin, Email: pierrette.melin@chu.ulg.ac.be.
Patrick De Mol, Email: P.DeMol@ulg.ac.be.
Marie-Pierre Hayette, Email: mphayette@chu.ulg.ac.be.
References
- 1.WHO . World malaria report 2014. Geneva: World Health Organization; 2014. [Google Scholar]
- 2.Wellems T, Plowe C. Chloroquine-resistance malaria. J Infect Dis. 2001;184:770–776. doi: 10.1086/322858. [DOI] [PubMed] [Google Scholar]
- 3.WHO. Antimalarial drug combination therapy. Report of a WHO technical consultation. Geneva: World Health Organization; 2001.
- 4.Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM. Evidence of artemisinin-resistant malaria in Western Cambodia. N Engl J Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 5.Phyo AP, Nkhoma S, Stepniewska K, Ashley EA, Nair S, McGready R, et al. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet. 2012;379:1960–1966. doi: 10.1016/S0140-6736(12)60484-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kyaw MP, Nyunt MH, Chit K, Aye MM, Aye KH, Lindegardh N, et al. Reduced susceptibility of Plasmodium falciparum to artesunate in southern Myanmar. PLoS One. 2013;8:e57689. doi: 10.1371/journal.pone.0057689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.WHO. Assessment and monitoring of antimalarial drug efficacy for the treatment of uncomplicated falciparum malaria. WHO/HTM/RBM/2003.50. Geneva: World Health Organization; 2003.
- 10.WHO . Global report on antimalarial drug efficacy and drug resistance: 2000–2010. Geneva: World Health Organization; 2010. [Google Scholar]
- 11.Kazadi WM, Vong S, Makina BN, Mantshumba JC, Kabuya W, Kebela B, et al. Assessing the efficacy of chloroquine and sulfadoxine–pyrimethamine for treatment of uncomplicated Plasmodium falciparum malaria in the Democratic Republic of Congo. Trop Med Int Health. 2003;8:868–875. doi: 10.1046/j.1365-3156.2003.01098.x. [DOI] [PubMed] [Google Scholar]
- 12.Andriantsoanirina V, Ménard D, Rabearimanana S, Hubert V, Bouchier C, Tichit M, et al. Association of microsatellite variations of Plasmodium falciparum Na+/H+ exchanger (Pfnhe-1) gene with reduced in vitro susceptibility to quinine: lack of confirmation in clinical isolates from Africa. Am J Trop Med Hyg. 2010;82:782–787. doi: 10.4269/ajtmh.2010.09-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Severini C, Menegon M, Sannella AR, Paglia MG, Narciso P, Matteelli A, et al. Prevalence of pfcrt point mutations and level of chloroquine resistance in Plasmodium falciparum isolates from Africa. Infect Genet Evol. 2006;6:262–268. doi: 10.1016/j.meegid.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Wilson PE, Kazadi W, Kamwendo DD, Mwapasa V, Purfield A, Meshnick SR. Prevalence of pfcrt mutations in Congolese and Malawian Plasmodium falciparum isolates as determined by a new Taqman assay. Acta Trop. 2005;93:97–106. doi: 10.1016/j.actatropica.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 15.Alker AP, Kazadi WM, Kutelemeni AK, Boland PB, Tshefu AK, Meshnick SR. dhfr and dhps genotype and sulfadoxine–pyrimethamine treatment failure in children with falciparum malaria in the Democratic Republic of Congo. Trop Med Int Health. 2008;13:1384–1391. doi: 10.1111/j.1365-3156.2008.02150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohuet S, Bonnet M, van Herp M, van Overmeir C, d’Allessandro U, Guthmann JP. Molecular markers associated with Plasmodium falciparum resistance to sulfadoxine–pyrimethamine in the Democratic Republic of Congo. Am J Trop Med Hyg. 2006;75:152–154. [PubMed] [Google Scholar]
- 17.Swarthout TD, van den Broek IV, Kayembe G, Montgomery J, Pota H, Roper C. Artesunate + amodiaquine and artesunate + sulphadoxine–pyrimethamine for treatment of uncomplicated malaria in Democratic Republic of Congo: a clinical trial with determination of sulphadoxine and pyrimethamine-resistant haplotypes. Trop Med Int Health. 2006;11:1503–1511. doi: 10.1111/j.1365-3156.2006.01710.x. [DOI] [PubMed] [Google Scholar]
- 18.Taylor SM, Antonia AL, Parobek CM, Juliano JJ, Janko M, Emch M, et al. Plasmodium falciparum sulfadoxine resistance is geographically and genetically clustered within the DR Congo. Sci Rep. 2013;3:1165. doi: 10.1038/srep01165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antonia AL, Taylor SM, Janko M, Emch M, Tshefu AK, Meshnick SR. A cross-sectional survey of Plasmodium falciparum pfcrt mutant haplotypes in the Democratic Republic of Congo. Am J Trop Med Hyg. 2014;90:1094–1097. doi: 10.4269/ajtmh.13-0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mobula L, Lilley B, Tshefu AK, Rosenthal PJ. Resistance-mediating polymorphisms in Plasmodium falciparum infections in Kinshasa, Democratic Republic of the Congo. Am J Trop Med Hyg. 2009;80:555–558. [PubMed] [Google Scholar]
- 21.Taylor SM, Parobek CM, DeConti DK, Kayentao K, Coulibaly SO, Greenwood BM, et al. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in sub-Saharan Africa: a molecular epidemiologic study. J Infect Dis. 2015;211:680–688. doi: 10.1093/infdis/jiu467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juliao PC, Sosa S, Gonzalez LD, Padilla M, Ortiz L, Goldman I, et al. Importation of chloroquine-resistant Plasmodium falciparum by Guatemalan peacekeepers returning from the Democratic Republic of the Congo. Malar J. 2013;12:344. doi: 10.1186/1475-2875-12-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mvumbi DM, Boreux R, Sacheli R, Mvumbi L, Bobanga L, Situakibanza NT, et al. Assessment of pfcrt 72-76 haplotypes eight years after chloroquine withdrawal in Kinshasa, Democratic Republic of Congo. Malar J. 2013;12:459. doi: 10.1186/1475-2875-12-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamau E, Campino S, Amenga-Etego L, Drury E, Ishengoma D, Johnson K, et al. K13-propeller polymorphisms in Plasmodium falciparum parasites from sub-Saharan Africa. J Infect Dis. 2015;211:1352–1355. doi: 10.1093/infdis/jiu608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–871. doi: 10.1016/S1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beshir K, Sutherland CJ, Merinopoulos I, Durrani N, Leslie T, Rowland M, et al. Amodiaquine resistance in Plasmodium falciparum malaria in Afghanistan is associated with the pfcrt SVMNT allele at codons 72 to 76. Antimicrob Agents Chemother. 2010;12:3714–3716. doi: 10.1128/AAC.00358-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang P, Read M, Sims PF, Hyde JE. Sulfadoxine resistance in the human malaria parasite Plasmodium falciparum is determined by mutations in dihydropteroate synthetase and an additional factor associated with folate utilization. Mol Microbiol. 1997;23:979–986. doi: 10.1046/j.1365-2958.1997.2821646.x. [DOI] [PubMed] [Google Scholar]
- 28.Peterson DS, Walliker D, Wellems TE. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc Natl Acad Sci USA. 1988;85:9114–9118. doi: 10.1073/pnas.85.23.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim HS, Okuda Y, Begum K, Nagai Y, Wataya Y, Kimura M, et al. Analysis of Pfmdr 1 gene in mefloquine-resistant Plasmodium falciparum. Nucleic Acids Res Suppl. 2001;1:231–232. doi: 10.1093/nass/1.1.231. [DOI] [PubMed] [Google Scholar]
- 30.Sisowath C, Ferreira PE, Bustamante LY, Dahlström S, Mårtensson A, Björkman A, et al. role of pfmdr1 in Plasmodium falciparum tolerance to artemether-lumefantrine in Africa. Trop Med Int Health. 2007;12:736–742. doi: 10.1111/j.1365-3156.2007.01843.x. [DOI] [PubMed] [Google Scholar]
- 31.Chavchich M, Gerena L, Peters J, Chen N, Cheng Q, Kyle DE. Role of pfmdr1 amplification and expression in induction of resistance to artemisinin derivatives in Plasmodium falciparum. Antimicrob Agents Chemother. 2010;54:2455–2464. doi: 10.1128/AAC.00947-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sidhu AB, Uhlemann AC, Valderramos SG, Valderramos JC, Krishna S, Fidock DA. Decreasing pfmdr1 copy number in Plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J Infect Dis. 2006;194:528–535. doi: 10.1086/507115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The Demographic and Health Surveys Program-Democratic Republic of Congo. 2007. http://dhsprogram.com/publications/publication-FR208-DHS-Final-Reports.cfm. Accessed 17 Nov 2014.
- 34.World Health Organization (WHO). Surveillance de la résistance aux antipaludiques, Rapport d’une consultation de l’OMS, Genève, Suisse, 3–5 décembre; 2001.
- 35.Gama BE, Pereira-Carvalho GA, Lutucuta Kosi FJ, Almeida de Oliveira NK, Fortes F, Rosenthal PJ, et al. Plasmodium falciparum isolates from Angola show the SVMNT haplotype in the pfcrt gene. Malar J. 2010;9:174. doi: 10.1186/1475-2875-9-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alifrangis M, Dalgaard MB, Lusingu JP, Vestergaard LS, Staalsoe T, Jensen AT, Enevold A, Ronn AM, Khalil IF, Warhurst DC, Lemnge MM, Theander TG, Bygbjerg IC. Occurrence of the Southeast Asian/South American SVMNT haplotype of the chloroquine-resistance transporter gene in Plasmodium falciparum in Tanzania. J Infect Dis. 2006;193:1738–41. doi: 10.1086/504269. [DOI] [PubMed] [Google Scholar]


