Figure 2.
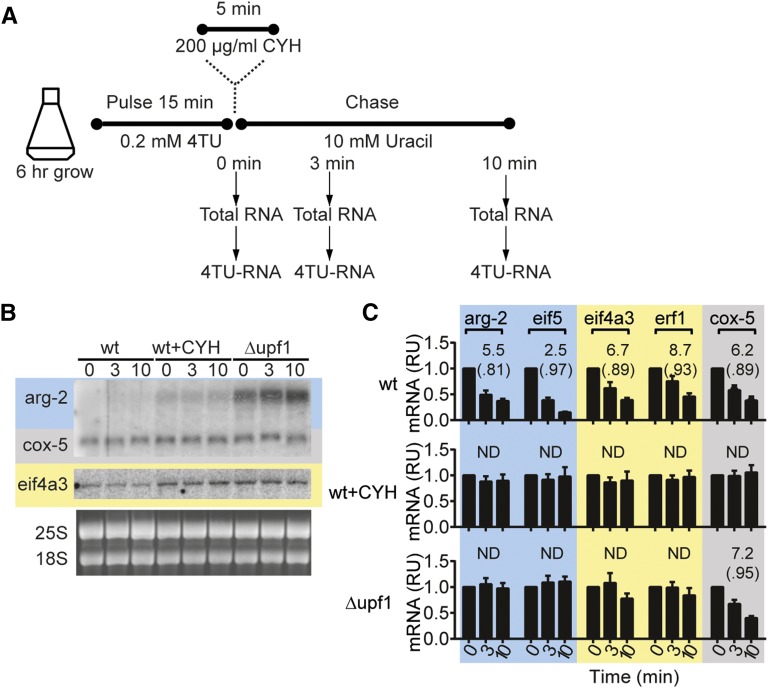
mRNA levels and mRNA stability are affected by growth in cycloheximide and by Δupf1. (A) Strategy for pulse–chase analyses of mRNA stability. N. crassa conidia were germinated and grown for 6 hr; 0.2 mM 4TU was then added. After 15 min incubation with 4TU, 10 mM uracil was added to the culture, and total RNA and 4TU–RNA was purified from samples taken at 0, 3, and 10 min. In one experiment, cycloheximide (CYH) was added after the 15-min 4TU-pulse; following 5 min of incubation with CYH, the uracil-chase was then performed. (B) Total RNA from wt, Δupf1, and wt cells treated with 200 μg/ml CYH and subjected to 4TU pulse–chase were analyzed by Northern blotting as in Figure 1B; the arg-2 and cox-5 probes were added together. (C) Pulse–chase analyses of 4TU RNA from wt, Δupf1, and CYH-treated wt. 4TU RNA was purified and quantified by RT–qPCR and levels of 4TU mRNAs for arg-2, eif5, eif4a3, erf1, and cox-5 were normalized to levels of 4TU-labeled 25S rRNA in each sample (4TU-labeled 25S rRNA is stable during a 10-min chase) and then normalized to the level at time 0. Differences in expression are shown in relative units (RUs). The results are the average of three independent experiments. The mRNA half-lives (in minutes), calculated by fitting data to first order-decay parameters, are shown, as are R2 values for each calculation (in parentheses). ND, not determined. See also Figure S3.