Abstract
The yeast Candida albicans can mate. However, in the natural environment mating may generate progeny (fusants) fitter than clonal lineages too rarely to render mating biologically significant: C. albicans has never been observed to mate in its natural environment, the human host, and the population structure of the species is largely clonal. It seems incapable of meiosis, and most isolates are diploid and carry both mating-type-like (MTL) locus alleles, preventing mating. Only chromosome loss or localized loss of heterozygosity can generate mating-competent cells, and recombination of parental alleles is limited. To determine if mating is a biologically significant process, we investigated if mating is under selection. The ratio of nonsynonymous to synonymous mutations in mating genes and the frequency of mutations abolishing mating indicated that mating is under selection. The MTL locus is located on chromosome 5, and when we induced chromosome 5 loss in 10 clinical isolates, most of the resulting MTL-homozygotes could mate with each other, producing fusants. In laboratory culture, a novel environment favoring novel genotypes, some fusants grew faster than their parents, in which loss of heterozygosity had reduced growth rates, and also faster than their MTL-heterozygous ancestors—albeit often only after serial propagation. In a small number of experiments in which co-inoculation of an oral colonization model with MTL-homozygotes yielded small numbers of fusants, their numbers declined over time relative to those of the parents. Overall, our results indicate that mating generates genotypes superior to existing MTL-heterozygotes often enough to be under selection.
Keywords: Candida albicans, mating, parasexual cycle, cryptic sexual cycle
SEX is costly and disrupts well-adapted allele combinations. It can also be advantageous by speeding up adaptation or by purging deleterious mutations. It has been difficult to establish how, precisely, these and other benefits outweigh the cost of sex. Indeed, abandoning sex in favor of clonal reproduction can be advantageous—asexual species arise frequently. Their life spans are short, however, indicating that sex may be essential for the long-term survival of species (Otto and Lenormand 2002; Rice 2002).
Sexual cycles have not been observed for ∼20% of fungal species (Carlile et al. 2001). Whether these species are truly asexual or merely restrict the frequency of sex—a strategy believed to maximize its benefits (Heitman 2006)—is difficult to determine. Genetic marker distributions in such species often suggest limited recombination (Carlile et al. 2001). However, clonal reproduction will initially copy the genetic marker distributions that were generated by sex, and new mutations and genetic drift will only slowly erase evidence of past recombination (Schmid et al. 2004; Cox et al. 2013). With rare exceptions, fungi without observable sexual cycles are derived from recent sexual ancestors (Carlile et al. 2001; Schmid et al. 2004; Butler 2007). Thus genetic marker distributions may reflect limited ongoing recombination or be an “echo” of recombination events that occurred in sexual ancestors.
The yeast Candida albicans, a commensal colonizer and occasional opportunistic pathogen of humans (Odds 1988), can mate, but it is a convoluted process. Most isolates are diploid and carry both mating-type-like (MTL) locus alleles, preventing mating. Apparently incapable of meiosis, loss of an entire copy of chromosome 5 (chr 5), on which the MTL locus is located, or localized loss of heterozygosity (LOH), generate MTL-homozygotes, which can mate. To do so they must, in addition, undergo an epigenetic switch from the “white” phenotype to the mating-competent “opaque” phenotype (Miller and Johnson 2002; Alby et al. 2009; Heitman 2010; Xie et al. 2013). A small fraction of mating encounters generates fusants with genetic markers from both parents (Lockhart et al. 2003; Bennett et al. 2005). These subsequently return to diploidy by chromosome loss (Bennett and Johnson 2003) and, in the process, intrachromosomal recombination can generate true recombinants (Forche et al. 2008). Alternatively, mating can be induced between haploids, which spontaneously arise from diploids by concerted chromosome loss, resulting immediately in diploid progeny (Hickman et al. 2013).
However, mating has never been observed in the human host, which is likely to be the natural environment of strains found on humans (Edelmann et al. 2005; Jacobsen et al. 2008; Wrobel et al. 2008) and the predominant environment of the species as a whole (Skinner and Fletcher 1960; Odds 1988). That mating is very rare is also indicated by the largely clonal population structure of the species (Gräser et al. 1996; Tibayrenc 1997; Tavanti et al. 2004; Bougnoux et al. 2008).
The existing data are consistent with two hypotheses: The first is that, while rare, mating in the natural environment produces progeny (fusant) lineages that surpass clonal lineages in fitness often enough to render the process biologically significant. If so, mutations that have turned mating into a convoluted process may have been selected because, for C. albicans, the benefits of mating are maximized by restricting its frequency (Heitman 2006). Alternatively, it is possible that mating so rarely generates fusants that are superior to clonal lineages that the process is no longer biologically significant. In that case, the mutations that make mating convoluted would indicate progressive mutational decay of a process that is no longer protected by selection—indeed mutations rendering strains entirely incapable of mating are not uncommon (Legrand et al. 2004). To distinguish between these two hypotheses we have investigated whether mating is under selection in C. albicans and, if so, why.
Materials and Methods
Analyses of dn/ds
The dn/ds analyses for the C. albicans mating genes MTLα1, MTLα2, MTLa1, and MTLa2 and their orthologs in other species were carried out using PAML (Yang 2007) as described previously (Zhang et al. 2010). Sequences were aligned and the M2 model in the codeml program from PAML v. 4.5 was used to model their evolution, allowing different dn/ds ratios for groups or species with full and cryptic sexual cycles in different parts of a phylogenetic tree based on ITS sequences (Supporting Information, Figure S1; the tree is consistent with relationships between these species established by other authors; Tavanti et al. 2005b; Wang et al. 2009; Butler 2010). The group dn/ds ratios reported in Results are those extracted from these models. We also used PAML to calculate dn/ds ratios from pairwise ortholog sequence comparisons between C. albicans and each of the three fully sexual species.
Strains, culture conditions, and growth rate comparisons
All strains used are listed in Tables 1-3. The 10 strains used for mating experiments are listed in Table 1. They were chosen (Holland and Schmid 2005) from a collection of 266 infection-causing isolates from 12 geographical regions in 6 countries (Schmid et al. 1999) so that 5 represented the GPG group (equivalent to clade 1; Schmid et al. 1999; Tavanti et al. 2005a; Odds et al. 2007) and 5 the remainder of the species. The strains had not been extensively subcultured after their isolation from patients and were revived from glycerol stocks for these experiments. Twenty-three MTL-homozygous clinical isolates used for growth-rate determinations were WO-1 (Slutsky et al. 1987; Miller and Johnson 2002); L26, GC75, p37005, 19F, p87, 12C, p60002, p78048, p57072, p94015 (Wu et al. 2007); 85/005, AM2005/0377, T101, M97105, 81/139, SCS103353G, SCS103354N, AM2003/018, AM2003/0165, AM2002/087, RIHO9, 81/196 (Schmid et al. 1999; Odds et al. 2007).
Table 1. Strains used for mating experiments: Clinical isolates.
| Strain | Ca3 typea | Geographical origin | Body site |
|---|---|---|---|
| AU35 | C | New Zealand | Sputum |
| AU7 | GPG A2 | New Zealand | Urine |
| Au90 | GPG A2 | New Zealand | Skin wound |
| RIHO11 | C | United States | Bloodstream |
| W43 | GPG A2 | New Zealand | Oral |
| FJ11 | C | Fiji | Throat |
| HUN97 | GPG A2 | Britain | Bloodstream |
| OD8916 | GPG A2 | Britain | Oral |
| W17 | C | New Zealand | Throat |
| YSU63 | C | Malaysia | Urine |
Source: Schmid et al. (1999).
Based on Ca3 fingerprinting; GPG, general-purpose genotype, equivalent to major group A, which is subdivided in to subgroups A1 and A2 (Schmid et al. 1999)
Table 3. Fusants used in growth experiments.
| Fusant | Fusant |
|---|---|
| AU35a × HUN97α.1 | AU90a × YSU63α.3 |
| AU35a × OD8916α.1 | RIHO11a × OD8916α.1 |
| AU35a × OD8916α.2 | RIHO11a × OD8916α.2 |
| AU35a × W17α.1 | RIHO11a × OD8916α.3 |
| AU35a × W17α.2 | RIHO11a × W17α.1 |
| AU35a × W17α.3 | RIHO11a × W17α.2 |
| AU35a × YSU63α.1 | RIHO11a × W17α.3 |
| AU35a × YSU63α.2 | RIHO11a × YSU63α.1 |
| AU35a × YSU63α.3 | W43a × OD8916α.1 |
| AU7a × YSU63α.1 | W43a × OD8916α.2 |
| AU90a × HUN97α.1 | W43a × OD8916α.3 |
| AU90a × HUN97α.2 | W43a × W17α.1 |
| AU90a × OD8916α.1 | W43a × W17α.2 |
| AU90a × W17α.1 | W43a × W17α.3 |
| AU90a × W17α.2 | W43a × YSU63α.1 |
| AU90a × YSU63α.1 | W43a × YSU63α.2 |
| AU90a × YSU63α.2 | W43a × YSU63α.3 |
All of the fusants are (MPAR) (NATR) MTLa/α. Names of fusants used under harsh conditions are underlined.
Serial transfers and growth-rate determinations were carried out under two conditions: Either (i) in YPD (1% yeast extract, 2% Bacto-peptone (BactoTM, Becton Dickinson, Sparks, MD) and 2% glucose (Asia Pacific Specialty Chemical Limited, NSW, Australia)) at 37° or (ii) in a defined medium containing 0.17% yeast nitrogen base without amino acids and ammonium sulfate (Becton and Dickinson), 0.5% ammonium sulfate, 0.08% glucose, 0.2 M NaCl (Goddard et al. 2005) at 42°.
During serial transfers, cultures were grown overnight, to saturation. Then 3.0 × 106 cells (from YPD medium) or 2 × 105 cells (from defined medium) were transferred for further propagation. The number of generations was calculated from the increase in cell number. Growth rates of exponentially growing cultures were determined by measuring optical density at 600 nm (OD600) at ≥ 8 time points. The growth rate was determined as the slope of the line of best fit in plots of ln(OD600) vs. time (r2 values >0.99). Growth rates that were to be compared with each other were, as far as possible, determined in the same experiments. See File S1 for details.
Selection of MTL-homozygous derivatives
MTL-homozygous derivatives were identified by PCR amplification of MTLa and MTLα alleles, either after inducing chr 5 loss on sorbose medium (Rustchenko et al. 1994) or from red sectors in colonies grown on YPD + phloxine B medium (Lockhart et al. 2002). See File S1 for details.
Resistance cassettes for selection of fusants
Strains to be used in mating experiments were transformed with a mycophenolic acid resistance (MPAr) cassette or a nourseothricin resistance cassette (NATr), by lithium acetate heat shock (Beckerman et al. 2001) or electroporation (De Backer et al. 1999), to allow selection of fusants on the basis of resistance to both drugs.
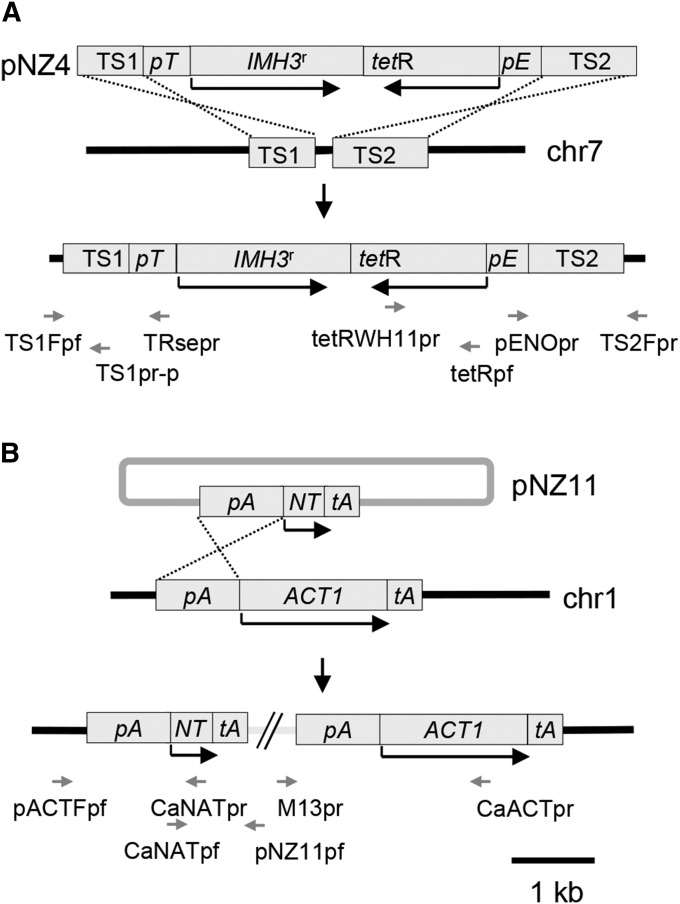
The MPA-resistance vector, pNZ4 (Figure 1A), contained the IMH3r gene under the control of a tetracycline-responsive promoter, the ORF encoding the tetracycline activator tetR–ScHAP4–WH11 under the control of the ENO1 promoter and two flanking sequences targeting its integration to a noncoding region of chr 7. It was constructed as follows: A fragment containing pENO plus tetR–ScHAP4–WH11 was amplified by PCR from the plasmid pCAITHE5 (Nakayama et al. 2000) with primers ENOpf and tetRWH11pr (the sequences of all primers used are given in Table S1). The fragment was cloned into SmaI-linearized plasmid pBSKS(+) to produce plasmid pNZ1. The tetracycline-responsive promoter (pTR) was PCR amplified from plasmid p99RLU (Nakayama et al. 2000) with primers TRpf and TRpr. The IMH3r ORF was amplified from plasmid p3408 (Beckerman et al. 2001) using primers IMH3pfatg and IMH3pr. Both fragments were purified using a PCR cleanup kit (Roche) and used as templates for recombinant PCR to combine the TR promoter and the IMH3r ORF using primers TRpf, TRpIMHp, and IMH3pr. The recombinant PCR product was cloned into EcoRV-linearized plasmid pBSKS(+) to produce plasmid pNZ2. The PCR reactions were carried out as described in File S1. Plasmid pNZ1 was cut with BamHI and treated with Klenow enzyme (Roche) to generate blunt ends. The cassette containing pTR and IMH3r was then removed from pNZ2 with SmaI and cloned into linearized pNZ1 resulting in plasmid pNZ3. To direct the integration of the resistance cassette to a noncoding region of the C. albicans genome, two targeting sequences, TS1 and TS2 (bp 367394–368202 and 366038–367194, respectively, on chr 7 of strain SC5314 as per genome assembly 21; http://www.candidagenome.org/) were PCR amplified with primers TS1pf/TS1pr and TS2pf/TS2pr and cloned into plasmid pGemT-Easy (Promega). TS1 and TS2 were removed from pGemT using NotI and HindIII, respectively, and cloned into pNZ3 to obtain pNZ4.
Figure 1.
Resistance cassettes used to mark parental strains by integration following transformation. (A) The pNZ4 cassette contains: two noncoding sequences, TS1 and TS2, from the region of chr 7 to which integration of the cassette is targeted; the MPA resistance-conferring IMH3 allele (IMH3r) under the control of a tetracycline-responsive promoter (pT); and the gene encoding the tetracycline transactivator (tetR) under control of the ENO1 promoter (pE). In the absence of tetracycline, the tetracycline activator induces the tetracycline-responsive promoter controlling IMH3r expression thus conferring resistance to MPA. Integration occurs by a double crossover in TS1 and TS2. (B) Plasmid pNZ11 contains, in a pBlueScript KS(+) backbone (the latter not drawn to scale): the CaNAT ORF (NT) under control of the ACT1 promoter (pA), followed by the ACT1 terminator region (tA). It integrates by a single crossover in the ACT1 promoter region on chr 1. Black arrows indicate ORFs, and grey arrows indicate the position of primers.
The NAT-resistance vector pNZ11 (designed so that integration would occur in chr 1; Figure 1B) was constructed as follows: The cassette containing a CaNAT-resistance gene flanked by an ACT1 promoter and the ACT1 terminator was PCR amplified from plasmid pJK850 (Shen et al. 2005) using primers M13pr and Sacpf1. The PCR product was purified and then digested with both KpnI and SacI and inserted into KpnI/SacI-digested vector pBlueScript KS(+) to obtain pNZ11.
All constructs were verified by DNA sequencing. The sequences of the pNZ4 and pNZ11 cassettes were submitted to GenBank (accession nos. FJ804172.1 and FJ804173.1).
Mating in laboratory culture
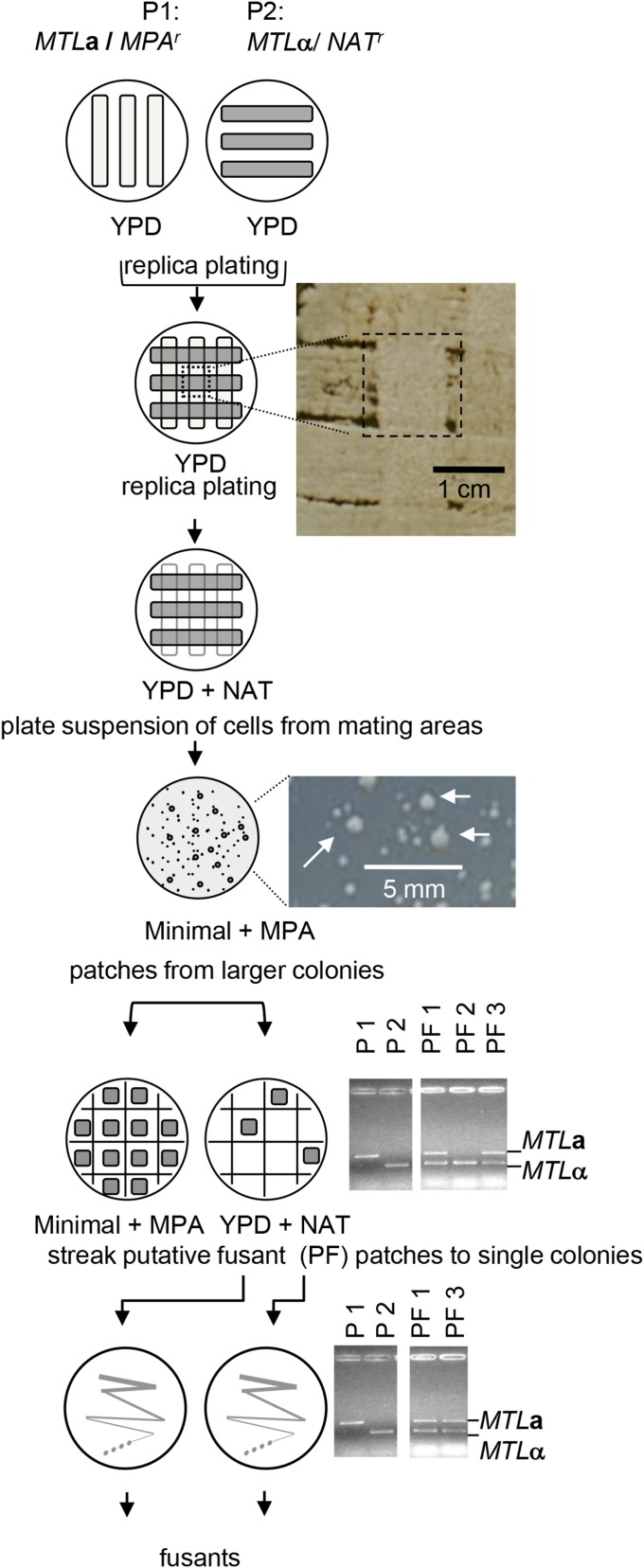
The mating protocol (Figure 2) was a modification of the method of Legrand et al. (2004). In brief, both parents were grown as broad bands across YPD plates and replica plated at right angles to each other on a single YPD or spider medium (Bennett et al. 2005) plate. The cross-replicated plate was incubated for 7 days at room temperature. We did not find a growth medium in which both MPA and NAT were effective when C. albicans was plated at high cell density and therefore we sequentially exposed cells to each drug to select for fusants. We first replica plated from the mating plate onto YPD + NAT (200 µg/ml). After incubation at 30° for 2 days, all cells from areas where mating could have occurred (the intersections of the bands of cells) were scraped off the plate with a loop, suspended in 1 ml water, and 100 µl aliquots of this suspension were plated on 10 minimal medium plates containing MPA (5 µg/ml) and incubated at 30° for 5 days. Large colonies were patch plated on both minimal medium + MPA and on YPD + NAT plates. After incubation at 30° for 2 days, the patches from the YPD + NAT plates, whose counterparts on minimal medium + MPA also grew, were tested by PCR for the presence of both MTL alleles as described above. If both MTL alleles could be amplified, cells from the patches were streaked on YPD agar plates and the PCR assay was repeated for single colonies. These precautions were necessary because MPA selection has a high false-positive background; cells without the resistance marker can still grow, albeit more slowly. Only single colonies with both MTL alleles from the final YPD plates were considered as true fusants and stored at −80°. In some cases, PCR analysis to detect the MPAr and NATr markers (using primer sets TS2Fpr/pENOpr and pACTFpf/CaNATpr respectively) was used to confirm fusants.
Figure 2.
Overview of mating procedure and selection of fusants from two parent strains P1 and P2. Photographs are included to illustrate key steps, namely the area of mating on YPD plates, marked by a dashed rectangle, examples of large colonies, marked with arrows, on minimal medium plus MPA and gel images of MTL allele-based PCR products of parents and three putative fusants, PF1, PF2, and PF3. PF2 is an example of a putative fusant, which was not confirmed as a fusant by PCR (false positive).
Mating of clinical isolates with universal mating tester strains was performed to validate the mating method. The MTLa derivatives of unmarked clinical isolates were mated with the universal mating tester strain 3685 (His−, Arg−, MPAr, MTLα), and the MTLα derivatives were mated with the tester strain 3710 (His−, Arg−, MPAr, MTLa) as described above; fusants were selected on minimal medium plus MPA as described by Magee et al. (2002).
Flow cytometry analysis of DNA content
Cells from glycerol stocks were patched on YPD agar and, after overnight incubation at 30°, the patch was used to inoculate liquid YPD medium to an initial OD600 of approximately 0.2. This culture was incubated at 30° with shaking (150 rpm) and cells were harvested when the OD600 was between 1 and 2. The cells were then fixed in 70% ethanol at 4° for 1 hr to 4 days. They were then treated for 1 hr with RNAse A (Sigma) (2 mg/ml in 50 mM Tris-HCl pH 8.0, 5 mM EDTA) and then for 1 hr with Pepsin (Sigma) (5 mg/ml in 55 mM HCl) and were subsequently washed with 50 mM Tris-HCl pH 7.5, 5 mM EDTA, and stained with 1 mM Sytox green (Invitrogen) in 50 mM Tris-HCl pH 7.5, 5 mM EDTA, as described by Hull et al. (2000). Flow cytometry analysis was performed on a BD FACSCalibur (BD Biosciences), using an excitation wavelength of 488 nm (15 mW argon-ion laser). Emission from Sytox green was measured using a 530/30-nm band pass filter. The sample was collected at a rate of 12 µl/min equating to a rate of >500 events per second. A minimum of 50,000 events was collected. See Figure S2 for examples. DNA content of a fusant relative to that of parents was calculated by first determining the median fluorescence in its M1 and M3 peaks, followed by dividing the fusant’s M1 median by the sum of its parents’ M1 medians, and the fusant’s M3 median by the sum of its parents’ M3 medians, followed by averaging these two ratios.
Nuclear staining
Cells from patches grown on YPD plates at 37° overnight were fixed in 70% ethanol at 4° for at least 1 hr and stained with DAPI (4’,6’-diamidino-2-phenylindole) dissolved in water (1 μg/ml) (Legrand et al. 2004). Cells were visualized using a DFC320 camera and a FM Olympus BX-51 microscope using a U-MWU2 filter set (excitation 330-385 nm, emission 420 nm, dichromatic mirror cut-off 400 nm) with a 100× Magna File objective (see Figure S3 for examples).
In vivo mating and competition
Details of these experiments are given in File S1. In brief, immunosuppressants (doxycycline and dexamethasone) were added to the drinking water of male Sprague-Dawley rats (∼200 g in weight) and oral cavities were co-inoculated with mating-compatible pairs of MTL-homozygotes. Thereafter the tongues of rats were swabbed weekly with sterile swabs moistened in sterile saline. Swabs were vortex mixed in 1 ml sterile saline. Portions of the suspension (5 µl and 50 µl) were plated on YPD agar containing chloramphenicol to determine the total number of C. albicans cells present, and portions (100 µl) were plated on a double selection medium (DSM; 0.67% yeast nitrogen base with amino acids, 2% glucose, 1.5% bacteriological agar, 5 μg MPA/ml, 100 µg NAT/ml) selecting for fusants. Individual colonies were checked by PCR amplification of MTLa or MTLα markers to determine the number of fusants and parents present.
Animal experiments were carried out in facilities approved by the New Zealand statutory body for animal experiments, ANZCCART (Animal Assurance no. A5608-01) in accordance with the ANZCCART Code of Practice for the Use of Veterinary and Human Medicines in Research, Testing and Teaching Organizations and the Animal Welfare Act 1999, New Zealand. Procedures were approved by the University of Otago Animal Ethics Committee (approval 60/2007).
Results
Mutations in mating genes indicate that mating is under selection
The C. albicans MTL locus contains four genes (MTLα1, MTLα2, MTLa1, MTLa2) that are required for mating. They are part of an evolutionarily conserved regulatory circuit, the primary function of which is the control of mating (Tsong et al. 2003; Butler et al. 2009). Expression of genes required for mating is costly, and mutations that abolish mating ability accumulate rapidly in the absence of selection for mating, as has been demonstrated in the yeast Saccharomyces cerevisiae (Lang et al. 2011). Mating competency also carries a cost in C. albicans (Ibrahim et al. 2005; Lockhart et al. 2005; Wu et al. 2007; see also below). The biological significance of the mating process should thus be deducible from an analysis of mutations in these genes.
A commonly used indicator of selection of gene function is the ratio of nonsynonymous to synonymous mutations (dn/ds) (Yang and Nielsen 2002; Spielman and Wilke 2015). If MTLα1, MTLα2, MTLa1, and MTLa2 have retained their original function—mating—and if the ability to mate is under selection, then nonsynonymous mutations in these genes would tend to be eliminated by selection because many such mutations would interfere with mating. Selectively neutral synonymous mutations should be retained more often; as a result the ratio between the two (dn/ds) should be <1.0. If the genes have lost their function, nonsynonymous mutations will not be selected against and would be as likely to be retained as synonymous mutations (dn/ds = 1). Finally, it is conceivable that the genes are functional, but are in the process of acquiring a new primary function, different from mating. In this scenario they could still retain some marginal functionality in regard to mating even though mating ability itself is no longer under selection. If so, nonsynonymous, function-altering mutations may be favored by selection and be more likely to be retained than synonymous mutations (dn/ds > 1) (Yang and Nielsen 2002).
We calculated dn/ds for MTLα1, MTLα2, MTLa1, and MTLa2 in C. albicans and their orthologs in six closely related yeasts. Two of these species, Candida dubliniensis and C. tropicalis, have limited (para) sexual abilities that resemble those of C. albicans (Pujol et al. 2004; Porman et al. 2011). One species, C. orthopsilosis, has a defective pheromone response and ineffective MTLa1 splicing. The occurrence of MTL-homozygous and -heterozygous C. orthopsilosis strains suggest that it may, nevertheless, mate (Sai et al. 2011). Three species (Pichia stipitis, C. lusitaniae, and Debaromyces hansenii) are capable of sexual recombination resembling that of the fully sexual S. cerevisiae. Haploid cells are easily induced to mate and generate diploids, which then return to haploidy by meiosis or processes which lead to meiosis-like marker distributions (Breuer and Harms 2006; Reedy et al. 2009; Bajwa et al. 2010; Butler 2010).
An analysis of the dn/ds ratios indicated that the C. albicans genes have retained their function in mating. First, even though dn/ds ratios were slightly higher in the parasexual than in the fully sexual group, they were well below 1.0 (ranges 0.099–0.252 vs. 0.023–0.188; Table 4), and the difference between the two groups was not statistically significant. We established this by simulating (with PAML; Yang 2007) the evolution of the aligned sequences, using two models, one in which dn/ds was allowed to differ between the parasexual group and other species and one in which dn/ds was constant. We found, using a likelihood test (Zhang et al. 2010), that allowing dn/ds to alter did not lead to models that fit the data significantly better than models with constant dn/ds. This analysis was carried out for each the three genes present in all species (MTLα1, MTLa1, MTLa2). MTLα2 orthologs are absent in the fully sexual group (Butler et al. 2009) and for this gene we established that a model in which dn/ds was free to vary fits the data better than one in which dn/ds was set to 1.0—indicating that the ratio was significantly <1.0. In addition, all nine dn/ds ratios for mutations distinguishing C. albicans MTLα1, MTLa1, and MTLa2 from their orthologs in the three sexual species were <1.0 (range 0.15–0.56); i.e., there was no indication that the C. albicans genes lost their function in mating as they and their orthologs in each of the fully sexual species diverged. These analyses were carried out on one reference genome per species, including the C. albicans SC5314 genome. We therefore cannot exclude that the results may not apply to the entire species. However, for one of the four genes, MTLa1, we found in GenBank the complete amino acid sequences it encodes in two other C. albicans strains, P94015 and P60002. These were identical to the sequence of the SC5314 protein.
Table 4. Ratio of nonsynonymous to synonymous mutations in mating genes.
| Genea | dn/ds parasexual groupb | dn/ds sexual groupb | dn/ds C. ortholopsisb | fraction of alignment usedc |
|---|---|---|---|---|
| MTLa1 | 0.144 | 0.067 | 0.141 | 312/834 |
| MTLa2d (i) | 0.252 | 0.095 | — | 264/834 |
| (ii) | 0.077 | 0.030 | 0.058 | 276/450 |
| MTLα1 | 0.057 | 0.023 | 0.188 | 432/663 |
| MTLα2 | 0.099 | — | — | 555/618 |
The sequences are from strains SC5314 (C. albicans), CD 36 (C. dubliniensis), 90–125 (C. orthopsilosis), MYA-3404 (C. tropicalis), CL143 (C. lusitaniae), CBS767 (D. hansenii), CBS 6054 (P. stipitis).
The ratio of nonsynonymous to synonymous mutations was estimated using the M2 model in the codeml program from PAML v. 4.5. This model allows a different dn/ds ratio in different parts of the tree and is optimized using maximum likelihood. We fit three different ratios: one for the sub-tree including C. tropicalis, C. albicans, and C. dubliniensis; one for the sub-tree including Pichia stipitis, C. lusitaniae, and D. hansenii; and one for the branch leading to C. ortholopsis.
Any deletions and insertions have to be removed from the alignment prior to the analysis.
The C. dubliniensis and the C. tropicalis coding sequences were deduced from nonannotated genomic sequence provided by Gary Moran and Derek Sullivan and, since the gene contained an intron and parts of it are poorly conserved, the intron boundary was difficult to establish. To overcome this problem, the two species were either eliminated from the analysis (i), or the part of the alignment that could contain intron sequences in the two species, was removed prior to analysis (ii).
Numerous mutations must occur to alter the dn/ds ratio of a gene significantly, but a single mutation can destroy mating ability. We therefore also assessed if mating is under selection by estimating the selection coefficient of mutations that prevent mating, i.e., how detrimental such mutations are to survival. This is possible because the frequency (p) of a deleterious mutation in a population is determined by the balance between the rate at which mutations of this type arise (m), and the rate at which individuals carrying them are eliminated. The latter frequency is the selection coefficient s, which indicates by how much the probability of survival is reduced by a deleterious mutation (s = m/p) (Ridley 1996). The estimate is not entirely accurate because chance events (genetic drift) also influence the frequency of mutations in a population. However, genetic drift plays only a minor role in microbial species, given their large effective population sizes (Lynch and Conery 2003; Tsai et al. 2008; Charlesworth 2009).
We are aware of two naturally occurring mutations that abolish mating, one found in an analysis of 12 MTL homozygotes by Legrand et al. (2004) and one in the 10 strains tested in this study (see below; both render MTLα1 dysfunctional). Thus our best estimate of the frequency of mutations that abolish mating (p) is 2/22, i.e., 0.09 (binominal 95% confidence interval 0.011–0.292).
How frequently mutations that abolish mating arise can be estimated by multiplying (i) the genome-wide mutation rate [3.3 × 10−10; point mutation rate determined in S. cerevisiae (Lynch et al. 2008); similar to the C. albicans rate (Gomez-Raja and Larriba 2013)] with (ii) the probability that any given mutation will affect function (0.107), (iii) the number of genes required for mating (25–80), and (iv) the average size of these genes (1539 bp; see File S1 for details on how the latter three estimates were derived). Based on the upper and lower estimate of the number of genes required for mating, the resulting estimates of m range from 1.7 × 10−6 to 5.5 × 10−6 per division. This results in a selection coefficient (s) between 5.9 × 10−6 and 4.9 × 10−4. In other words, carrying a mutation that interferes with mating reduces the fitness of a strain by an estimated 0.0006–0.0490%.
These values for s are small, but nevertheless indicate that mating is under selection. Selection is not overwhelmed by genetic drift as long as selection coefficients are ≥4 times the inverse of effective population size (Charlesworth 2009) and our lowest estimate of s is greater than 4 times the inverse of typical effective population sizes of unicellular eukaryotic species (107–108; Lynch and Conery 2003; Tsai et al. 2008).
Mating of MTL-homozygotes derived from genetically distinct clinical isolates often yielded fusant offspring
To investigate why mating is under natural selection we wanted to assess directly the benefits of mating for naturally occurring strains, using 10 clinical isolates from our international collection (Table 1). None of the isolates had been extensively subcultured and they should thus represent naturally occurring genotypes. All isolates were MTL-heterozygous. We used sorbose selection (Magee and Magee 2000) to induce the loss of one copy of chr 5, and thus one MTL allele, to generate mating-competent MTL-homozygous derivatives. Most strains lost one of their MTL alleles much more frequently than the other, and for several we could recover only either MTLα or MTLa derivatives (Table 5). We chose frequently recovered types of MTL-homozygous derivatives for further experiments.
Table 5. MTL status of colonies recovered from sorbose plates.
| Strain | % MTLa/MTLαa (no. of colonies) | % MTLaa (no. of colonies) | % MTLαa (no. of colonies) |
|---|---|---|---|
| AU35 | 71 (17) | 29 (7) | 0 (0) |
| AU7 | 94 (45) | 6 (3) | 0 (0) |
| Au90 | 25 (6) | 75 (18) | 0 (0) |
| RIHO11 | 96 (23) | 4 (1) | 0 (0) |
| W43 | 4 (1) | 25 (6) | 71 (17) |
| FJ11 | 4 (1) | 17 (4) | 79 (19) |
| HUN97 | 50 (6) | 0 (0) | 50 (6) |
| OD8916 | 79 (19) | 0 (0) | 21 (5) |
| W17 | 42 (10) | 0 (0) | 58 (14) |
| YsU63 | 25 (6) | 0 (0) | 75 (18) |
determined by colony PCR as described in File S1.
A likely reason why mating is under selection would be that it can generate advantageous genotypes, not otherwise achievable through the frequent genome rearrangements in individual cells (Chibana et al. 2000; Forche et al. 2005; Rustchenko-Bulgac et al. 1990), by mating between the unrelated commensal strains which often cocolonize the same individual (Soll et al. 1991; Jacobsen et al. 2008). We therefore investigated the potential of genetically distinct MTL-homozygotes to generate fusants.
Only a small percentage of encounters between mating-compatible MTL-homozygous cells actually generate fusants (Lockhart et al. 2003; Bennett et al. 2005). We therefore marked five of our sorbose selection-derived MTLa strains with a mycophenolic acid-resistance cassette (Beckerman et al. 2001) and five MTLα strains with a nourseothricin-resistance cassette (Shen et al. 2005) (Table 2), allowing selection of fusants after mating on the basis of dual drug resistance (see Materials and Methods for details).
Table 2. Strains used for mating experiments: MTL-homozygous derivatives.
| MTL-homozygote | Genotype | Drug-resistant derivative | Genotype |
|---|---|---|---|
| AU35a | MTLa | AU35a-pNZ4 | MPAR, MTLa |
| AU7a | MTLa | AU7a-pNZ4 | MPAR, MTLa |
| AU90a | MTLa | AU90a-pNZ4 | MPAR, MTLa |
| RIHO11a | MTLa | RIHO11a-pNZ4 | MPAR, MTLa |
| W43a | MTLa | W43a-pNZ4 | MPAR, MTLa |
| FJ11α | MTLα | FJ11α-pNZ11 | NATR, MTLα |
| HUN97α | MTLα | HUN97α-pNZ11 | NATR, MTLα |
| OD8916α | MTLα | OD8916α-pNZ11 | NATR, MTLα |
| W17α | MTLα | W17α-pNZ11 | NATR, MTLα |
| W17α-pNZ11.2 | NATR, MTLα | ||
| YSU63α | MTLα | YSU63α-pNZ11 | NATR, MTLα |
| 3685a | MTLα | 3685a | His−, Arg−, MPAr, MTLα |
| 3710a | MTLa | 3710a | His−, Arg−, MPAr, MTLa |
Source: Magee et al. (2002); MTL-homozygous derivatives of MPA-resistant strains.
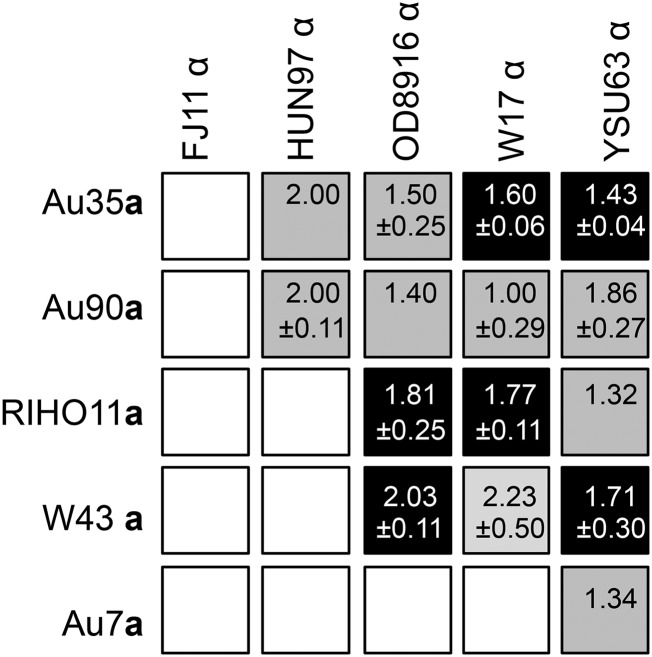
We obtained one or more fusants from 15 of 25 possible combinations of parents, although in some cases only after several attempts (Figure 3). One strain, FJ11α, did not mate with any of the five MTLa derivatives, because of a mutation in the MTLα1 gene essential for mating (Tsong et al. (2003); File S2). All remaining strains could mate with at least one of the five strains of the opposing mating type.
Figure 3.
Matrix of outcomes of matings between MTL-homozygous derivatives. Black boxes indicate combinations yielding seven or more fusants per experiment and in which ≥10% of all colonies tested were fusants. Grey boxes indicate combinations in which fewer fusants were obtained and open boxes indicate combinations yielding no fusants in three or more independent experiments (some matings involving FJ11α were attempted only once after it was discovered that a genetic defect prevented it from mating). Numbers in boxes indicate the DNA content of fusants (mean ±SD where multiple fusants were analyzed) relative to the mean DNA content of the parents. Fusants were selected and identified, and their DNA content assessed by flow cytometry as described in Materials and Methods. All fusants were mononucleate apart from a small percentage of W17α cells and a small percentage of W17α offspring, which were multinucleate (see Figure S3).
The DNA content of fusants was often less than that of both parents combined (Figure 3), indicating chromosome loss during the selection procedure. Thus loss of the markers that we used to select and identify fusants (drug resistance and MTL alleles) may have prevented us from recovering fusants from additional matings. However, there was no correlation between the reduction of DNA content of fusants and the frequency with which fusants were obtained (Figure 3). Furthermore, there was no indication of selection for loss-of-resistance cassettes even in the absence of the drugs and we never observed loss of MTL-heterozygosity when serially propagating fusants (Figure S4 and File S2). It thus seems more likely that, apart from one case of mutational decay of a key gene (in FJ11), it was incompatibility of genetic backgrounds that prevented, or severely curtailed, the generation of fusants in 10 of the 25 pairwise combinations.
Fusants often grew better than their parents in laboratory culture
To test if mating can generate genotypes that are fitter than parental genotypes, we assessed the growth rates of 34 fusants (3 per parent combination unless fewer fusants were obtained; listed in Table 3) and their 9 MTL-homozygous parents.
C. albicans’ largely clonal population structure (Gräser et al. 1996; Tibayrenc 1997; Tavanti et al. 2004; Bougnoux et al. 2008) suggests that mating may only rarely increase fitness in the host. We therefore tested fitness under conditions that should favor fusants. We assessed growth rates during serial propagation, for 100–120 generations, in YPD medium at 37°, and under harsh conditions under which not all MTL-homozygotes could grow, namely at 42° in a low-glucose minimal medium containing 0.2 M NaCl. Under the latter conditions we assessed only 11 fusants that were close to tetraploid upon their isolation. In other fusants, parental alleles beneficial to the harsh environment may have been lost, because the conditions under which fusants were isolated differed substantially from the extreme conditions (see Table 3 for a list of fusants tested).
Both conditions represent novel environments in which (i) the benefits of mating in the form of accelerated adaptation should be high, (especially under harsh conditions under which the strains could barely survive) and (ii) the cost of breaking up compatible parental allele combinations should be lower than in the environment in which they were selected (Goddard et al. 2005; de Visser and Elena 2007; Morran et al. 2011). Reproductive success in culture will depend on maximizing growth rates in the novel environment and reducing the cost of features that enhanced fitness in the host but are now superfluous (Agrawal et al. 2010; Hill et al. 2015). Unlike their parents, fusants initially contain two entire parental genomes. Chromosome loss subsequently generates a variety of aneuploid, and eventually diploid, lineages from which the best-adapted combination of parental alleles can be selected. If some fusant lineages were to become MTL-homozygous as they reduce their genome size, their mating could further speed up adaptation to the new environment. However, we never observed MTL allele loss in serially propagated fusants (Figure S4).
Mating clearly enhanced reproductive success, over that of continued clonal reproduction, for the mating-competent MTL-homozygous isolates (Table 6). Even prior to serial propagation about one-third of fusants grew faster than their parents and some faster than any of the MTL-homozygotes. After serial propagation fusants’ growth rates had increased more than those of their parents and most fusants grew better than parents. Under harsh conditions most fusants grew better than any of the MTL-homozygotes.
Table 6. Comparison of the growth rates of fusants and the MTL-homozygotes that parented fusants.
| Initial | After serial propagation for 100-120 generations | |||||||
|---|---|---|---|---|---|---|---|---|
| n | Average growth rate (doublings/hr) | % of fusants with rates higher than rates of both parents/higher than parental average | No. of fusants growing faster than all MTL-homozygotesa | n | Average growth rate (doublings/hr) | % of fusants with rates higher than rates of both parents/higher than parental average | No. of fusants growing faster than all MTL-homozygotesa | |
| YPD at 37° | ||||||||
| MTL-homozygotes | 9 | 0.960 | n/a | n/a | 9 | 0.972 | n/a | n/a |
| Fusants | 34 | 0.964 | 32/44 | 5 | 34 | 1.037 | 47/79 | 10 |
| Harsh conditions | ||||||||
| MTL-homozygotes | 8 | 0.502 | n/a | n/a | 8 | 0.481 | n/a | n/a |
| Fusants | 11 | 0.566 | 27/36 | 0 | 11 | 0.655 | 82/91 | 8 |
All MTL-homozygotes with resistance cassettes, which parented one or several of the fusants tested.
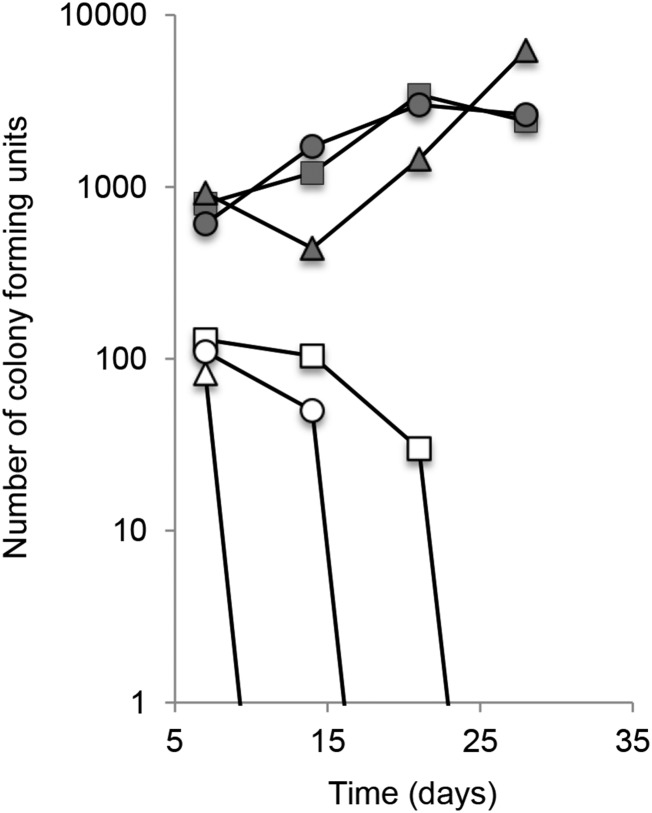
We also assessed the benefits of mating under conditions more closely resembling C. albicans’ natural environment, in a model of oral commensal colonization. The outcome indicated that mating can occur during commensal colonization, but also that it confers less benefit in C. albicans’ normal environment than in culture. In 13 of 51 rats, orally inoculated with equal numbers of cells of mating-compatible parents, fusants were initially detectable (Table S2). However, even though colonization levels in the oral cavity increased over time, fusant numbers declined (Figure 4), as did the frequency of one parent (Table S2), and the percentage of mating-competent opaque cells (data not shown). This suggests that fusants formed initially when equal numbers of mating-competent MTLa and MTLα cells were present, but could not compete well with their parents. Thus while in laboratory culture between 47% (YPD) and 82% (harsh conditions) of fusants eventually grew faster than both parents (Table 6), fusants could compete with their parents in <8% (<1/13) of animals.
Figure 4.
Number of fusants detected over 28 days in a rat model of oral colonization. The figure shows the total number of colony forming units (solid symbols) and the number of fusants (open symbols) recovered by swabbing the oral cavities of a set of three rats (each represented by a differently shaped symbol) co-inoculated orally with opaque cells of strains W43a–pNZ4 and OD8916α–pNZ11. This is one of several experiments with similar outcomes (see Table S2 for outcomes of all experiments and more details).
Many fusants also grow faster than their MTL-heterozygous ancestors
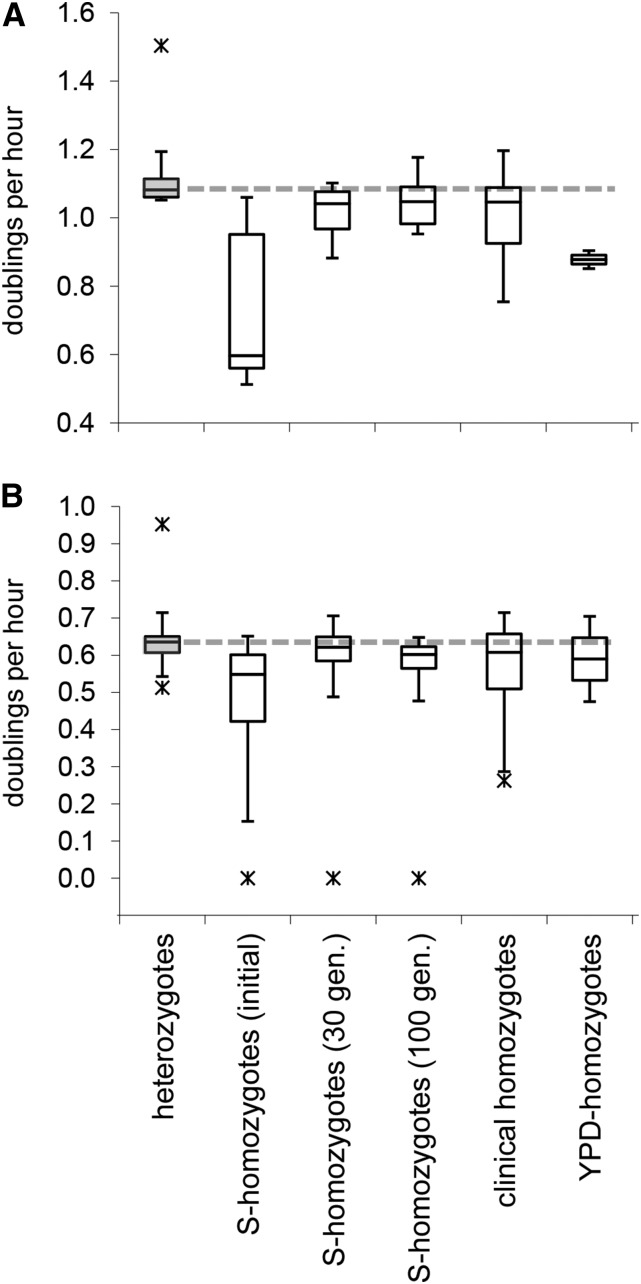
One likely reason why fusants grew faster than their parents in laboratory culture was that mating reversed LOH at the MTL locus and the remainder of chr 5. That such LOH events reduce fitness is indicated by the low prevalence (∼10%) of MTL-homozygous C. albicans clinical isolates (Legrand et al. 2004) and their reduced virulence (Ibrahim et al. 2005; Lockhart et al. 2005; Wu et al. 2007). We also observed that MTL/chr 5 LOH resulted in a significantly reduced fitness under our growth conditions. When first isolated after sorbose selection, the median growth rate of MTL-homozygotes was 45% lower than that of their heterozygous ancestors in YPD (Figure 5A) and 15% lower under harsh conditions (Figure 5B; one MTL-homozygote, W17α, could not grow under harsh conditions at all - not included when calculating the median). Growth rates recovered within 30 generations of serial propagation but remained ∼3% below those of the heterozygous ancestors. The rates measured after 30 generations were close to those of a set of 23 naturally occurring MTL-homozygous clinical isolates (Figure 5). Two MTL-homozygotes that spontaneously arose from isolates OD8916 and W43 on YPD medium grew 20% slower than their MTL-heterozygous ancestors (Figure 5). Their growth rates did not improve over time.
Figure 5.
Impact of MTL-homozygosity on growth rates. Box and whisker plots of growth rates of the 10 clinical MTL-heterozygous clinical isolates used in this study (shaded boxes), their MTL-homozygote derivatives, selected on sorbose (S-homozygotes) initially and after 30 and 100 generations of serial propagation, MTL-homozygous clinical isolates (clinical homozygotes) and spontaneous MTL-homozygotes isolated on YPD medium (YPD-homozygotes); serial propagation for 30 generations of the latter did not increase the growth rates over the initial rates depicted in the figure. Asterisks, outliers. A dashed line indicates the median growth rate of the MTL-heterozygous clinical isolates. (A) Strains grown on YPD, (B) strains grown under harsh conditions (W17α could not grow under harsh conditions).
A comparison of fusants with their MTL-heterozygous ancestors (Table 7) indicated that mating could lead to fitness increases beyond those explicable by restoration of chr 5 heterozygosity. After ≥100 generations of serial propagation in YPD, 15% of fusants grew faster than both MTL-heterologous ancestors, and 27% after serial propagation under harsh conditions—although no fusant grew faster than the fastest growing of all MTL-heterologous ancestor strains. Approximately 10% of fusants already grew faster than either ancestor prior to serial propagation.
Table 7. Comparison of the growth rates of fusants and their MTL- heterozygous ancestors.
| Initial | After serial propagation for 100-120 generations | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | Average rate adjustment factor ± SDa | Average growth rate (doublings/hr) | % of fusants with rates higher than both ancestors/higher than ancestral average | No. of fusants growing faster than all MTL-hetero-zygotesb | n | Average rate adjustment factor ± SDa | Average growth rate (doublings/hr) | % of fusants with rates higher than both ancestors/higher than ancestral average | No. of fusants growing faster than all MTL-hetero-zygotesb | |
| YPD at 37° | ||||||||||
| MTL-heterozygotes | 9 | n/a | 1.133 | n/a | n/a | 9 | n/a | 1.152 | n/a | n/a |
| fusants | 34 | n/a | 0.936 | 12/15 | 0 | 34 | n/a | 1.037 | 15/29 | 0 |
| fusants adjusteda | 34 | 0.92 ± 0.11 | 1.063 | 38/38 | 0 | 34 | 0.91 ± 0.10 | 1.158 | 47/50 | 1 |
| adjusted w/o W17αa,c | 24 | 0.99 ± 0.04 | 0.961 | 17/21 | 0 | 24 | 0.96 ± 0.03 | 1.063 | 8/25 | 0 |
| Harsh conditions | ||||||||||
| MTL-heterozygotes | 8 | n/a | 0.626 | n/a | n/a | 8 | n/a | 0.642 | n/a | n/a |
| fusants | 11 | n/a | 0.566 | 9/27 | 0 | 11 | n/a | 0.655 | 27/64 | 0 |
| fusants adjusteda,d | 11 | 0.89 ± 0.05 | 0.638 | 45/55 | 1 | 11 | 0.93 ± 0.06 | 0.704 | 73/91 | 4 |
| adjusted w/o W17αa,e | 8 | 0.90 ± 0.06 | 0.606 | 38/38 | 0 | 8 | 0.94 ± 0.05 | 0.679 | 63/75 | 1 |
In addition to data based on actual growth rates, the table includes data based on fusant growth rates that were adjusted to compensate for the impact of transformation on their parents. To generate the adjusted rates, the actual growth rate of each fusant was divided by an adjustment factor. The adjustment factor was calculated as follows: The average of the rates of a fusant’s actual parents was determined. This was then divided by the average of the rates of the same strains prior to their transformation with the resistance cassettes.
All MTL-heterozygotes, whose MTL-homozygous derivatives parented one or several of the fusants tested.
Values excluding W17α-derived fusants. Strain W17α was the fastest growing of all initially recovered MTL-homozygotes. After each of two transformation attempts, W17α’s growth rate was reduced by almost 40%, compared to 0–5% in other MTL-homozygotes strains (Table S3). This made the cassette-bearing W17α derivative the slowest growing of all transformed parent strains. However, the strain parented the fastest-growing fusants, indicating that the growth defect was not passed on to fusants and that a 40% adjustment of growth rate was excessive.
The normal procedure for growth rate adjustment (described in footnote a) could not be applied to all fusants. It is based on growth rates of parents, but neither W17α nor its resistance cassette-bearing-transformed derivative could grow under harsh conditions. Growth rates of fusants parented by W17α were therefore adjusted assuming that W17α’s growth rate reduction after transformation equaled the average reduction in other MTL-homozygotes.
Values excluding rates for W17α-derived fusants, since these had to be adjusted using a different procedure, as explained in footnote d.
These results may underestimate the true fitness gain achievable by mating in culture, because the fusants’ parents, but not the MTL-heterozygous ancestors, had been transformed with drug-resistance cassettes. The cassettes themselves apparently had little effect on fusants’ fitness since they were retained during serial propagation in the absence of drug selection (File S2 and Figure S4). However, transformation did, in many cases, reduce the fitness of MTL-homozygotes (Table S3). This suggests that spurious genetic defects acquired by parents during their transformation (rather than the cassettes) might be inherited by fusants and reduce their fitness.
To compensate for the potential reduction in fusant fitness caused by transformation-related parental genetic defects, we computationally increased the growth rates of fusants according to the fitness differential between transformed and original MTL-homozygotes (see Table 7 footnote a for details on the computational adjustment). This provides an upper estimate of the benefits of mating, since it assumes that mating cannot ameliorate these genetic defects by complementation or their elimination through chromosome loss in fusants. Indeed there was, at least in YPD, a reasonably good correlation between the extent by which transformation had reduced parents’ growth rates and the extent by which the rate of their fusant progeny exceeded it, as expected if mating eliminated these defects (Figure S5). The most striking example was strain W17α. Transformation reduced its growth rate by almost 40%, compared to 0–5% in other MTL-homozygotes (Table S3), making its transformed derivative the slowest growing of all transformed parent strains. Nevertheless this strain parented the fastest-growing fusants (Table 7).
Applying the computational adjustment, 47% of fusants grew better than both of their ancestors after serial propagation in YPD and 73% after serial propagation under harsh conditions. Four grew faster than the fastest growing of the MTL-heterologous ancestors (Table 7).
We limited propagation to 100–120 generations, and it is possible that fusants can eventually reach higher growth rates. One likely cause would be further reduction in ploidy. Indeed a decrease of DNA content of fusants during serial propagation was associated with an increase in growth rate (tested in YPD; Figure S6A). Nevertheless after 100 generations only ∼20% of fusants tested had a DNA content indicative of diploidy. Also, the DNA content after 100 generations did not predict how well fusants grew, relative to their parents or MTL-heterologous ancestors (Figure S6, B and C). This suggests that euploidy may be less important in determining fitness than other factors, such as two copies of a chromosome homolog containing fitness-enhancing alleles. Maximal fitness and diploidy may be attainable only after a lengthy series of mitotic recombination events.
Discussion
To date, mating in C. albicans has been observed only in the laboratory. Judging by our analysis of mutations in mating genes, the ability to mate is under selection, indicating that C. albicans mates in its natural environment and that mating is a biologically relevant process. To be under selection, mating must generate genotypes of increased fitness. Even if it does so rarely these genotypes will increase the reproductive success of the species because their progeny will, over time, replace or reduce in frequency other less-fit genotypes. Furthermore, even if the fitness increase is small, compounded over many generations it can have a substantial impact on the reproductive success of the species. However, each mating also involves costs, diminishing the species’ reproductive success. To be under selection, C. albicans mating must thus generate fitter genotypes often enough to outweigh the cost of mating.
Our laboratory growth rate data show that mating can generate fusants that are fitter than their parents and their MTL-heterologous ancestors. One possible reason is that mating can assist in adaptation to novel environments. Goddard et al. (2005) showed that mating led to faster adaptation of a long-cultured (Tauro and Halvorson 1966) S. cerevisiae laboratory strain to novel unusual growth conditions than clonal reproduction. In normal growth medium, mating did not increase fitness (Goddard et al. 2005). Similarly, selective advantages, in terms of growth rate, of C. albicans were observable in many of our culture experiments but not, in terms of survival, in a small number of oral model colonization experiments, conditions more similar to C. albicans’ normal environment. Mating probably also increased fitness in our experiments by removal, or complementation, of deleterious mutations present in parental genomes. The latter is suggested by the correlation between mating-associated increase in fitness and the reduction of fitness associated with genetic modification of parental strains (Figure S5) and also by the observation that some fusants already grew faster than ancestors prior to serial propagation in the novel environment.
A likely category of frequent natural genetic defects that mating, and only mating, can repair are LOH events that occur frequently throughout the C. albicans genome and that can “expose” heterologous deleterious mutations (Diogo et al. 2009; Forche et al. 2009, 2011; Hickman et al. 2013). The immediate fitness increase of some fusants, over that of their MTL-homozygous parents, may appear as striking examples of benefits of mating in the form LOH correction, but a significant part of the fitness reduction associated with chr 5 LOH may be caused by the expression of genes required for mating in mating-competent MTL-homozygous cells. In S. cerevisiae mating competency is associated with a 2% growth rate reduction attributed to expression of such genes (Lang et al. 2009), equivalent to more than half of the ∼3% growth rate difference between chr 5 homozygotes and chr 5 heterozygotes that we measured.
The fitness reduction associated with mating competency is one of many factors that contribute to the cost of mating. Slow-growing mating-competent cells produce fewer progeny, decreasing their reproductive success relative to that of continued clonal reproduction of faster-growing MTL-heterozygotes—and the longer it takes for them to mate after reaching mating competency, the greater this cost becomes. In the host, MTL-homozygotes may even become extinct when competing for limited resources with faster-growing MTL-heterozygotes, eliminating any chance to eventually recoup this cost in the form of fast-reproducing fusants. The act of mating itself reduces reproductive success, yielding fewer new cells than could be generated in the same time period by continued clonal propagation (Lockhart et al. 2003). Other costs of mating include matings that “fail” in that they do not yield fusants fitter than parents or MTL-heterologous ancestors. The parental alleles combined in some fusants may never lead to genotypes fitter than those of ancestors or parents, even after prolonged periods of fusant growth. In the natural host, competition with parents and ancestors for limited niches and resources in the presence of host clearance mechanisms can also eliminate, before they have a chance to do so, fusants that could, over time, become fitter than parents and ancestors. Even fusants that are fitter than the parents or ancestors may thus often become extinct and contribute to the number of “failed” matings. If mating is rare in the host, a small number of resulting fusants will be at risk of extinction regardless of their fitness, among the more abundant parents or MTL-heterozygous ancestor cells (Kimura and Ohta 1969). This may be part of the reason, with a reduced probability of mating to generate genotypes better than existing genotypes in an organism’s normal environment (Goddard et al. 2005; De Visser and Elena 2007; Morran et al. 2011), why fusants apparently struggled to survive in competition with their parents in the animal model. Similarly, a low probability of survival of fusants in the human host would explain why mating, although under selection, has so little impact on the population structure of the species, which is largely clonal (Gräser et al. 1996; Tibayrenc 1997; Tavanti et al. 2004; Bougnoux et al. 2008).
While fusants that survive but only eventually grow faster than parents or ancestors do not contribute to the failed matings, the time it takes them to reach a higher growth rate is also a cost. It reduces the differential between the number of their progeny generated over time compared to that generated by continued clonal reproduction.
A difficulty in deducing from our findings how the benefits of mating outweigh its costs in C. albicans’ natural environment is that mating has never been observed in the human host and may occur via different scenarios. We have focused on one of these, the mating of genetically different strains after chr 5 loss. While chr 5 loss is the most common mechanism of spontaneous loss of MTL-heterozygosity (Wu et al. 2005), in most MTL-homozygous clinical isolates MTL-homozygosity has arisen by localized recombination events (Wu et al. 2007). It seems unlikely though that the net benefit of mating is significantly affected by the mechanism of MTL-heterozygosity loss. The growth rates of clinical MTL-homozygous isolates were indistinguishable from those we generated by chr 5 loss (Figure 5); i.e., the fitness of the two is comparable. Also, while parents heterozygous for large parts of chr 5 bring more alleles to the initial tetraploids, because of selection for MTL-heterozygosity (our data; Ibrahim et al. 2005; Lockhart et al. 2005; Wu et al. 2007), the final diploid products of mating will contain one copy of chr 5 from each parent—except possibly when rare circumstances allow mating of strains of the same mating type (Alby and Bennett 2011). The costs and benefits of these two scenarios are thus probably fundamentally similar (although the prevalence of isolates with partial chr 5 LOH indicates they may have better chances of long-term survival (Wu et al. 2007) while chr 5 LOH, stress induced (Forche et al. 2011) in pairs of cocolonizing strains, could more often allow mating shortly after mating competency is reached).
Mating of diploid or aneuploid MTL-homozygotes derived from the same strain is possible (Magee and Magee 2000) at least in some strains (Table 5). Such matings offer benefits in terms of restoring MTL- and/or chr5-heterozygosity but cannot correct other LOHs or generate better-adapted allele combinations, beyond those also achievable by genome rearrangements during clonal reproduction (Chibana et al. 2000; Forche et al. 2005; Rustchenko-Bulgac et al. 1990).
Mating is also possible between C. albicans haploids spontaneously arising by chromosome loss (Hickman et al. 2013), immediately yielding diploid fusants and restoring chr 5 heterozygosity. However, the mating of haploids should have a lower probability of generating genotypes of improved fitness than mating between diploids. Fusants derived from haploid parents inherit one of two chromosome homologs from each parent, and the future compatibility of parental alleles in fusants plays no role in selecting which homolog a parent passes on. In contrast, when diploids mate, the merger of two diploid genomes followed by chromosome loss allows selection for the best pairs of parental homologs, including the option of retaining only homologs from one parent. Fusants generated by mating of haploid derivatives of the same strain grow poorly (Hickman et al. 2013).
Thus, among these possible scenarios, the mating of strains of different ancestry following chr 5 LOH or LOH at the MTL locus appears to offer the greatest net benefit. However, with the exception of haploid mating, all are likely to offer some net benefits that could contribute to the selection of mating ability.
After subtracting the estimated costs, the remaining net benefit of C. albicans mating appears considerably lower than that of a fully functional sexual cycle as exemplified by the yeast S. cerevisiae. One important reason is the absence of meiosis in C. albicans. In the absence of meiosis all of the initial fusants arising from the mating of two strains are genetically more or less identical tetraploids. Subsequent competition among clonally reproducing fusant lineages can only slowly improve fitness by removing allelic incompatibilities and reducing ploidy through chromosome loss and mitotic recombination. In S. cerevisiae, in contrast, each offspring is immediately diploid and each has a different combination of parental alleles, having inherited only one of two homologous parental chromosomes, with parental alleles on these partially unlinked by chiasma formation (Carlile et al. 2001). S. cerevisiae’s sexual cycle further increases efficacy of selection of the best allele combinations by allowing frequent return to the mating-competent state, not by spontaneous LOH but in response to environmental cues (Merlini et al. 2013), increasing the likelihood that mating-competent cells will quickly find a compatible partner and reducing the cost of the slower growth of mating competent cells.
A fully functional sexual cycle offers powerful protection against deleterious mutations (Felsenstein 1974). It is thus difficult to conceive why mutations reducing sexual ability swept through the species in the past if such loss was deleterious. Possibly loss of sex was advantageous at the time, as a means of protecting rare allele combinations (Vrijenhoek and Parker 2009) that allowed C. albicans to become one of the very few Candida species that colonize the human intestinal tract (Skinner and Fletcher 1960; Odds 1988). Bottlenecks and genetic drift among small numbers of initial colonizers of humans could also have contributed to the fixation of mutations that reduced the ability to mate, even if mating was advantageous (Lynch et al. 1995). Since then, long periods of predominantly clonal reproduction are likely to have generated clonal lines whose allele combinations are often poorly compatible (Carlile et al. 2001); this would also explain why only 15/25 pairwise combination of mating-capable strains yielded fusants in our experiments.
The mutational decay of the sexual cycle in C. albicans, while increasing the cost and reducing the benefits of mating for strains that engage in mating, has also effectively eliminated the cost of mating as far as the species as a whole is concerned and this may be pivotal to why mating is under selection. Fitness-decreasing LOH events are frequent in C. albicans (Diogo et al. 2009; Forche et al. 2009, 2011; Hickman et al. 2013) and the strains affected are doomed to become extinct unless these defects are corrected, which is achievable quickly only by mating. Thus mating is restricted to those members of the species that otherwise are destined for extinction, offering them a small chance of survival by mating if the LOHs include MTL-LOH. Any instance in which mating averts their extinction, or leads to fitness increases beyond restoration of their initial fitness, constitutes a net benefit of mating to the species.
Supplementary Material
Acknowledgments
We thank Fran Wolber for assistance with flow cytometry and Richard Bennett for providing MTL-homozygous clinical isolates. We thank Judith Berman, Hamish Spencer, Joseph Heitman, Rosie Bradshaw, Murray Cox, and anonymous referees for insightful comments on earlier versions of the manuscript. We thank Gary Moran and Derek Sullivan for providing sequences for dn/ds analyses and the curators of the Candida genome database for the annotations that allowed us to estimate the mutation rate across mating genes. This work was supported by Marsden grant MAU0603.
Footnotes
Communicating editor: A. P. Mitchell
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.177170/-/DC1.
Sequence data from this article have been deposited with the GenBank Data Libraries under accession nos. FJ804172.1, FJ804173.1, and JN099704.
Literature Cited
- Agrawal A. A., Conner J. K., Rasmann S., 2010. Tradeoffs and negative correlations in evolutionary ecology, pp. 243–268 in Evolution after Darwin: The First 150 Years, edited by Bell G., Eanes W. F., Futuyma D. J., Levinton J. S. Sinauer, Sunderland, MA. [Google Scholar]
- Alby K., Bennett R. J., 2011. Interspecies pheromone signaling promotes biofilm formation and same-sex mating in Candida albicans. Proc. Natl. Acad. Sci. USA 108: 2510–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alby K., Schaefer D., Bennett R. J., 2009. Homothallic and heterothallic mating in the opportunistic pathogen Candida albicans. Nature 460: 890–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajwa P. K., Pinel D., Martin V. J. J., Trevors J. T., Lee H., 2010. Strain improvement of the pentose-fermenting yeast Pichia stipitis by genome shuffling. J. Microbiol. Methods 81: 179–186. [DOI] [PubMed] [Google Scholar]
- Beckerman J., Chibana H., Turner J., Magee P. T., 2001. Single-copy IMH3 allele is sufficient to confer resistance to mycophenolic acid in Candida albicans and to mediate transformation of clinical Candida species. Infect. Immun. 69: 108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R. J., Johnson A. D., 2003. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 22: 2505–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R. J., Miller M. G., Chua P. R., Maxon M. E., Johnson A. D., 2005. Nuclear fusion occurs during mating in Candida albicans and is dependent on the KAR3 gene. Mol. Microbiol. 55: 1046–1059. [DOI] [PubMed] [Google Scholar]
- Bougnoux M.-E., Pujol C., Diogo D., Bouchier C., Soll D. R., et al. , 2008. Mating is rare within as well as between clades of the human pathogen Candida albicans. Fungal Genet. Biol. 45: 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer U., Harms H., 2006. Debaryomyces hansenii: an extremophilic yeast with biotechnological potential. Yeast 23: 415–437. [DOI] [PubMed] [Google Scholar]
- Butler G., 2007. The evolution on MAT: the Ascomycetes, pp. 3–18 in Sex in Fungi: Molecular Determination and Evolutionary Implications, edited by Heitman J., Kronstad J. W., Taylor J. W., Casselton L. A. ASM Press, Washington, DC. [Google Scholar]
- Butler G., 2010. Fungal sex and pathogenesis. Clin. Microbiol. Rev. 23: 140–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G., Rasmussen M. D., Lin M. F., Santos M. A., Sakthikumar S., et al. , 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459: 657–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile, M., S. Watkinson, and G. Gooday, 2001 Genetic variation and evolution, pp. 245–296 in The Fungi. Academic Press, San Diego. [Google Scholar]
- Charlesworth B., 2009. Fundamental concepts in genetics: effective population size and patterns of molecular evolution and variation. Nat. Rev. Genet. 10: 195–205. [DOI] [PubMed] [Google Scholar]
- Chibana H., Beckerman J. L., Magee P. T., 2000. Fine-resolution physical mapping of genomic diversity in Candida albicans. Genome Res. 10: 1865–1877. [DOI] [PubMed] [Google Scholar]
- Cox M. P., Holland B. R., Wilkins M. C., Schmid J., 2013. Reconstructing historic changes in locus-specific recombination rates. BMC Genet. 14: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Backer M. D., Maes D., Vandoninck S., Logghe M., Contreras R., et al. , 1999. Transformation of Candida albicans by electroporation. Yeast 15: 1609–1618. [DOI] [PubMed] [Google Scholar]
- de Visser J. A., Elena S. F., 2007. The evolution of sex: empirical insights into the roles of epistasis and drift. Nat. Rev. Genet. 8: 139–149. [DOI] [PubMed] [Google Scholar]
- Diogo D., Bouchier C., d’Enfert C., Bougnoux M.-E., 2009. Loss of heterozygosity in commensal isolates of the asexual diploid yeast Candida albicans. Fungal Genet. Biol. 46: 159–168. [DOI] [PubMed] [Google Scholar]
- Edelmann A., Schmid J., Krüger M., 2005. Genetic relationships between human and animal isolates of Candida albicans. J. Clin. Microbiol. 43: 6164–6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J., 1974. The evolutionary advantage of recombination. Genetics 78: 737–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A., May G., Magee P. T., 2005. Demonstration of loss of heterozygosity by single-nucleotide polymorphism microarray analysis and alterations in strain morphology in Candida albicans strains during Infection. Eukaryot. Cell 4: 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A., Alby K., Schaefer D., Johnson A. D., Berman J., et al. , 2008. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol. 6: e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A., Magee P. T., Selmecki A., Berman J., May G., 2009. Evolution in Candida albicans populations during a single passage through a mouse host. Genetics 182: 799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A., Abbey D., Pisithkul T., Weinzierl M. A., Ringstrom T., et al. , 2011. Stress alters rates and types of loss of heterozygosity in Candida albicans. MBio 2: e00200–e00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard M. R., Godfray H. C., Burt A., 2005. Sex increases the efficacy of natural selection in experimental yeast populations. Nature 434: 636–640. [DOI] [PubMed] [Google Scholar]
- Gomez-Raja J., Larriba G., 2013. Comparison of two approaches for identification of haplotypes and point mutations in Candida albicans and Saccharomyces cerevisiae. J. Microbiol. Methods 94: 47–53. [DOI] [PubMed] [Google Scholar]
- Gräser Y., Volovsek M., Arrington J., Schönian G., Presber W., et al. , 1996. Molecular markers reveal that population structure of the human pathogen Candida albicans exhibits both clonality and recombination. Proc. Natl. Acad. Sci. USA 93: 12473–12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J., 2006. Sexual reproduction and the evolution of microbial pathogens. Curr. Biol. 16: R711–R725. [DOI] [PubMed] [Google Scholar]
- Heitman J., 2010. Evolution of eukaryotic microbial pathogens via covert sexual reproduction. Cell Host Microbe 8: 86–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman M. A., Zeng G., Forche A., Hirakawa M. P., Abbey D., et al. , 2013. The ’obligate diploid’ Candida albicans forms mating-competent haploids. Nature 494: 55–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J., O’Meara T. R., Cowen E. L., 2015. Fitness trade-offs associated with the evolution of resistance to antifungal drug combinations. Cell Reports 10: 809–819. [DOI] [PubMed] [Google Scholar]
- Holland, B. R., and J. Schmid, 2005 Selecting representative model micro-organisms. BMC Microbiol. 5: paper 26. [DOI] [PMC free article] [PubMed]
- Hull C. M., Raisner R. M., Johnson A. D., 2000. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science 289: 307–310. [DOI] [PubMed] [Google Scholar]
- Ibrahim A. S., Magee B. B., Sheppard D. C., Yang M., Kauffman S., et al. , 2005. Effects of ploidy and mating type on virulence of Candida albicans. Infect. Immun. 73: 7366–7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen M. D., Bougnoux M. E., d’Enfert C., Odds F. C., 2008. Multilocus sequence typing of Candida albicans isolates from animals. Res. Microbiol. 159: 436–440. [DOI] [PubMed] [Google Scholar]
- Kimura M., Ohta T., 1969. The average number of generations until fixation of a mutant gene in a finite population. Genetics 61: 763–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G. I., Murray A. W., Botstein D., 2009. The cost of gene expression underlies a fitness trade-off in yeast. Proc. Natl. Acad. Sci. USA 106: 5755–5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G. I., Botstein D., Desai M. M., 2011. Genetic variation and the fate of beneficial mutations in asexual populations. Genetics 188: 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand M., Lephart P., Forche A., Mueller F. M., Walsh T., et al. , 2004. Homozygosity at the MTL locus in clinical strains of Candida albicans: karyotypic rearrangements and tetraploid formation. Mol. Microbiol. 52: 1451–1462. [DOI] [PubMed] [Google Scholar]
- Lockhart S. R., Pujol C., Daniels K. J., Miller M. G., Johnson A. D., et al. , 2002. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics 162: 737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart S. R., Daniels K. J., Zhao R., Wessels D., Soll D. R., 2003. Cell biology of mating in Candida albicans. Eukaryot. Cell 2: 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart S. R., Wu W., Radke J. B., Zhao R., Soll D. R., 2005. Increased virulence and competitive advantage of a/α over a/a or α/α offspring conserves the mating system of Candida albicans. Genetics 169: 1883–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Conery J. S., 2003. The origins of genome complexity. Science 302: 1401–1404. [DOI] [PubMed] [Google Scholar]
- Lynch M., Conery J., Burger R., 1995. Mutational meltdowns in sexual populations. Evolution 49: 1067–1080. [DOI] [PubMed] [Google Scholar]
- Lynch M., Sung W., Morris K., Coffey N., Landry C. R., et al. , 2008. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc. Natl. Acad. Sci. USA 105: 9272–9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee B. B., Magee P. T., 2000. Induction of mating in Candida albicans by construction of MTLa and MTLalpha strains. Science 289: 310–313. [DOI] [PubMed] [Google Scholar]
- Magee B. B., Legrand M., Alarco A. M., Raymond M., Magee P. T., 2002. Many of the genes required for mating in Saccharomyces cerevisiae are also required for mating in Candida albicans. Mol. Microbiol. 46: 1345–1351. [DOI] [PubMed] [Google Scholar]
- Merlini L., Dudin O., Martin S. G., 2013. Mate and fuse: how yeast cells do it. Open Biol 3: 130008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M., Johnson A., 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110: 293. [DOI] [PubMed] [Google Scholar]
- Morran L. T., Schmidt O. G., Gelarden I. A., Parrish R. C., Lively C. M., 2011. Running with the Red Queen: host-parasite coevolution selects for biparental sex. Science 333: 216–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama H., Mio T., Nagahashi S., Kokado M., Arisawa M., et al. , 2000. Tetracycline-regulatable system to tightly control gene expression in the pathogenic fungus Candida albicans. Infect. Immun. 68: 6712–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds F. C., 1988. Candida and Candidosis. Bailliere Tindall, London. [Google Scholar]
- Odds F. C., Bougnoux M. E., Shaw D. J., Bain J. M., Davidson A. D., et al. , 2007. Molecular phylogenetics of Candida albicans. Eukaryot. Cell 6: 1041–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto S. P., Lenormand T., 2002. Resolving the paradox of sex and recombination. Nat. Rev. Genet. 3: 252–261. [DOI] [PubMed] [Google Scholar]
- Porman A. M., Alby K., Hirakawa M. P., Bennett R. J., 2011. Discovery of a phenotypic switch regulating sexual mating in the opportunistic fungal pathogen Candida tropicalis. Proc. Natl. Acad. Sci. USA 108: 21158–21163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol C., Daniels K. J., Lockhart S. R., Srikantha T., Radke J. B., et al. , 2004. The closely related species Candida albicans and Candida dubliniensis can mate. Eukaryot. Cell 3: 1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reedy J. L., Floyd A. M., Heitman J., 2009. Mechanistic plasticity of sexual reproduction and meiosis in the Candida pathogenic species complex. Curr. Biol. 19: 891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice W. R., 2002. Experimental tests of the adaptive significance of sexual recombination. Nat. Rev. Genet. 3: 241–251. [DOI] [PubMed] [Google Scholar]
- Ridley M., 1996. Evolution. Blackwell Science, Cambridge, MA. [Google Scholar]
- Rustchenko-Bulgac E. P., Sherman F., Hicks J. B., 1990. Chromosomal rearrangements associated with morphological mutants provide a means for genetic variation of Candida albicans. J. Bacteriol. 172: 1276–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustchenko E. P., Howard D. H., Sherman F., 1994. Chromosomal alterations of Candida albicans are associated with the gain and loss of assimilating functions. J. Bacteriol. 176: 3231–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sai S., Holland L. M., McGee C. F., Lynch D. B., Butler G., 2011. Evolution of mating within the Candida parapsilosis species group. Eukaryot. Cell 10: 578–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid J., Herd S., Hunter P. R., Cannon R. D., Yasin M. S. M., et al. , 1999. Evidence for a general-purpose genotype in Candida albicans, highly prevalent in multiple geographic regions, patient types and types of infection. Microbiology 145: 2405–2414. [DOI] [PubMed] [Google Scholar]
- Schmid J., Cannon R. D., Holland B., 2004. A futile act?: thoughts on the reproductive biology of Candida albicans. Mycologist 18: 158–163. [Google Scholar]
- Shen J., Guo W., Kohler J. R., 2005. CaNAT1, a heterologous dominant selectable marker for transformation of Candida albicans and other pathogenic Candida species. Infect. Immun. 73: 1239–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner C. E., Fletcher D. W., 1960. A review of the genus Candida. Bacteriol. Rev. 24: 397–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slutsky B., Staebell M., Anderson J., Risen L., Pfaller M., et al. , 1987. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J. Bacteriol. 169: 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll D. R., Galask R., Schmid J., Hanna C., Mac K., et al. , 1991. Genetic dissimilarity of commensal strains of Candida spp. carried in different anatomical locations of the same healthy women. J. Clin. Microbiol. 29: 1702–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman S. J., Wilke C. O., 2015. The relationship between dN/dS and scaled selection coefficients. Mol. Biol. Evol. 32: 1079–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauro P., Halvorson H. O., 1966. Effect of gene position on the timing of enzyme synthesis in synchronous cultures of yeast. J. Bacteriol. 92: 652–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavanti A., Gow N. A., Maiden M. C., Odds F. C., Shaw D. J., 2004. Genetic evidence for recombination in Candida albicans based on haplotype analysis. Fungal Genet. Biol. 41: 553–562. [DOI] [PubMed] [Google Scholar]
- Tavanti A., Davidson A. D., Fordyce M. J., Gow N. A. R., Maiden M. C. J., et al. , 2005a Population structure and properties of Candida albicans, as determined by multilocus sequence typing. J. Clin. Microbiol. 43: 5601–5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavanti A., Davidson A. D., Gow N. A., Maiden M. C., Odds F. C., 2005b Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis groups II and III. J. Clin. Microbiol. 43: 284–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc M., 1997. Are Candida albicans natural populations subdivided? Trends Microbiol. 5: 253–257. [DOI] [PubMed] [Google Scholar]
- Tsai I. J., Bensasson D., Burt A., Koufopanou V., 2008. Population genomics of the wild yeast Saccharomyces paradoxus: quantifying the life cycle. Proc. Natl. Acad. Sci. USA 105: 4957–4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsong A. E., Miller M. G., Raisner R. M., Johnson A. D., 2003. Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell 115: 389–399. [DOI] [PubMed] [Google Scholar]
- Vrijenhoek, R. C., and E. J. Parker, 2009 Geographical parthogenesis: general purpose genotypes and frozen niche variation, pp. 99–132 in Lost Sex: The Evolutionary Biology of Parthenogenesis, edited by K. M. Isa Schön, Peter J. Dijk, Peter van Dijk. Springer, New York. [Google Scholar]
- Wang H., Xu Z., Gao L., Hao B., 2009. A fungal phylogeny based on 82 complete genomes using the composition vector method. BMC Evol. Biol. 9: 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel, L., J. K. Whittington, C. Pujol, S.-H. Oh, M. O. Ruiz et al., 2008 Molecular phylogenetic analysis of a geographically and temporally matched collection of Candida albicans isolates from humans and non-migratory wildlife in central Illinois. Eukaryot. Cell, EC.00162–00108. [DOI] [PMC free article] [PubMed]
- Wu W., Pujol C., Lockhart S. R., Soll D. R., 2005. Chromosome loss followed by duplication is the major mechanism of spontaneous mating-type locus homozygosis in Candida albicans. Genetics 169: 1311–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Lockhart S. R., Pujol C., Srikantha T., Soll D. R., 2007. Heterozygosity of genes on the sex chromosome regulates Candida albicans virulence. Mol. Microbiol. 64: 1587–1604. [DOI] [PubMed] [Google Scholar]
- Xie J., Tao L., Nobile C. J., Tong Y., Guan G., et al. , 2013. White-opaque switching in natural MTLa/alpha isolates of Candida albicans: evolutionary implications for roles in host adaptation, pathogenesis, and sex. PLoS Biol. 11: e1001525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., 2007. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z., Nielsen R., 2002. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 19: 908–917. [DOI] [PubMed] [Google Scholar]
- Zhang N., Cannon R. D., Holland B., Patchett M., Schmid J., 2010. Impact of genetic background on allele selection in a highly mutable Candida albicans gene, PNG2. PLoS ONE 5: e9614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.