Abstract
The Kaiser Permanente Research Program on Genes, Environment, and Health (RPGEH) Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort includes DNA specimens extracted from saliva samples of 110,266 individuals. Because of its relationship to aging, telomere length measurement was considered an important biomarker to develop on these subjects. To assay relative telomere length (TL) on this large cohort over a short time period, we created a novel high throughput robotic system for TL analysis and informatics. Samples were run in triplicate, along with control samples, in a randomized design. As part of quality control, we determined the within-sample variability and employed thresholds for the elimination of outlying measurements. Of 106,902 samples assayed, 105,539 (98.7%) passed all quality control (QC) measures. As expected, TL in general showed a decline with age and a sex difference. While telomeres showed a negative correlation with age up to 75 years, in those older than 75 years, age positively correlated with longer telomeres, indicative of an association of longer telomeres with more years of survival in those older than 75. Furthermore, while females in general had longer telomeres than males, this difference was significant only for those older than age 50. An additional novel finding was that the variance of TL between individuals increased with age. This study establishes reliable assay and analysis methodologies for measurement of TL in large, population-based human studies. The GERA cohort represents the largest currently available such resource, linked to comprehensive electronic health and genotype data for analysis.
Keywords: relative telomere length, GERA cohort, saliva DNA, robotic assay, quantitative PCR
TELOMERES are the protective DNA–protein complexes that cap the ends of eukaryotic chromosomes and are required for genome stability. The essential telomeric DNA consists of a tract of a tandemly repeated short sequence specified and maintained by the highly regulated reverse transcriptase action of the cellular enzyme telomerase. Telomeric DNA is susceptible to natural terminal erosion through a variety of processes including the end replication problem of linear chromosomal DNA, which causes telomeres to get shorter each time a somatic cell divides (Olovnikov 1973; Blackburn 2005), and other processes in cells that can act to diminish telomere length (TL) including nuclease action, replication fork stalling through telomeric DNA repeat tracts, DNA recombination, and oxidative damage (Jain and Cooper 2010). While telomerase can counteract shortening by elongating and protecting telomeres, telomerase activity is generally down-regulated in normal human cells (Blackburn 1997). When telomeres become too short, cells become senescent, losing the ability to divide and function normally (Allsopp et al. 1992; Blackburn 2000; Armanios and Blackburn 2012). Mutations that decrease telomerase and cause short telomeres in humans lead to a spectrum of premature-onset diseases and conditions collectively termed “telomere syndromes,” which share many features of the common diseases of aging in the human population (Armanios and Blackburn 2012). Multiple independent studies have found impaired human telomeric DNA length maintenance to be associated with a wide range of diseases and for several age-related diseases to predict future risks and outcomes including mortality (Von Zglinicki et al. 2000; Samani et al. 2001; Cawthon et al. 2003; Panossian et al. 2003; Valdes et al. 2005; Bischoff et al. 2006; Harris et al. 2006; Martin-Ruiz et al. 2006; Bakaysa et al. 2007; Brouilette et al. 2007; Fitzpatrick et al. 2007, 2011; Aubert and Lansdorp 2008; Farzaneh-Far et al. 2008; Kimura et al. 2008; Epel et al. 2009; Njajou et al. 2009; Astrup et al. 2010; Codd et al. 2010, 2013; Salpea et al. 2010; Willeit et al. 2010a,b; Zee et al. 2010; Strandberg et al. 2011; Wentzensen et al. 2011; Yaffe et al. 2011; Honig et al. 2012; Lee et al. 2012; Weischer et al. 2012; Bojesen 2013; Muezzinler et al. 2013; Gardner et al. 2014; Haycock et al. 2014; Walsh et al. 2014).
Most studies associating TL with human clinical and other states have been done by measuring TL in white blood cells, typically peripheral blood mononuclear cells (PBMC TL) or leukocytes, using either Southern blotting or a well-validated PCR-based method that has been shown to have strong concordance with Southern blotting measurements (Aviv et al. 2011). In multiple independent cohort studies, TL in PBMCs or leukocyte samples has been correlated with sex (longer in females) and negatively correlated with age (Muezzinler et al. 2013; Gardner et al. 2014). A recent study incorporating data from the National Health and Nutrition Examination Survey (NHANES) 1999–2002, a large, nationally representative, ethnically and socioeconomically diverse sample, reported that sex, race-ethnicity, and socioeconomic status (SES) moderated the associations between TL and mortality in older individuals (Needham et al. 2012). However, while some human cohort studies have found shorter PBMC TL or LTL is associated with increased mortality (Cawthon et al. 2003; Martin-Ruiz et al. 2006; Bakaysa et al. 2007; Farzaneh-Far et al. 2008; Kimura et al. 2008; Epel et al. 2009; Astrup et al. 2010; Willeit et al. 2010b; Fitzpatrick et al. 2011; Honig et al. 2012; Lee et al. 2012; Weischer et al. 2012), others have not (Bischoff et al. 2006; Harris et al. 2006; Njajou et al. 2009; Strandberg et al. 2011). These inconsistencies in the literature may be due to differences in study cohorts, sample sizes, assay methods, and other methodological differences (Bojesen 2013).
It is known that mean TL is under both cell type-specific and systemic controls. Specifically, cell-subtype differences in mean TL are observed in white blood cells (Lin et al. 2010). Thus, in the same individual, granulocyte and B-lymphocyte mean TL is typically longer than TL of T-lymphocytes, and in turn both CD4+ and CD8+CD28+ T-lymphocytes have longer mean TL than CD8+CD28− T-cells (Lin et al. 2010). However, comparing between different individuals, mean TL in all these cell types also shifts up and down in concert in any given individual, indicative of systemic regulation of TL in addition to cell-type differences. The existence of systemic effects on TL in different cell types is further reinforced by analyses showing that the correlation of mean PBMC or leukocyte TL with TL of buccal cells or fibroblasts is much greater within an individual than between individuals (Graakjaer et al. 2006; Gadalla et al. 2010).
Here we describe novel high-throughput automated methods and analytics used for measurement of mean TL of cells derived from saliva samples from >100,000 individual human subjects in the Kaiser Permanente Northern California (KPNC) Research Program on Genes, Environment, and Health (RPGEH) Genetic Epidemiology on Adult Health and Aging (GERA) cohort. As up to 74% of the DNA in saliva derives from leukocytes (Thiede et al. 2000), we anticipated that results obtained here from saliva samples would demonstrate characteristics comparable to blood-derived DNA. One way to evaluate this is to compare our results obtained using saliva DNA with previous human studies, in which associations between TL and demographic parameters (age, sex) were studied using leukocytes or peripheral blood mononuclear cells prepared from blood-drawn samples.
As members of the KPNC health plan, GERA cohort members have available extensive electronic health data covering diagnoses, laboratory tests, pathology reports, and other clinical assessments that can be analyzed for relationship with the derived TL measurements. This also represents the largest, single cohort with TL measurements available.
Materials and Methods
Study subjects
The GERA cohort is composed of 110,266 adult male and female members of the KPNC health plan. It is a component of the KPNC Research Program on Genes, Environment and Health (RPGEH). The detailed description of the cohort and study design can be found in the database of Genotypes and Phenotypes (dbGaP) (study accession: phs000674.v1.p1). Beginning in July 2008, study subjects who completed consent forms were mailed saliva collection kits (Oragene), with the final specimens for the GERA cohort being obtained by February 2011.
Experimental methods
DNA was extracted at the KPNC RPGEH biorepository, and samples were shipped in 96-well plates to the Blackburn lab at UCSF for TL analysis. Well position C04 was left empty for all plates as a control. DNA sample concentration, quantified by the Picogreen method, was normalized to 35 ng/μl. All DNA samples were heat sealed and stored in their original plates in a −80° freezer upon arrival. DNA samples were thawed at 4° on the day of the assay and transferred to 384-well plates.
Principles of the assay
TL is measured as the ratio of telomeric product/single copy gene product (T/S) obtained using quantitative PCR, as shown in Figure 1 (Cawthon 2002). The rationale of the method is that the longer the telomeres are in a given sample, the more PCR product will be generated in PCR reactions using primers specific for the telomeric DNA. This can be quantified by quantitative PCR using a serially diluted standard DNA and the standard curve method. To normalize the quantity of the input DNA, a single copy gene is amplified in parallel. The ratio of the telomeric product vs. the single copy gene product reflects the average length of the telomeres.
Figure 1.
Measurement of telomere length (TL) using quantitative PCR. For each sample, T and S values are measured in separate reactions using the standard curve method. The T/S ratio of each sample is calculated from T and S values.
Hardware design
As this project was supported by a 2-year grant, it was critical that we develop an approach for analyzing a large number of samples in a period of at most 6 months. Hence, to assay the 110,266 GERA samples in a reliable and high-throughput fashion, we designed an integrated robotic system that had the following components:
Biomek FXP liquid handler with a 96-channel pipetting head and a Span-8 module
Thermo VAL 50-inch robotic arm
Three Thermo Multidrop Combi reagent dispensers
Temperature-controlled reagent reservoirs
Two Thermo ALPS 3000 microplate plate sealers
BioRad PTC-100 Thermal Cycler
Carousels for tips and plate storage
Thermo Cytomat 2 C4 Automated Incubator
Four Roche LightCycler 480 real-time PCR machines
Agilent Vspin Automated Centrifuge
Thermo Tower of Power de-lidder and 2D barcode reader.
The system was integrated by Thermo CRS and controlled by the Thermo Momentum scheduling software. The computer systems were accessible via Virtual Network Computing (VNC) or Remote Desktop Protocol (RDP), and Microsoft LifeCam video cameras enabled video monitoring via iCam iPhone/iPad application. These systems in conjunction allowed 24/7 operations with remote control and monitoring, with an overall daily throughput of 1920 patient samples (10- to 20-fold increase in throughput over assays carried out manually).
Reagents
Primers:
The primer assay was adopted from Cawthon (2002) with modifications from Lin et al. (2010). Primers for the telomere PCR (T runs) are tel1b [5′-CGGTTT(GTTTGG)5GTT-3′], used at a final concentration of 100 nM, and tel2b [5′-GGCTTG(CCTTAC)5CCT-3′], used at a final concentration of 900 nM. The primers for the single-copy gene (human beta-globin) PCR (S runs) are hbg1 [5′-GCTTCTGACACAACTGTGTTCACTAGC-3′], used at a final concentration of 300 nM, and hbg2 [5′-CACCAACTTCATCCACGTTCACC-3′], used at a final concentration of 700 nM. The primers only bind within the repeat tracts, but it should be noted that they are designed to have a base mutation for every telomeric repeat sequence (e.g., TTTGGG vs. the wild-type telomere sequence TTAGGG). This design allows binding and amplification of the target telomere sequence, while eliminating the formation of primer dimers. All primers were purchased from IDT (www.idtdna.com) in standard desalted form.
Standard reference DNA and control DNAs:
We included eight control DNA samples from various cancer cell lines in every assay plate. These DNAs are: C1, Hela; C2, 293T; C3, H1299; C4, UMUC3 infected with hTER-population doubling (PD) 7; C5, UMUC3 infected with hTER-PD 11; C6, UMUC3 infected with hTER-PD 15; C7, UMUC3 infected with hTER-PD 19; and C8, UMUC3.
Each control DNA was diluted threefold three times with DI buffer, resulting in four dilution points for each control DNA. The highest concentration is 16 ng/μl in the PCR reaction. These control DNA samples allowed us to create eight standard curves, which were then integrated into a composite standard curve, used for T and S concentration calculations. The diluted control DNA samples were prepared as a single batch for the entire project, aliquoted, and stored at −80°.
Having eight control DNA samples, each with four points, allows calculation of a composite standard curve that is not solely dependent on the accuracy of one standard curve; hence this method is more robust than one based on a single standard curve. The tradeoff is that assaying six points for each standard curve would result in too many wells being used. To get around this problem, the normalized 35 ng/μl concentration was chosen for the input DNA. As a consequence, the range of input DNA concentration is very narrow and falls within the four-point threefold dilution range.
E. coli DNA:
E. coli DNA was included in the assay to reduce well-to-well assay variability; neither the T primers nor the S primers amplify Escherichia coli DNA. E. coli DNA stock (600 ng/μl): lyophilized E. coli DNA (Sigma-Aldrich, cat. no. D2001) was resuspended in sterile Millipore H2O to 600 ng/μl. The resuspended DNA was rotated at room temperature for 2 hr and stored at −20° until further use. Denaturation buffer (25 mM Tris-HCl, pH 8.4; 62.5 mM KCl, 3.8 ng/μl of E. coli DNA) was made by using 10× PCR buffer (200 mM Tris-HCl, pH 8.4; 500 mM KCl) supplied in the Platinum Taq kit (Invitrogen, cat no. 10966-083) and 600 ng/μl of E. coli DNA stock. Denaturation buffer for the entire study was made as a single batch and stored at −80° as individual aliquots for each batch of 60 plates.
U, T, and S mixes:
U mix contains Tris-HCl, PH 8.4; KCl, MgCl2, dNTPs, and DMSO. U mix was made from 10× PCR buffer and 50 mM MgCl2 (from Platinum Taq kit), 100 mM dNTP (Roche Applied Science, cat. no. 03622614001) and DMSO (Sigma-Aldrich, cat. no. 154938). T mix contains tel 1b and tel 2b primers. S mix contains hbg1 and hbg2 primers and MgCl2. U, T, and S mixes were made as a single batch for the entire study and stored as individual aliquots at −80°.
SYBR Green I:
Working stocks of SYBR Green I (10×) were made from 10,000× in DMSO (Invitrogen, cat. no. S7585) with ddH2O, wrapped in aluminum foil and stored at −80° as individual aliquots.
Platinum Taq polymerase:
Platinum Taq polymerase was purchased from Invitrogen and stored at −20°. The entire study used a single lot of Platinum Taq. The final reaction mix contained 20 mM Tris-HCl, pH 8.4; 50 mM KCl; 200 μM each dNTP; 1% DMSO; 0.5× SRBR Green I; 16.5 ng E. coli DNA per reaction; 0.44 units of Platinum Taq DNA polymerase per 11-μl reaction.
Assay plate design
The 96 DNA samples from each sample plate were placed in 96 wells of a quadrant of a 384-well plate. Three of the four quadrants contained samples from three different original sample plates. The fourth quadrant contained control samples as described below. In this way, three sample plates were assayed along with the controls (Figure 2). A computer-generated, randomized plate-testing schedule was developed so that any specific source DNA plate was never tested more than once with another source DNA plate. Each of the eight control DNA samples was serially diluted threefold three times; the serial dilution was carried out three times independently. The resulting 96 control samples (8 controls × 4 dilutions × 3 replicate dilutions) were placed in the fourth quadrant. The 384-well plate arrayed with samples from the three original sample plates and 96 controls is called a superplate. Each DNA sample was placed in a congruent pair of superplates for the T assay and similarly in a congruent pair of superplates for the S assay. The same DNA was also placed in two other pairs of T superplates, with different source plate configurations as well as in two other pairs of S superplates. Thus each sample was assayed in six T wells and six S wells.
Figure 2.
Assay plate design. Locations indicated by S1, S2, and S3 indicate tested samples from plates 1, 2, and 3, and locations with C indicate controls.
Experimental procedures
Thermo Multidrop Combi was used to dispense 30 μl of denaturation buffer into a 384-well plate (Biorad cat no. HSP-3901), and 1.5 μl of DNA samples were mixed with the denaturation buffer using the Biomek 96-channel head. Similarly, 1.5 μl of control DNA dilution standards were pipetted into the same 384-well plate. The 384-well plate layout is shown in Figure 2. This 384-well plate was heated at 95° for 10 min in a Thermal cycler (Biorad PTC-100).
A complete master mix was made by combining U mix, SYBR Green, Taq polymerase, and T mix (for T run) or S mix (for S run). Thermo Multidrop Combi reagent dispenser was used to aliquot 7.5 μl of the master mix into each well of a 384-well PCR plate (Roche Applied Science, cat. no. 0510243001), and 3.5 μl of the denatured DNA samples were mixed into each well using the 96-channel head on the Biomek deck. The PCR plate was heat sealed with an optically clear seal (Thermo cat. no. AB3799) with Thermo ALPS 3000, briefly spun down on Agilent Vspin, and then loaded onto Roche LightCycler 480 for qPCR.
The Thermal cycling profiles were as follows for the T run: denature at 96° for 1 min for 1 cycle, denature 96° for 1 sec, anneal/extend at 54° for 60 sec, with fluorescence data collection, 30 cycles; and for the S run: denature at 96° for 1 min for 1 cycle, denature at 95° for 15 sec, anneal at 58° for 1 sec, extend at 72° for 20 sec, 8 cycles, followed by denature at 96° for 1 sec, anneal at 58° for 1 sec, extend at 72° for 20 sec, hold at 83° for 5 sec with data collection, 35 cycles.
Data analysis
In computing relative TL from the raw assay data, the overall goal was to obtain TL of uniformly high quality across the entire cohort while minimizing batch effects and experimental nonstationarities across the duration of the experiment. These goals were accomplished by creating extensions of the standard curve analysis to control for nonbiological sources of noise.
The first step was to filter out poor control sets. This was done by first creating a linear fit standard curve of the samples in each control set on each superplate. The median of the derived standard curve slopes from the dilution sequences for all the control sets was calculated and used to detect poorly behaving control sets; if abs(slope-median) > 0.2, a control set was flagged. Recovery of a flagged control set was attempted by removing the most outlying data point and recalculating the slope. If the slope still deviated from the median by greater than the threshold amount, the control set was removed from further analysis.
For each sample, a standard curve estimate was obtained by using the control set(s) physically closest to the sample well on the superplate. Because each sample was subjected to two telomere measurements (T) and two measurements of the single copy gene (S), four measures were created by contrasting each of the two T measurements with each of the two S measurements. To obtain a more Gaussian distribution, we formed the T/S ratio and log-transformed it (log base 2) to create the quantity log2(T/S) (denoted as LTS).
Each sample was run at least three times on different plates. Therefore, a typical sample had 12 measurements, although the actual number varied from 4 to 20, with occasionally fewer due to failed qPCR reactions. The LTS measures were then fit to a mixed effects model to control for nonbiological sources of variation, such as overall superplate plate variation and overall superwell variation.
The residuals from this model became the final sets of LTS measurements for each sample. The relative TL for a sample was computed as the median of the LTS residuals.
Results
Control sample properties
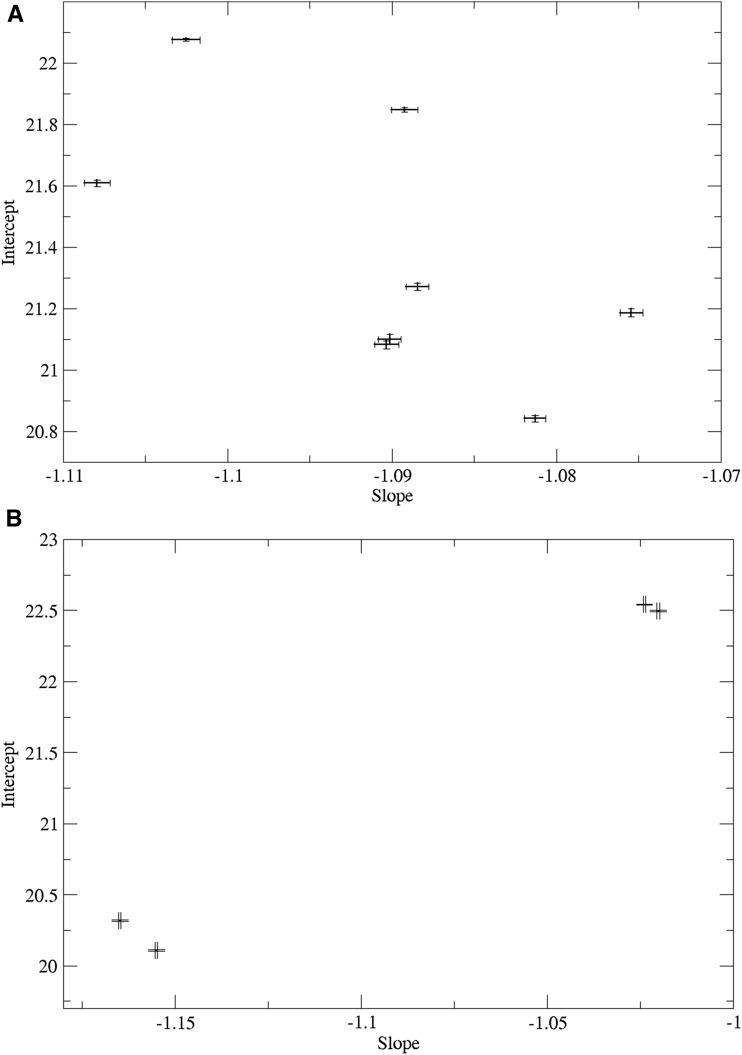
Standard curves were calculated for all control sample dilution sets on all superplates and those failing QC thresholds (2.4% of control sets) were filtered out. Figure 3A shows the mean standard curve slope and intercept for the eight sets of control samples. The sets of control samples are well clustered, but ANOVA tests showed significant differences among control samples for both slope (P < 2e-16) and intercept (P < 2e-16).
Figure 3.
(a) Mean standard curve slope and intercept for the eight sets of control samples. Error bars represent ±1 standard error. (b) Mean standard curve slope and intercept for control samples on the four LightCycler qPCR systems.
Figure 3B shows the mean standard curve slope and intercept for control sample sets grouped by the four LightCycler qPCR systems used for the assay. ANOVA tests showed significant differences among qPCR systems for both slope (P < 2e-16) and intercept (P < 2e-16).
A linear regression of slope vs. control sample produced an adjusted R^2 correlation of 0.013. A linear regression of slope vs. qPCR system produced an adjusted R^2 correlation of 0.658. Together, both factors explain about 2/3 of the slope variation. qPCR system heterogeneity was controlled for by matching control sets on different systems and normalizing LTS values to a single effective system. Control sample heterogeneity was controlled for through the superplate well factor in mixed-effects model.
Overall sample success rate
A total of 106,902 individuals were initially assayed for TL. We note that with the robotic system, laboratory processing was completed within a 4-month time frame. Such rapid throughput, required for this project, would have been completely infeasible without the automation.
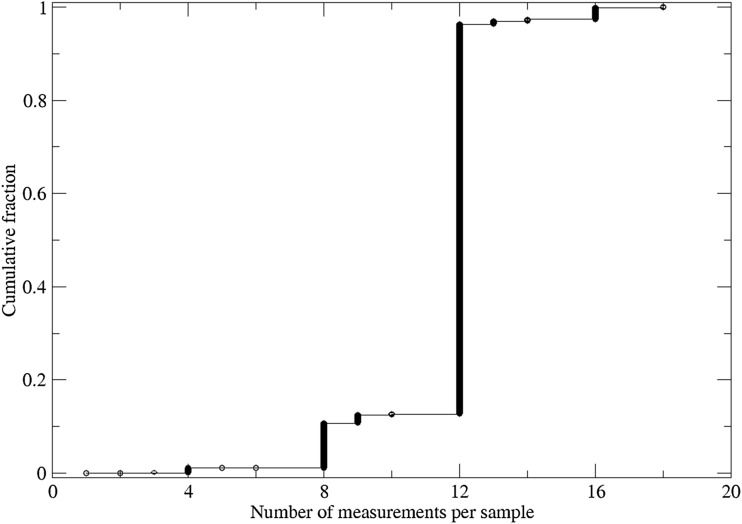
Figure 4 provides the distribution of the number of replicate measurements per individual. A large majority of individuals, 87%, had 12 or more replicate measures available. A smaller proportion, 12%, had between 8 and 12 replicates available, while 11 subjects had fewer than 4 replicates; 1.2% had fewer than 8 replicates and were filtered out.
Figure 4.
Cumulative distribution of number of TL measurements per sample.
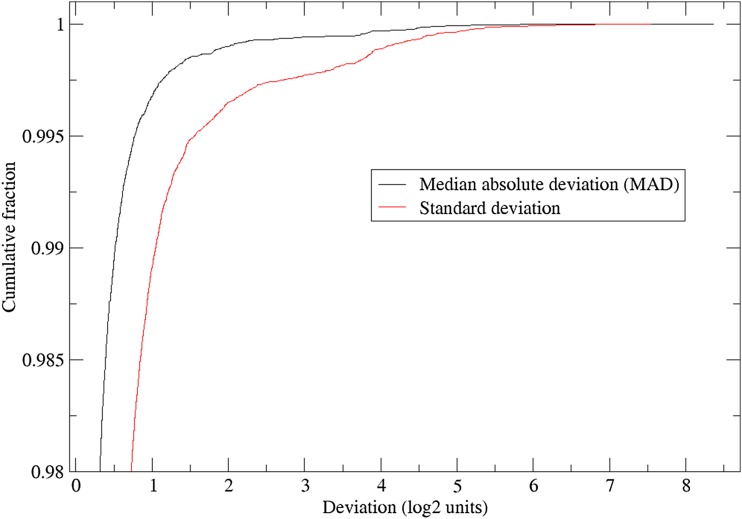
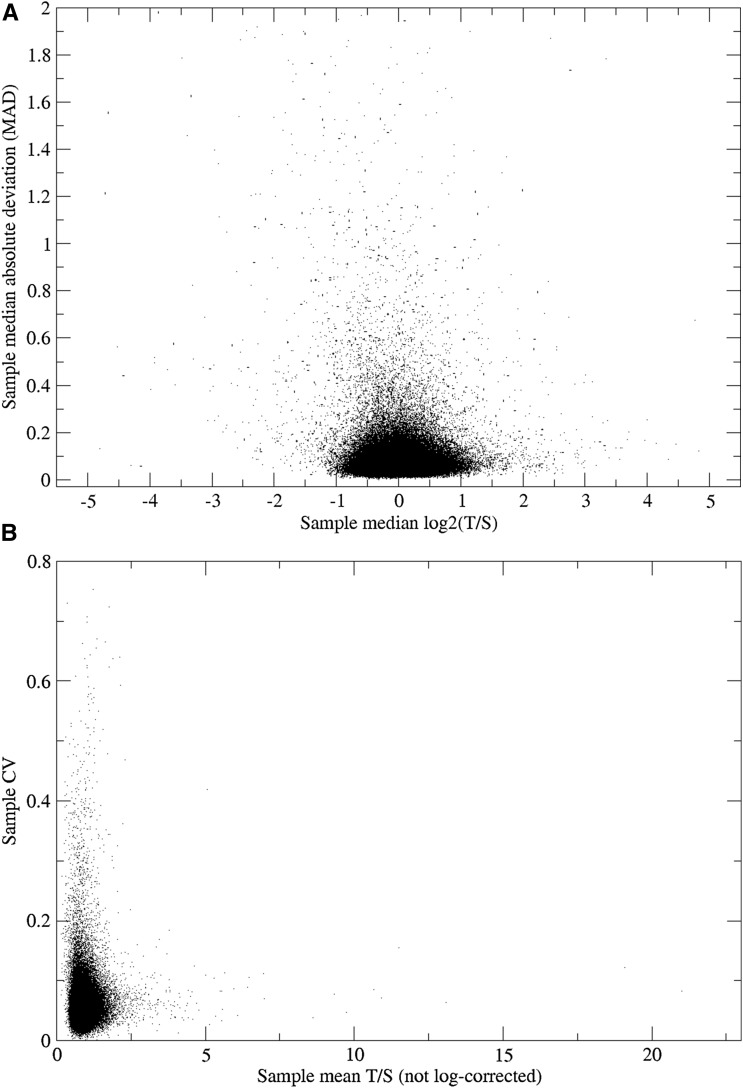
We were also concerned about the variability among the different replicates within an individual, as a high level of variation would suggest the presence of faulty measurements. Figure 5 shows the cumulative distribution of both the within-subject standard deviation and the robust median absolute deviation from the sample median. Figure 6A presents the median absolute deviation plotted against the sample median. As seen in both figures, the within-subject variability is skewed, with a subset of individuals having large within-sample variability for their replicates. Figure 6B shows the within-subject CV vs. the non-log-corrected mean (T/S) telomere length. Because these samples are likely to include unreliable measures, guided by these figures, we further removed individuals with an absolute median deviation at a threshold value of >0.4. This led to the elimination of an additional 1372 subjects (1.3% of the total). Hence, the final sample for subsequent analysis included a total of 105,539 subjects with summary telomere measurements.
Figure 5.
Cumulative distribution of within individual TL measurement deviation.
Figure 6.
(a) Within individual transformed TL [log2(T/S)] median absolute deviation (MAD) vs. median TL value (MAD values truncated at a value of 2). (b) Within individual CV vs. non-log-corrected TL (T/S).
The CV of this assay from our lab with blood-derived leukocyte DNA samples is 3–4%; for this cohort of saliva-derived DNA, the median CV across all samples is 5.1% and the mean CV is 5.6% ± 3.1%. The median sample CV vs. the number of sample replicate measurements is given in Table 1. The sample CV does not significantly vary with number of replicates; nonetheless, samples with fewer than eight replicates were considered undersampled and were removed.
Table 1. Median sample CV vs. number of sample replicate measurements.
| Sample replicates | Number of samples | Median sample CV | Standard error |
|---|---|---|---|
| 2 | 40 | 0.03 | 0.003 |
| 4 | 1,135 | 0.03 | 0.001 |
| 6 | 65 | 0.05 | 0.01 |
| 7 | 31 | 0.05 | 0.004 |
| 8 | 10,689 | 0.05 | 0.0003 |
| 9 | 25 | 0.06 | 0.03 |
| 10 | 551 | 0.06 | 0.003 |
| 12 | 86,974 | 0.05 | 0.0001 |
| 14 | 21 | 0.07 | 0.04 |
| 16 | 4,799 | 0.06 | 0.0004 |
Relationship of TL with age and sex
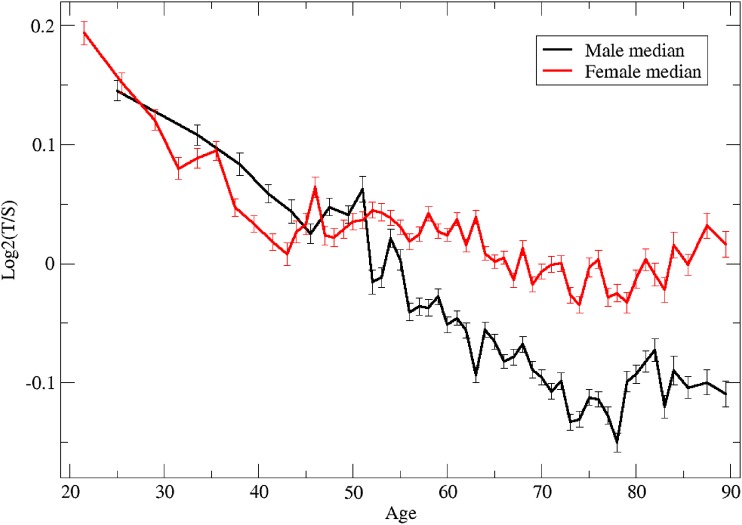
We next examined the relationship between TL and age, separately for males and females (Figure 7). Several characteristics of this relationship are notable, in particular the sex dimorphism. While, overall, males have shorter TLs than females, the difference is age dependent. For males, there is a continuous linear decline from young adulthood to late adulthood (approximately age 75), at which point the decline ends, and ultimately rises slightly at around age 80 (t-test on males age 70–78 vs. 80–90: P = 0.0210). By contrast, in females, there is a parallel linear decline in TL from young adulthood to middle age; however, this pattern changes at around age 50, at which point the decline diminishes and eventually reverses, again at around age 75 (t-test on females age 70–78 vs. 80–90: P = 0.000796). Prior to age 40, TLs appear to be slightly but not significantly shorter in women (t-test on males vs. females age < 40: P = 0.287); however, after age 50, the opposite occurs, with a gradually increasing difference in TL favoring females (t-test on males vs. females age > 50: P < 2.2e-16).
Figure 7.
Median TL as a function of age, in males and females.
TL variance with age
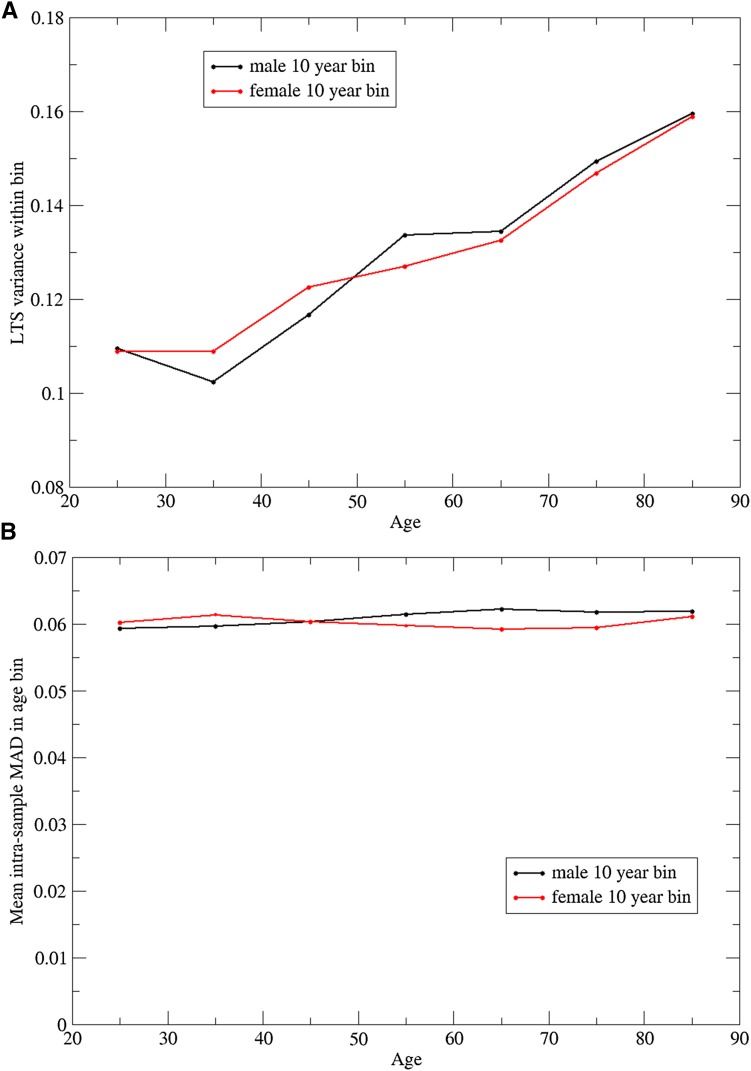
Finally, we also examined the variability of TL as a function of age. We calculated the variance in TL among individuals within age decade for each sex separately (Figure 8A). There is a clear pattern of linear increase in variation in TL by decade that starts to occur at age 35 in both sexes. A linear regression model was fit to these data with variance measured at each integral age as the dependent variable and age as the independent variable, for each sex separately. For males, the regression slope coefficient was 0.00102 ± 0.00010 (t-test for null slope: P < 10−12); similarly, for females, the regression slope coefficient was 0.000920 ± 0.000098 (t-test for null slope: P < 10−12).
Figure 8.
Variation in TL measurements between individuals (a) and within individuals (b) as a function of age, for males and females.
For comparison, we examined the variability of the TL measurements for an individual, as determined by the median absolute deviation. If we bin the individual (MAD) values by age decade and plot the mean MAD (Figure 8B), it is apparent that the variation among measurements within an individual does not increase with age.
These results suggest that systematic differences in the parameters of the PCR reaction, that might have reflected different quality of the sample or DNA preparations with age, could not explain the observed increased variance of TL with age.
Discussion
We have presented the largest study to create TL data in a single population sample to date. The development of these data over a short time period required the development of novel automation for the assay. Many of the steps, traditionally done by hand, were implemented through robotic systems to enable high throughput. Also notable among the methods, were procedures for assuring quality during the process, including the replication of the assays, randomization of the placement of samples, and real-time assessment of the experiment to detect problems as they emerged.
It is instructive to compare our results, regarding the relationship of sex and age with TL, to prior studies, most of which used blood leukocytes and other cell types as the source of telomeric DNA. In the current study, we observed an overall declining pattern with age and a sex difference, as has been observed in other studies. In our data, the declining trend with age was highly dimorphic in men and women, with a continuing decline in men but not women after the age of 50. These results are consistent with a number of other studies that have documented a stronger decline in TL with age in men compared to women (Iwama et al. 1998; Unryn et al. 2005; Vasa-Nicotera et al. 2005; De Meyer et al. 2007; Nordfjall et al. 2010), but not others (Gardner et al. 2014). These results are also consistent with a 5-year follow-up study of leukocyte TL that showed more rapid shortening in middle-aged men than women (Farzaneh-Far et al. 2010).
In both sexes, we saw a slight reversal in TL at late age, starting at approximately age 75. The change in relationship between age and TL is also reminiscent of the observation, in a smaller study, that in the general Costa Rican population, the earlier age-related decline in leukocyte TL was diminished between ages ∼80 and 100 years (Rehkopf et al. 2013).
The nonmonotonic relationship between TL and age we report in the large GERA cohort may explain inconsistencies between previous reports on relationships of TL with chronological age. The reversal of TL occurs at a time of increasing mortality and hence may reflect a survival advantage for those with longer TLs. This speculation can be subsequently confirmed directly by relating TL to mortality as this cohort ages.
Our finding of increasing variance of TL with advancing age is also intriguing and to our knowledge not previously reported. We eliminated methodological or technical reasons that might have explained this increase of variance with age. It suggests that environmental factors influencing TL may become more prominent with age. Alternatively, it may represent a selection bias in that older individuals who participated in providing a saliva sample were “healthier” than typical individuals of the same age. One approach to further exploring this question is by examination of correlations of TL between relatives, stratified by age of the relatives. Because of the large size of the GERA cohort and the presence of related individuals, we will be able to report results from such analyses in a future publication.
The GERA cohort contains a wealth of longitudinal electronic health data, as well as demographic and behavioral data from self-report surveys and environmental exposures obtained from geocoded linkage of environmental databases to study subject residences. In addition, the cohort has full genome-wide genotype data available. Bringing these other sources of information together with the TL data described here will enable a broad array of analyses addressing important questions regarding the genetic and environmental factors that determine TL and the role of telomere maintenance in the aging process, onset of various diseases, and mortality.
Conclusion
We have performed the largest single relative-length telomere length assay to date. For this saliva-based cohort, the median sample CV was 5.1%, vs. 3–4% for blood-derived leukocyte assays on other sample sets in the same lab. It was found that overall, telomere length decreased for both men and women with increasing age. Telomere length decreased more for men than women from age 50 to 75, but TL increased modestly for the age 80 to 90 group for both men and women. The variation of TL among individuals increased with age, but the variation of TL measurements within an individual did not.
Supplementary Material
Acknowledgments
We thank Judith Millar for her assistance in preparing the manuscript for publication. We are grateful to the Kaiser Permanente Northern California members who have generously agreed to participate in the Kaiser Permanente Research Program on Genes, Environment, and Health. This work was supported by grant RC2 AG036607 from the National Institutes of Health. Participant enrollments and survey and sample collection for the RPGEH were supported by grants from the Robert Wood Johnson Foundation, the Ellison Medical Foundation, the Wayne and Gladys Valley Foundation, and Kaiser Permanente. Information about data access can be obtained at: http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000674.v1.p1 and https://rpgehportal.kaiser.org.
Note added in proof: See Kvale et al. 2015 (pp. 1051–1060) and Banda et al. 2015 (pp. 1285–1295) in this issue for related works.
Footnotes
Communicating editor: N. R. Wray
The accession number for the RPGEH Parent Level/Study on dbGaP is: phs00078. The title of the dbGaP Parent study is: The Kaiser Permanente Research Program on Genes, Environment, and Health (RPGEH). GO Project IRB: CN-09CScha-06-H.
Literature Cited
- Allsopp R. C., Vaziri H., Patterson C., Goldstein S., Younglai E. V., et al. , 1992. Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl. Acad. Sci. USA 89: 10114–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios M., Blackburn E. H., 2012. The telomere syndromes. Nat. Rev. Genet. 13: 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astrup A. S., Tarnow L., Jorsal A., Lajer M., Nzietchueng R., et al. , 2010. Telomere length predicts all-cause mortality in patients with type 1 diabetes. Diabetologia 53: 45–48. [DOI] [PubMed] [Google Scholar]
- Aubert G., Lansdorp P. M., 2008. Telomeres and aging. Physiol. Rev. 88: 557–579. [DOI] [PubMed] [Google Scholar]
- Aviv A., Hunt S. C., Lin J., Cao X., Kimura M., et al. , 2011. Impartial comparative analysis of measurement of leukocyte telomere length/DNA content by Southern blots and qPCR. Nucleic Acids Res. 39: e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakaysa S. L., Mucci L. A., Slagboom P. E., Boomsma D. I., McClearn G. E., et al. , 2007. Telomere length predicts survival independent of genetic influences. Aging Cell 6: 769–774. [DOI] [PubMed] [Google Scholar]
- Bischoff C., Petersen H. C., Graakjaer J., Andersen-Ranberg K., Vaupel J. W., et al. , 2006. No association between telomere length and survival among the elderly and oldest old. Epidemiology 17: 190–194. [DOI] [PubMed] [Google Scholar]
- Blackburn E. H., 1997. The telomere and telomerase: nucleic acid-protein complexes acting in a telomere homeostasis system. A review. Biochemistry (Mosc.) 62: 1196–1201. [PubMed] [Google Scholar]
- Blackburn E. H., 2000. Telomere states and cell fates. Nature 408: 53–56. [DOI] [PubMed] [Google Scholar]
- Blackburn E. H., 2005. Telomeres and telomerase: their mechanisms of action and the effects of altering their functions. FEBS Lett. 579: 859–862. [DOI] [PubMed] [Google Scholar]
- Bojesen S. E., 2013. Telomeres and human health. J. Intern. Med. 274: 399–413. [DOI] [PubMed] [Google Scholar]
- Brouilette S. W., Moore J. S., McMahon A. D., Thompson J. R., Ford I., et al. , 2007. Telomere length, risk of coronary heart disease, and statin treatment in the West of Scotland Primary Prevention Study: a nested case-control study. Lancet 369: 107–114. [DOI] [PubMed] [Google Scholar]
- Cawthon R. M., 2002. Telomere measurement by quantitative PCR. Nucleic Acids Res. 30: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthon R. M., Smith K. R., O’Brien E., Sivatchenko A., Kerber R. A., 2003. Association between telomere length in blood and mortality in people aged 60 years or older. Lancet 361: 393–395. [DOI] [PubMed] [Google Scholar]
- Codd V., Mangino M., van der Harst P., Braund P. S., Kaiser M., et al. , 2010. Common variants near TERC are associated with mean telomere length. Nat. Genet. 42: 197–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codd, V., C. P. Nelson, E. Albrecht, M. Mangino, J. Deelen et al., 2013 Identification of seven loci affecting mean telomere length and their association with disease. Nat. Genet. 45: 422–427, 427e1–2. [DOI] [PMC free article] [PubMed]
- De Meyer T., Rietzschel E. R., De Buyzere M. L., De Bacquer D., Van Criekinge W., et al. , 2007. Paternal age at birth is an important determinant of offspring telomere length. Hum. Mol. Genet. 16: 3097–3102. [DOI] [PubMed] [Google Scholar]
- Epel E. S., Merkin S. S., Cawthon R., Blackburn E. H., Adler N. E., et al. , 2009. The rate of leukocyte telomere shortening predicts mortality from cardiovascular disease in elderly men. Aging (Albany, N.Y.) 1: 81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzaneh-Far R., Cawthon R. M., Na B., Browner W. S., Schiller N. B., et al. , 2008. Prognostic value of leukocyte telomere length in patients with stable coronary artery disease: data from the Heart and Soul Study. Arterioscler. Thromb. Vasc. Biol. 28: 1379–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzaneh-Far R., Lin J., Epel E., Lapham K., Blackburn E., et al. , 2010. Telomere length trajectory and its determinants in persons with coronary artery disease: longitudinal findings from the heart and soul study. PLoS One 5: e8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick A. L., Kronmal R. A., Gardner J. P., Psaty B. M., Jenny N. S., et al. , 2007. Leukocyte telomere length and cardiovascular disease in the cardiovascular health study. Am. J. Epidemiol. 165: 14–21. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick A. L., Kronmal R. A., Kimura M., Gardner J. P., Psaty B. M., et al. , 2011. Leukocyte telomere length and mortality in the Cardiovascular Health Study. J. Gerontol. A Biol. Sci. Med. Sci. 66: 421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadalla S. M., Cawthon R., Giri N., Alter B. P., Savage S. A., 2010. Telomere length in blood, buccal cells, and fibroblasts from patients with inherited bone marrow failure syndromes. Aging (Albany, N.Y.) 2: 867–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M., Bann D., Wiley L., Cooper R., Hardy R., et al. , 2014. Gender and telomere length: systematic review and meta-analysis. Exp. Gerontol. 51: 15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graakjaer J., Londono-Vallejo J. A., Christensen K., Kolvraa S., 2006. The pattern of chromosome-specific variations in telomere length in humans shows signs of heritability and is maintained through life. Ann. N. Y. Acad. Sci. 1067: 311–316. [DOI] [PubMed] [Google Scholar]
- Harris S. E., Deary I. J., MacIntyre A., Lamb K. J., Radhakrishnan K., et al. , 2006. The association between telomere length, physical health, cognitive ageing, and mortality in non-demented older people. Neurosci. Lett. 406: 260–264. [DOI] [PubMed] [Google Scholar]
- Haycock P. C., Heydon E. E., Kaptoge S., Butterworth A. S., Thompson A., et al. , 2014. Leucocyte telomere length and risk of cardiovascular disease: systematic review and meta-analysis. BMJ 349: g4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honig L. S., Kang M. S., Schupf N., Lee J. H., Mayeux R., 2012. Association of shorter leukocyte telomere repeat length with dementia and mortality. Arch. Neurol. 69: 1332–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwama H., Ohyashiki K., Ohyashiki J. H., Hayashi S., Yahata N., et al. , 1998. Telomeric length and telomerase activity vary with age in peripheral blood cells obtained from normal individuals. Hum. Genet. 102: 397–402. [DOI] [PubMed] [Google Scholar]
- Jain D., Cooper J. P., 2010. Telomeric strategies: means to an end. Annu. Rev. Genet. 44: 243–269. [DOI] [PubMed] [Google Scholar]
- Kimura M., Hjelmborg J. V., Gardner J. P., Bathum L., Brimacombe M., et al. , 2008. Telomere length and mortality: a study of leukocytes in elderly Danish twins. Am. J. Epidemiol. 167: 799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Sandford A. J., Connett J. E., Yan J., Mui T., et al. , 2012. The relationship between telomere length and mortality in chronic obstructive pulmonary disease (COPD). PLoS One 7: e35567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Epel E., Cheon J., Kroenke C., Sinclair E., et al. , 2010. Analyses and comparisons of telomerase activity and telomere length in human T and B cells: insights for epidemiology of telomere maintenance. J. Immunol. Methods 352: 71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Ruiz C., Dickinson H. O., Keys B., Rowan E., Kenny R. A., et al. , 2006. Telomere length predicts poststroke mortality, dementia, and cognitive decline. Ann. Neurol. 60: 174–180. [DOI] [PubMed] [Google Scholar]
- Muezzinler A., Zaineddin A. K., Brenner H., 2013. A systematic review of leukocyte telomere length and age in adults. Ageing Res. Rev. 12: 509–519. [DOI] [PubMed] [Google Scholar]
- Needham B. L., Fernandez J. R., Lin J., Epel E. S., Blackburn E. H., 2012. Socioeconomic status and cell aging in children. Soc. Sci. Med. 74: 1948–1951. [DOI] [PubMed] [Google Scholar]
- Njajou O. T., Hsueh W. C., Blackburn E. H., Newman A. B., Wu S. H., et al. , 2009. Association between telomere length, specific causes of death, and years of healthy life in health, aging, and body composition, a population-based cohort study. J. Gerontol. A Biol. Sci. Med. Sci. 64: 860–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordfjall K., Svenson U., Norrback K. F., Adolfsson R., Roos G., 2010. Large-scale parent-child comparison confirms a strong paternal influence on telomere length. Eur. J. Hum. Genet. 18: 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olovnikov A. M., 1973. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 41: 181–190. [DOI] [PubMed] [Google Scholar]
- Panossian L. A., Porter V. R., Valenzuela H. F., Zhu X., Reback E., et al. , 2003. Telomere shortening in T cells correlates with Alzheimer’s disease status. Neurobiol. Aging 24: 77–84. [DOI] [PubMed] [Google Scholar]
- Rehkopf D. H., Dow W. H., Rosero-Bixby L., Lin J., Epel E. S., et al. , 2013. Longer leukocyte telomere length in Costa Rica’s Nicoya Peninsula: a population-based study. Exp. Gerontol. 48: 1266–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salpea K. D., Talmud P. J., Cooper J. A., Maubaret C. G., Stephens J. W., et al. , 2010. Association of telomere length with type 2 diabetes, oxidative stress and UCP2 gene variation. Atherosclerosis 209: 42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samani N. J., Boultby R., Butler R., Thompson J. R., Goodall A. H., 2001. Telomere shortening in atherosclerosis. Lancet 358: 472–473. [DOI] [PubMed] [Google Scholar]
- Strandberg T. E., Saijonmaa O., Tilvis R. S., Pitkala K. H., Strandberg A. Y., et al. , 2011. Association of telomere length in older men with mortality and midlife body mass index and smoking. J. Gerontol. A Biol. Sci. Med. Sci. 66: 815–820. [DOI] [PubMed] [Google Scholar]
- Thiede C., Prange-Krex G., Freiberg-Richter J., Bornhauser M., Ehninger G., 2000. Buccal swabs but not mouthwash samples can be used to obtain pretransplant DNA fingerprints from recipients of allogeneic bone marrow transplants. Bone Marrow Transplant. 25: 575–577. [DOI] [PubMed] [Google Scholar]
- Unryn B. M., Cook L. S., Riabowol K. T., 2005. Paternal age is positively linked to telomere length of children. Aging Cell 4: 97–101. [DOI] [PubMed] [Google Scholar]
- Valdes A. M., Andrew T., Gardner J. P., Kimura M., Oelsner E., et al. , 2005. Obesity, cigarette smoking, and telomere length in women. Lancet 366: 662–664. [DOI] [PubMed] [Google Scholar]
- Vasa-Nicotera M., Brouilette S., Mangino M., Thompson J. R., Braund P., et al. , 2005. Mapping of a major locus that determines telomere length in humans. Am. J. Hum. Genet. 76: 147–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zglinicki T., Serra V., Lorenz M., Saretzki G., Lenzen-Grossimlighaus R., et al. , 2000. Short telomeres in patients with vascular dementia: an indicator of low antioxidative capacity and a possible risk factor? Lab. Invest. 80: 1739–1747. [DOI] [PubMed] [Google Scholar]
- Walsh K. M., Codd V., Smirnov I. V., Rice T., Decker P. A., et al. , 2014. Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nat. Genet. 46: 731–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischer M., Bojesen S. E., Cawthon R. M., Freiberg J. J., Tybjaerg-Hansen A., et al. , 2012. Short telomere length, myocardial infarction, ischemic heart disease, and early death. Arterioscler. Thromb. Vasc. Biol. 32: 822–829. [DOI] [PubMed] [Google Scholar]
- Wentzensen I. M., Mirabello L., Pfeiffer R. M., Savage S. A., 2011. The association of telomere length and cancer: a meta-analysis. Cancer Epidemiol. Biomarkers Prev. 20: 1238–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willeit P., Willeit J., Brandstatter A., Ehrlenbach S., Mayr A., et al. , 2010a Cellular aging reflected by leukocyte telomere length predicts advanced atherosclerosis and cardiovascular disease risk. Arterioscler. Thromb. Vasc. Biol. 30: 1649–1656. [DOI] [PubMed] [Google Scholar]
- Willeit P., Willeit J., Mayr A., Weger S., Oberhollenzer F., et al. , 2010b Telomere length and risk of incident cancer and cancer mortality. JAMA 304: 69–75. [DOI] [PubMed] [Google Scholar]
- Yaffe K., Lindquist K., Kluse M., Cawthon R., Harris T., et al. , 2011. Telomere length and cognitive function in community-dwelling elders: findings from the Health ABC Study. Neurobiol. Aging 32: 2055–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zee R. Y., Castonguay A. J., Barton N. S., Germer S., Martin M., 2010. Mean leukocyte telomere length shortening and type 2 diabetes mellitus: a case-control study. Transl. Res. 155: 166–169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.